Introduction
Gastric carcinoma (GC) is the fourth most prevalent
type of cancer and is the second highest contributor to global
cancer-related mortality (1). GC
remains the second most common cause of cancer-associated mortality
and health problems in China, despite the lower incidence of GC
observed over the past several years (2). Although gastric cancer can be
surgically excised and inhibited by chemotherapy, gastric tumor
cells continue to grow and metastasize elsewhere in the body, which
leads to a poor prognosis. Uncontrolled tumor cell proliferation
and migration are two important features of GC. Therefore,
inhibition of cancer cell proliferation and migration, and
promotion of cancer cell apoptosis are promising therapeutic
targets for GC. Investigation of the pathological characteristics
of GC and associated mechanisms will provide novel insights for the
discovery of potential therapeutic targets.
C1q and tumor necrosis factor superfamily are
involved in several biological processes, including inflammation,
apoptosis and cell differentiation. C1q/tumor necrosis
factor-related proteins (CTRP proteins) have been revealed to serve
a role in carcinogenesis and cancer progression (3). The C1q/TNF-related protein family is
comprised of 16 CTRP members, CTRP1-9, 9B, 10-15. Among these,
CTRP3, CTRP4 and C1QTNF6 have been revealed to be associated with
tumor promotion. All CTRP members are secreted proteins, and are
widely expressed in various tissues and cell types (4-8).
CTRP4 was demonstrated to function as a tumor-promoting
inflammatory regulator, and to promote tumor cell survival and
reduce drug-induced apoptosis (3). These findings strongly suggested
that CTRP4 is a potential therapeutic target. CTRP8 was reported to
be involved in brain cancer (9).
It was also demonstrated to enhance motility and matrix invasion by
human glioblastoma cells (10).
CTRP8-induced migration of human glioma cells was revealed to be
inhibited by a small competitor peptide derived from C1QTNF6
(11). Western blotting
experiments have demonstrated that C1QTNF6 is highly expressed in
human hepatocellular carcinoma tissues. An immunohistochemistry
assay indicated that C1QTNF6 is mainly localized in hepato-cellular
carcinoma cells and endothelial cells in tumor tissues. High
expression of C1QTNF6 was revealed to activate the Akt signaling
pathway, increase tumor angiogenesis and reduce the necrosis of
HepG2 cells (12).
C1QTNF6-interference was revealed to inhibit the Erk1/2 signaling
pathway in 3T3-L1 adipocytes (13). C1QTNF6 was also revealed to serve
as an endogenous complement regulator that exhibited a prominent
therapeutic effect in arthritis (14). It was also demonstrated that
C1QTNF6 inhibited fibrogenesis by TGF-β1-stimulated human dermal
fibroblasts (15).
The present study investigated the expression and
localization of C1QTNF6 in GC specimens. Furthermore, the
pathological functions of C1QTNF6 in GC, including cell growth,
proliferation, cycle, migration and apoptosis, were studied by
transfection of AGS cells with lentivirus expressing
siRNA-C1QTNF6.
Materials and methods
Cell culture and regents
All cell lines were purchased from American Type
Culture Collection (American Type Culture Collection, Manassas, VA,
USA). SGC-7901, AGS, BGC-823 and MGC-803 cells were cultured in
Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), supplemented with 10% fetal
bovine serum (FBS) and 1% penicillin/streptomycin in 5%
CO2 at 37°C. Cell growth was monitored using Celigo
instrument (Nexcelom Corporation, Lawrence, MA, USA). The siRNA
targeting C1QTNF6 (target sequence site: TGT GTG AGA TCC CTA TGG T)
was purchased from Shanghai GeneChem (Shanghai, China). SYBR-Green
quantitative polymerase chain reaction (qPCR) master mix was
obtained from Takara Biotechnology Co., Ltd., Dalian, China (cat.
no. DRR041B). BrDU kit was obtained from Roche Diagnostics, Basel,
Switzerland. FITC-Annexin V apoptosis detection kit was purchased
from eBioscience; Thermo Fisher Scientific, Inc. (cat. no.
88-8007).
Patient selection
A total of 64 patients (34 males and 18 females)
aged between 27 and 85 years (mean, 62.8 years) undergoing surgical
resection of primary GC between March 2012 and June 2015 at the
Qingdao Municipal Hospital (Qingdao, China) were selected for the
present study. Gene expression profiles were generated from gastric
biopsy specimens of 12 patients using GeneChip. Immunohistochemical
analysis for C1QTNF6 was performed on the tumors and non-neoplastic
gastric mucosal specimens (located >2 cm away from the margins
of the tumor) obtained from the other 52 patients. None of the
patients received chemotherapy or radiation therapy prior to
surgery. The pathological diagnosis was made by 2 pathologists
based on the depth of infiltration, differentiation, lymph node
metastasis, microvascular invasion and nerve invasion (Table I). The Institutional Review Board
at the Qingdao Municipal hospital approved the study protocol, and
all patients provided written informed consent.
 | Table IPathological characteristics of
gastric carcinoma (n=52). |
Table I
Pathological characteristics of
gastric carcinoma (n=52).
| Pathological
characteristic | Number | % |
|---|
|
Differentiation | | |
| Well | 2 | 3.8 |
| Moderate | 11 | 21.2 |
| Poor | 39 | 75 |
| Depth | | |
| Mucosa and
submucosa | 11 | 21.1 |
| Muscularis | 7 | 13.5 |
| Serosa | 34 | 65.4 |
| Lymph node
metastasis | | |
| (+) | 38 | 73.1 |
| (−) | 14 | 26.9 |
| Micro vessel
invasion | | |
| (+) | 30 | 57.7 |
| (−) | 22 | 42.3 |
| Nerve invasion | | |
| (+) | 30 | 57.7 |
| (−) | 22 | 42.3 |
Gene microarray analysis
Total RNA samples were extracted from tumor and
peri-tumoral normal gastric tissues using TRIzol reagent (Thermo
Fisher Scientific, Inc.). RNA microarray was conducted by Shanghai
GeneChem. In brief, the RNA was reverse transcribed at 37°C for 60
min and then at 95°C for 5 min using the GeneChip IVT Express kit
(Agilent Technologies, Inc., Santa Clara, CA, USA). The product
from each reverse transcription reaction was pre-amplified and then
the RNA expression was profiled using human GeneChip primeview
array (Affymetrix; Thermo Fisher Scientific, Inc.), according to
the manufacturer’s protocol.
