Introduction
The intestinal epithelium forms a physical barrier
that separates the luminal contents of the gut from the internal
milieu (1). Intestinal epithelial
cells (IECs) also perform several other functions, such as the
sampling of the intestinal microenvironment, the sensing of both
beneficial and harmful microbes and the secretion of numerous
factors in response to positive or negative signals, which are
crucial for intestinal homeostasis (2). To fulfill these diverse functions,
the intestinal epithelium has unique anatomical and cellular
adaptations, and IECs comprise several specialized cell types with
distinct function (2,3). The dynamic crosstalk between IECs
and local immune cells represents one of the basic features of
intestinal homeostasis (2,3).
These interactions are not only essential for maintaining
homeostasis, but are also critical for the pathogenesis of
inflammatory bowel disease (IBD) and intestinal epithelium
restitution following injury.
Interleukin (IL)-22 belongs to the IL-10 cytokine
family and is expressed by several types of lymphocytes, including
CD4+ T cells, Th17 cells, natural killer (NK) cells and
neutrophils (4-14). IL-22 binds to a heterodimeric
receptor consisting of IL-22RA1 and IL10-RB. In contrast to the
wide expression pattern of IL-10RB, IL-22RA1 is expressed by
non-hematopoietic cells, such as epithelial cells of the
gastrointestinal tract and skin (15-17). Thus, IL-22 can function in both
lymphocytes and epithelial cells. In the intestine, IL-22 has been
shown to prevent prolonged inflammation in models of dextran sodium
sulfate (DSS)-mediated colitis and graft versus host disease
(13,18,19). On the contrary, IL-22 can induce
inflammatory responses in the other intestine models (20). These findings indicate that IL-22
has dual functions of promoting and antagonizing inflammation,
which likely depends on the specific context. To date, these
complex roles of IL-22 are not yet fully understood.
It is well known that intestinal stem cells (ISCs)
are located at the crypt base and specifically express markers,
such as leucine-rich repeat-containing G-protein coupled receptor 5
(Lgr5) (21). They play
significant roles in intestinal homeostasis and regeneration. An
ex vivo 3D culture system of intestinal organoids
(epithelial mini-guts), highly mimicking the growth in vivo,
has been successfully established by Clever’s laboratory. This was
done by culturing intestinal epithelial organoids in Matrigel and
supplementing epidermal growth factor (EGF), activators of the Wnt
and Notch pathways, and an inhibitor of the bone morphogenetic
protein (BMP) pathway (22). This
powerful system has been widely used for the study of intestinal
epithelium homeostasis (22-26).
In this study, we applied DSS and X-ray induced
colitis mouse models, and an ex vivo organoid culture system
to explore the role of IL-22 in regulating the homeostasis of the
intestinal epithelium during inflammation. We found that IL-22 was
dynamically expressed with the development of intestinal
inflammation. IL-22 promoted cell proliferation, but inhibited the
differentiation of intestinal organoids. It also suppressed the
self-renewal capacity of ISCs and accordingly resulted in the
destruction of organoids. Mechanistically, IL-22 activated signal
transducer and activator of transcription 3 (Stat3) activity, but
suppressed the Wnt and Notch signaling pathways, in which the
change in Wnt signaling was more dominant.
Materials and methods
Ethics statement
All animal experiments conformed to the regulations
drafted by the Association for Assessment and Accreditation of
Laboratory Animal Care in Shanghai and were approved by the East
China Normal University Center for Animal Research.
Animal experiments
For the model of IBD, a total of 35 male (n=5/group)
C57BL/6 mice (8 weeks old, weighing approximately 20 g) were
obtained from Shanghai SLAC Laboratory Animal Co., Ltd. (Shanghai,
China) and maintained under SPF experimental animal conditions
(temperature, 23±2°C; humidity, 65±5%) in a 12-h light/dark cycle
and fed a standard laboratory diet and water in East China Normal
University Center for Animal Research. Acute colitis was induced at
day 0 by the administration of 2.5% (w/v) DSS (molecular weight,
36-50 kDa; MP Biomedicals, Santa Ana, CA, USA) in their drinking
water and this was then changed to normal drinking water at day 5.
The mice were sacrificed at different time points as indicated
(days 0, 2, 4, 6, 8, 10 and 12) and intestinal samples were
collected for RNA extraction or histological analysis.
For in vivo regeneration assay, a total of 20
male (n=5/group) C57BL/6 mice (8 weeks old, weighing approximately
20 g) were obtained from Shanghai SLAC Laboratory Animal Co., Ltd.
and kept under SPF experimental animal conditions (temperature,
23±2°C; humidity, 65±5%) in a 12-h light/dark cycle and fed a
standard laboratory diet and water in East China Normal University
Center for Animal Research. The mice were irradiated by 9.0 Gy
X-ray using a Biological X-ray irradiator (RS 2000 series; Rad
Source Technologies, Inc., Suwanee, GA, USA) operated at 250 kVp,
15 mA. The mice were sacrificed every other day after irradiation
by cervical dislocation.
Organoid culture
Small intestinal crypt isolation and culture were
performed as previously described (21,22). Briefly, the small intestine was
washed in cold PBS. The tissue was then chopped into approximately
3-5-mm-thick sections, washed with cold PBS and incubated in 5 mM
EDTA in PBS for 30 min on ice. The tissue fragments were suspended
vigorously, and the supernatants, including most of the villi were
discarded. The sediment was further resuspended with cold PBS; the
yielded supernatants enriched in crypts were collected and filter
through a 70 μm cell mesh (BD Biosciences, San Jose, CA,
USA). The crypts were then pelleted by centrifugation (132 × g, 5
min), suspended in ADF medium (advanced DMEM F12) (Thermo Fisher
Scientific, Waltham, MA, USA) containing 1% GlutaMAX (Thermo Fisher
Scientific). The crypts were centrifuged at low speed (33 × g, 3
min) for 2-3 times to remove single cells. The final crypts were
resuspended with growth factor reduced to Matrigel (356231; BD
Biosciences) at 5-10 crypts per μl, followed by seeding on a
96-well plate (5 μl per well) or 48-well plate (30 μl
per well). Culture medium was made by ADF supplemented with
penicillin/streptomycin, 10 mM HEPES, 1X GlutaMAX, 1X N2, 1X B27
(all from Thermo Fisher Scientific) and 1 μM
N-acetylcysteine (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)
containing 50 ng/ml Murine EGF, 100 ng/ml Murine Noggin and 1
μg/ml Human R-spondin-1 (all from PeproTech, Rocky Hill, NJ,
USA). The medium was changed every other day.
For single cell culture, 4-6-day cultured organoids
were dissociated with TrypLE express (Thermo Fisher Scientific)
including 2,000 U/ml DNase (Roche Diagnostics, Indianapolis, IN,
USA) for 10 min at 37°C. Dissociated cells were passed through a 20
μm cell mesh (BD Biosciences) and embedded in Martrigel
containing 1 μM jag1 (AnaSpec, Fremont, CA, USA) at up to
5,000-10,000 cells per 30-50 μl. The culture medium was
regular organoid culture medium supplemented with 10 μM ROCK
inhibitor Y27632 (Sigma-Aldrich; Merck KGaA).
For rescue experiments, 100 ng/ml IL-22 (R&D
Systems, Minneapolis, MN, USA), 200 ng/ml Wnt3a (R&D Systems),
1 μM Jag1 (AnaSpec), 5 μM WP1066 (Stat3 inhibitor;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA), 3 μM
Stattic (Stat3 inhibitor; Santa Cruz Biotechnology, Inc.,) were
used at different combinations in oranoid culture medium: Vehicle
(without IL22), Stattic, WP1066, Wnt3a, Jag1, IL-22, IL-22 plus
Stattic, IL-22 plus WP1066, IL-22 plus Wnt3a, IL-22 plus Jag1.
