Introduction
High-resolution magic angle spinning (HRMAS) nuclear
proton magnetic resonance spectroscopy (1H NMR) is a
novel non-destructive technique that substantially improves
spectral line-widths and allows high-resolution spectra to be
obtained from intact cells, cultured tissues (1,2)
and unprocessed tissues (3–7).
HRMAS 1H NMR enables us to investigate relationships
between metabolites and cell processes. For example, choline
(Cho)-containing compounds involved in phospholipid metabolism and
lipids, such as triglycerides, that are involved in apoptosis have
been studied (8–11). Nevertheless, HRMAS
1H-NMR has only been performed ex vivo thus
far.
Studies combining in vivo 1H NMR
with ex vivo HRMAS 1H NMR have demonstrated an
important functional role of intramyocellular lipids (IMCLs) in
rodent burn biology (11,12), while other ex vivo HRMAS
1H NMR studies have focused on lipid metabolism
(13). Szczepaniak et al
demonstrated that IMCL stores could be quantified accurately in a
clinical setting by in vivo 1H NMR (14). In a recently published
1H NMR study, Van der Graaf et al found an
inverse correlation between IMCL content in human calf muscle and
local glycogen synthesis rate (15). Jacob et al emphasized the
importance of these resonances as biomarkers of insulin resistance
in type-2 diabetes patients and their offspring (16). Additionally, IMCL content was
found to be increased in the soleus muscle of insulin-resistant
elderly patients, providing support for the hypothesis that an
age-associated decline in mitochondrial function contributes to
insulin resistance (17).
In vivo HRMAS 1H NMR is a
potentially useful tool in Drosophila since in vitro
NMR studies have shown the metabolic effects of hypoxia (18) and temperature stress (19) in flies. Although Drosophila
is a distinctively useful model organism that can be employed to
investigate genetics, physiology, and metabolism (20), with the exception of a recent
feasibility report (21), in
vivo NMR studies in Drosophila are lacking. Thus, we
attempted to implement an in vivo HRMAS 1H NMR
method that we developed in Drosophila (22), with the aim of investigating the
metabolism of Drosophila mutants. Such a study would be
particularly useful for assessing the biomarkers of pathophysiology
with the long-term goal of providing critical information that may
direct novel therapeutic development.
State-of-the art, in vivo NMR techniques are
used to elucidate metabolic patterns in Drosophila
melanogaster as a model organism of interest owing to the
notable parallels in the metabolism between Drosophila and
mammals (23,24). Indeed, the study of
Drosophila metabolism is an emerging field that can
potentially elucidate conserved metabolic mechanisms. Furthermore,
the powerful genetic tools available in Drosophila research
render the fruit fly a particularly tractable model organism in
which to probe metabolic pathways and lead to a better
understanding of human metabolic disorders.
Drosophila melanogaster glutathione
S-transferase (GST2, also known as DmGSTS1-1) was recognized
originally as an indirect flight muscle-associated protein with no
known catalytic properties. In relation to mammalian GSTs,
Drosophila GST2 is most similar to the sigma class of GSTs,
and the mammalian GSTA4 gene is an ortholog of Drosophila
GST2. In the present study, we investigated mutant flies that
do not express the GST2 gene in skeletal muscle. We examined
the feasibility of a novel, in vivo HRMAS 1H NMR
approach towards the investigation of the metabolic derangements in
these GST2 mutant flies and compared them to isogenic
control flies.
Materials and methods
Drosophila flies
Male Gst2 gene deletion flies (25), designated as GstS1M38 were used,
and compared to male wild-type (wt) isogenic strain C5 flies. The
two strains were kindly provided by Helen Benes (University of
Arkansas). At the time of the experiments, all flies were 5–8 days
of age and weighed 0.7–1.0 mg (n=6 per group). Prior to insertion
in the spectrometer, each fly was anesthetized by being placed on
ice for <1 min. Flies were kept at 4°C while in the
spectrometer.
In vivo HRMAS 1H NMR
spectroscopy
All HRMAS 1H NMR experiments were
performed on a wide-bore Bruker Bio-Spin Avance NMR spectrometer
(600.13 MHz) using a 4-mm triple resonance (1H,
13C, 2H) HRMAS probe (Bruker, Billerica, MA,
USA). The flies were placed into a zirconium oxide rotor tube (4 mm
diameter, 50 μl), and 8 μl of external standard
trimethylsilyl-propionic-2,2,3,3-d4 acid (TSP) (molecular mass =
172 Da, d = 0.00 ppm, 50 mM in D2O) was introduced. TSP
functioned as a reference for both resonance chemical shift and
quantification. Each fly was placed in the rotor using the insert,
which was sealed with a screw and covered with parafilm to prevent
contact between the fly and the TSP/D2O (Fig. 1). The samples were secured and
tightened in the rotors with a top cap (Bruker). The HRMAS
1H NMR was performed at 4°C with 2 kHz MAS.
