Introduction
Despite its declining incidence, gastric cancer
remains a worldwide health problem that is the second leading cause
of cancer-related death with little improvement of long-term
survival during the past decades. In order to improve patient
prognosis and therapy, it is now mandatory to further understand
the molecular features involved in the pathogenesis. Although it
has been under intensive investigation, the molecular mechanism of
gastric cancer is not well understood and needs further
exploring.
In an attempt to identify new factors involved in
gastric cancer, we previously performed a comprehensive analysis of
differentially expressed genes in gastric cancer by SSH (1). RPS12 was identified as one of
the genes overexpressed in gastric cancer compared with the
corresponding non-tumorous tissue, which was further confirmed by
RT-PCR and DOT blot analysis in more samples. Clinicopathological
analysis revealed that the enhanced RPS12 expression was
related to metastasis of gastric cancer (1). Subsequent cDNA microarray studies
showed that RPS12 expression in lymph node metastases was
much higher than that in the primary gastric cancers (2). All the above results suggested that
RPS12 might play important roles in the progression and
metastasis of gastric cancer, which led us to investigate the
function of RPS12 in gastric cancer cells. Our previous
study showed that suppression of RPS12 expression by RNAi
led to increased apoptosis of gastric cancer cells (3). In the current study, we focused on
further investigating the effects of RPS12 on the
proliferation and migration of gastric cancer cells, and the
underlying mechanisms by exploring the downstream effector of
RPS12.
Materials and methods
Cell culture
The human gastric cancer cell line BGC823 and
SGC7901 were cultured in RPMI-1640 medium (Invitrogen, Carlsbad,
CA, USA) supplemented with 10% fetal bovine serum in a humidified
37°C incubator with 5% CO2.
Construction and transfection of
RPS12-specific short hairpin RNA (shRNA) expression vector
Two different RPS12 hairpin oligonucleotides
(oligos) were designed by selecting appropriate sequences from the
human RPS12 mRNA according to the online software of Ambion
Inc. (Austin, TX, USA). The first one named RPS12-shRNA-A
oligo (sense:
5′-GATCCagtggttggttgcagttgtTTCAAGAGAacaactgcaaccaaccactttA-3′;
anti sense:
5′-AGCTTaaagtggttggttgcagttgtTCTCTTGAAacaactgcaaccaaccactG-3′)
contained a region specific to bases 386–406 of RPS12 mRNA
(marked in bold); the second one named RPS12-shRNA-B oligo
(sense:
5′-GATCCgctgccaaagccttagacaTTCAAGAGAtgtctaaggctttggcagcttA-3′;
antisense: 5′-AGCTTaagctgccaaagccttagacaTCTCTT
GAAtgtctaaggctttggcagcG-3′) contained a region specific
to bases 192–212 of RPS12 mRNA (marked in bold). The
RPS12-shRNA-A or RPS12-shRNA-B oligo contained a
hairpin loop region (italicized), and 5′ and 3′ linker sequences
for subcloning into the BamHI and HindIII sites of
the pSliencer 4.1-CMV neo vectors. Each oligo was annealed with its
complementary strand, and the resulting double-stranded shRNA oligo
was cloned into the pSilencer 4.1-CMV neo vector (Ambion Inc.)
making pRPS12-shRNA-A or pRPS12-shRNA-B according to
the manufacturer’s instructions. The control shRNA expression
vector (pControl-shRNA) was the same as previously reported
(4). Competent JM109 bacteria
(Takara, Japan) were transformed with pRPS12-shRNA-A/B or
pControl-shRNA. The plasmids were prepared from individual
bacterial colonies, and confirmed by sequencing analysis.
pRPS12-shRNA-A/B or pControl-shRNA was
transfected into BGC823 cells respectively, using Lipofectamine™
2000. We selected the more efficient one in inhibiting RPS12
mRNA expression and renamed it as pRPS12-shRNA, this was
then used to inhibit RPS12 expression in BGC823 and SGC7901
cells in the subsequent studies.
The cells transfected with pRPS12-shRNA or
pControlshRNA were referred to as
BGC823/SGC7901/pRPS12-shRNA cells or
BGC823/SGC7901/pControl-shRNA cells, respectively.
RNA extraction and semi-quantitative
RT-PCR
Total RNA of the cells was extracted using TRIzol
reagent (Invitrogen). The reverse transcription reaction was
performed using the First-Strand cDNA synthesize kit (Promega,
Madison, WI, USA) with 1 μg of RNA in a final volume of 20
μl. The newly synthesized cDNA was amplified by PCR. Primers
specific for human RPS12, S100A4 or β-actin
were designed as follows: RPS12 sense
5′-ggCTTgggTgCgTTCAAgAT-3′, antisense 5′-ggCCT gAgACTCCTTgCCA-3′;
S100A4 sense 5′-gATgT gATggTgTC CACCTT-3′, antisense
5′-ATTTCTTCCTgggCTgCTTA-3′; β-actin sense
5′-CTCTTCCagCCTTCCTTCCT-3′, antisense 5′-CACCTTCACCgTTCCAgTTT-3′.