Reverse transcription-quantitative PCR
(RT-qPCR)
RNA from the organoids, intestines (IECs) and immune
cells [macrophages, monocytes, CD4+ T cells and
dendritic cells (DCs) were sorted from mouse peripheral blood by
corresponding magnetic beads; Miltenyi Biotec, Bergisch Gladbach,
Germany] was obtained by tissue homogenization using TRIzol reagent
(Takara Bio, Inc., Otsu, Japan), followed by RNA precipitation in
isopropanol, resuspension in DEPC-treated water and verification as
DNA free by PCR. cDNA was synthesized with the Superscript RNase H2
Reverse Transcriptase kit (Takara Bio, Inc.) using 2.5 mM random
hexamers (Takara Bio, Inc.). Quantitative (real-time) PCR (qPCR)
was performed using the SYBR-Green PCR kit (Takara Bio, Inc.) on a
Mx3005P thermal cycler (Stratagene, San Diego, CA, USA). The
amplification conditions included 3 processes, namely hold stage (1
cycle): 95°C for 1 min; PCR stage (40 cycles): 95°C for 5 sec, 55°C
for 30 sec; melt curve stage (1 cycle): 95°C for 5 sec, 55°C for 30
sec and 95°C for 30 sec. The sequences of the main primers were as
follows: β-actin sense, CAGCCTTCCTTCTTGGGTAT and antisense,
TGATCTTGATCTTCATGGTGC; Il-22 sense, ACATCGTCAACCGCACCTTT and
antisense, CAGCCTTCTGACATTCTTCTGGAT; Il-22ra1 sense,
CTACGTGTGCCGAGTGAAGA and antisense, GGTAGGTTGCAGGATGGAGA;
Il-10rb sense, TTCGGAGTGGGTCAATGTCA and antisense,
GACCTCAGAGTCGTACGGAG; Lgr5 sense, CCTACTCGAAGACTTACCCAG T
and antisense, GCATTGGGGTGAATGATAGCA; olfactomedin 4 (Olfm4)
sense, TCTTGGGCAGAAGGTGGGACT and antisense, GGACCGTCAGGTTCAGGAGC;
achaete-scute family bHLH transcription factor 2 (Ascl2)
sense, AAGCACACCTTGACTGGTACG; and anti-sense,
AAGTGGACGTTTGCACCTTCA; zinc and ring finger 3 (Znrf3) sense,
CCAGTGGTGTATGTGAAGGGT G and antisense, AATTCTGACTACGACGTTGCT; WD
repeat domain 43 (Wdr43) sense, GCATACAGTGGCATCAAGAC and
antisense, GTGAACCTAAGCGACGAAA; Prominin 1 (Prom1) sense,
CTGAAGATTGCCCTCTATGA and anti-sense, AGTTTCTGGGTCCCTTTGAT; Axin2
sense, TGACTCTCCTTCCAGATCCCA and antisense, TGCCCACACTAGGCTGACA;
and EPH receptor B3 (Ephb3) sense, GACCTTGCTGCCCGAAACAT and
antisense, CCCACATGACAATCCCATAGCT; Mus musculus mucin 2
(Muc2) sense, ATGACGTCTGGTGGAATGGT and antisense,
TGTTCTGACAGTTGCACGTG; Mus musculus SAM pointed domain
containing ets transcription factor (Spdef) sense,
TGAACATCACAGCAGACCCT and antisense, ACTTCCAGATGTCCAGGTGG; Chga
chromogranin A (Chga) sense, ACTTCAAGACCTGGCTCT CC and
antisense, CCTAGGTCCCTCTGTGGTTG; Mus musculus neurogenin 3
(Nuero3) sense, ACACAACAACCTTTTCCCGG and antisense,
CCCGATCATTGGCCTTCTTG; Mus musculus caudal type homeobox 1
(Cdx1) sense, AAGGTTCCAGAGTGAGGTG and anti-sense,
TAGGGCAGGTGAAAGTGAGG; Mus musculus caudal type homeobox 2
(Cdx2) sense, CTCTCCTCCTACCCACGAAC and antisense,
TGGGGCAAAAGAGGCTGATA; Mus musculus lysozyme 1 (Lyz1)
sense, GCTTCTACTGCAGCCCATTC and antisense, GCTGACTGACAAGGGAGACT;
Mus musculus SRY (sex determining region Y)-box 9
(Sox9) sense, CAAGAACAAGCCACACGT CA and antisense,
GTGGTCTTTCTTGTGCTGCA; Mus musculus defensin, alpha, 29
(Defa29 or Crs1c) sense, AAAGAAGGTTTCCGTGGTGC and
antisense, TGAGGCTAAGCACAATGGGA; Mus musculus defensin,
alpha, 5 (Defa5) sense, ATTTGTCCTCCTCTCTGCCC and antisense,
ACGCGTTCTCTTCTTTTGCA; Axin2 sense, TGACTCTCCTTCCAGATCCCA and
antisense, TGCCCACACTAGGCTGACA; cyclin D1 (Ccnd1) sense,
AGTTCATTTCCAACCCACCC and antisense, AGACCAGCCTCTTCCTCCAC;
transcriptional regulator Myc-like (C-myc) sense,
CCCTACCCGCTCAACGACAG and antisense, TTGCCTCTTCTCCACAGACACC; Eph
receptor B3 (Ephb3) sense, GACCTTGCTGCCCGAAACAT and
antisense, CCCACATGACAATCCCATAGCT; Eph receptor B2 (Ephb2)
sense, GGCTTTACCTCTTTC GAC G and antisense, CAATCCCCTTTTCCTTCC;
wingless-type MMTV integration site family, member 3A
(Wnt3a) sense, GCACCACCGTCAGCAACAGC and antisense,
TCCCTGGCATCGGCAAA CTC; Notch1 sense, AATGGAGGGAGGTGCGAA GT
and antisense, CAGAGGTGTCAGGCAGAGGG; hes family bHLH transcription
factor 1(Hes1) sense, GGAGAAGAGGCGAAGGGCAAG A and antisense,
CGTGGACAGGAAGCGGGTCA; jagged 1 (Jag1) sense,
AATGGTTATCGCTGTATCTGTCC and antisense, AGTTCTTGCCCTCATAGTCCTC;
delta like canonical Notch ligand 1 (Dll-1) sense,
CCGATGACCTCGCAACAGAA and antisense, CCAGGGTCGCACATCTTCTC. β-actin
served as the control gene, all reactions were performed in
triplicate. The relative expression was calculated using the
2-ΔΔCq method (27).
Histological analysis
The intestines were dissected and flushed gently
with cold PBS. They were then immediately fixed by 4% PFA in PBS or
snap-frozen on dry ice. Frozen samples were stored at -80°C until
cut into frozen slices using a cryostat microtome. Fixed intestines
were dehydrated by gradient alcohols, embedded in paraffin, and cut
into 4-μm-thick sections which were then subjected to
H&E staining or immunohistochemistry (IHC).
Organoid histological staining was carried out with
the sections or whole-mount sections. For the sections, organoids
were collected after Matrigel was melted by Cell Recovery Solution
(BD Biosciences). The organoids were then dehydrated and embedded
in paraffin. The sections were deparaffinized in xylene and
rehydrated in gradient alcohols. Endogenous peroxidase activity was
quenched with 3% H2O2 in methanol for 20 min
and washed in PBS. Antigen retrieval was performed by boiling the
slides in 10 mM sodium citrate buffer, pH 6.0 or 10 mM Tris-base, 1
mM EDTA solution, pH 9.0. The sections were blocked with 1% BSA and
incubated with the primary antibodies as listed below overnight at
4°C. For IHC staining, following incubation with secondary antibody
(HRP labeled polymer, GK500710; Shanghai Genetech Co., Shanghai,
China) for 30 min, the sections were developed with DAB and
counterstained with hematoxylin, then dehydrated and mounted in
neutral resins. For BrdU staining, before fixture, the organoids
were treated with 3 μg/ml BrdU for 2 h. A similar procedure
was performed with the addition of a 30-min treatment with 2 N HCl
after antigen retrieval. For immunofluorescence (IF), the sections,
following primary antibody as listed below incubation overnight at
4°C were washed and incubated with secondary Alexa Fluor 488 (1:500
dilution; cat. no. ab150077; Abcam, Cambridge, MA, USA) or 647
antibodies (1:500 dilution; cat. no. ab150115; Abcam) for 2 h at
room temperature with DAPI (1:10,000 dilution; cat. no. ab104139;
Abcam) counterstaining for 5 min at room temperature (Thermo Fisher
Scientific) and subjected to immunofluorescence microscopy (BX53;
Olympus Corporation, Tokyo, Japan).
For the whole-mount sections, the organoids were
fixed with 4% PFA in a plate without melting the Matrigel, then
permeabilized with 0.2% Triton in PBS and incubated overnight with
primary antibodies as listed below. Fluorescence was detected as
mentioned above.
The primary antibodies were as follows: Cleaved
caspase-3 (1:200; cat. no. 9661; Cell Signaling Technology,
Danvers, MA, USA), Ki67 (1:2,000; cat. no. ab873; Abcam), Lysozyme
(Paneth cell marker; 1:2,000; cat. no. ab108508; Abcam),
Chromogranin A (endocrine cell marker; 1:2,000; cat. no. 20085;
ImmunoStar, Hudson, WI, USA), Muc2 (goblet cell marker; 1:100; cat.
no. sc-59859; Santa Cruz Biotechnology, Inc.,), phosphor-Stat3 at
Tyr705 (1:200; cat. no. 4113; Cell Signaling Technology), BrdU
(1:500; cat. no. B8434; Sigma-Aldrich; Merck KGaA) and IL-22RA1
(1:400; cat. no. ab211675; Abcam).
Flow cytometry
Lgr5-EGFP-IRES-creERT2 mice (Stock no.
008875) were obtained from The Jackson Laboratory (Bar Harbor, ME,
USA) and a total of 5 male, Lgr5-EGFP-IRES-creERT2 mice (8
weeks old, weighing approximately 20 g) were kept under SPF
experiment animal conditions (temperature,23±2°C; humidity, 65±5%)
in a 12-h light/dark cycle and fed a standard laboratory diet and
water in East China Normal University Center for Animal Research.
Endogenous GFP (green) driven by Lgr5 promoter indicates crypt base
columnar stem cells in the small intestine. Crypts from the
Lgr5-EGFP-IRES- CreERT2 mice were isolated as described
above after the mice were sacrificed and organoid culture was
performed. Following treatment with 100 ng/ml recombinant mouse
IL-22 (R&D Systems, Inc., Minneapolis, MN, USA), organoids were
collected and dissociated with TypLE and 2,000 U/ml DNase for 15
min at 37°C. The dead cells were then excluded by scatter
characteristics and propidium iodide. Lgr5+ISCs
were identified by their endogenous GFP expression.