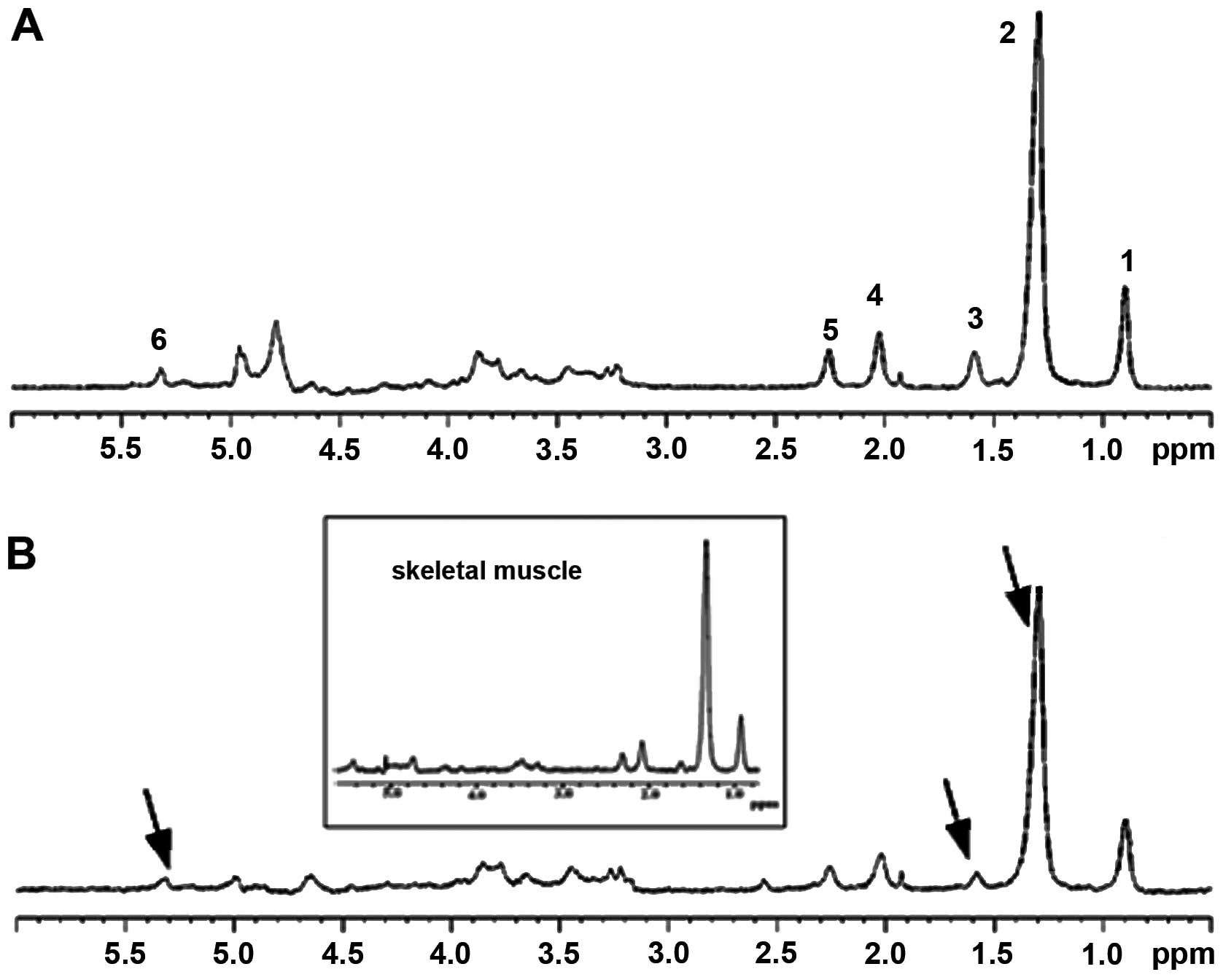 | Figure 1In vivo 1D HRMAS 1H
CPMG spectra of: (A) GST2 and (B) young wt flies. Lipid
components were: 1, CH3 (0.89 ppm); 2, (CH2)n
(1.33 ppm, putative IMCLs); 3, CH2C-CO (1.58 ppm,
putative EMCLs), acetate (Ac, 1.92 ppm); 4, CH2C=C (2.02
ppm); 5, CH2C=O (2.24 ppm); and 6, CH=CH (5.33 ppm).
Other spectral components included: β-alanine (β-Ala, 2.55 ppm),
phosphocholine (PC, 3.22 ppm), phosphoethanolamine (PE, 3.22 ppm),
and glycerol (4.10, 4.30 ppm 1,3-CH; 5.22 ppm 2-CH2).
The spectra in the inset are from the thorax of dissected flies and
thus represent primarily skeletal muscle. Note the similarity of
spectra for the dissected and whole flies. The spectra shown were
normalized to TSP at each echo time and therefore do not exhibit a
T2 decay. HRMAS, high-resolution magic angle spinning; wt,
wild-type; IMCLs, intramyocellular lipids; EMCLs, extramyocellular
lipids. |
One dimensional (1D) water-suppressed spin-echo
Carr-Purcell-Meiboom-Gill (CPMG) pulse sequencing [90° − (τ − 180°
−τ)n − acquisition] (26) was performed on single flies. CPMG
is a methodological improvement of particular interest in
developing ex vivo 1D HRMAS of intact tissue samples since
it suppresses broad signals that destroy the linear baseline in
typical Free Induction Decay (FID) spectra. Thus, the CPMG proton
NMR spectra are free from the broad component that contributes to
the baseline of simple FID spectra. The CPMG sequence has also been
applied to two-dimensional sequences for the same reason.
Additional parameters for the CPMG sequence included
an inter-pulse delay of τ = 2π/ωr = 250 msec, a total
spin-echo delay of 30 msec, two total 180° cycles, 256 transients,
a spectral width of 7.2 kHz, 32,768 (32k) data points, and a 3-sec
relaxation time. A spin-echo delay of 30 msec was chosen based on
the observation that at this echo time, line broadening without
loss of signal from triglycerides was avoided. When the spin-echo
delay was increased, all the lipid signals were affected, but not
in favor of other metabolites.
In vivo 1H HRMAS NMR data
processing
MR spectra of specimens were analyzed using MestReC
software (Mestrelab Research, www.mestrec.com). A
0.5-Hz line-broadening apodization function was applied to CPMG
HRMAS 1H FIDs prior to Fourier transformation. MR
spectra were referenced with respect to TSP at δ = 0.0 ppm
(external standard), manually phased, and a Whittaker baseline
estimator was applied to subtract the broad components of the
baseline.
Quantification of metabolites from 1D
1H CPMG HRMAS spectra
For metabolite quantification from 1D 1H
CPMG HRMAS spectra, we used the highly accurate ‘external
standard’ technique. Metabolite concentrations were calculated
using MestReC software. An automated fitting routine based on the
Levenberg-Marquardt (27,28) algorithm was applied after manual
peak selection; peak positions, intensities, line widths, and
Lorentzian/Gaussian ratios were adjusted until the residual
spectrum was minimized. Metabolite concentration (mol/kg) was
calculated using the equation (29): massTSP/PMTSP
× Met(area)/TSP(area) ×
NTSP/NMet × 1/wt(sample), where
massTSP was constant (0.069 mg), PMTSP was
the molar mass of TSP (172.23 g/mol), Met signifies metabolites,
NTSP was the TSP proton number (9 1H),
NMet was the metabolite proton number, and
wt(sample) was the sample weight in mg (29).