Reverse transcription (RT)-qPCR
analysis
Total RNA was extracted from cells and tissues using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer’s protocol. The RNA was reverse
transcribed at 42°C for 60 min using the SYBR-Green master mix in a
20-μl reaction. The primer sequences were as follows
(designed and synthesized by Shanghai GeneChem): GAPDH forward,
5′-TGA CTT CAA CAG CGA CAC CCA-3′ and reverse, 5′-CAC CCT GTT GCT
GTA GCC AAA-3′; and C1QTNF6 forward, 5′-GAA AGG GTC TTT GTG AAC CTT
GA-3′ and reverse, 5′-CTG CGC GTA CAG GAT GAC AG-3′. The qPCR
reaction cycle was performed using SYBR green master mix as
follows: 10 min at 95°C and 40 cycles of 15 sec at 95°C and 60 sec
at 60°C. Quantification for each sample was performed using the
2-∆∆Cq method (16).
Immunohistochemistry (IHC)
Following the tissues being fixed in 10% formalin
for 24 h at room temperature and embedded in paraffin, sections
from tumor and peri-tumoral normal gastric tissues were
deparaffinized in xylene (15 min at room temperature, twice),
rehydrated in a descending alcohol series (100%, 5 min twice; 95,
85 and 75%, 2 min each), and washed in distilled water. Prior to
antigen retrieval, the tissues were treated with sodium citrate
buffer (pH 6.0) at 100°C for 20 min, and endogenous hydrogen
peroxidase activity was inhibited by 0.3% hydrogen peroxide.
Following microwave antigen retrieval at 95-100°C, the sections
were incubated with 5% FBS for 10 min at room temperature to
inhibit non-specific staining. Next, the slides were incubated
overnight with anti-C1QTNF6 antibody (1:1,000; ab36900; Abcam,
Cambridge, UK) in a moist chamber at 4°C, followed by incubating
with anti-rabbit immunoglobulin G as secondary antibody (1:2,000;
ab205718; Abcam) at room temperature for 1 h. The antigen was
visualized using a light microscope (magnification, ×200 or
×400).
Construction of AGS si-C1QTNF6 cell
line
AGS cell lines stably expressing siRNAs were
generated by transduction of 5 μg of GV115-si-C1QTNF6 or
empty vector (Shanghai GeneChem) in a 100 ul transfection mix and
10 μl lipo-fectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) to 1 ml of medium without serum. At 6 h following
transfection, full medium was used for cell culture. Stable
knockdown of the gene was determined by RT-qPCR.
Cell cycle assay
Cells were plated in 6-cm culture plates and grown
to 80% confluence. Subsequently, the cells were harvested by
centrifugation at 179 x g for 5 min, washed twice with PBS, fixed
overnight with 75% cold ethanol at 4°C and stained with 0.05 mg/ml
propidium iodide (PI) solution (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) containing 2.5 μg/ml RNase A for 40 min
at 37°C. The cell cycle distribution was detected using a FACScan
flow cytometer (EMD Millipore, Billerica, MA, USA) and the data was
analyzed with FlowJo software 10.5.2 (Tree Star, Inc., Ashland, OR,
USA).
Wound healing assay
Cells (3×104) were seeded onto 96-well
plates and grown to 90% confluence. Next, the cell culture DMEM
medium was changed to DMEM containing 0.5% FBS, and cells were
scratched with a 96 Wounding Replicator (V&P Scientific, Inc.,
San Diego, CA, USA; cat. no. VP408FH) to create a mechanical wound.
Images were obtained at 0, 8 and 24 h using a phase-contrast
microscope (magnification, ×100). The distance of wound closure was
calculated and the associated P-values calculated.
Transwell chamber invasion assay
Transwell chamber invasion assay was conducted using
a Transwell kit (Corning Incorporated, Corning, NY, USA), as
previously described (17). In
brief, following infection with shRNA lentivirus for 3 days, the
cells were harvested and suspended at a density of 1×105
cells/ml in serum-free DMEM medium. A total of 100 μl cells
were placed in the upper Matrigel-coated chambers, and 600
μl DMEM medium containing 30% FBS was placed in the lower
chambers. Additionally, cell suspension containing 5,000 cells was
seeded into 96-well plates, and was subsequently used for
measurement of OD570. After 24-h incubation at 37°C, the
non-invaded cells were removed using a cotton swab, while the
invaded cells were stained with 2-3 drops of Giemsa solution for
3-5 min at room temperature. The invaded cells on the lower surface
of the membrane filter were counted under an inverted microscope
(magnification, ×100). The data are presented as the mean number of
migratory cells per field.
Colony assay
Cells (400-1,000 cells/well) were seeded into 6-well
plates with three replicates. A total of 14 days after seeding, the
cells were washed with PBS and fixed with 4% paraformaldehyde for
30-60 min at room temperature. The fixed cells were stained with
Giemsa dyes for 10-20 min at room temperature. The colonies were
visualized and counted using an inverted microscope (magnification,
×100; Shanghai Caikon Optical Instrument Co., Ltd., Shanghai,
China; cat. no. XDS-100).
Proliferation assay
Cell proliferation was assessed by MTT assay or with
the use of Roche BrDU kit (cat. no. 11647229001), according to the
manufacturer’s protocols. For the MTT assay, 2,000 cells were
seeded into 96-well plates. On the next day, 5 mg/ml MTT solution
(GenView SA, Lausanne, Switzerland; cat. no. JT343) was added to
each well. Four hours later, 100 μl DMSO was added to
dissolve formazan particles, with votex for 2-5 min. OD490/570 of
each well was read using a microplate spectrophotometer (Tecan
Group Ltd., Mannedorf, Switzerland; cat. no. M2009PR). For the BrDU
assay, cells were seeded into 96-well plates, and the OD450 of each
well was recorded.
Apoptosis assay
Flow cytometry was used to detect the apoptotic
rate. AGS cells (5×105) cells/well with or without
siC1QTNF6 lentivirus infection were plated into 6-well plates and
grown to 90% confluence. Following incubation, the AGS cells were
harvested by centrifugation at 179 x g for 5 min and incubated with
10 μl Annexin V-APC for 10-15 min. Next, the cells were
analyzed using a flow cytometer (EMD Millipore) and FlowJo software
10.5.2.
Statistical analysis
Fisher’s exact test and Student’s t-test were used
to analyze the relative expression of C1QTNF6 protein in human
gastric carcinoma and normal tissues. Two-way analysis of variance
with a post hoc Bonferroni’s test was used to analyze the relative
expression of C1QTNF6 in SGC-7901, AGS, BGC-823 and MGC-803 cell
lines, and cell proliferation, migration, invasion and apoptosis.
Data are presented as the mean ± standard error of the mean (n=3).
All these experiments were conducted in triplicate. P<0.05 was
considered to indicate a statistically significant difference.
Results were analyzed using GraphPad 5.0 (GraphPad Software, Inc.,
La Jolla, CA, USA).