Western blot analysis
Organoids were collected after Matrigel melting. The
cells were then lysed with 0.5% NP-40 lysis buffer [50 mM Tris-base
(pH 7.4), 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate,
0.1% SDS and Complete Mini Protease Inhibitor mixture (Roche
Diagnostics)] for western blot analysis. The protein concentration
was determined using a BCA Protein Assay kit (cat. no. p0011;
Beyotime, Shanghai, China). The proteins (30 μg each sample)
were resolved on 12% SDS-PAGE gels, transferred onto PVDF membranes
(EMD Millipore, Billerica, MA, USA). Following blocking with PBS
containing 3% skim milk, the membranes were incubated overnight at
4°C with primary antibodies as follows: Anti-cleaved caspase-3
(1:1,000; cat. no. 9661), anti-Bax (1:1,000; cat. no. 2772),
anti-Bcl2 (1:1,000; cat. no. 3498), anti-Bcl-xL (1:1,000; cat. no.
2762), anti-Stat3 (1:1,000; cat. no. 9139), anti-phospho-Stat3
(Tyr705; 1:1,000; cat. no. 9145) (all from Cell Signaling
Technology) and anti-β-actin (1:1,000; cat. no. A1978;
Sigma-Aldrich; Merck KGaA). Following incubation with peroxidase
AffiniPure secondary antibodies (goat anti-mouse IgG, cat. no.
115-035-003; goat anti-rabbit IgG, cat. no. 111-035-003; Jackson
ImmunoResearch Laboratories, Inc., West Grove, PA, USA) for 1 h at
room temperature, the blots were then analyzed using the ECL
detection system or scanned with an Odyssey Infrared Imager (LI-COR
Biosciences, Lincoln, NE, USA).
Enzyme-linked immunosorbent assay
(ELISA)
The colons from mice with DSS-induced colitis were
obtained and completely homogenized in PBS containing proteinase
inhibitors. Following centrifugation at 825 × g/min for 20 min, the
supernatant was collected. The level of secreted IL-22 from IECs
was measured using a mouse IL-22 ELISA kit (Abcam) following the
manufacturer’s instructions. The experiment was repeated 3
times.
Statistical analysis
Data were statistically analyzed by one-way ANOVA or
the Student’s t-test. A value of P<0.05 considered to indicate a
statistically significant difference. Results are presented as the
means ± standard error of the means (SEM).
Results
IL-22 is dynamically expressed during
intestinal inflammation
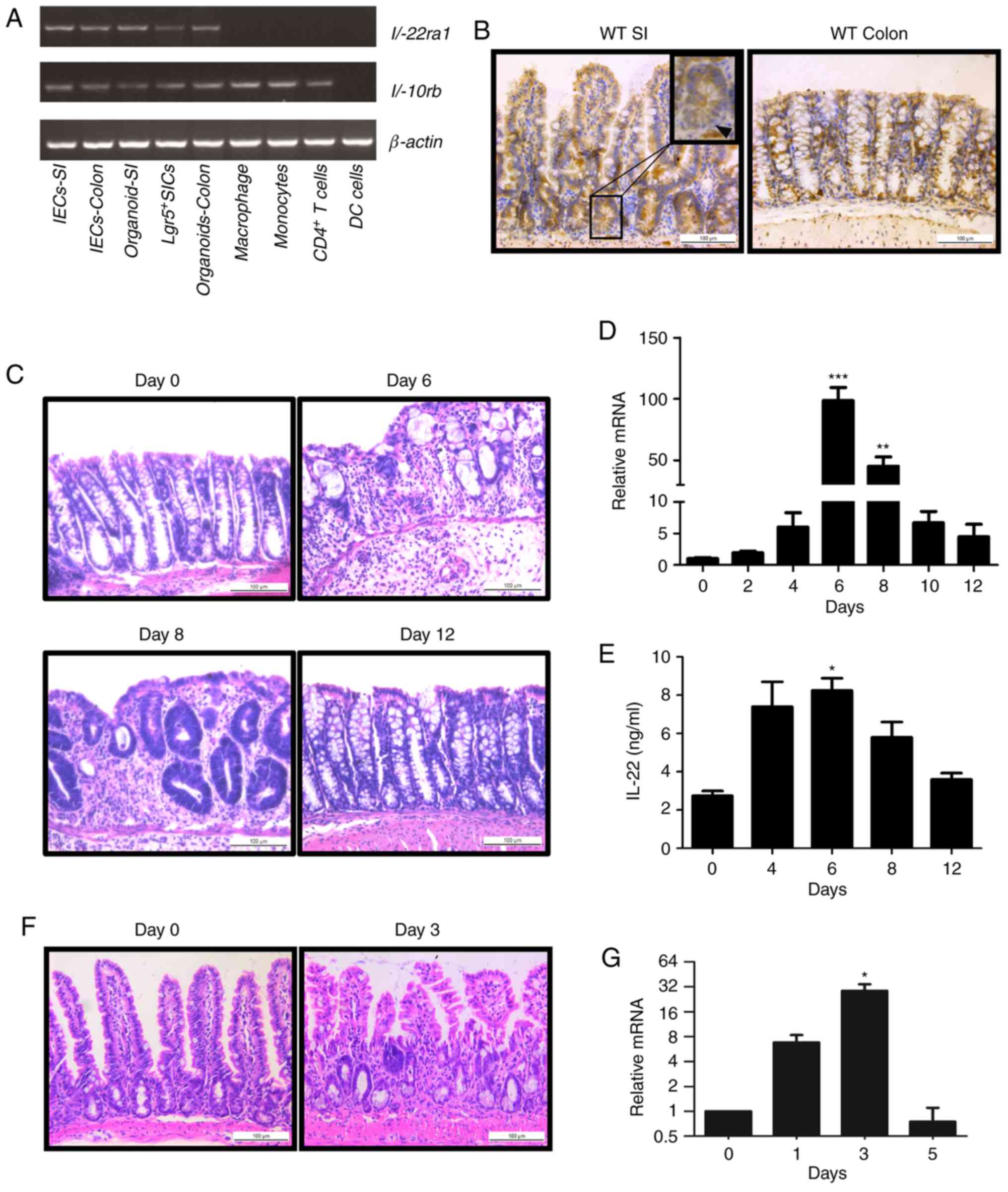
We first detected the gene transcription of
IL-22RA1 and IL-10RB, heterodimeric receptors of
IL-22, in both immune cells and IECs by RT-qPCR. While
IL-22RA1 was specifically expressed in epithelial cells,
IL-10RB expression was found in both immune and IECs
(Fig. 1A). The results of
immunohistochemistry revealed that IL-22RA1 was expressed in
the epithelium of the colon and intestine (Fig. 1B). These data suggest IL-22
signaling functions in the intestinal epithelium.
We then examined IL-22 expression in the colon in a
mouse model of DSS-induced colitis. Histological analysis revealed
that damage to the intestinal epithelium gradually increased, being
most severe on day 6 (1 day after DSS administration), and were
recovered from day 8 to day 12 (3rd day to the 7th day after DSS
administration) (Fig. 1C).
Notably, IL-22 expression was consistently altered with the
development of colitis. It reached maximum levels on day 6
following DSS administration and decreased during the regeneration
of the intestinal epithelium (Fig. 1D
and E). In another mouse model of irradiation, we observed
similar results (Fig. 1F and G).
Taken together, our results reveal that IL-22 expression is
associated with the state of intestinal damage.
IL-22 regulates the formation of
intestinal organoids
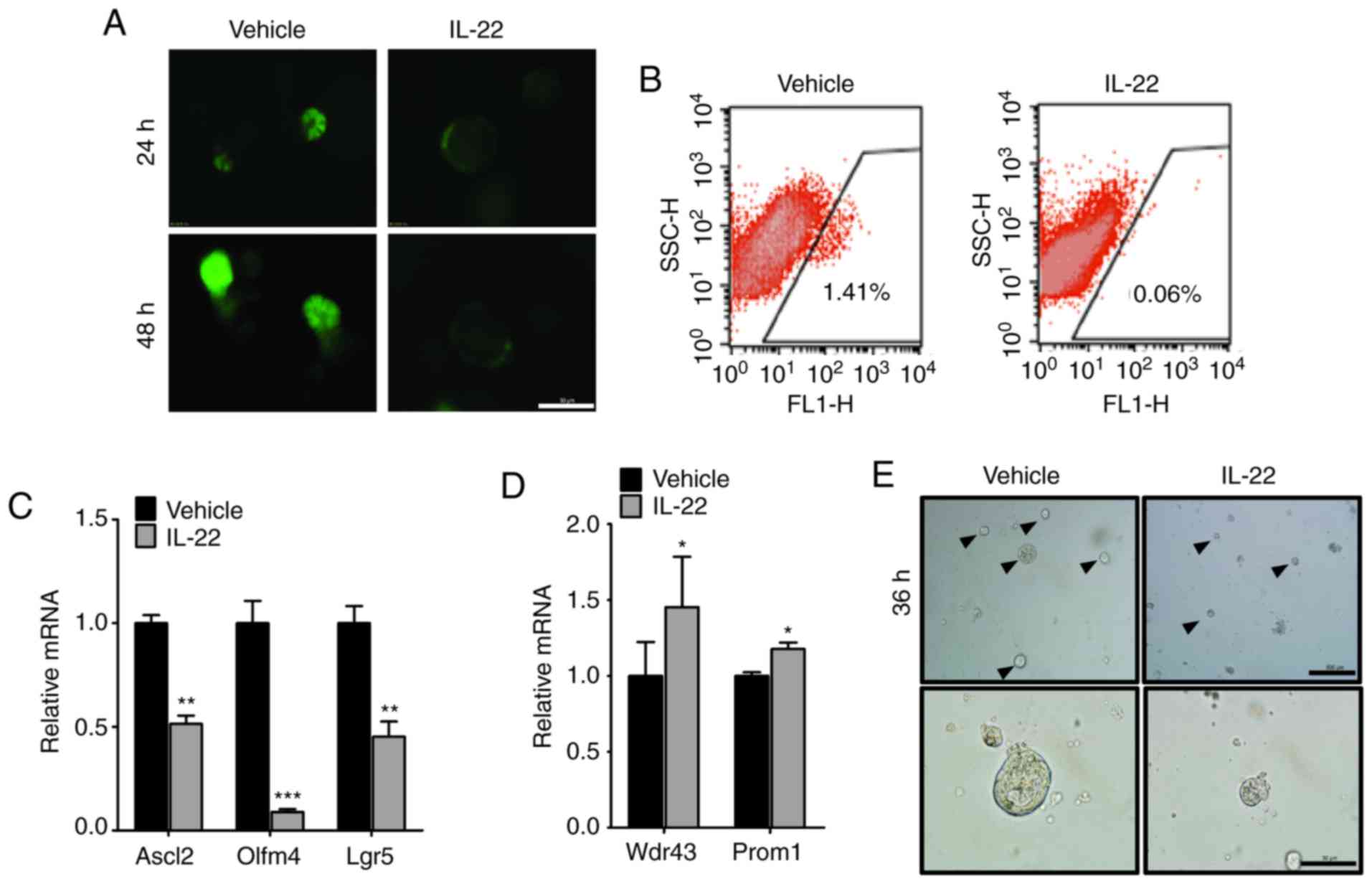
As it is well known that IL-22 promotes cell
proliferation during tissue repair, we wished to explore the direct
function of IL-22 in IECs. In an extra in vitro organoid
culture system, we found that IL-22 treatment led the organoids
acquiring a rounded shape and they began to degenerate after 3 days
(Fig. 2A). On day 4, the buds of
the organoids were significantly decreased and almost all organoids
were disrupted in the IL-22-treated group (Fig. 2B-D).