Statistical analysis
Group data were compared with the Student’s t-test.
A P-value of 0.05 (corrected) was accepted as significant and all
P-values are reported to two significant digits. Calculations were
performed using SPSS (SPSS 12, SPSS Inc).
Results and Discussion
In the present study, we detected and quantified
lipids and small metabolites in live Drosophila using
1H HRMAS NMR at 14.1 T (Fig. 1) (30). All the flies survived the
procedure, which was completed in ~45 min per fly. Our results
confirmed our expectations in that we were able to reduce
acquisition time, and thereby achieve zero mortality. We employed a
novel in vivo HRMAS 1H NMR approach in
Drosophila to examine the hypothesis that the GST2
mutation results in insulin resistance, due to a phylogenetically
conserved pathway for the regulation of glucose and lipid
metabolism between flies and mammals (31,32).
Drosophila was utilized in this study
because, relative to other animal models, flies are inexpensive and
easy to maintain and manipulate, and they have a well-known genome
as well as numerous available mutants. These characteristics make
Drosophila an ideal genetically amenable model organism with
which to investigate the physiology of biomedical paradigms. To
this end, invertebrate Drosophila models have already
provided powerful experimental systems for muscle developmental
biology investigations (33–35), age-related decline in function
(36), such as neurodegeneration
(37) and loss of immune
(38,39) and cardiac (40) functions, and specifically,
regarding muscle degeneration, for the investigation of protein
synthesis (41,42), sarcomere integrity (43–45), apoptosis (46), mitochondrial function and
morphology (44,45,47–50), stress response (48,51), glycogen content (45), muscle function and morphology
(52,53), flight ability (54) (flight) myofiber stiffness and
power (44), and protein
modifications (55,56) and related transcriptional changes
(48,57,58). The conservation of insulin
signaling between flies and mammals (31) renders Drosophila a
particularly interesting model organism for metabolism studies. The
focus of the present study on Drosophila as a model organism
distinguishes this study from traditional metabolism experiments.
The findings of this study are supported by findings in mammals
showing evidence of insulin resistance and mitochondrial
dysfunction in mGsta4 null mice (59).
The in vivo fly spectra (see representative
spectra in Fig. 1) of this study
compare well to other published in vivo skeletal muscle
spectra (11,60,61). All of these studies have shown
high amounts of lipids in skeletal muscle, particularly
triglycerides. Other HRMAS reports involving skeletal muscle showed
spectra with more metabolites than those of the present study
(8,62). The samples and set conditions in
our experiments differed from those of prior studies in that we had
a smaller quantity of sample (0.6–1.1 mg) and we performed the
experiment with a lower spin rate, which may have limited spectral
resolution. The NMR-visible non-lipid components are expected to
contribute only a small percentage in the total signal from sample
flies, which are of extremely small size (0.7–0.8 mg total body
weight), with concomitantly low sensitivity of detection. Even
spectra from the thorax of dissected flies, which is highly
enriched in skeletal muscle, are similar to whole fly spectra
(inset of Fig. 1). Nevertheless,
we were still able to detect certain metabolites from the 1D
experiment (Fig. 1).
From a biomedical perspective, the principal finding
of our experiments was that the GST2 mutation was associated
with an accumulation of mobile lipids in muscle tissue. The
quantitative data of selected components (triglycerides) detected
in live Drosophila with HRMAS 1H NMR are
summarized in Table I. Fig. 2 (30) shows a bar graph of the amounts of
the same selected components. There was a marked and significant
increase in (CH2)n (1.33 ppm) in the mutants relative to
the wt controls. Additionally, we observed a trend towards more
CH2C-CO lipids (1.58 ppm) in the mutants (Table I).
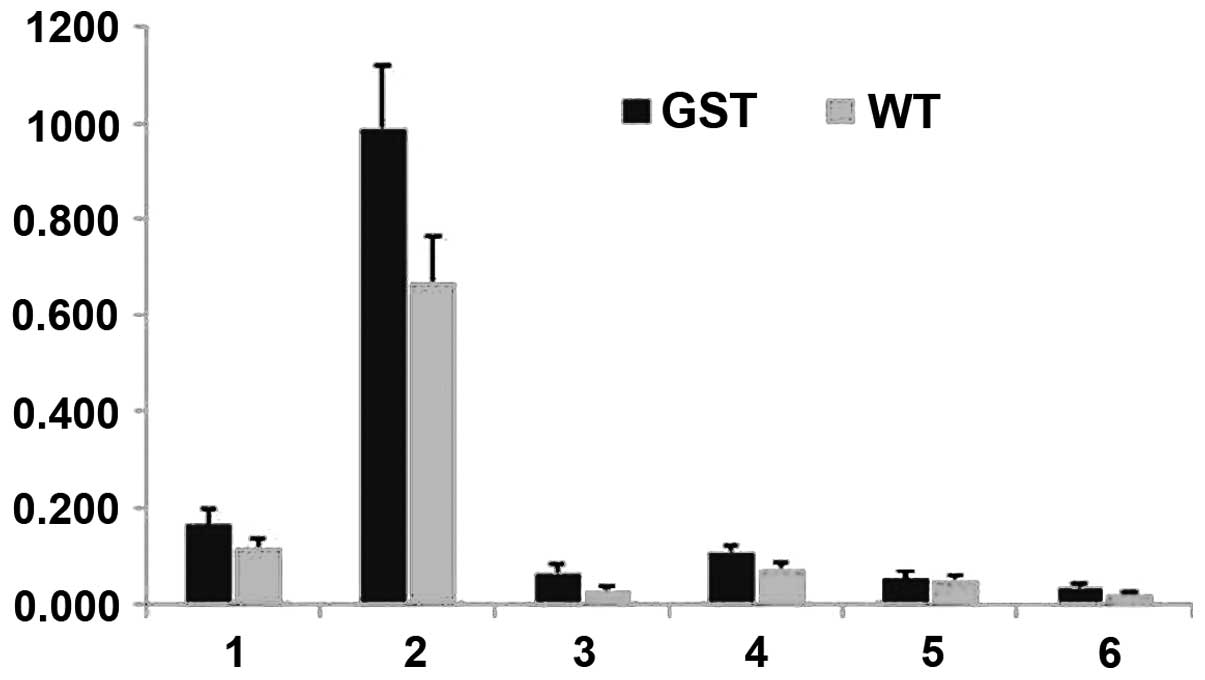 | Figure 2Lipid quantities calculated from
in vivo 1D HRMAS 1H CPMG spectra of young wt
flies (light gray) and GST2 mutant flies (black). 1,
CH3 (0.89 ppm); 2, (CH2)n (1.33 ppm) or
IMCLs; 3, CH2C-CO (1.58 ppm) or EMCLs; 4,
CH2C=C (2.02 ppm); 5, CH2C=O (2.24 ppm); 6,
CH=CH (5.33 ppm). HRMAS, high-resolution magic angle spinning; wt,
wild-type; IMCLs, intramyocellular lipids; EMCLs, extramyocellular
lipids. |
 | Table IQuantity of selected lipid components
in live Drosophila according to 1H HRMAS NMR
(n=6/group). |
Table I
Quantity of selected lipid components
in live Drosophila according to 1H HRMAS NMR
(n=6/group).