Amplication cycles were: 95°C for 5 min, then 30 cycles at 95°C for
30 sec, 58°C for 30 sec and 72°C for 30 sec, followed by 72°C for
10 min. Aliquots of PCR product were checked by electrophoresis on
1.5% agarose gels, and the fragments were visualized by ethidium
bromide staining. The experiments were performed three times.
Western blot analysis
Cells were washed with ice-cold PBS after being
trypsinized, and then centrifuged at 1,000 rpm for 5 min at 4°C.
The pellet containing 106 cells was lysed in 100
μl of lysis buffer (50 mM Tris, pH 7.2, 1% Triton X-100,
0.5% sodium deoxycholate, 0.1% SDS, 500 mM NaCl, 10 mM
MgCl2 with 10 μg/ml leupeptin, 10 μg/ml
aprotinin and 1 mM PMSF) and quantified by Bradford method. Total
protein (100 μg) was separated by SDS-PAGE (12%
polyacrylamide gel) and transferred to polyvinylidene difluoride
(PVDF) membranes (Bio-Rad). The membranes were incubated in
blocking solution consisting of 5% non-fat milk in TBST (10 mM
Tris-HCl (pH 8.0), 150 mM NaCl, and 0.1% Tween-20) at room
temperature for 3 h, then immunoblotted with primary antibodies
such as anti-RPS12 (1:800 dilution; Proteintech), anti-S100A4
(1:1,000 dilution; Lab Vision) and anti-β-actin (1:1,500 dilution;
Santa Cruz) respectively, followed by incubation with a
peroxidaseconjugated second antibody (goat anti-rabbit IgG for
RPS12, S100A4 and β-actin). The reagent for enhanced
chemiluminescence (Amersham, Freiburg, Germany) was used for
detection and developed on X-ray film. The experiments were
performed three times.
MTT assay
BGC823/SGC7901/pRPS12-shRNA cells and
BGC823/SGC7901/pControl-shRNA cells at 12 h to 3 days after
transfection were plated in 96-well plates at a density of
5×103 cells per well and incubated at 37°C in 5%
CO2. At different time points (day 3, 4, 5, 6, 7 and 8
after transfection), the proliferation ability was assayed by using
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
as previously reported (4). The
absorbance was read at a wavelength of 570 nm in Multiscan MC
microplate reader. Each experiment was performed in triplicate and
repeated three times.
Transwell assay
For cell migration assays, 5×104
BGC823/SGC7901/pRPS12-shRNA cells and
BGC823/SGC7901/pControl-shRNA cells at 4th day after transfection
were trypsinized, washed, resuspended in serum-free RPMI-1640, and
plated in the upper chamber of 8-μm pore transwells (Costar,
USA), respectively. The lower chamber was filled with RPMI-1640
supplemented with 10% FBS as a chemoattractant. Cells were
incubated at 37°C for 24 h. Non-migrated cells on the upper surface
of the insert membrane were removed with a cotton swab. Migrated
cells on the lower surface were washed with PBS, fixed with 10%
formaldehyde, stained, photographed and counted under high-power
magnification. Five random fields were analyzed for each chamber.
The experiments were performed three times.
Luciferase reporter assay
The plasmid pGL3-S100A4-luc vector containing
a regulatory fragment located in the promoter region and the first
intron of S100A4 gene (from −387 to +550), was constructed
in a previous study by our group (5). On day 4 after transfection of
RPS12-specific shRNA, BGC823/RPS12-shRNA cells at a
density of 1.5×104 cells/well in a 24-well plate were
transfected with 0.8 μg of the pGL3-S100A4-luc or
pGL3-basic-luc constructs, with Renilla luciferase (Promega) as the
internal control (5). After
incubation for 24 h, the cells were harvested, and luciferase
activity was measured using a Dual-Luciferase Reporter Assay System
(Promega) and normalized by the internal control values. The
experiments were performed three times.
Rescue assay after ectopic expression of
S100A4 in BGC823/pRPS12-shRNA cells
To investigate the effect of S100A4 on
gastric cancer cells after RPS12 suppression, at 48 h after
RPS12-sepcific shRNA transfection, BGC823/RPS12-shRNA
cells were transfected with 4 μg
pcDNA3.1-S100A4(6) using
Lipofectamine 2000, with pcDNA3.1-empty vector transfected cells as
a control. At different time points, cells were harvested for
determining the proliferation, and migration by MTT and transwell
assay, respectively, as described above.
Statistical analysis
The data of absorbance in MTT, cell numbers in
transwell and luciferase activities in luciferase reporter assay
are presented as the means ± SD. Statistical analyses were carried
out using the Statistical Package for the Social Sciences (SPSS
Inc., Chicago, IL, USA), where p<0.05 was considered
significant.