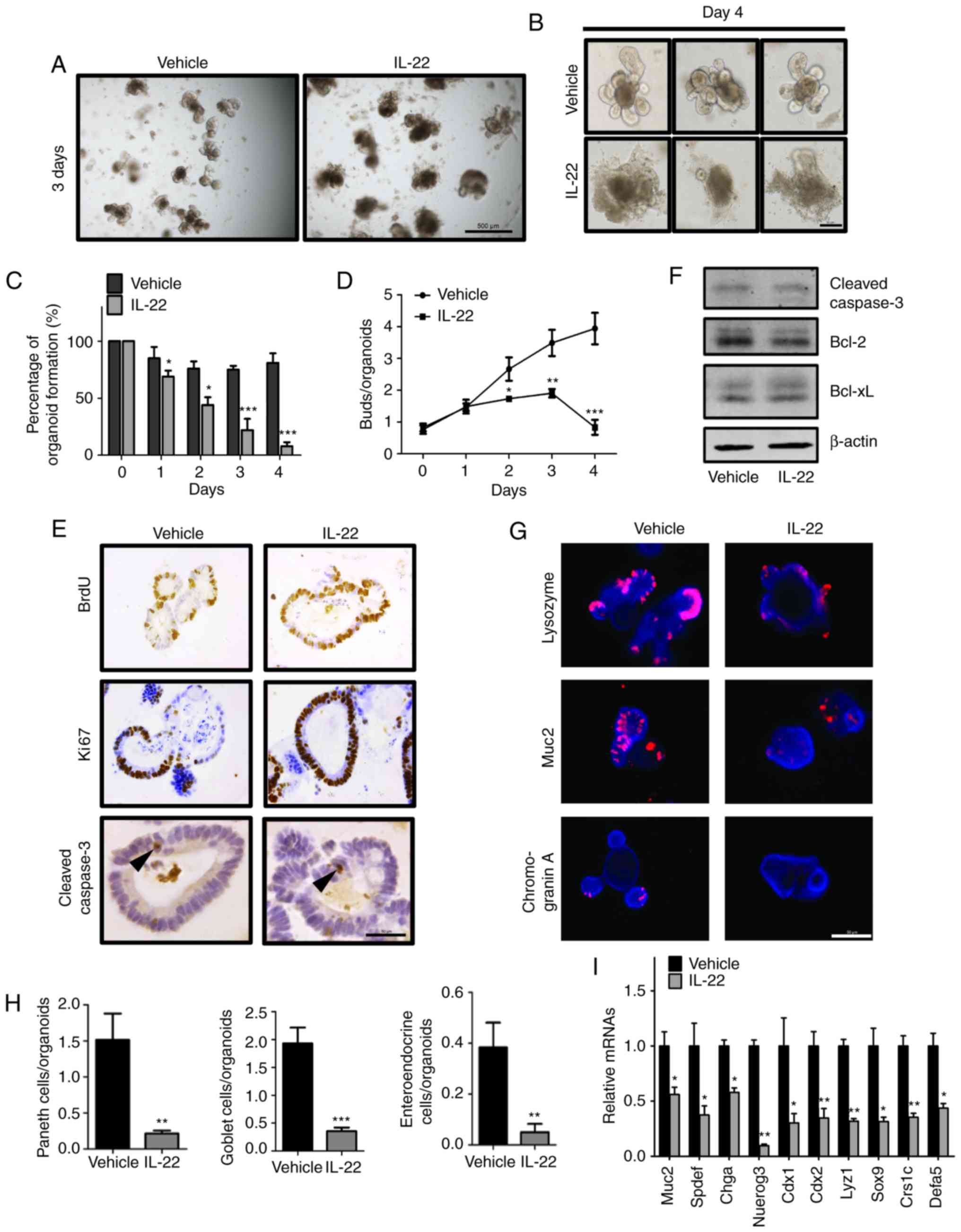 | Figure 2Interleukin (IL)-22 regulates growth
of intestinal organoids. (A) Organoids were cultured for 2 days
followed by treatment with the vehicle (the organoid cultured
without IL-22, left panel) or IL-22 (100 ng/ml treatment group,
right panel); representative images are shown on day 3 after
treatment. Scale bar, 500 μm. (B) Organoids were cultured
for 2 days followed by treatment with the vehicle (upper panels) or
IL-22 (100 ng/ml, bottom panels); representative 3 images are shown
on day 4 after treatment. Scale bar, 50 μm. (C and D)
Organoids were cultured for 2 days followed by treatment with the
vehicle or IL-22 (100 ng/ml). The percentage of (C) organoid
formation and (D) the number of buds are shown at different time
points after treatment. Data are the means ± SEM;
*P<0.05, **P<0.01, and
***P<0.001 vs. the vehicle at the indicated time
points; analyzed by t-test. (E) Organoids were cultured for 2 days
followed by treatment with the vehicle or IL-22 (100 ng/ml). The
expression of BrdU, Ki67 and cleaved caspase-3 was detected by
immunohistochemical staining. Arrowheads indicate apoptotic cells.
Scale bar, 50 μm. (F) Immunoblotting of cleaved caspase-3,
Bcl2 and Bcl-xL in the organoids treated with or without IL-22 for
2 days (100 ng/ml). (G) The expression of Lysozyme (Paneth cell
marker), Muc2 (goblet cell marker) and chromogranin A (endocrine
cell marker) was determined by immunofluorescence. Scale bar, 50
μm. (H) Quantitative analysis of the data in (G) are
presented. Data are the means ± SEM; **P<0.01 and
***P<0.001 vs. the vehicle; analyzed by t-test. (I)
Organoids were cultured for 2 days followed by treatment with the
vehicle or IL-22 (100 ng/ml). The expression of Muc2, Spdef (goblet
cell marker), Chga, Nuerog3 (enteroendocrine cell marker), Cdx1,
Cdx2 (columnar absorptive cell marker), Lyz1, Sox9, Crs1c and Defa5
(Paneth cell marker) was detected by RT-qPCR. Data are the means ±
SEM; *P<0.05, **P<0.01, and vs.
vehicle; analyzed by t-test. |
Subsequently, to determine the mechanisms through
which IL-22 regulates organoid formation, we assessed the
proliferation and apoptosis of IECs following IL-22 treatment. IHC
staining for BrdU and Ki67 revealed that the majority of the cells
were in a proliferative state (Fig.
2E). By contrast, we failed to detect a marked difference in
cell apoptosis between the vehicle (the organoid cultured without
IL-22) and the IL-22-treated samples, as determined by measuring
the levels of apoptosis-related markers (Fig. 2E and F).
We then wished determine whether IL-22 affects the
differentiation of IECs. The results of IF staining revealed that
the numbers of different differentiated cell types, including
Paneth cells, goblet cells and enteroendocrine cells were
significantly decreased following IL-22 treatment (Fig. 2G and H). In addition, the
expression of specific markers of differentiated cells was
suppressed by IL-22, as shown by RT-qPCR (Fig. 2I). Collectively, these results
indicated that IL-22 regulates organoid formation through
activation of cell proliferation and inhibition of IECs
differentiation.
IL-22 counteracts the self-renewal
ability of ISCs
Paneth cells provide EGF, Notch and Wnt ligands to
maintain ISC self-renewal ability, which is essential for organoid
formation. Since IL-22 was found to inhibit the differentiation of
Paneth cells, we wished to determine whether it also affects ISCs
self-renewal. By isolating the organoids from
Lgr5-EGFP-IRES-creERT2 mice, we found that the number of
ISCs, positive for Lgr5-EGFP, was markedly decreased by IL-22
(Fig. 3A and B). Consistently,
the results of RT-qPCR revealed that the expression of stem cell
markers, including Ascl2, Olfm4 and Lgr5 was significantly lower in
the group treated with IL-22 (Fig.