| | Mean quantity ± SE
(μmol/g) | | |
|---|
| |
| | |
|---|
| Peak no.
(ppm)a | Lipid | wt | GST2 | % difference | P-value |
|---|
| 1 (0.89) | CH3 | 0.12±0.02 | 0.17±0.03 | +41.7 | 0.1342 |
| 2 (1.33) |
(CH2)n | 0.67±0.09 | 0.99±0.13 | +47.7 | 0.0444b |
| 3 (1.58) |
CH2CCO | 0.03±0.01 | 0.07±0.02 | +133.3 | 0.0748 |
| 4 (2.02) |
CH2C=C | 0.07±0.01 | 0.11±0.01 | +57.1 | 0.0276b |
| 5 (2.24) |
CH2CO=O | 0.05±0.01 | 0.05±0.02 | 0 | 0.4676 |
| 6 (5.33) | CH=CH | 0.02±0.004 | 0.04±0.005 | +100 | 0.0251b |
Although determining the source of these accumulated
lipids is beyond the scope of this study, it should be considered
that extramyocellular lipids (EMCLs), IMCLs, and triglycerides can
all contribute to cellular lipid peaks (14,63,64). Specifically, EMCLs and IMCLs can
be distinguished by in vivo NMR by differences in their bulk
magnetic susceptibilities and geometric arrangements (65), with 1.33 ppm lipids,
(CH2)n, being attributed to IMCLs and 1.58 ppm lipids,
CH2C-CO, being attributed to EMCLs. However,
discrimination is not likely in the present study. Spinning a
sample at the magic angle (HRMAS) with respect to the static field
direction averages the second-order tensors of the anisotropic
chemical shift, the dipolar interaction, and the susceptibility
variations in heterogeneous samples (66–68). Garroway (67) indicated that MAS eliminates the
broadening effect produced by magnetic susceptibility, and
eliminates the shift itself. In their study, Chen et al
(69) clarified that,
irrespective of system geometry, MAS eliminates only the
anisotropic contribution of bulk susceptibility inside a
homogeneous susceptibility region. Inspecting the isotropic part of
the susceptibility tensors available for IMCLs and EMCLs (63,70), we can deduce that IMCLs and EMCLs
have an identical chemical shift under MAS conditions due to bulk
susceptibility.
IMCLs probably serve as an energy substrate for
oxidative metabolism (71), and
can be mobilized and utilized with turnover times in the range of
several hours (72). In insects,
triglycerides are located in body fat. Triglycerides in insect body
fat (73–75) are used for storage of both energy
and fatty acid precursors, such as transported lipids,
phospholipids (membrane structure), hydrocarbons, and wax esters
(that minimize water loss from the cuticle due to evaporation)
(76). In our study, mobility of
fat body contents may have been affected by trauma or immune
status, leading to strong IMCL and EMCL signals (77). However, this suggestion is only
hypothetical as the intracellular signaling cascade mediating
mobilization of triglycerides has not been as fully elucidated in
insects as it has in mammals (78). Nevertheless, we suggest that there
was mobilization of triglycerides in the GST2−/−
flies because the peaks indicative of triglycerides at 1.33 and
1.58 ppm were increased (79).
The significantly greater triglyceride signals (both IMCLs and
EMCLs) detected in the GST2−/− mutants vs. wt
controls (Figs. 1 and 2, Table
I) resembles the metabolic profile of akhr flies with an
obese phenotype and abnormal accumulation of both lipids and
carbohydrates (79).
Specifically, elevated IMCL levels are associated with insulin
resistance, a major metabolic dysfunction of diabetes, aging
(80,81), burn trauma (82,83) and obesity (84–87).
Moreover, our observations of increased peaks
indicative of triglycerides at 1.33 ppm in
GST2−/− flies agree with prior findings in
chico flies (79) with
disrupted insulin signaling. Chico flies have a mutated
insulin receptor substrate (IRS) gene, a Drosophila homolog
of vertebrate IRS1-4. Chico flies have a small stature and
show abnormally high triglyceride levels (88,89) that are attributable to a
dysfunctional mutated insulin signaling pathway (31), resulting in insulin resistance.
The high 1.33 ppm peak in chico flies is clearly due to
IMCLs and not to EMCLs since these flies are not obese.