Results
Downregulation of RPS12 expression in
gastric cancer cells by RNAi
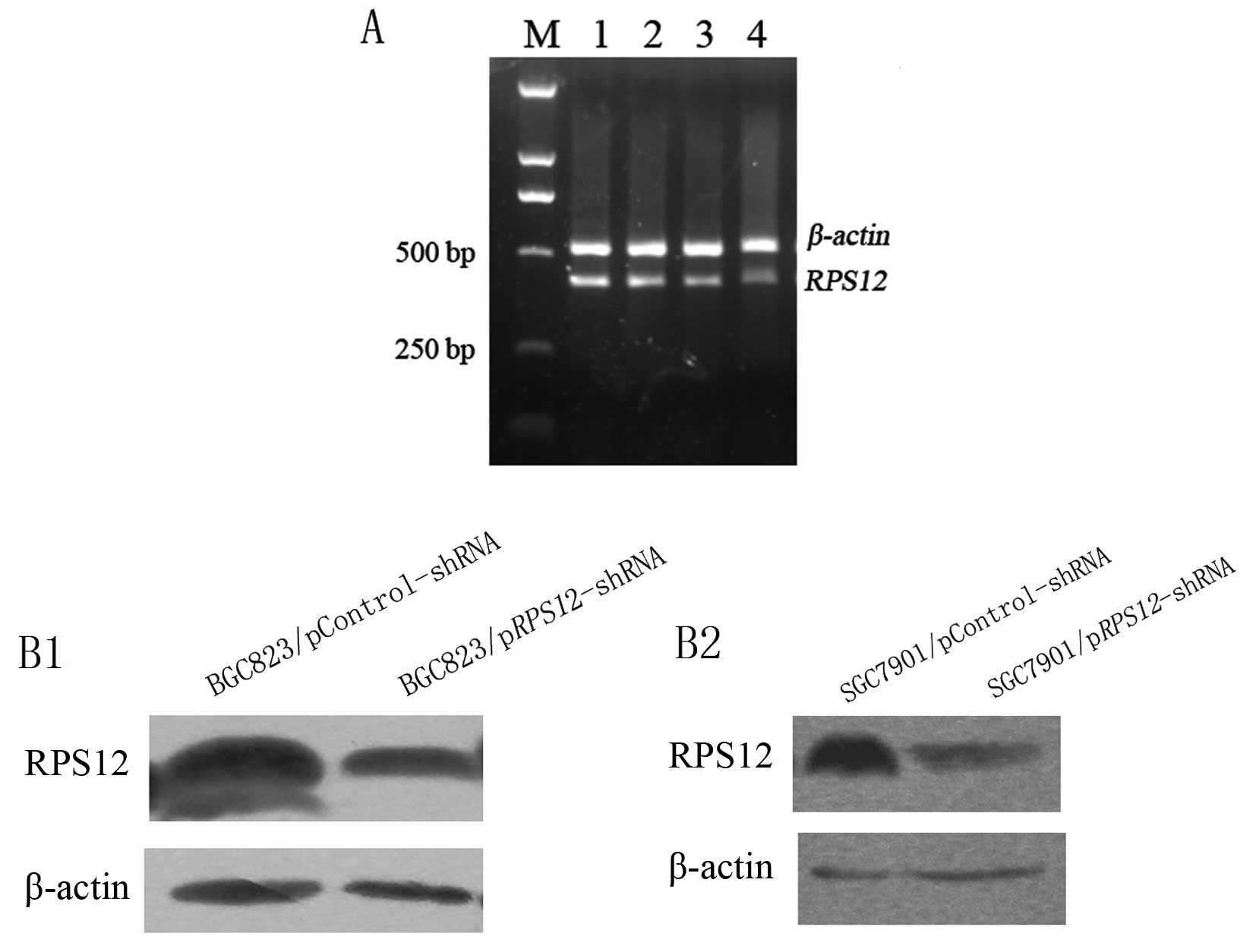
We first evaluated the efficiency of the two shRNAs
specific to different regions of RPS12 in silencing
RPS12 expression on day 5 after transfection by using
semi-quantitative RT-PCR (Fig.
1A). The results showed that RPS12 mRNA expression in
pRPS12-shRNA-B transfected BGC823 cells was obviously lower
than that in pRPS12-shRNA-A transfected cells indicating
that pRPS12-shRNA-B can suppress RPS12 expression
more efficiently than pRPS12-shRNA-A in BGC823 cells.
Therefore, we selected RPS12-shRNA-B (renamed
RPS12-shRNA) to inhibit RPS12 expression in BGC823
and SGC7901 cells in the subsequent studies, and observed the
cellular effects mainly at/or around day 5 after RPS12-shRNA
transfection.
The results of western blot analysis showed that
RPS12 protein expression in BGC823/SGC7901/pRPS12-shRNA
cells at day 5 after transfection was obviously lower than that in
BGC823/SGC7901/pControl-shRNA cells (Fig. 1B).