3C). Moreover, the expression of Wdr43 and Prom1, the
transit-amplifying (TA) cells markers, was slightly upregulated by
IL-22. This suggested that IL-22 promotes the differentiation into
TA cells from Lgr5+ISCs (Fig. 3D).
To further validate the effects of IL-22 on the
maintenance of ISC self-renewal, we disassociated the organoids
into single cells and assessed the growth ability. We observed that
the single cells could individually grow to a cyst-like structure
and finally formed an organoid structure (Fig. 3E, arrowheads). On the contrary,
these cells could not survive following culture for 3-4 days with
the addition of IL-22 (Fig. 3E,
bottom panels). Taken together, these data suggest that IL-22
counteracts the self-renewal ability of ISCs.
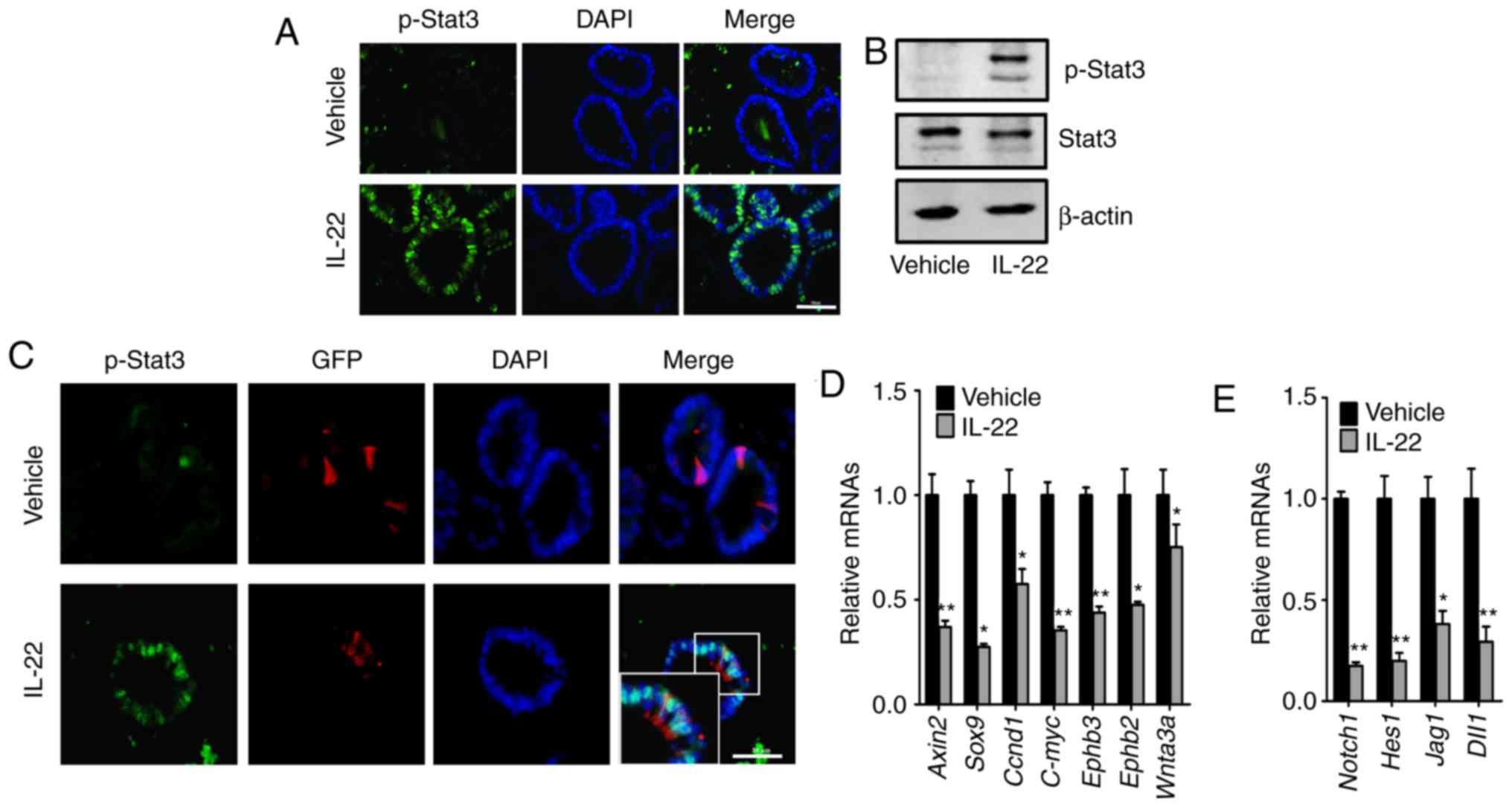
IL-22 promotes the phosphorylation of
Stat3 and suppresses the Wnt and Notch signaling pathways
It has been reported that IL-22 modulates the
phosphorylation of Stat3 (18,28). In this regard, we assessed the
changes in the levels of phosphorylated Stat3 following treatment
of the organoids with IL-22. In the assay of immunofluorescence, we
observed that the addition of IL-22 increased the level of
phosphorylated Stat3 in the nucleus of the organoids (Fig. 4A and C). Consistently, similar
results were got by immunoblotting (Fig. 4B). Moreover, in the organoids of
Lgr5-EGFP-IRES-creERT2 mice, increased levels of nuclear
phosphorylated Stat3 were observed in the Lgr5-positive ISCs
following treatment with IL-22 (Fig.
4C).
We then examined the activity of the Wnt and Notch
signaling pathways, which are essential for ISC self-renewal and
differentiation. It was found that both the Wnt and Notch signaling
pathways were markedly suppressed by IL-22 (Fig. 4D and E). In summary, these results
demonstrate that IL-22 activates the Stat3, and inhibits the Wnt
and Notch signaling pathways.
IL-22 leads to defects in organoid
formation mainly through the Wnt pathway
To understand the importance of signaling pathway
changes caused by IL-22, we carried out rescue experiments. We
applied WP1066 and Stattic as Stat3 specific inhibitors, and added
Wnt3a and Jag1 as respective ligands for the Wnt and Notch
pathways.
In the absence of IL-22, these supplements did not
affect the formation and survival of organoids (Fig. 5A and B). Notably, only the
addition of Wnt3a recovered the defects in the organoids induced by
the addition of IL-22 (43% vs. 7%; Fig. 5A and B). Furthermore, we found
that the number of Paneth cells was retrieved by Wnt3a, but not by
Stattic, WP1066 and Jag1 (Fig.
5D). Notably, Stattic treatment significantly restrained
IL-22-induced cell hyper-proliferation (Fig. 5C). This observation suggested that
IL-22 promotes cell proliferation mainly through Stat3 activation.
Consistently, the upregulation of Wnt target genes, including Axin2
and Ephb3, was confirmed by RT-qPCR following the addition of Wnt3a
in IL-22-treated organoids (Fig.
5E). Moreover, the levels of ISC markers (Lgr5, Olfm4 and
Znrf3) were elevated as well. Taken together, these data suggest
that IL-22 caused the deficiency of organoid formation mainly by
suppressing the Wnt signaling pathway.