Accordingly, chico flies do not exhibit significantly
increased 1.58 ppm peaks, which are frequently attributed to EMCLs
(79). Thus, despite the
theoretical considerations of HRMAS, it remains likely that the
lipids that produce the peak at 1.33 ppm are primarily IMCLs,
whereas the lipids that yield a peak at 1.58 ppm are primarily
EMCLs. Thus, chico flies are a suitable comparison strain
for GST2 flies, which also exhibit increased triglycerides,
evidently due to increased IMCLs since they are not obese, and thus
not expected to have increased EMCLs. Conversely, akhr flies
exhibit a metabolic profile with significantly increased peaks in
all assigned lipids, agrees with their obese phenotype (79).
Another principal finding of our experiments was
that peaks 4 (CH2C=C at 2.02 ppm) and 6 (CH=CH at 5.33
ppm), which includes protons from ceramide, were also significantly
increased in the mutant flies compared to wt controls (Table I and Fig. 2). Ceramide accumulation decreases
insulin-stimulated GLUT4 translocation to the plasma membrane and,
consequently, reduces glucose transport (90), resulting in insulin resistance.
Paumen and co-workers demonstrated that saturated fatty acids such
as palmitoleic acid at 2.02 ppm in our study, induce de novo
synthesis of ceramide and programmed cell death (90). They suggested that inhibition of
carnitine palmitoyltransferase I activity induces both sphingolipid
synthesis and palmitate-induced cell death. Ruddock et al
(91) suggested that long-chain
saturated fatty acids (palmitoleic acid C16:0) attenuate insulin
signal transduction in hepatoma cell lines. Their study suggests
that an increase in palmitoleic acid signifies insulin resistance.
If this is the case, then the signal at 2.02 ppm in our study may
also be a biomarker of insulin resistance; this peak was elevated
in our GST2−/− flies (CH2C=C at 2.02
ppm, peak 4 in Figs. 1 and
2) and in chico flies
(79).
From a biomedical perspective, the findings of this
study support the hypothesis that the GST2 mutation is
associated with insulin signaling and suggest that the IMCL level
may be a biomarker of insulin resistance in
GST2−/− flies. However, whether IMCLs are
directly involved in the development of insulin resistance simply
serve as an indirect marker is currently a topic of debate
(92). Insulin resistance has not
been demonstrated previously in flies with currently available
assays. Furthermore, direct links between GST2 mutation (the
Drosophila ortholog of the GSTA4 gene in mammals) and
insulin resistance, as suggested in this study, have not been made
previously. The common characteristics shared among innate immunity
activation, obesity, and insulin resistance, as recently described
(79), support the findings of
this study.
In conclusion, findings of the present study have
demonstrated that a novel solid-state HRMAS NMR method is a
sensitive tool for the molecular characterization of metabolic
perturbations in Drosophila. We observed increased levels of
triglycerides in GST2−/− Drosophila mutant
that may be indicative of insulin resistance. These findings may
thus be directly relevant to the mitochondrial dysfunction that
occurs in a wide range of metabolically disruptive conditions, such
as trauma, aging, and immune system deficiencies, that lead to
elevated susceptibility to infection. Our findings advance the
development of novel in vivo non-destructive research
approaches in Drosophila strains, offers biomarkers to
investigate biomedical paradigms, and thus may direct novel
therapeutic development.
Acknowledgements
This study was supported in part by a grant from
DM103014 of the Defense Medical Research and Development Program
(DMRDP) to Laurence G. Rahme (A. Aria Tzika, co-investigator). We
would like to thank Professor Helen Benes of the University of
Arkansas for Medical Sciences for providing the Drosophilas
used in this study. We would also like to thank Dr Ann Power Smith
(Write Science Right, Las Vegas, NV, USA) for editorial
assistance.
Abbreviations:
|
Ac
|
acetate
|
|
akhr
|
adipokinetic hormone receptor
|
|
β-Ala
|
β-alanine
|
|
CPMG
|
Carr-Purcell-Meiboom-Gill
|
|
Cho
|
choline
|
|
EMCLs
|
extramyocellular lipids
|
|
FFA
|
free fatty acids
|
|
HRMAS
|
high-resolution magic angle
spinning
|
|
IMCLs
|
intramyocellular lipids
|
|
Lip
|
lipids
|
|
PUFA
|
poly-unsaturated fatty acid
|
|
TGA
|
triglycerides
|
|
wt
|
wild-type
|
References
|
1
|
Weybright P, Millis K, Campbell N, Cory DG
and Singer S: Gradient, high-resolution, magic angle spinning
1H nuclear magnetic resonance spectroscopy of intact
cells. Magn Reson Med. 39:337–345. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Blankenberg FG, Storrs RW, Naumovski L,
Goralski T and Spielman D: Detection of apoptotic cell death by
proton nuclear magnetic resonance spectroscopy. Blood.
87:1951–1956. 1996.PubMed/NCBI
|
|
3
|
Cheng LL, Ma MJ, Becerra L, et al:
Quantitative neuropathology by high resolution magic angle spinning
proton magnetic resonance spectroscopy. Proc Natl Acad Sci USA.
94:6408–6413. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cheng LL, Newell K, Mallory AE, Hyman BT
and Gonzalez RG: Quantification of neurons in Alzheimer and control
brains with ex vivo high resolution magic angle spinning proton
magnetic resonance spectroscopy and stereology. Magn Reson Imaging.
20:527–533. 2002. View Article : Google Scholar
|
|
5
|
Millis KK, Maas WE, Cory DG and Singer S:
Gradient, high-resolution, magic-angle spinning nuclear magnetic
resonance spectroscopy of human adipocyte tissue. Magn Reson Med.
38:399–403. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Millis K, Weybright P, Campbell N, et al:
Classification of human liposarcoma and lipoma using ex vivo proton
NMR spectroscopy. Magn Reson Med. 41:257–267. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Barton SJ, Howe FA, Tomlins AM, et al:
Comparison of in vivo 1H MRS of human brain tumours with
1H HR-MAS spectroscopy of intact biopsy samples in
vitro. MAGMA. 8:121–128. 1999.
|
|
8
|
Griffin JL, Williams HJ, Sang E and
Nicholson JK: Abnormal lipid profile of dystrophic cardiac tissue
as demonstrated by one- and two-dimensional magic-angle spinning
(1)H NMR spectroscopy. Magn Reson Med. 46:249–255. 2001. View Article : Google Scholar
|
|
9
|
Tzika AA, Cheng LL, Goumnerova L, et al:
Biochemical characterization of pediatric brain tumors by using in
vivo and ex vivo magnetic resonance spectroscopy. J Neurosurg.