Results
C1QTNF6 gene expression was upregulated
in GC tissues compared with that in peritumoral normal gastric
tissues
To investigate the alterations of mRNAs, total RNA
samples were isolated from tumor and the peritumoral normal gastric
tissues to conduct microarray analysis. Of the ~49,395 detectable
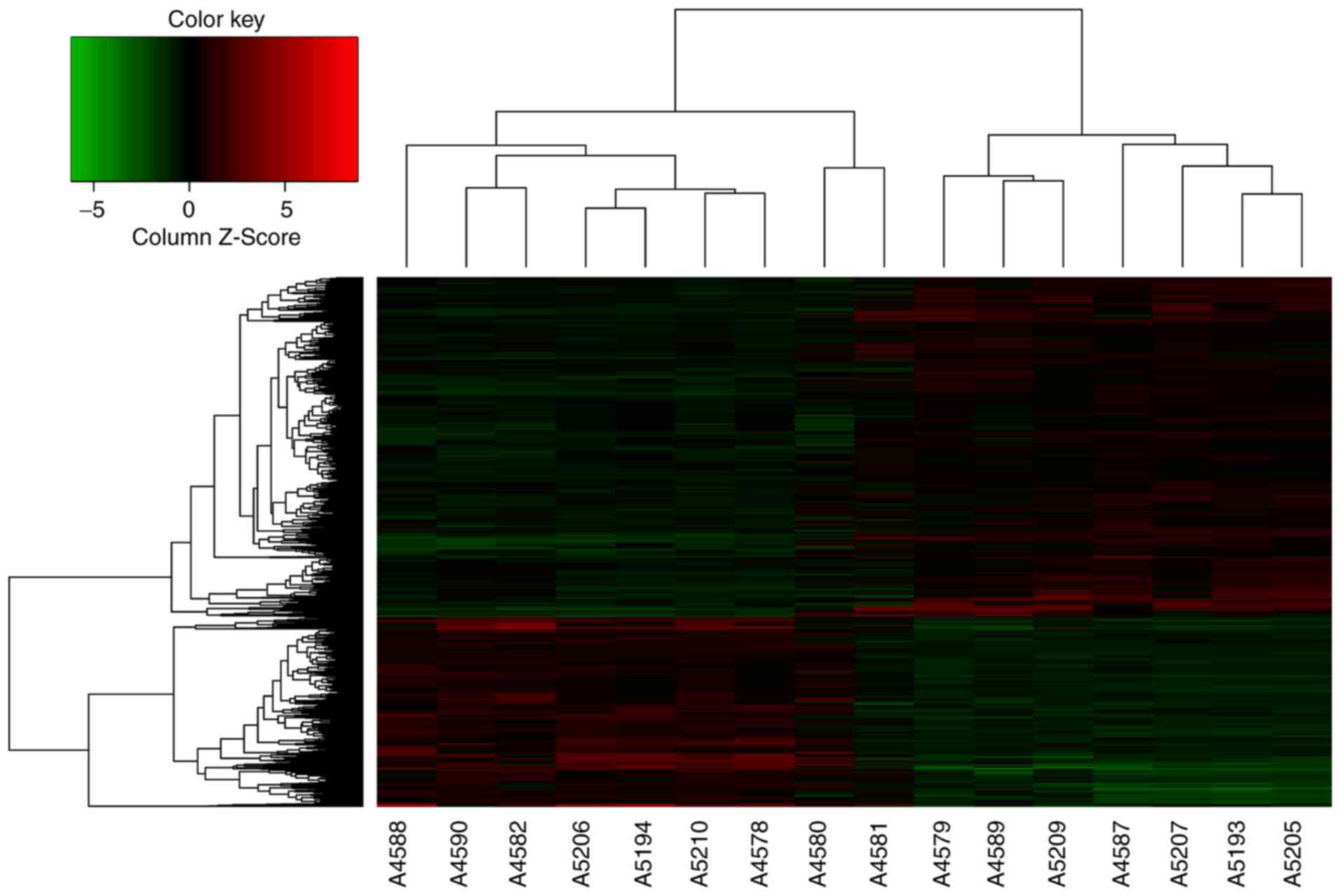
mRNAs, the expression of 238 genes had changed by >5 times. The
microarray measurements are also displayed as a heat-map matrix
(Fig. 1). Compared with the
peritu-moral normal gastric tissues, the expression of 109 mRNAs
was upregulated, while the expression of 129 mRNAs was
downregulated in GC tissues. C1QTNF6 was significantly upregulated
by 1.59-fold in GC tissues. The influence of the C1QTNF6 gene on GC
cell proliferation, migration and invasion in vitro was
investigated in the present study.
C1QTNF6 was highly expressed in GC
Next, the present study aimed to determine the
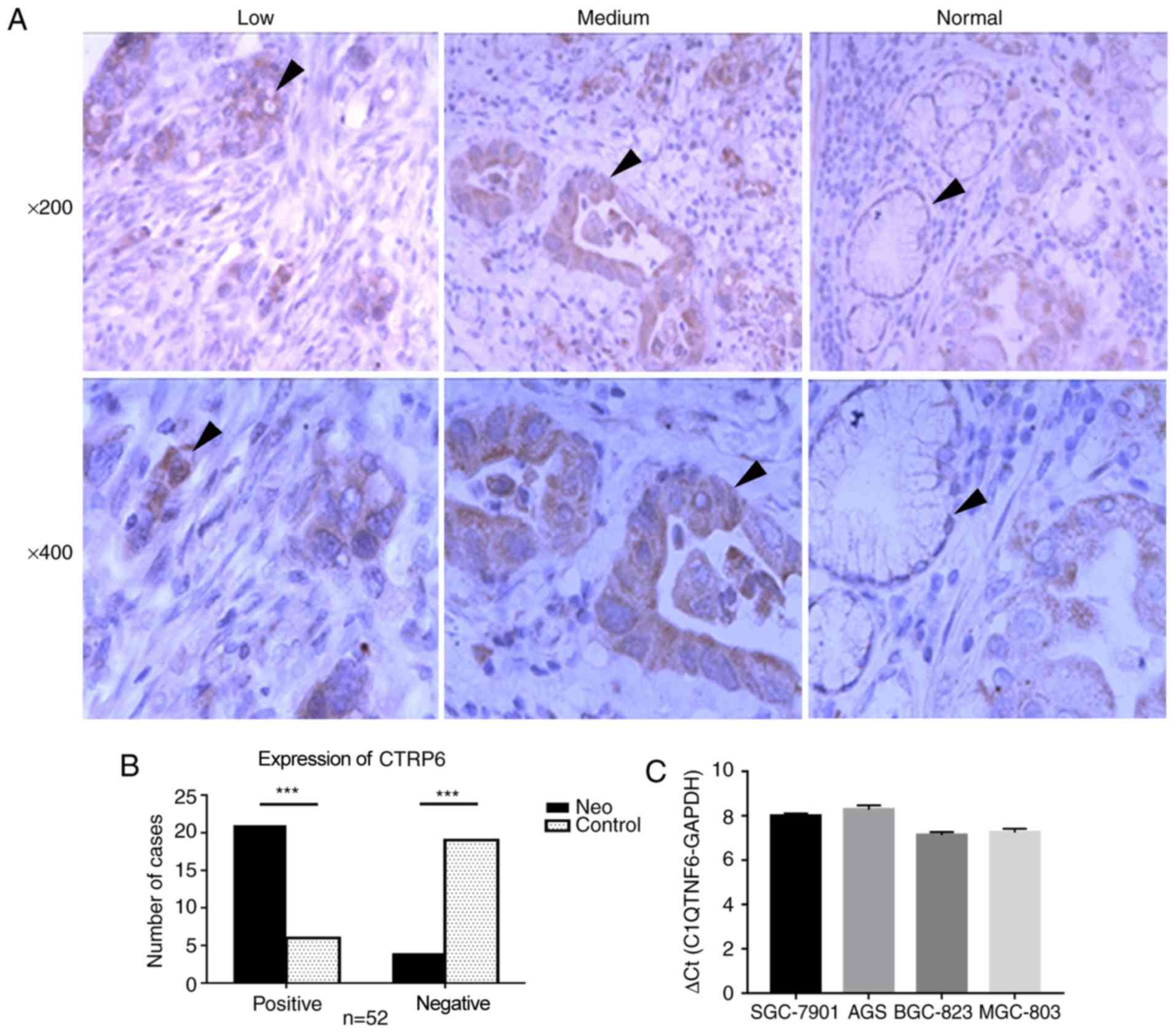
expression status of C1QTNF6 in GC. Initially, the protein
expression of C1QTNF6 in 52 primary GC samples and paratumoral
normal gastric tissues was examined by IHC (Fig. 2A). Notably, the protein expression
of C1QTNF6 was observed among GC tissues with different degrees of
differentiation, including in poorly- and moderately-differentiated
adenocarcinoma tissues. C1QTNF6 was primarily expressed in the
cytoplasm and exhibited a diffuse granular distribution. The
expression rate of C1QTNF6 [80.7% (42/52)] in GC tissues was
significantly higher than that in the normal gastric tissues [23.1%
(12/52); P<0.05; Fig. 2B].