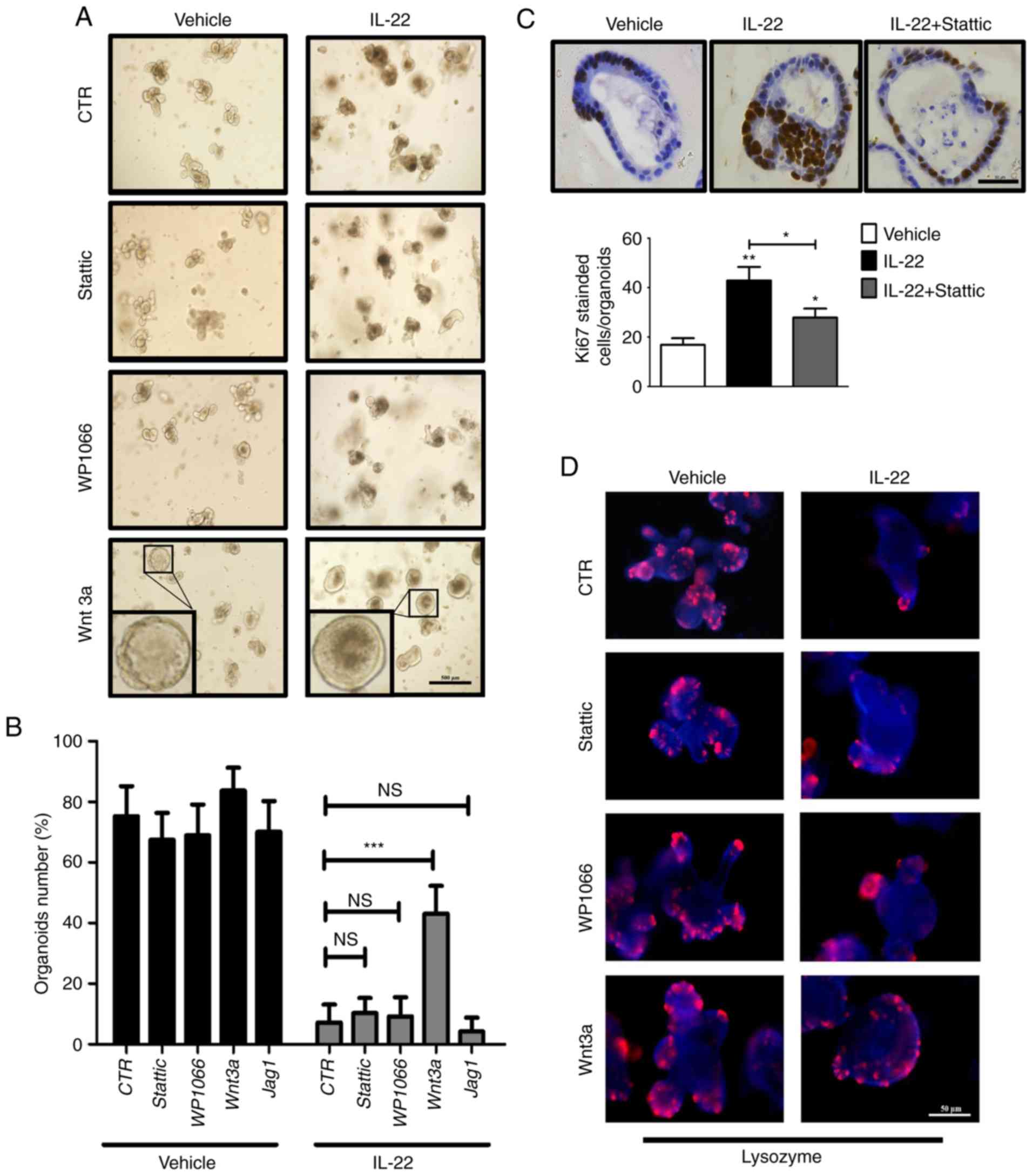 | Figure 5Interleukin (IL)-22 causes defects in
organoid formation mainly through the Wnt pathway. (A) Organoids
were cultured for 2 days followed by treatment with various
combinations of IL-22 (100 ng/ml), Wnt3a (200 ng/ml), WP1066 (5
μM) and Stattic (3 μM). Scale bar, 500 μm. The
inset is the magnification of one representative organoid. (B) The
number of organoids after 2 days of treatment with various
combinations of IL-22, Wnt3a, WP1066, Stattic and Jag1 was counted.
Data are the means ± SEM; and ***P<0.001 vs. Control
(100 ng/ml IL-22 treatment group); NS, not significant; analyzed by
one-way ANOVA. (C) Organoids were cultured for 2 days followed by
treatment with various combinations of IL22 and Stattic. The
expression of Ki67 was detected by immunohistochemical staining.
Scale bar, 50 μm. The quantitative analysis is presented at
the bottom panel. Data are the means ± SEM; *P<0.05,
**P<0.01 and vs. the vehicle; analyzed by one-way
ANOVA. (D) Organoids were cultured for 2 days followed by treatment
with various combinations of IL-22, Wnt3a, WP1066 and Stattic. The
expression of Lysozyme (Paneth cell marker) was determined by
immunofluorescence. Scale bar, 50 μm. (E) Organoids were
cultured for 2 days followed by treatment with various combinations
of IL-22 and Wnt3a. The expression of ISC markers (Lgr5, Olfm4 and
Znrf3) and Wnt target genes (Axin2 and Ephb3) was examined by
RT-qPCR. Data are the means ± SEM; *P<0.05,
**P<0.01 and vs. Control (the organoid cultured
without IL-22) or Control (100 ng/ml IL-22 treatment group);
analyzed by t-test. |
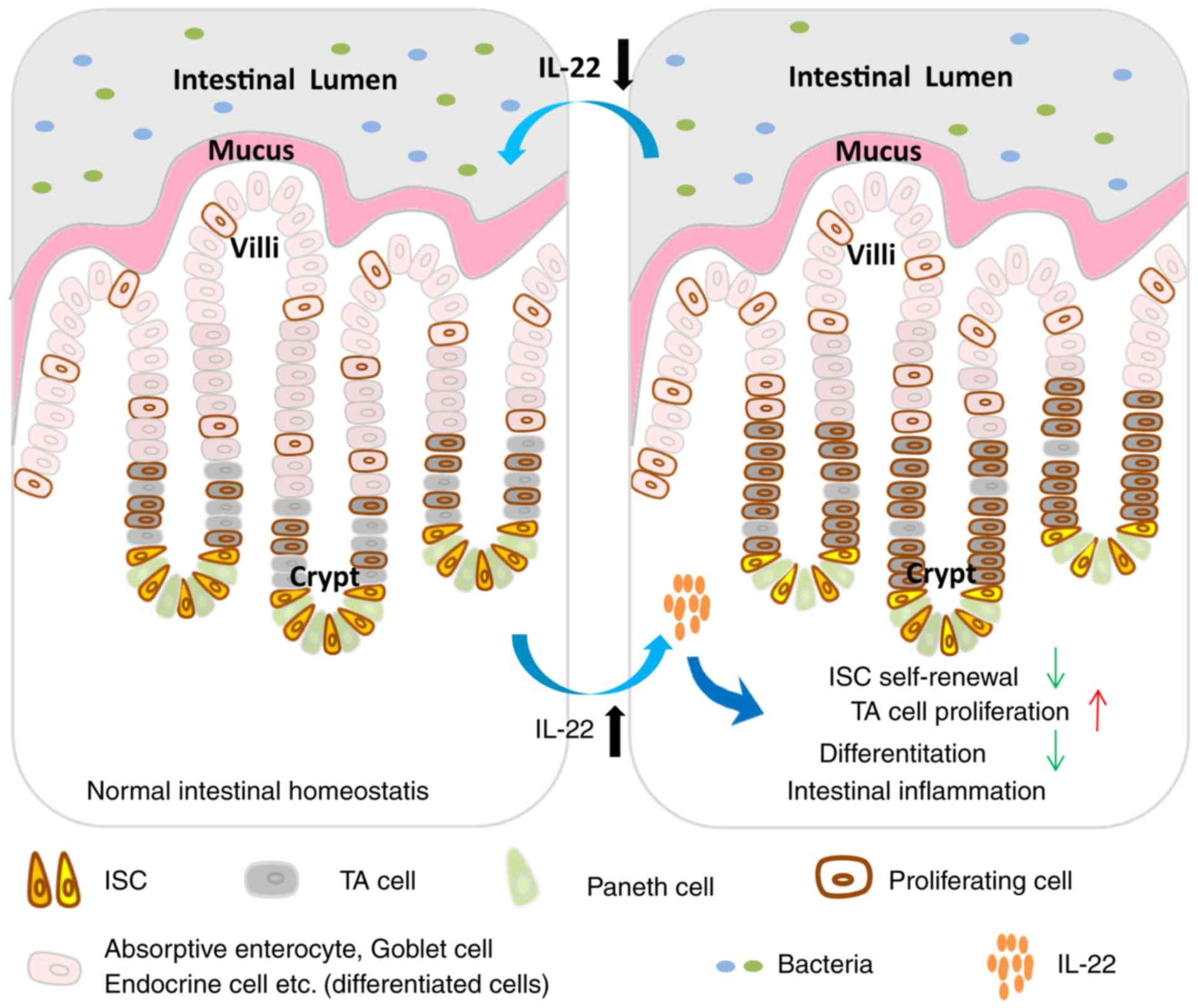
Taken together, in this study, we observed that the
IL-22 level initially increased and gradually decreased during
inflammation. Elevated IL-22 levels of modulated the Stat3, Wnt and
Notch pathways to accelerate the proliferation of TA cells,
suppressing cell differentiation and causing the deficiency of ISC
self-renewal (Fig. 6).
Discussion
It has been reported that IL-22 is a dual-natured
cytokine depending on the context of inflammation. It has either
pro-inflammatory or anti-inflammatory properties (13,29). In the majority of inflammatory
bowel disease animal models (14,18,28,30,31), infectious disease (32-34) and graft versus host disease (GVHD)
(35), IL-22 plays a protective
role against inflammation. By contrast, in a Treg cell transferring
animal model and during Toxoplasma gondii-induced ileitis,
IL-22 has been shown to play a pro-inflammatory function (20,36). In this respect, a more precise
association between IL-22 and epithelial cells needs to be
determined. In this study, we used an ex vivo organoid
culture system to investigate the role of IL-22 in the homeostasis
of intestinal epithelium during inflammation. The advantage of this
system is that it can rule out the effects of other cells
surrounding the IECs, such as mesenchyme cells and leukocytes. In
this manner, we discovered a previously unknown direct association
between IL-22 and the intestinal epithelium in that IL-22 promotes
the proliferation but inhibits the differentiation of intestinal
organoids. Moreover, it suppresses the self-renewal ability of ISCs
and consequently leads to the death of organoids. These data reveal
that IL-22 has more complex functions in IECs than what was
previously known.
Following IL-22 treatment, we observed that the
organ-oids became rounded without buds whose tips were enriched
with ISCs and Paneth cells (21,22,37). In addition, fewer terminal
differentiated cells were found in IL-22-treated organoids. It has
been known that the downregulation of Wnt signaling suppresses EphB
expression and thus impairs the ephrinB-EphB repulsion, which is
critical for the crypt-villi axis establishment (38,39). Thus, the rounded structure caused
by IL-22 treatment is likely attributed to the accelerated
proliferation of IECs and the loss of migration axis, causing the
cells to detach from the epithelial structures and to shed into the
lumen.