Downregulation of RPS12 expression leads
to decreased proliferation and migration of gastric cancer
cells
We observed the effects of RPS12 inhibition
on the proliferation and migration of BGC823 and SGC7901 cells. The
MTT assay showed that BGC823/SGC7901/pRPS12-shRNA cells
displayed significantly reduced proliferation, as compared to
BGC823/SGC7901/pControl-shRNA cells at different time points after
transfection of the corresponding shRNA (Fig. 2A). Transwell assays revealed that
the number of migrated BGC823/SGC7901/pRPS12-shRNA cells
(46.2±9.45, 54.68±8.43) was significantly fewer than that of their
corresponding pControl-shRNA cells (118.05±15.78, 78.4±27.49)
(p<0.05) (Fig. 2B and C).
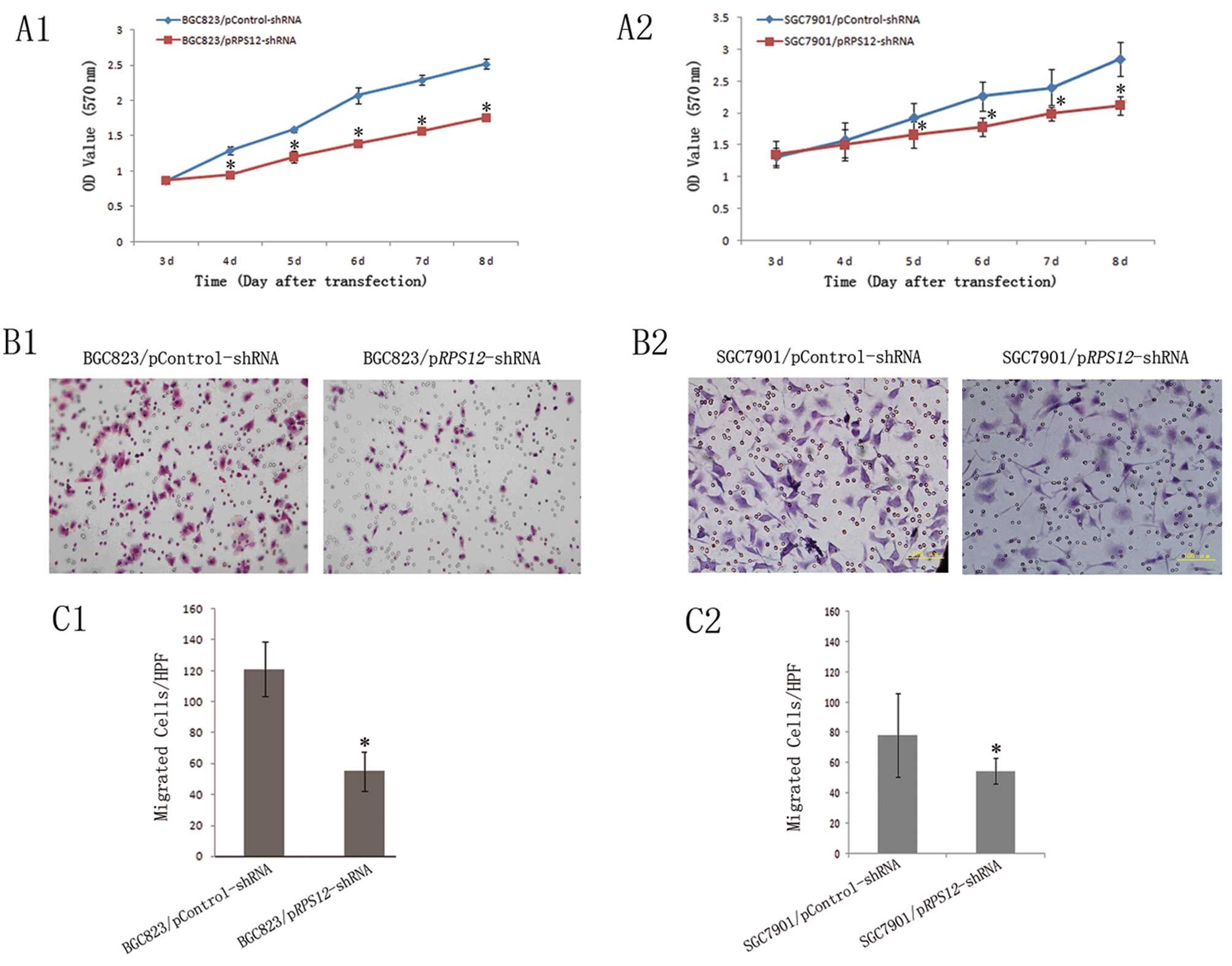 | Figure 2Downregulation of RPS12
expression inhibits proliferation and migration of gastric cancer
cell BGC823 and SGC7901. (A) Proliferation of
BGC823/SGC7901/pRPS12-shRNA and BGC823/SGC7901/pControl-shRNA cells
was analyzed by (A1–A2) MTT assay as described in Materials and
methods. On day 3, 4, 5, 6, 7 and 8 after transfection.
*p<0.05 vs the corresponding pControl-shRNA cells.