There was no significant difference in C1QTNF6 expression between
GC samples with different pathological grades, depth of
infiltration, lymph node metastasis, lymph vascular space
involvement or nerve infiltration.
Additionally, the mRNA expression profile of C1QTNF6
was also assessed in GC cell lines. The results revealed high mRNA
expression of C1QTNF6 in all four GC cell lines (SGC-7901, AGS,
SGC-823 and MGC-803; Fig. 2C). In
the subsequent experiments, in order to assess the in vitro
effect of C1QTNF6 on cell proliferation, migration, invasion and
apop-tosis, the AGS cell line was used for C1QTNF6-knockdown.
Protein detection of C1QTNF6-knockdown
cell lines
In order to determine whether C1QTNF6 serves a role
in the growth and proliferation of GC cells, the present study
first established the C1QTNF6-knockdown AGS cell line. RT-qPCR
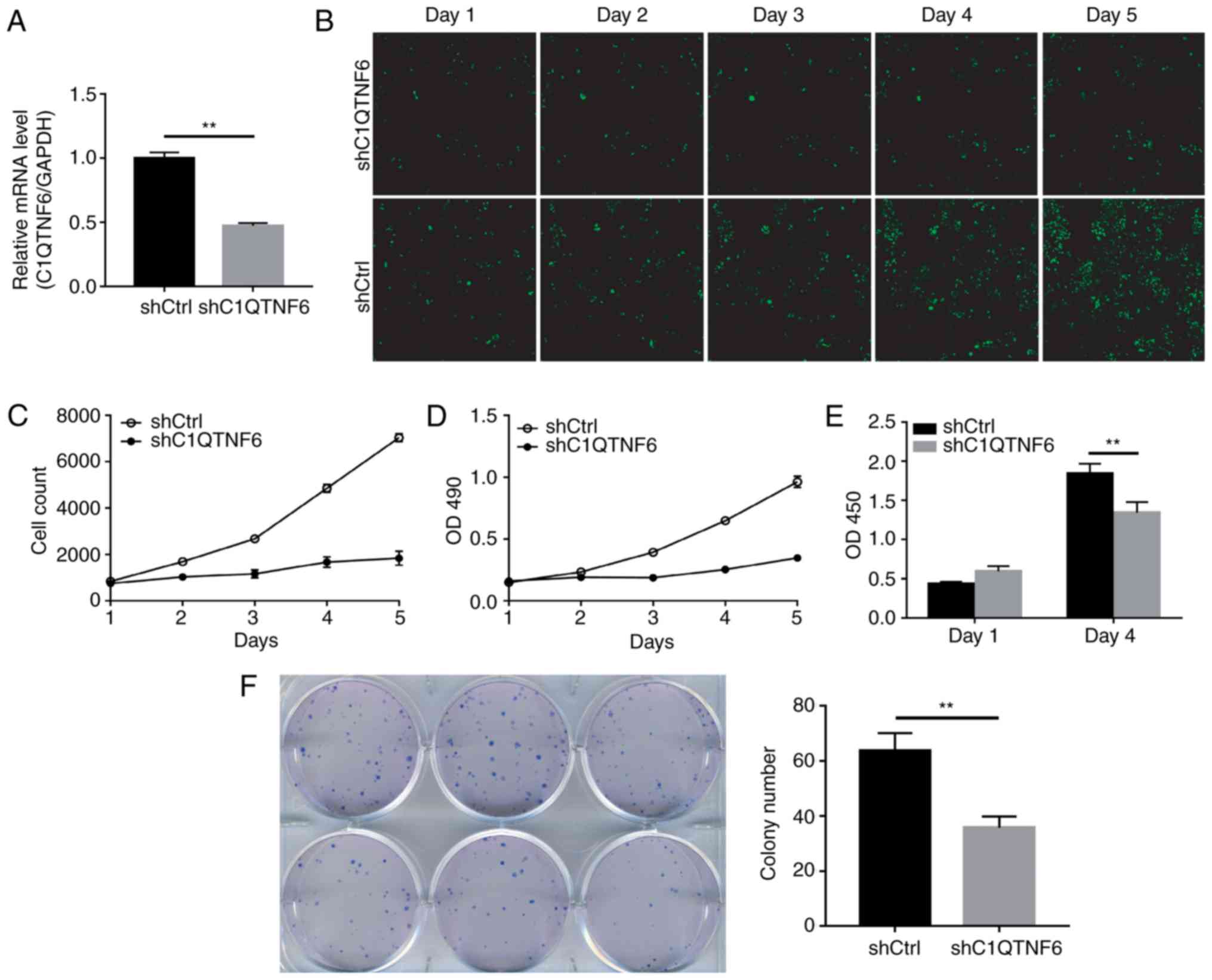
confirmed the efficiency of C1QTNF6-knockdown (Fig. 3A). The expression of C1QTNF6 in
knockdown cells was 52.8% lower than that in the shCtrl cells,
which indicated the successful establishment of the
C1QTNF6-knockdown cell line. The growth of siC1QTNF6 and shCtrl
cells from days 1 to 5 was monitored using Celigo instrument and
immunofluorescence microscopy. The fluorescence intensity (Fig. 3B) and cell number (Fig. 3C) of the C1QTNF6-knockdown AGS
cell line were weaker than that of the shCtrl cells.