In vivo, IL-22 expression was upregulated
within 6-8 days of DSS-induced inflammation and on the 3rd day of
X-ray radiation-induced inflammation; during this period, the
intestine incurs the most severe inflammatory response. We
postulate that high level of IL-22 promotes anti-microbial peptide
expression and alleviates the bacterial burden, which is critical
for the initiation of intestinal regeneration (32,40,41). However, following bacterial
clearance, the epithelium begins to recover and reconstitute; the
limitation of IL-22 is important for IEC differentiation. The
decreased IL-22 level is possible due to the reduced number of
adaptive immune cells, such as Th17 and Th22, which are the source
of IL-22, while responding the inflammatory stresses and bacterial
states (33,42-44).
The stem cell niche is indispensable for the
homeostasis of the intestinal epithelium. In this study, we
observed that IL-22 activates Stat3 signaling to promote the
hyper-proliferation of intestinal organoids. Moreover, IL-22
inhibited the Wnt and Notch signaling pathways in IECs. Notably,
only Wnt3a rescued the organoid defects caused by IL-22, which
suggests that the Wnt pathway is a key factor, downstream of IL-22,
to regulate intestinal homeostasis. A recent study indicated that
Wnt signaling synergizes with R-spondin to maintain ISC
self-renewal (45). However, the
mechanisms through which IL-22 influences the Wnt pathway remain
unclear. One possibility is that IL-22 directly modulates important
players controlling Wnt signaling. Another possibility is that
IL-22 suppresses the differentiation of Paneth cells which secret
Wnt ligands for ISCs and IECs.
Our results are not consistent with those from a
previous study, which demonstrated that IL-22 maintained the
self-renewal ability of Lgr5-positive stem cells by using an IL-22
knockout mouse strain (35).
IL-22 has been reported to have diverse functions in the intestine.
For example, it can promote antibacterial peptide secretion and
mucus production (18). We
hypothesized that the loss of IL-22 may cause global consequences,
such as impairing bacterial clearance, and therefore leads to the
destruction of the intestinal barrier and structure.
Inconsistently, another study recently demonstrated that IL-22
promotes the proliferation and expansion of ISCs in intestinal
organoids (46). Of note, we used
a much higher concentration of IL-22 (100 ng/ml vs. 5-10 ng/ml) in
this study. This is more coincident with the high level of IL-22
expression during inflammation (47). We reason that a greater amount of
IL-22 likely reaches a certain threshold that causes the
suppression of Wnt signaling. Additionally, given that IL-22
receptors are also bound to other cytokines (10,48-50), a greater amount of IL-22 may
competitively inhibit other signaling, such as IL-20 and IL-24
(48), which affect ISC
self-renewal.
Given that IL-22 is well documented to regulate
barrier immunity and anti-microbiota, it is considered to play a
protective role in IBD (51).
Agents to activate the IL-22 pathway are being developed for the
treatment of patients with IBD. For example, Indigo
naturalis has been shown to be beneficial for ulcerative
colitis in a clinical trial (52). However, a previous study
demonstrated that the continuous stimulation of IL-22 potentially
increases the risk of colitis-associated cancer (53). In this study, we found that the
dynamic alteration of IL-22 was important for the maintenance of
intestinal homeostasis during inflammation (Fig. 6), which sheds light into the role
of IL-22 in regulating the homeostasis of the intestinal epithelium
during inflammation. Thus, this study suggests that a prolonged
administration of agents to activate the IL-22 pathway for IBD
treatment may not be beneficial. Our findings further the current
understanding of IL-22 and may aid in the clinical therapy of
IBD.
In conclusion, in this study, we found that IL-22
treatment promoted cell proliferation, suppressed the
differentiation of intestinal organoids and led to self-renewal
defects of intestinal stem cells. As regards the underlying
mechanisms, IL-22 activated Stat3 phosphorylation and suppressed
the Wnt and Notch signaling pathways. This study reveals that IL-22
regulates the homeostasis of the intestinal epithelium and is
critical for the regeneration of the intestine during inflammation.
Thus, it deepens our current understanding of the regulation and
function of IL-22 and provides the theoretical basis for IBD
treatment.
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 81772622 to JZ) and the
Animal Research Project of Shanghai Science and Technology
Commision (grant no. 16140902000 to XuZ).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors’ contributions
XiZ, JZ, YC, XuZ designed the experiments. XiZ, SL,
YWa, HH, LL performed the experiments. SL DC and YWu performed the
data analysis. YWu contributed materials/facilities. XiZ, JZ, YC
and XuZ wrote the manuscript. All authors have read and approved
the final manuscript.
Ethics approval and consent to
participate
All animal experiments conformed to the regulations
drafted by the Association for Assessment and Accreditation of
Laboratory Animal Care in Shanghai and were approved by the East
China Normal University Center for Animal Research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Sturm A and Dignass AU: Epithelial
restitution and wound healing in inflammatory bowel disease. World
J Gastroenterol. 14:348–353. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Artis D: Epithelial-cell recognition of
commensal bacteria and maintenance of immune homeostasis in the
gut. Nat Rev Immunol. 8:411–420. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hooper LV and Macpherson AJ: Immune
adaptations that maintain homeostasis with the intestinal
microbiota. Nat Rev Immunol. 10:159–169. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kreymborg K, Etzensperger R, Dumoutier L,
Haak S, Rebollo A, Buch T, Heppner FL, Renauld JC and Becher B:
IL-22 is expressed by Th17 cells in an IL-23-dependent fashion, but
not required for the development of autoimmune encephalomyelitis. J
Immunol. 179:8098–8104. 2007. View Article : Google Scholar
|
|
5
|
Satoh-Takayama N, Vosshenrich CA,
Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention JJ, Thiam
K, Cerf-Bensussan N, Mandelboim O, et al: Microbial flora drives
interleukin 22 production in intestinal NKp46+ cells that provide
innate mucosal immune defense. Immunity. 29:958–970. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cella M, Fuchs A, Vermi W, Facchetti F,
Otero K, Lennerz JK, Doherty JM, Mills JC and Colonna M: A human
natural killer cell subset provides an innate source of IL-22 for
mucosal immunity. Nature. 457:722–725. 2009. View Article : Google Scholar
|
|
7
|
Cupedo T, Crellin NK, Papazian N, Rombouts
EJ, Weijer K, Grogan JL, Fibbe WE, Cornelissen JJ and Spits H:
Human fetal lymphoid tissue-inducer cells are interleukin
17-producing precursors to RORC+ CD127+
natural killer-like cells. Nat Immunol. 10:66–74. 2009. View Article : Google Scholar
|
|
8
|
Luci C, Reynders A, Ivanov II, Cognet C,
Chiche L, Chasson L, Hardwigsen J, Anguiano E, Banchereau J,
Chaussabel D, et al: Influence of the transcription factor
RORgammat on the development of NKp46+ cell populations
in gut and skin. Nat Immunol. 10:75–82. 2009. View Article : Google Scholar
|
|
9
|
Sanos SL, Bui VL, Mortha A, Oberle K,
Heners C, Johner C and Diefenbach A: RORgammat and commensal
microflora are required for the differentiation of mucosal
interleukin 22-producing NKp46+ cells. Nat Immunol.
10:83–91. 2009. View Article : Google Scholar
|
|
10
|
Ouyang W, Rutz S, Crellin NK, Valdez PA
and Hymowitz SG: Regulation and functions of the IL-10 family of
cytokines in inflammation and disease. Annu Rev Immunol. 29:71–109.
2011. View Article : Google Scholar
|
|
11
|
Sawa S, Lochner M, Satoh-Takayama N,
Dulauroy S, Bérard M, Kleinschek M, Cua D, Di Santo JP and Eberl G:
RORγt+ innate lymphoid cells regulate intestinal
homeostasis by integrating negative signals from the symbiotic
microbiota. Nat Immunol. 12:320–326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Spits H and Di Santo JP: Regulators and
effectors of immunity and tissue remodeling. Nat Immunol. 12:21–27.
2011. View Article : Google Scholar
|
|
13
|
Zenewicz LA and Flavell RA: Recent
advances in IL-22 biology. Int Immunol. 23:159–163. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zindl CL, Lai JF, Lee YK, Maynard CL,
Harbour SN, Ouyang W, Chaplin DD and Weaver CT: IL-22-producing
neutrophils contribute to antimicrobial defense and restitution of
colonic epithelial integrity during colitis. Proc Natl Acad Sci
USA. 110:12768–12773. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xie MH, Aggarwal S, Ho WH, Foster J, Zhang
Z, Stinson J, Wood WI, Goddard AD and Gurney AL: Interleukin
(IL)-22, a novel human cytokine that signals through the interferon
receptor-related proteins CRF24 and IL-22R. J Biol Chem.
275:31335–31339. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kotenko SV, Izotova LS, Mirochnitchenko
OV, Esterova E, Dickensheets H, Donnelly RP and Pestka S:
Identification of the functional interleukin-22 (IL-22) receptor
complex: The IL-10R2 chain (IL-10Rbeta) is a common chain of both
the IL-10 and IL-22 (IL-10-related T cell-derived inducible factor,
IL-TIF) receptor complexes. J Biol Chem. 276:2725–2732. 2001.