(B) The representative pictures of transwell assays for
BGC823/SGC7901/pRPS12-shRNA and
BGC823/SGC7901/pControl-shRNA cells (B1 and B2, BGC823 and SGC7901,
respectively), as described in Materials and methods. (C) The
numbers of migrated cells were plotted as mean from triplicate
experiments; *p<0.05 vs the corresponding
pControl-shRNA cells (C1 and C2, BGC823 and SGC7901, respectively).
Bars indicate SD. HPF, high power field (×200). |
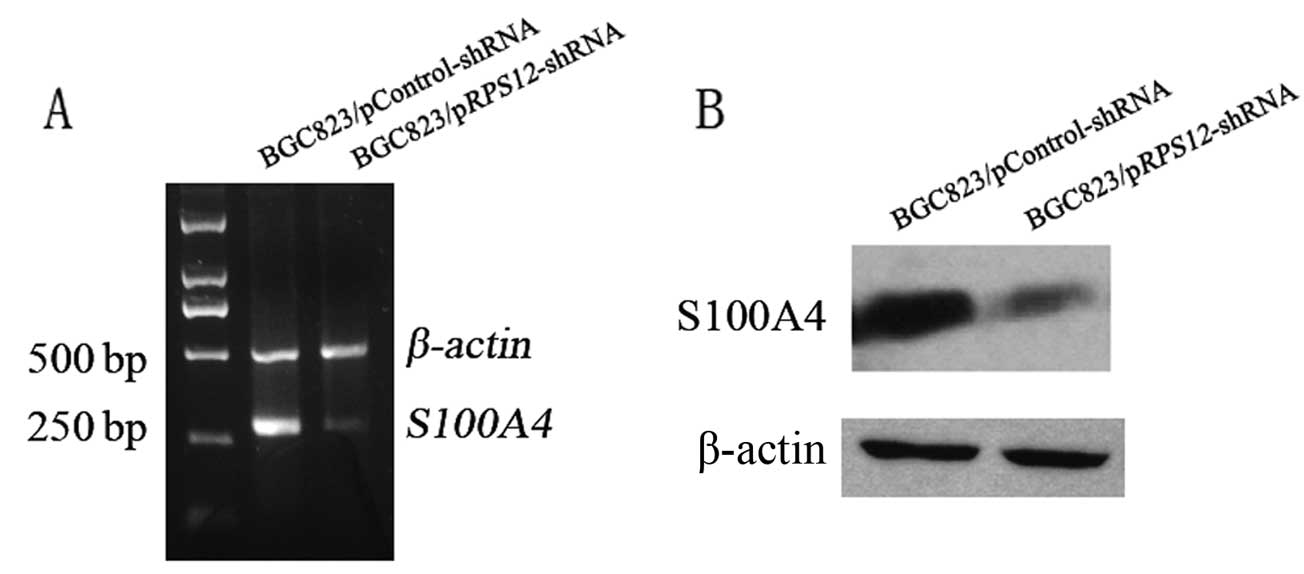
Effect of RPS12 inhibition on S100A4
expression in BGC8 3 cells
Having demonstrated that RPS12 plays
important roles in regulating proliferation and migration of
gastric cancer cells, we attempted to investigate the molecular
mechanism through which RPS12 exerts its functions. It has
been demonstrated by our group that S100A4 plays crucial
roles in regulating proliferation of gastric cancer cells (4), and also the migration of the cells
(unpublished). We hypothesized that S100A4 might mediate the
effect of RPS12 as its downstream target. To test our
hypothesis, S100A4 expression was determined by RT-PCR and
western blot analysis after RPS12 inhibition. We found that
BGC823/pRPS12-shRNA cells displayed an obviously decreased
S100A4 mRNA and protein expression compared with
BGC823/pControl-shRNA cells (Fig.
3).