The effect of C1QTNF6 on AGS cell proliferation was
further assessed by MTT assay or using the Roche BrDU kit. For the
MTT assay, 3 days after the shRNA lentivirus infection, ~2,000
cells were seeded into 96-well plates and were allowed to grow for
5 days. The AGS cell proliferation rate was significantly
suppressed following C1QTNF6 interference (Fig. 3D). The results of the BrDU assay
demonstrated that on the 4th day of culture, the proliferation
index of AGS cells was reduced by ~27% following
C1QTNF6-interference (Fig. 3E),
which suggested that the proliferative ability of cells was blocked
as a result of C1QTNF6-knockdown.
Subsequently, the colony formation ability of the
AGS cells was investigated; the results revealed that
C1QTNF6-knockdown markedly decreased the colony formation
efficiency of AGS cells (P=0.00329), compared with that of cells
transfected with negative control plasmid (Fig. 3F).
C1QTNF6-knockdown inhibits AGS cell
invasion and migration
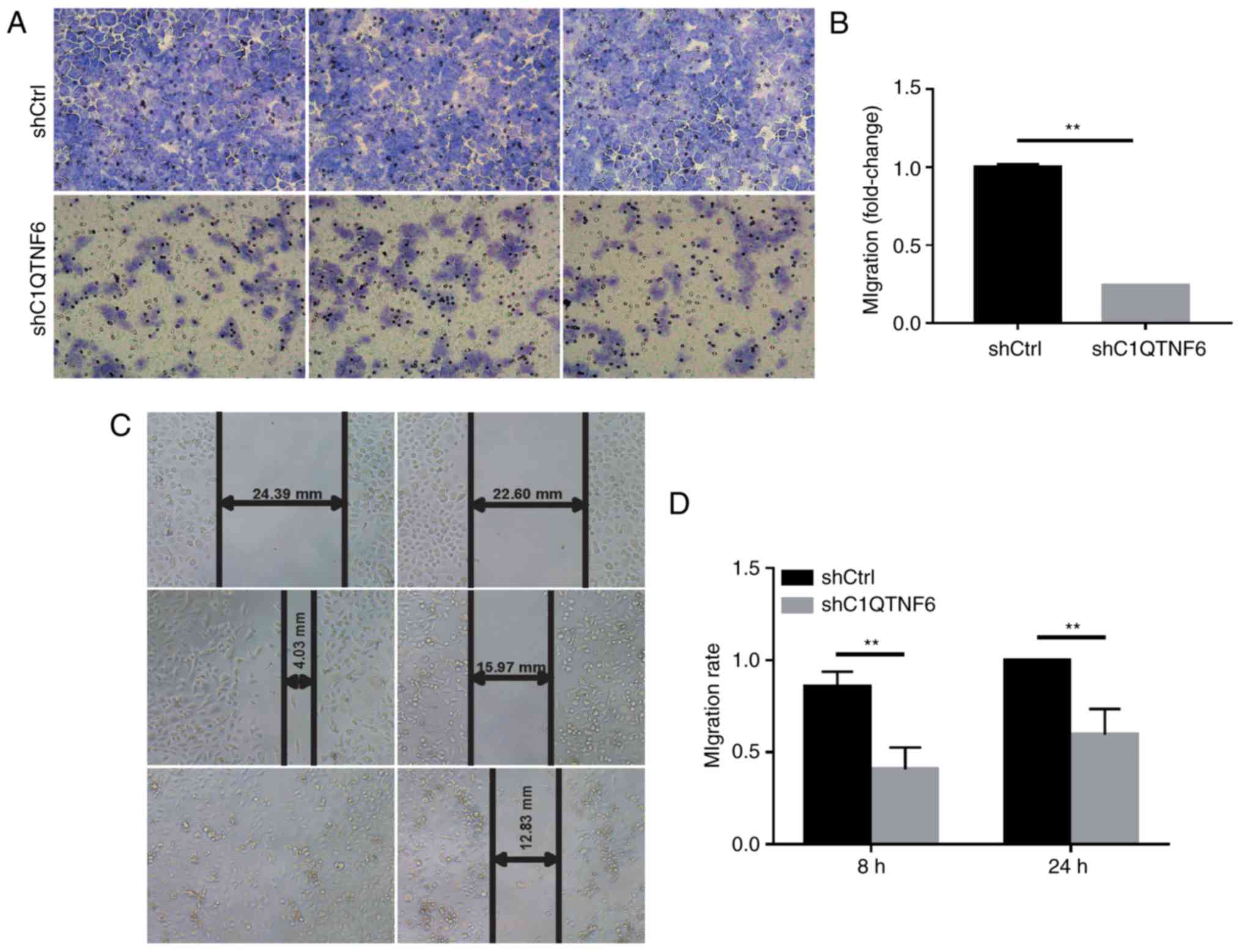
To detect whether C1QTNF6 affects the cell invasion
ability, a Transwell chamber migration assay was conducted using
AGS cells infected with siC1QTNF6 or siCtrl lentivirus. As
demonstrated in Fig. 4A,
infection with siC1QTNF6 notably decreased the invasive ability of
AGS cells. The inhibitory rate of siC1QTNF6 on AGS cell invasion
was ~76% (Fig. 4B).
Wound-healing assay revealed that the migration
ability of C1QTNF6-knockdown cells at 8 and 24 h of culture was
decreased by ~52.33% (P=0.0001) and ~40% (P=0.003), respectively
(Fig. 4C and D); these results
indicated a significant difference between the shCtrl and the
siC1QTNF6 groups.