96:1023–1031. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tugnoli V, Schenetti L, Mucci A, et al:
Ex vivo HR-MAS MRS of human meningiomas: a comparison with
in vivo 1H MR spectra. Int J Mol Med. 18:859–869.
2006.
|
|
11
|
Astrakas LG, Goljer I, Yasuhara S, et al:
Proton NMR spectroscopy shows lipids accumulate in skeletal muscle
in response to burn trauma-induced apoptosis. FASEB J.
19:1431–1440. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tzika AA, Astrakas LG, Cao H, et al:
Murine intramyocellular lipids quantified by NMR act as metabolic
biomarkers in burn trauma. Int J Mol Med. 21:825–832.
2008.PubMed/NCBI
|
|
13
|
Bollard ME, Garrod S, Holmes E, et al:
High-resolution (1)H and (1)H-(13)C magic angle spinning NMR
spectroscopy of rat liver. Magn Reson Med. 44:201–207. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Szczepaniak LS, Babcock EE, Schick F, et
al: Measurement of intracellular triglyceride stores by H
spectroscopy: validation in vivo. Am J Physiol. 276:E977–E989.
1999.PubMed/NCBI
|
|
15
|
van der Graaf M, Tack CJ, de Haan JH,
Klomp DW and Heerschap A: Magnetic resonance spectroscopy shows an
inverse correlation between intramyocellular lipid content in human
calf muscle and local glycogen synthesis rate. NMR Biomed.
23:133–141. 2009.
|
|
16
|
Jacob S, Machann J, Rett K, et al:
Association of increased intramyocellular lipid content with
insulin resistance in lean nondiabetic offspring of type 2 diabetic
subjects. Diabetes. 48:1113–1119. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Petersen KF, Befroy D, Dufour S, et al:
Mitochondrial dysfunction in the elderly: possible role in insulin
resistance. Science. 300:1140–1142. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Feala JD, Coquin L, McCulloch AD and
Paternostro G: Flexibility in energy metabolism supports hypoxia
tolerance in Drosophila flight muscle: metabolomic and
computational systems analysis. Mol Syst Biol. 3:992007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pedersen KS, Kristensen TN, Loeschcke V,
et al: Metabolomic signatures of inbreeding at benign and stressful
temperatures in Drosophila melanogaster. Genetics.
180:1233–1243. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bharucha KN: The epicurean fly: using
Drosophila melanogaster to study metabolism. Pediatr Res.
65:132–137. 2009.PubMed/NCBI
|
|
21
|
Null B, Liu CW, Hedehus M, Conolly S and
Davis RW: High-resolution, in vivo magnetic resonance imaging of
Drosophila at 18.8 Tesla. PLoS One. 3:e28172008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Righi V, Apidianakis Y, Rahme LG and Tzika
AA: Magnetic resonance spectroscopy of live Drosophila
melanogaster using magic angle spinning. J Vis Exp.
38:17102010.
|
|
23
|
Baker KD and Thummel CS: Diabetic larvae
and obese flies-emerging studies of metabolism in
Drosophila. Cell Metab. 6:257–266. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Leopold P and Perrimon N:
Drosophila and the genetics of the internal milieu. Nature.
450:186–188. 2007. View Article : Google Scholar
|
|
25
|
Singh SP, Coronella JA, Benes H, Cochrane
BJ and Zimniak P: Catalytic function of Drosophila
melanogaster glutathione S-transferase DmGSTS1–1 (GST-2) in
conjugation of lipid peroxidation end products. Eur J Biochem.
268:2912–2923. 2001.PubMed/NCBI
|
|
26
|
Meiboom S and Gill D: Modified spiin-echo
method for measuring nuclear relaxation time. Rev Sci Instrum.
29:688–691. 1958. View Article : Google Scholar
|
|
27
|
Levenberg K: A method for the solution of
certain non-linear problems in least squares. Q Appl Math.
2:164–168. 1944.
|
|
28
|
Marquardt D: An algorithm for
least-squares estimation of nonlinear parameters. SIAM J Appl Math.
11:431–441. 1963. View Article : Google Scholar
|
|
29
|
Swanson MG, Zektzer AS, Tabatabai ZL, et
al: Quantitative analysis of prostate metabolites using
1H HR-MAS spectroscopy. Magn Reson Med. 55:1257–1264.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Righi V, Apidianakis Y, Psychogios N,
Rahme LG, Tompkins RG and Tzika AA: In vivo high-resolution magic
angle spinning proton NMR spectroscopy of Drosophila
melanogaster flies as a model system to investigate
mitochondrial dysfunction in Drosophila mutants. Intl Soc
Mag Reson Med. 1460:192011.
|
|
31
|
Garofalo RS: Genetic analysis of insulin
signaling in Drosophila. Trends Endocrinol Metab.
13:156–162. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Saltiel AR and Kahn CR: Insulin signalling
and the regulation of glucose and lipid metabolism. Nature.
414:799–806. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Abmayr SM, Zhuang S and Geisbrecht ER:
Myoblast fusion in Drosophila. Methods Mol Biol. 475:75–97.
2008. View Article : Google Scholar
|
|
34
|
Richardson B, Beckett K and Baylies M:
Visualizing new dimensions in Drosophila myoblast fusion.
Bioessays. 30:423–431. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rochlin K, Yu S, Roy S and Baylies MK:
Myoblast fusion: when it takes more to make one. Dev Biol.
341:66–83. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Partridge L and Tower J: Yeast, a feast:
The fruit fly Drosophila as a model organism for research
into aging. The Molecular Biology of Aging. Guarente L and
Partridge L: Cold Spring Harbor Laboratory Press; pp. 267–308.