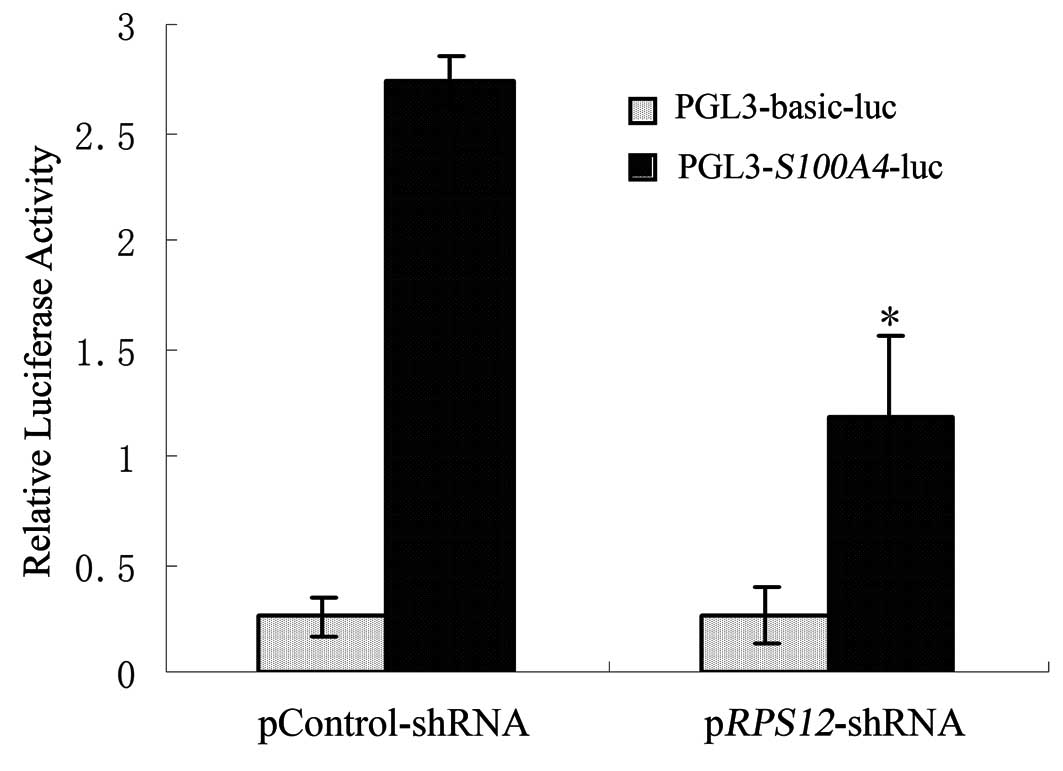
Effect of RPS12 inhibition on the
transcriptional activity of S100A4 promoter in BGC823 cells
In order to investigate whether RPS12
regulates the transcriptional activity of S100A4 promoter,
pGL3-S100A4-luc plasmid was transfected into
BGC823/pRPS12-shRNA cells or BGC823/pControlshRNA cell,
respectively. The results of Dual-Luciferase Reporter Assay showed
that luciferase activity of pGL3-S100A4-luc plasmid in