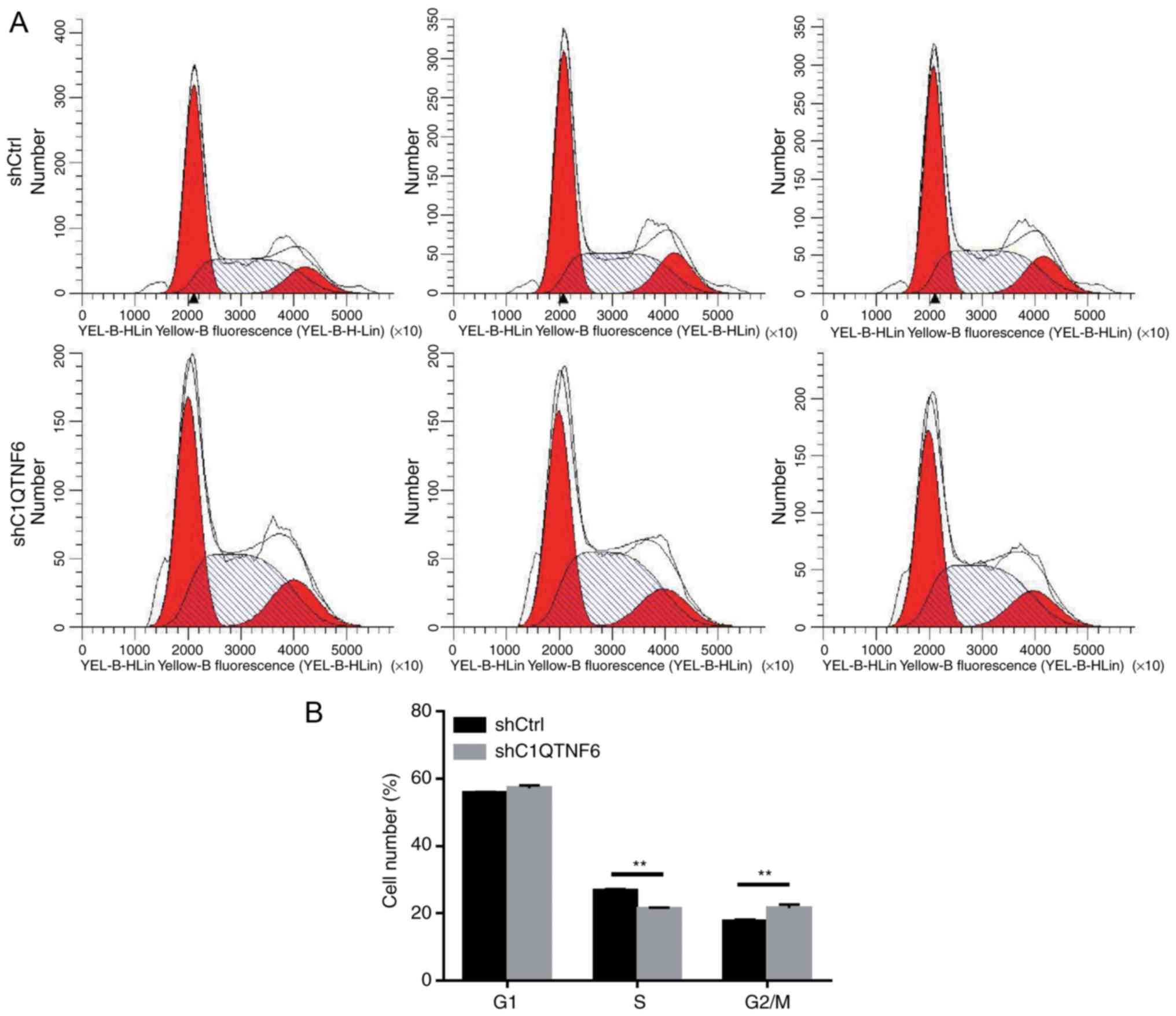
C1QTNF6-knockdown inhibits the AGS cell
cycle
To investigate whether cell cycle alteration leads
to decreased proliferation of siC1QTNF6 AGS cells, the cell cycle
distribution was detected using a FACScan flow cytometer. Following
infection with lentivirus for 5 days, there were fewer AGS cells in
the S phase and more cells in the G2/M phase following
C1QTNF6-knockdown (Fig. 5A and
B). No significant difference was observed between the
siC1QTNF6 and shCtrl groups with respect to the distribution of
cells in the G1 phase, which suggested that the C1QTNF6
gene is associated with AGS cell cycle distribution.
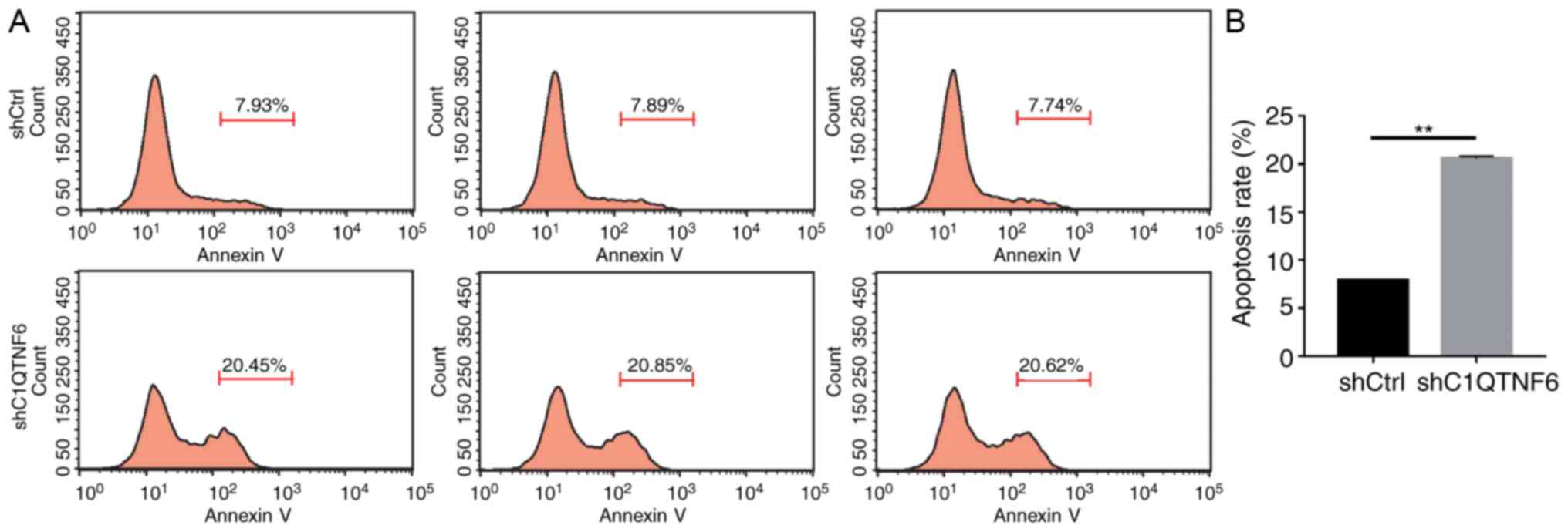
C1QTNF6-knockdown promotes AGS cell
apoptosis
In order to test the effect of C1QTNF6 on AGS cell
apoptosis, flow cytometry was used to detect the apoptotic rate.
Five days after transfection with lentivirus, more early apoptotic
cells were observed in the siC1QTNF6 AGS group, compared with that
in the shCtrl group (Fig. 6A).
The apoptotic rate of siC1QTNF6 AGS cells was increased by ~62%,
compared with that of the shCtrl cells (Fig. 6B), which suggested that the
C1QTNF6 gene is significantly associated with AGS cell
apoptosis.