View Article : Google Scholar
|
|
17
|
Pestka S, Krause CD, Sarkar D, Walter MR,
Shi Y and Fisher PB: Interleukin-10 and related cytokines and
receptors. Ann Rev Immunol. 22:929–979. 2004. View Article : Google Scholar
|
|
18
|
Sugimoto K, Ogawa A, Mizoguchi E,
Shimomura Y, Andoh A, Bhan AK, Blumberg RS, Xavier RJ and Mizoguchi
A: IL-22 ameliorates intestinal inflammation in a mouse model of
ulcerative colitis. J Clin Invest. 118:534–544. 2008.PubMed/NCBI
|
|
19
|
Honda K: IL-22 from T cells: Better late
than never. Immunity. 37:952–954. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kamanaka M, Huber S, Zenewicz LA, Gagliani
N, Rathinam C, O’Connor W Jr, Wan YY, Nakae S, Iwakura Y, Hao L and
Flavell RA: Memory/effector (CD45RB(lo)) CD4 T cells are controlled
directly by IL-10 and cause IL-22-dependent intestinal pathology. J
Exp Med. 208:1027–1040. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sato T, Vries RG, Snippert HJ, van de
Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters
PJ and Clevers H: Single Lgr5 stem cells build crypt-villus
structures in vitro without a mesenchymal niche. Nature.
459:262–265. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sato T, van Es JH, Snippert HJ, Stange DE,
Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M
and Clevers H: Paneth cells constitute the niche for Lgr5 stem
cells in intestinal crypts. Nature. 469:415–418. 2011. View Article : Google Scholar
|
|
23
|
Dekkers JF, Wiegerinck CL, de Jonge HR,
Bronsveld I, Janssens HM, de Winter-de Groot KM, Brandsma AM, de
Jong NW, Bijvelds MJ, Scholte BJ, et al: A functional CFTR assay
using primary cystic fibrosis intestinal organoids. Nat Med.
19:939–945. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yilmaz ÖH, Katajisto P, Lamming DW,
Gültekin Y, Bauer-Rowe KE, Sengupta S, Birsoy K, Dursun A, Yilmaz
VO, Selig M, et al: mTORC1 in the Paneth cell niche couples
intestinal stem-cell function to calorie intake. Nature.
486:490–495. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yui S, Nakamura T, Sato T, Nemoto Y,
Mizutani T, Zheng X, Ichinose S, Nagaishi T, Okamoto R, Tsuchiya K,
et al: Functional engraftment of colon epithelium expanded in vitro
from a single adult Lgr5(+) stem cell. Nat Med. 18:618–623. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sato T and Clevers H: Growing
self-organizing mini-guts from a single intestinal stem cell:
Mechanism and applications. Science. 340:1190–1194. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
28
|
Pickert G, Neufert C, Leppkes M, Zheng Y,
Wittkopf N, Warntjen M, Lehr HA, Hirth S, Weigmann B, Wirtz S, et
al: STAT3 links IL-22 signaling in intestinal epithelial cells to
mucosal wound healing. J Exp Med. 206:1465–1472. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sonnenberg GF, Fouser LA and Artis D:
Border patrol: Regulation of immunity, inflammation and tissue
homeostasis at barrier surfaces by IL-22. Nat Immunol. 12:383–390.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zenewicz LA, Yancopoulos GD, Valenzuela
DM, Murphy AJ, Stevens S and Flavell RA: Innate and adaptive
interleukin-22 protects mice from inflammatory bowel disease.
Immunity. 29:947–957. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cox JH, Kljavin NM, Ota N, Leonard J,
Roose-Girma M, Diehl L, Ouyang W and Ghilardi N: Opposing
consequences of IL-23 signaling mediated by innate and adaptive
cells in chemically induced colitis in mice. Mucosal Immunol.
5:99–109. 2012. View Article : Google Scholar
|
|
32
|
Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa
SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ and
Ouyang W: Interleukin-22 mediates early host defense against
attaching and effacing bacterial pathogens. Nat Med. 14:282–289.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Basu R, O’Quinn DB, Silberger DJ, Schoeb
TR, Fouser L, Ouyang W, Hatton RD and Weaver CT: Th22 cells are an
important source of IL-22 for host protection against
enteropathogenic bacteria. Immunity. 37:1061–1075. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zenewicz LA, Yin X, Wang G, Elinav E, Hao
L, Zhao L and Flavell RA: IL-22 deficiency alters colonic
microbiota to be transmissible and colitogenic. J Immunol.
190:5306–5312. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hanash AM, Dudakov JA, Hua G, O’Connor MH,
Young LF, Singer NV, West ML, Jenq RR, Holland AM, Kappel LW, et
al: Interleukin-22 protects intestinal stem cells from
immune-mediated tissue damage and regulates sensitivity to graft
versus host disease. Immunity. 37:339–350. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Muñoz M, Heimesaat MM, Danker K, Struck D,
Lohmann U, Plickert R, Bereswill S, Fischer A, Dunay IR, Wolk K, et
al: Interleukin (IL)-23 mediates Toxoplasma gondii-induced
immu-nopathology in the gut via matrixmetalloproteinase-2 and IL-22
but independent of IL-17. J Exp Med. 206:3047–3059. 2009.
View Article : Google Scholar
|
|
37
|
Farin HF, Van Es JH and Clevers H:
Redundant sources of Wnt regulate intestinal stem cells and promote
formation of Paneth cells. Gastroenterology. 143:1518–1529.e7.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Batlle E, Henderson JT, Beghtel H, van den
Born MM, Sancho E, Huls G, Meeldijk J, Robertson J, van de Wetering
M, Pawson T and Clevers H: Beta-catenin and TCF mediate cell
positioning in the intestinal epithelium by controlling the
expression of EphB/ephrinB. Cell. 111:251–263. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cortina C, Palomo-Ponce S, Iglesias M,
Fernández-Masip JL, Vivancos A, Whissell G, Humà M, Peiró N,
Gallego L, Jonkheer S, et al: EphB-ephrin-B interactions suppress
colorectal cancer progression by compartmentalizing tumor cells.
Nat Genet. 39:1376–1383. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wolk K, Kunz S, Witte E, Friedrich M,
Asadullah K and Sabat R: IL-22 increases the innate immunity of
tissues. Immunity. 21:241–254. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Raffatellu M, George MD, Akiyama Y,
Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL,
Berger T, et al: Lipocalin-2 resistance confers an advantage to
Salmonella enterica serotype Typhimurium for growth and survival in
the inflamed intestine. Cell Host Microbe. 5:476–486. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chung Y, Yang X, Chang SH, Ma L, Tian Q
and Dong C: Expression and regulation of IL-22 in the
IL-17-producing CD4+ T lymphocytes. Cell Res.
16:902–907. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liang SC, Tan XY, Luxenberg DP, Karim R,
Dunussi-Joannopoulos K, Collins M and Fouser LA: Interleukin
(IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively
enhance expression of antimicrobial peptides. J Exp Med.
203:2271–2279. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zheng Y, Danilenko DM, Valdez P, Kasman I,
Eastham-Anderson J, Wu J and Ouyang W: Interleukin-22, a T(H)17
cytokine, mediates IL-23-induced dermal inflammation and
acanthosis. Nature. 445:648–651. 2007. View Article : Google Scholar
|
|
45
|
Yan KS, Janda CY, Chang J, Zheng GXY,
Larkin KA, Luca VC, Chia LA, Mah AT, Han A, Terry JM, et al:
Non-equivalence of Wnt and R-spondin ligands during
Lgr5+ intestinal stem-cell self-renewal. Nature.
545:238–242. 2007. View Article : Google Scholar
|
|
46
|
Lindemans CA, Calafiore M, Mer telsmann A
M, O’Connor MH, Dudakov JA, Jenq RR, Velardi E, Young LF, Smith OM,
Lawrence G, et al: Interleukin-22 promotes
intestinal-stem-cell-mediated epithelial regeneration. Nature.
528:560–564. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Andoh A, Zhang Z, Inatomi O, Fujino S,
Deguchi Y, Araki Y, Tsujikawa T, Kitoh K, Kim-Mitsuyama S,
Takayanagi A, et al: Interleukin-22, a member of the IL-10
subfamily, induces inflammatory responses in colonic subepithelial
myofibroblasts. Gastroenterology. 129:969–984. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dumoutier L, Leemans C, Lejeune D, Kotenko
SV and Renauld JC: Cutting edge: STAT activation by IL-19, IL-20
and mda-7 through IL-20 receptor complexes of two types. J Immunol.
167:3545–3549. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sabat R: IL-10 family of cytokines.
Cytokine Growth Factor Rev. 21:315–324. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang M, Tan Z, Zhang R, Kotenko SV and
Liang P: Interleukin 24 (MDA-7/MOB-5) signals through two
heterodimeric receptors, IL-22R1/IL-20R2 and IL-20R1/IL-20R2. J
Biol Chem. 277:7341–7347. 2002. View Article : Google Scholar
|
|
51
|
Li LJ, Gong C, Zhao MH and Feng BS: Role
of interleukin-22 in inflammatory bowel disease. World J
Gastroenterol. 20:18177–18188. 2014. View Article : Google Scholar
|
|
52
|
Sugimoto S, Naganuma M, Kiyohara H, Arai
M, Ono K, Mori K, Saigusa K, Nanki K, Takeshita K, Takeshita T, et
al: Clinical efficacy and safety of oral Qing-Dai in patients with
ulcerative colitis: A single-center open-label prospective study.
Digestion. 93:193–201. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Low D, Mino-Kenudson M and Mizoguchi E:
Recent advancement in understanding colitis-associated
tumorigenesis. Inflamm Bowel Dis. 20:2115–2123. 2014. View Article : Google Scholar : PubMed/NCBI
|