BGC823/pRPS12-shRNA cells was significantly lower than that
in BGC823/pControl-shRNA cells (Fig.
4).
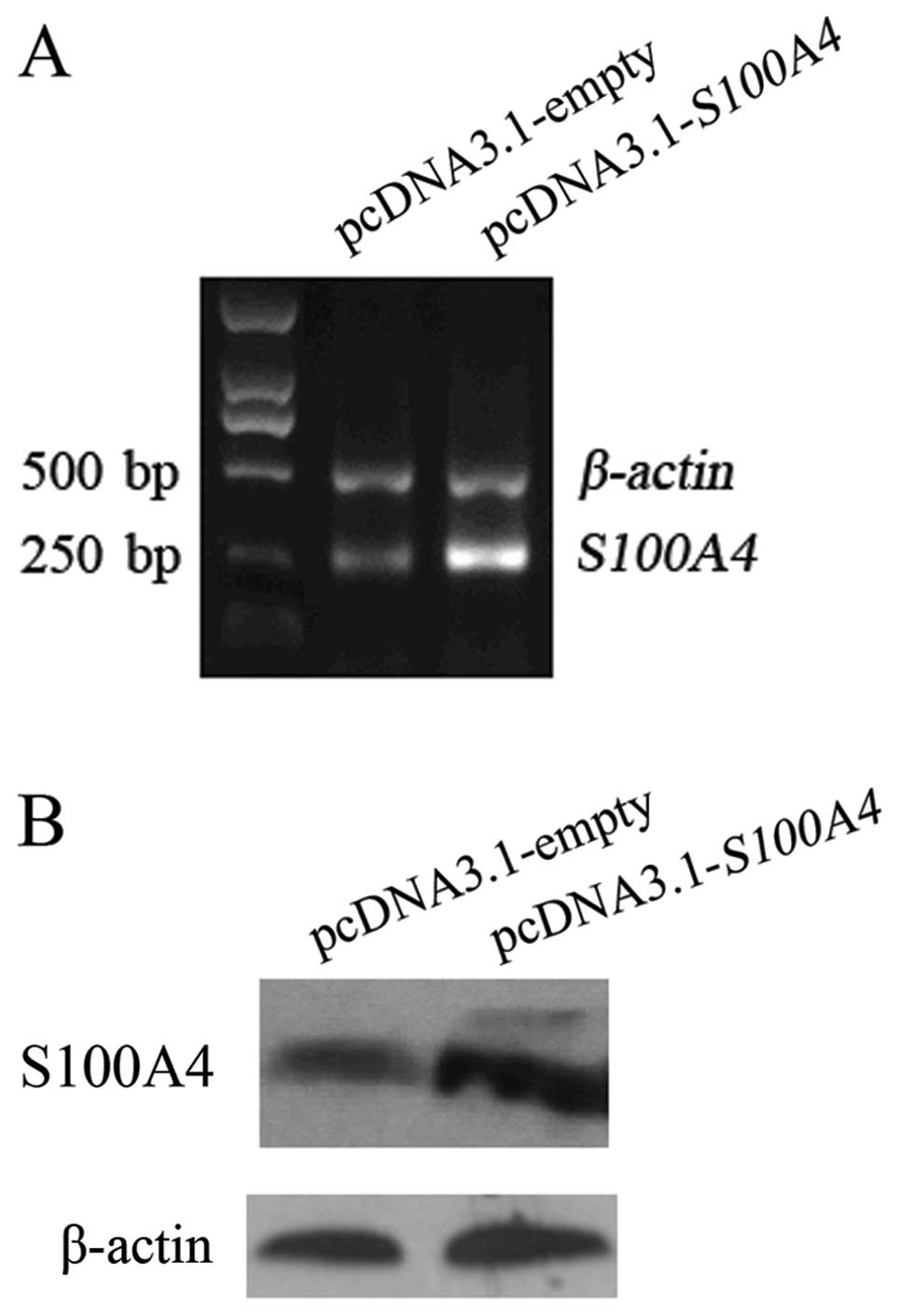
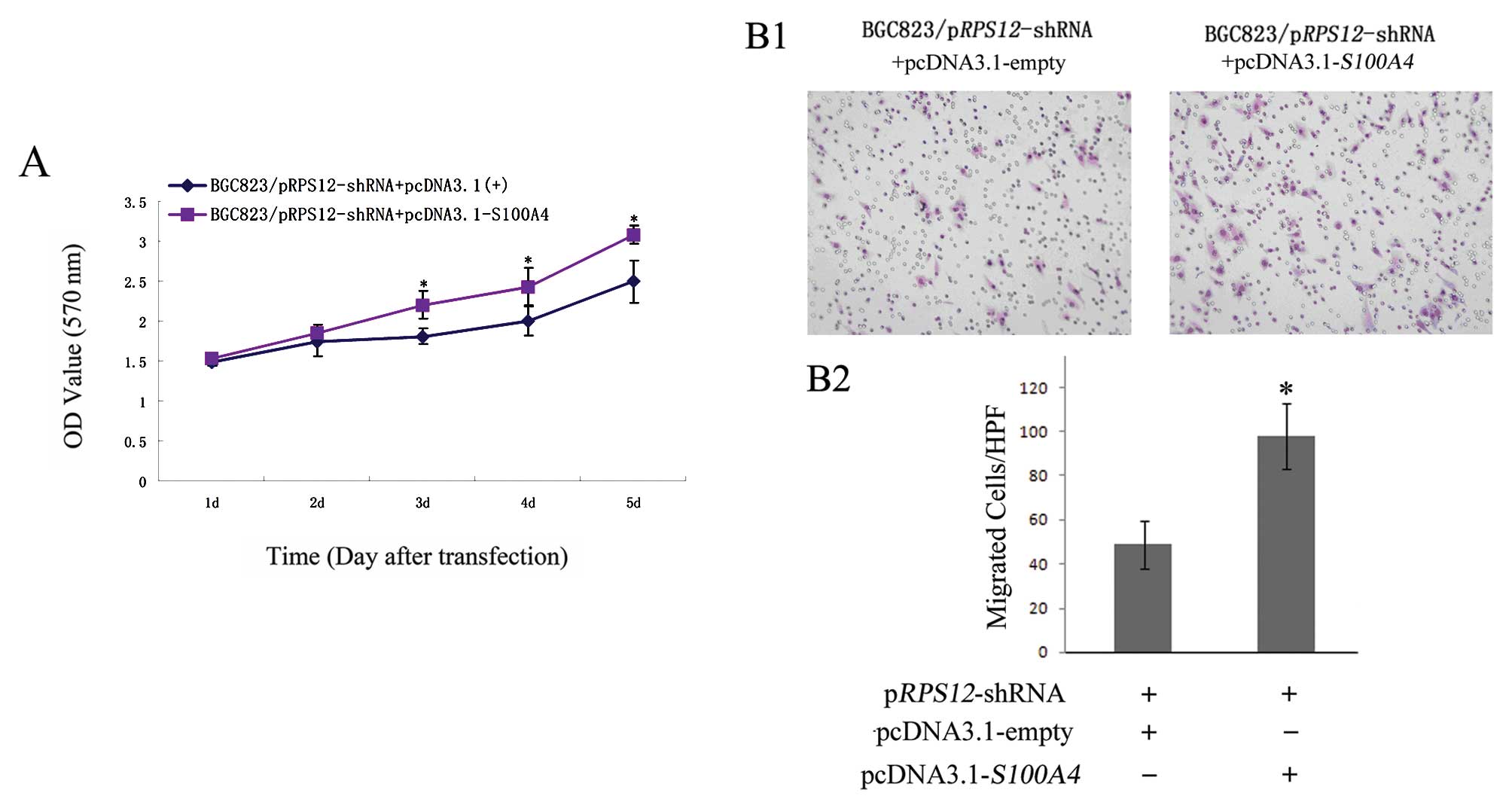
Ectopic expression of S100A4 reverses the
effects of RPS12 inhibition on the proliferation, migration of BGC8
3 cells
Having found that S100A4 is a molecular
target of RPS12, we further determined whether S100A4
could mediate the cellular effects of RPS12 by rescue assay.