Discussion
A previous study demonstrated that, under
physiological conditions, CTRPs function as molecular mediators
connecting inflammatory and metabolic diseases (18). In addition, CTRPs also function in
tumor tissues. C1QTNF6 was revealed to be highly expressed in
hepatocellular carcinoma (12),
and associated with clinicopathological parameters, which suggested
that C1QTNF6 is likely to be a diagnostic biomarker for
hepatocellular carcinoma. Furthermore, CTRP8-induced human glioma
cell migration can be inhibited by a small competitor peptide
derived from C1QTNF6 (11). Taken
together, these findings suggested an important role of C1QTNF6 in
physiological and pathological states. The present study reported
high expression levels of C1QTNF6 in GC, and C1QTNF6 was revealed
to promote the proliferation and migration of GC cells in
vitro and to serve a role in cell apoptosis. These results
provided crucial evidence for further studies on reproduction and
metastasis of GC cells in vivo. The present study provided
novel evidence for the design of small molecular drugs targeting
C1QTNF6 to treat GC. Furthermore, microarray assay was conducted,
and numerous differentially expressed proteins were detected, which
established the basis to determine the molecular mechanism of
C1QTNF6 in the proliferation and migration of GC cells.
The present study demonstrated that C1QTNF6
expression in GC tissues and cell lines has an impact on cell
proliferation, migration and invasion, which suggested that
alternative expression of C1QTNF6 altered cancer cell life
processes. However, Wang et al (19) demonstrated that downregulation of
C1QTNF6 mRNA contributed toward invasiveness of the human breast
cancer MCF-7 cell line. In this aforementioned study, C1QTNF6 was
revealed to be directly targeted by miR-29b, which may be
considered as a tumor promoter or suppressor, depending on the type
of cell or tissue. Therefore, C1QTNF6 may also serve different
roles in different cell types with respect to cell invasion or
migration. In the present study, inhibition of C1QTNF6 induced G2-M
cell cycle arrest and cell apoptosis, which suggested that C1QTNF6
sensitizes GC cells to apoptosis.
The GC AGS cells exhibited a notable decrease in
wound healing ability following C1QTNF6-knockdown following 8 or 24
h culture. This may have been caused by inhibition of the
expression of the potent anti-inflammatory cytokine, IL10, since
C1QTNF6 can induce IL-10 expression in Raw264.7 cells (20). IL-10 was revealed to enhance wound
healing of cutaneous flaps following ischemia reperfusion injury
(21), and to promote
regenerative healing in an adult model of scar formation (22). Additionally, numerous other
factors may also be involved in regulating C1QTNF6-mediated wound
healing.
The human C1QTNF family has 16 members that consist
of 4 distinct domains, i.e., an N-terminal signal peptide, a short
variable domain, a collagen-like domain, and a C-terminal C1q-like
globular domain (23-25). A number of protein members are
involved in the regulation of cell apoptosis. Administration of
CTRP9 was demonstrated to attenuate cardiomyocyte apoptosis
following acute myocardial infarction (26) or myocardial ischemia reperfusion
(27), and to decrease the
apoptotic rate of primary pulmonary artery epithelial cells due to
pulmonary arterial hypertension (28). CTRP3 protected bone marrow derived
mesenchymal stem cells from hypoxia-induced apoptosis through the
phosphoinositide 3-kinase/protein kinase B signaling pathway
(29). CTRP3 also serves a
protective role in cardiac infarction through its anti-apoptotic
effect in cardiomyocytes (30).
CTRP4 was demonstrated to protect against apoptosis induced by
chemotherapeutics and thereby promote tumor cell survival (3). The present study was the first to
report the anti-apoptotic effect of C1QTNF6 in GC AGS cells; the
results are consistent with those of previous studies (28-30), which suggested that C1QTNF family
proteins primarily perform cytoprotective and anti-apoptotic
functions. In addition, the results of the present study may be
applied to a C1QTNF6-knockout animal model to extend and confirm
the results derived from a cell culture model. A limitation of the
present study is that the cellular protection mechanism of C1QTNF6
remains unclear. Further transcriptomic analysis in GC cells
following inhibition of C1QTNF6 expression may provide further
insight into the anti-apoptotic mechanism of this protein.
In conclusion, the imbalance of cell proliferation
and apoptosis may contribute toward tumorigenesis, and changes in
cell motility may lead to metastasis of cancer cells. C1QTNF6
affects the proliferation, migration, invasion and apoptosis of GC
cells. With respect to the association between GC and C1QTNF6, the
present study demonstrated the following: i) The C1QTNF6 gene is
upregulated in GC tissues and cell lines; and ii) interference of
C1QTNF6 expression influenced the proliferation, migration and
invasion ability of GC cells in vitro. Therefore, the
results of the present study provided novel evidence for GC
therapy.
Acknowledgments
Not applicable.
Funding
The present study was supported by grants from
Qingdao Municipal Science and Technology Bureau (grant no.
17-3-3-35-nsh).
Availability of data and materials
The datasets generated and/or analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors’ contributions
X-JJ, DW and H-XQ designed the study. H-XQ, LC,
X-YM, Z-JW, Y-XC and Y-PY collected and analyzed the data. H-XQ, LC
and X-YM drafted and wrote the manuscript. X-JJ and DW revised the
manuscript critically for intellectual content. All authors gave
intellectual input to the study and approved the final version of
the manuscript.
Ethics approval and consent to
participate
The Institutional Review Board of Qingdao Municipal
Hospital approved the study protocol. All procedures involving
human participants were performed in accordance with the ethical
standards of the Institutional and National Research Committee, and
the 1964 Declaration of Helsinki and its later amendments or
comparable ethical standards. Written informed consent was obtained
from all patients.
Patient consent for publication
Written informed consent was obtained from all
patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chung HW and Lim JB: Role of the tumor
microenvironment in the pathogenesis of gastric carcinoma. World J
Gastroenterol. 20:1667–1680. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Qiu MZ and Xu RH: The progress of targeted
therapy in advanced gastric cancer. Biomark Res. 1:322013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li Q, Wang L, Tan W, Peng Z, Luo Y, Zhang
Y, Zhang G, Na D, Jin P, Shi T, et al: Identification of
C1qTNF-related protein 4 as a potential cytokine that stimulates
the STAT3 and NF-kappaB pathways and promotes cell survival in
human cancer cells. Cancer Lett. 308:203–214. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Akiyama H, Furukawa S, Wakisaka S and
Maeda T: CTRP3/cart-ducin promotes proliferation and migration of
endothelial cells. Mol Cell Biochem. 304:243–248. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Park SY, Choi JH, Ryu HS, Pak YK, Park KS,
Lee HK and Lee W: C1q tumor necrosis factor alpha-related protein
isoform 5 is increased in mitochondrial DNA-depleted myocytes and
activates AMP-activated protein kinase. J Biol Chem.