2008
|
|
37
|
Marsh JL and Thompson LM:
Drosophila in the study of neurodegenerative disease.
Neuron. 52:169–178. 2006. View Article : Google Scholar
|
|
38
|
Ramsden S, Cheung YY and Seroude L:
Functional analysis of the Drosophila immune response during
aging. Aging Cell. 7:225–236. 2008.
|
|
39
|
Zerofsky M, Harel E, Silverman N and Tatar
M: Aging of the innate immune response in Drosophila
melanogaster. Aging Cell. 4:103–108. 2005.PubMed/NCBI
|
|
40
|
Ocorr K, Akasaka T and Bodmer R:
Age-related cardiac disease model of Drosophila. Mech Ageing
Dev. 128:112–116. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Smith JM, Bozcuk AN and Tebbutt S: Protein
turnover in adult Drosophila. J Insect Physiol. 16:601–613.
1970. View Article : Google Scholar
|
|
42
|
Webster GC, Beachell VT and Webster SL:
Differential decrease in protein synthesis by microsomes from aging
Drosophila melanogaster. Exp Gerontol. 15:495–497. 1980.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Gartner LP: Aging and the visceral
musculature of the adult fruitfly: an ultrastructural
investigation. Trans Am Microsc Soc. 96:48–55. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Miller MS, Lekkas P, Braddock JM, et al:
Aging enhances indirect flight muscle fiber performance yet
decreases flight ability in Drosophila. Biophys J.
95:2391–2401. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Takahashi A, Philpott DE and Miquel J:
Electron microscope studies on aging Drosophila
melanogaster. 3 Flight muscle. J Gerontol. 25:222–228. 1970.
View Article : Google Scholar
|
|
46
|
Zheng J, Edelman SW, Tharmarajah G, Walker
DW, Pletcher SD and Seroude L: Differential patterns of apoptosis
in response to aging in Drosophila. Proc Natl Acad Sci USA.
102:12083–12088. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ferguson M, Mockett RJ, Shen Y, Orr WC and
Sohal RS: Age-associated decline in mitochondrial respiration and
electron transport in Drosophila melanogaster. Biochem J.
390:501–511. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Girardot F, Lasbleiz C, Monnier V and
Tricoire H: Specific age-related signatures in Drosophila
body parts transcriptome. BMC Genomics. 7:692006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Magwere T, Goodall S, Skepper J, Mair W,
Brand MD and Partridge L: The effect of dietary restriction on
mitochondrial protein density and flight muscle mitochondrial
morphology in Drosophila. J Gerontol A Biol Sci Med Sci.
61:36–47. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sohal RS, Sohal BH and Orr WC:
Mitochondrial superoxide and hydrogen peroxide generation, protein
oxidative damage, and longevity in different species of flies. Free
Radic Biol Med. 19:499–504. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Goddeeris MM, Cook-Wiens E, Horton WJ, et
al: Delayed behavioural aging and altered mortality in
Drosophila beta integrin mutants. Aging Cell. 2:257–264.
2003.PubMed/NCBI
|
|
52
|
Miller BM, Zhang S, Suggs JA, et al: An
alternative domain near the nucleotide-binding site of
Drosophila muscle myosin affects ATPase kinetics. J Mol
Biol. 353:14–25. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kronert WA, Dambacher CM, Knowles AF,
Swank DM and Bernstein SI: Alternative relay domains of
Drosophila melanogaster myosin differentially affect ATPase
activity, in vitro motility, myofibril structure and muscle
function. J Mol Biol. 379:443–456. 2008.PubMed/NCBI
|
|
54
|
Kronert WA, Melkani GC, Melkani A and
Bernstein SI: Mutating the converter-relay interface of
Drosophila myosin perturbs ATPase activity, actin motility,
myofibril stability and flight ability. J Mol Biol. 398:625–632.
2010.PubMed/NCBI
|
|
55
|
Das N, Levine RL, Orr WC and Sohal RS:
Selectivity of protein oxidative damage during aging in
Drosophila melanogaster. Biochem J. 360:209–216. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Toroser D, Orr WC and Sohal RS:
Carbonylation of mitochondrial proteins in Drosophila
melanogaster during aging. Biochem Biophys Res Commun.
363:418–424. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wheeler JC, Bieschke ET and Tower J:
Muscle-specific expression of Drosophila hsp70 in response
to aging and oxidative stress. Proc Natl Acad Sci USA.
92:10408–10412. 1995.
|
|
58
|
Zhan M, Yamaza H, Sun Y, Sinclair J, Li H
and Zou S: Temporal and spatial transcriptional profiles of aging
in Drosophila melanogaster. Genome Res. 17:1236–1243. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Singh SP, Niemczyk M, Saini D, Awasthi YC,
Zimniak L and Zimniak P: Role of the electrophilic lipid
peroxidation product 4-hydroxynonenal in the development and
maintenance of obesity in mice. Biochemistry. 47:3900–3911. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Weis J, Johansson L, Ortiz-Nieto F and
Ahlstrom H: Assessment of lipids in skeletal muscle by LCModel and
AMARES. J Magn Reson Imaging. 30:1124–1129. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang L, Salibi N, Wu Y, Schweitzer ME and
Regatte RR: Relaxation times of skeletal muscle metabolites at 7T.
J Magn Reson Imaging. 29:1457–1464. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chen JH, Sambol EB, Decarolis P, et al:
High-resolution MAS NMR spectroscopy detection of the spin
magnetization exchange by cross-relaxation and chemical exchange in
intact cell lines and human tissue specimens. Magn Reson Med.
55:1246–1256. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Boesch C, Slotboom J, Hoppeler H and Kreis
R: In vivo determination of intra-myocellular lipids in human
muscle by means of localized 1H-MR-spectroscopy. Magn
Reson Med. 37:484–493. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Vermathen P, Kreis R and Boesch C:
Distribution of intramyocellular lipids in human calf muscles as
determined by MR spectroscopic imaging. Magn Reson Med. 51:253–262.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Havel RJ, Carlson LA, Ekelund LG and
Holmgren A: Turnover rate and oxidation of different free fatty
acids in man during exercise. J Appl Physiol. 19:613–618.