284:27780–27789. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Peterson JM, Wei Z and Wong GW:
C1q/TNF-related protein-3 (CTRP3), a novel adipokine that regulates
hepatic glucose output. J Biol Chem. 285:39691–39701. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chavali VR, Khan NW, Cukras CA, Bartsch
DU, Jablonski MM and Ayyagari R: A CTRP5 gene S163R mutation
knock-in mouse model for late-onset retinal degeneration. Hum Mol
Genet. 20:2000–2014. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hofmann C, Chen N, Obermeier F, Paul G,
Buchler C, Kopp A, Falk W and Schäffler A: C1q/TNF-related
protein-3 (CTRP-3) is secreted by visceral adipose tissue and
exerts antiinflammatory and antifibrotic effects in primary human
colonic fibroblasts. Inflamm Bowel Dis. 17:2462–2471. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Thanasupawat T, Glogowska A, Burg M, Wong
GW, Hoang-Vu C, Hombach-Klonisch S and Klonisch T: RXFP1 is
targeted by complement C1q tumor necrosis factor-related factor 8
in brain cancer. Front Endocrinol (Lausanne). 6:1272015. View Article : Google Scholar
|
|
10
|
Klonisch T, Glogowska A, Thanasupawat T,
Burg M, Krcek J, Pitz M, Jaggupilli A, Chelikani P, Wong GW and
Hombach-Klonisch S: Structural commonality of C1q TNF-related
proteins and their potential to activate relaxin/insulin-like
family peptide receptor 1 signalling pathways in cancer cells. Br J
Pharmacol. 174:1025–1033. 2017. View Article : Google Scholar
|
|
11
|
Glogowska A, Kunanuvat U, Stetefeld J,
Patel TR, Thanasupawat T, Krcek J, Weber E, Wong GW, Del Bigio MR,
Hoang-Vu C, et al: C1q-tumour necrosis factor-related protein 8
(CTRP8) is a novel interaction partner of relaxin receptor RXFP1 in
human brain cancer cells. J Pathol. 231:466–479. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Takeuchi T, Adachi Y and Nagayama T:
Expression of a secretory protein C1qTNF6, a C1qTNF family member,
in hepatocellular carcinoma. Anal Cell Pathol (Amst). 34:113–121.
2011. View Article : Google Scholar
|
|
13
|
Wu WJ, Mo DL, Zhao CZ, Zhao C, Chen YS,
Pang WJ and Yang GS: Knockdown of CTRP6 inhibits adipogenesis via
lipogenic marker genes and Erk1/2 signalling pathway. Cell Biol
Int. 39:554–562. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Murayama MA, Kakuta S, Inoue A, Umeda N,
Yonezawa T, Maruhashi T, Tateishi K, Ishigame H, Yabe R, Ikeda S,
et al: CTRP6 is an endogenous complement regulator that can
effectively treat induced arthritis. Nat Commun. 6:84832015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fan RH, Zhu XM, Sun YW, Peng HZ, Wu HL and
Gao WJ: CTRP6 inhibits fibrogenesis in TGF-beta1-stimulated human
dermal fibroblasts. Biochem Biophys Res Commun. 475:356–360. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
17
|
Chen Z, He T, Zhao K and Xing C:
Anti-metastatic activity of fangchinoline in human gastric cancer
AGS cells. Oncol Lett. 13:655–660. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schaffler A and Buechler C: CTRP family:
Linking immunity to metabolism. Trends Endocrinol Metab.
23:194–204. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang C, Gao C, Zhuang JL, Ding C and Wang
Y: A combined approach identifies three mRNAs that are
down-regulated by microRNA-29b and promote invasion ability in the
breast cancer cell line MCF-7. J Cancer Res Clin Oncol.
138:2127–2136. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim MJ, Lee W, Park EJ and Park SY:
C1qTNF-related protein-6 increases the expression of interleukin-10
in macrophages. Mol Cells. 30:59–64. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hu M, Ludlow D, Alexander JS, McLarty J
and Lian T: Improved wound healing of postischemic cutaneous flaps
with the use of bone marrow-derived stem cells. Laryngoscope.
124:642–648. 2014. View Article : Google Scholar
|
|
22
|
Peranteau WH, Zhang L, Muvarak N, Badillo
AT, Radu A, Zoltick PW and Liechty KW: IL-10 overexpression
decreases inflammatory mediators and promotes regenerative healing
in an adult model of scar formation. J Invest Dermatol.
128:1852–1860. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ahima RS, Qi Y, Singhal NS, Jackson MB and
Scherer PE: Brain adipocytokine action and metabolic regulation.
Diabetes. 55(Suppl 2): S145–S154. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Scherer PE, Williams S, Fogliano M,
Baldini G and Lodish HF: A novel serum protein similar to C1q,
produced exclusively in adipocytes. J Biol Chem. 270:26746–26749.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kopp A, Bala M, Weigert J, Buchler C,
Neumeier M, Aslanidis C, Scholmerich J and Schaffler A: Effects of
the new adiponectin paralogous protein CTRP-3 and of LPS on
cytokine release from monocytes of patients with type 2 diabetes
mellitus. Cytokine. 49:51–57. 2010. View Article : Google Scholar
|
|
26
|
Sun Y, Yi W, Yuan Y, Lau WB, Yi D, Wang X,
Wang Y, Su H, Wang X, Gao E, et al: C1q/tumor necrosis
factor-related protein-9, a novel adipocyte-derived cytokine,
attenuates adverse remodeling in the ischemic mouse heart via
protein kinase A activation. Circulation. 128(11 Suppl 1):
S113–S120. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bai S, Cheng L, Yang Y, Fan C, Zhao D, Qin
Z, Feng X, Zhao L, Ma J, Wang X, et al: C1q/TNF-related protein 9
protects diabetic rat heart against ischemia reperfusion injury:
Role of endoplasmic reticulum stress. Oxid Med Cell Longev.
2016:19020252016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li Y, Geng X, Wang H, Cheng G and Xu S:
CTRP9 ameliorates pulmonary arterial hypertension through
attenuating inflammation and improving endothelial cell survival
and function. J Cardiovasc Pharmacol. 67:394–401. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hou M, Liu J, Liu F, Liu K and Yu B: C1q
tumor necrosis factor-related protein-3 protects mesenchymal stem
cells against hypoxia- and serum deprivation-induced apoptosis
through the phosphoinositide 3-kinase/Akt pathway. Int J Mol Med.
33:97–104. 2014. View Article : Google Scholar
|
|
30
|
Yi W, Sun Y, Yuan Y, Lau WB, Zheng Q, Wang
X, Wang Y, Shang X, Gao E, Koch WJ, et al: C1q/tumor necrosis
factor-related protein-3, a newly identified adipokine, is a novel
antiapoptotic, proangiogenic, and cardioprotective molecule in the
ischemic mouse heart. Circulation. 125:3159–3169. 2012. View Article : Google Scholar : PubMed/NCBI
|