1964.PubMed/NCBI
|
|
66
|
Mehring M: High Resolution NMR in Solids.
Springer-Verlag; New York: 1982
|
|
67
|
Garroway AN: Magic-angle sample spinning
of liquids. J Magn Reson. 49:168–171. 1982.
|
|
68
|
Barbara TM: Cylindrical demagnetization
fields and microprobe design in high resolution NMR. J Magn Reson
A. 109:2651994. View Article : Google Scholar
|
|
69
|
Chen JH, Enloe BM, Xiao Y, Cory DG and
Singer S: Isotropic susceptibility shift under MAS: the origin of
the split water resonances in 1H MAS NMR spectra of cell
suspensions. Magn Reson Med. 50:515–521. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Chu SC, Xu Y, Balschi JA and Springer CS
Jr: Bulk magnetic susceptibility shifts in NMR studies of
compartmentalized samples: use of paramagnetic reagents. Magn Reson
Med. 13:239–262. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Kayar SR, Hoppeler H, Howald H, Claassen H
and Oberholzer F: Acute effects of endurance exercise on
mitochondrial distribution and skeletal muscle morphology. Eur J
Appl Physiol Occup Physiol. 54:578–584. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Canavoso LE, Jouni ZE, Karnas KJ,
Pennington JE and Wells MA: Fat metabolism in insects. Annu Rev
Nutr. 21:23–46. 2001. View Article : Google Scholar
|
|
73
|
Gilby AR: Lipids and their metabolism in
insects. Annu Rev Entomol. 10:141–160. 1965. View Article : Google Scholar
|
|
74
|
Fast PG: A comparative study of the
phospholipids and fatty acids of some insect lipids. Science.
155:1680–1681. 1967.
|
|
75
|
Stanley-Samuelson DW, Jurenka RA, Cripps
C, Blomquist GJ and deRenobales M: Fatty acids in insects:
composition, metabolism, and biological significance. Arch Insect
Biochem Physiol. 9:1–33. 1988. View Article : Google Scholar
|
|
76
|
Horne I, Haritos VS and Oakeshott JG:
Comparative and functional genomics of lipases in holometabolous
insects. Insect Biochem Mol Biol. 39:547–567. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Patel RT, Soulages JL, Hariharasundaram B
and Arrese EL: Activation of the lipid droplet controls the rate of
lipolysis of triglycerides in the insect fat body. J Biol Chem.
280:22624–22631. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Bharucha KN, Tarr P and Zipursky SL: A
glucagon-like endocrine pathway in Drosophila modulates both
lipid and carbohydrate homeostasis. J Exp Biol. 211:3103–3110.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Righi V, Apidianakis Y, Mintzopoulos D,
Astrakas L, Rahme LG and Tzika AA: In vivo high-resolution
magic angle spinning magnetic resonance spectroscopy of
Drosophila melanogaster at 14.1 T shows trauma in aging and
in innate immune-deficiency is linked to reduced insulin signaling.
Int J Mol Med. 26:175–184. 2010.
|
|
80
|
Machann J, Thamer C, Schnoedt B, et al:
Age and gender related effects on adipose tissue compartments of
subjects with increased risk for type 2 diabetes: a whole body
MRI/MRS study. MAGMA. 18:128–137. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Nakagawa Y, Hattori M, Harada K, Shirase
R, Bando M and Okano G: Age-related changes in intramyocellular
lipid in humans by in vivo H-MR spectroscopy. Gerontology.
53:218–223. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Muller MJ and Herndon DN: The challenge of
burns. Lancet. 343:216–220. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Ikezu T, Okamoto T, Yonezawa K, Tompkins
RG and Martyn JA: Analysis of thermal injury-induced insulin
resistance in rodents. Implication of postreceptor mechanisms. J
Biol Chem. 272:25289–25295. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Sinha R, Dufour S, Petersen KF, et al:
Assessment of skeletal muscle triglyceride content by (1)H nuclear
magnetic resonance spectroscopy in lean and obese adolescents:
relationships to insulin sensitivity, total body fat, and central
adiposity. Diabetes. 51:1022–1027. 2002. View Article : Google Scholar
|
|
85
|
Schrauwen-Hinderling VB, Hesselink MK,
Schrauwen P and Kooi ME: Intramyocellular lipid content in human
skeletal muscle. Obesity (Silver Spring). 14:357–367. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Consitt LA, Bell JA and Houmard JA:
Intramuscular lipid metabolism, insulin action, and obesity. IUBMB
Life. 61:47–55. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Johnson AB, Argyraki M, Thow JC, Cooper
BG, Fulcher G and Taylor R: Effect of increased free fatty acid
supply on glucose metabolism and skeletal muscle glycogen synthase
activity in normal man. Clin Sci (Lond). 82:219–226.
1992.PubMed/NCBI
|
|
88
|
Clancy DJ, Gems D, Harshman LG, et al:
Extension of life-span by loss of CHICO, a Drosophila
insulin receptor substrate protein. Science. 292:104–106. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Bohni R, Riesgo-Escovar J, Oldham S, et
al: Autonomous control of cell and organ size by CHICO, a
Drosophila homolog of vertebrate IRS1–4. Cell. 97:865–875.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Paumen MB, Ishida Y, Muramatsu M, Yamamoto
M and Honjo T: Inhibition of carnitine palmitoyltransferase I
augments sphingolipid synthesis and palmitate-induced apoptosis. J
Biol Chem. 272:3324–3329. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Ruddock MW, Stein A, Landaker E, et al:
Saturated fatty acids inhibit hepatic insulin action by modulating
insulin receptor expression and post-receptor signalling. J
Biochem. 144:599–607. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Fernandez-Real JM and Pickup JC: Innate
immunity, insulin resistance and type 2 diabetes. Trends Endocrinol
Metab. 19:10–16. 2008. View Article : Google Scholar
|
