As shown in Fig. 5, transfection
of pcDNA3.1-S100A4 into BGC823/pRPS12-shRNA cells led
to increased S100A4 expression in the cells at both mRNA and
protein level compared with pcDNA3.1-empty vector transfection. We
then found the following cellular effects. At day 3, 4 and 5 after
pcDNA3.1-S100A4 transfection, the proliferation of
BGC823/pRPS12-shRNA cells was significantly higher than that
after pcDNA3.1-empty vector transfection (p<0.05) (Fig. 6A). The number of migrated cells was
significantly increased in BGC823/pRPS12-shRNA cells after
pcDNA3.1-S100A4 transfection (97±11.37), as compared to that
in BGC823/pRPS12-shRNA cells after pcDNA3.1-empty vector
transfection (49±8.73) (p<0.05) (Fig. 6B).
Discussion
The ribosome is an organelle essential for protein
synthesis in cells, which is composed of ribosomal RNAs and
ribosomal proteins (RPs). At present, approximately 80 different
ribosomal proteins are found in eukaryotic ribosomes (7). It is very interesting that the
reports showing the involvements of RPs in various tumors are
emerging (8–14). Kasai et al(15) reported decreased expression of 10
RP genes and increased expression of 2 RP genes in colorectal
cancer compared with normal epithelia. Other reports have also
shown the overexpression of RP genes in different types of cancers,
such as S8, L12, L23a, L27 and
L30 mRNAs in human hepatocellular carcinoma (16), RPL13 in gastric cancer
(9), and RPL19 in
colorectal cancer (10).
Additionally, transcriptional loss of RPL14 was observed in
lung and oral cancers (17).
Reports on the association of abnormal expression of RPs with
tumorigenesis are still increasing. RPS12 gene, located in
6q23.2, encoding a ribosomal protein of 132 amino acids as a
component of the 40S subunit of ribosome, was isolated as a clone
overexpressed in gastric cancer in our previous studies by SSH.
Subsequent experiments showed that its overexpression was
associated with lymph node metastasis of gastric cancer (1,2). The
enhanced expression of RPS12 gene has also been reported in
colorectal cancer and squamous cell carcinoma of the uterine cervix
when compared with normal tissues (11,18).
O’Donohue et al reported depletion of RPS12 causes
delayed processing of 18S ribosomal RNA, although a less severe
phenotype is seen for RPS12 than for other small subunit
r-proteins (19). However, whether
or how the overexpression of RPS12 contributes to the
development and progression of gastric cancers is not clear.
We carried out the current study to investigate the
function of RPS12 in gastric cancer and demonstrated that
RPS12 suppression by RNAi led to reduced proliferation and
migration in gastric cancer cells BGC823 and SGC7901. The findings
indicate that RPS12 may take part in the development,
progression and metastasis of gastric cancer or prognosis of the
patients by regulating proliferation, migration of gastric cancer
cells. Similar to our findings on the cellular effects of
RPS12, it has been reported that inhibition of RPL15
expression suppressed the proliferation of gastric cancer cells
(20), while RPL44 enhanced
colony formation and cell proliferation of hepatocellular carcinoma
cells (13). Taken together, the
findings above suggest that aberrant expression of certain
ribosomal proteins contributes to tumorigenesis and progression of
certain types of cancers by regulating biological behavior of the
cancer cells.
Based on the important roles of RPS12 in
gastric cancer cells as described above, we try to decipher the
mechanisms through which RPS12 performs the functions.
Previous studies indicated that ribosomal proteins may exert their
roles by regulating the expression of downstream genes. Khanna
et al(21) demonstrated
that ribosomal protein S29 induced apoptosis in non-small
cell lung cancer cells by downregulating BCL-2,
BCL-XL and survivin, and also upregulating P53
and BAX expression. It was also reported that RPL6
inhibited cell cycle progression through downregulation of cyclin E
(22). Since we found that
RPS12 affected many biological characteristics of gastric
cancer cells, we investigated whether RPS12 might exert its
functions by regulating a multifunctional downstream target gene.
S100A4, also known as metastatin-1 (Mts-1), is a member of
the S100 family calcium-binding proteins. It is overexpressed in
gastric cancer and its expression correlates with poor prognosis of
the patients (23–25). It has also been shown that
S100A4 inhibition leads to decreased in vitro and
in vivo growth of gastric cancer BGC823 cells (4). We previously found that S100A4
suppression leads to decreased migration of BGC823 cells (data not
shown). These findings suggest that S100A4 can regulate many
biological characteristics of gastric cancer cells. We speculated
that S100A4 may be a downstream effector mediating the role
of RPS12. We examined and found that RPS12 inhibition
led to decreased expression of S100A4 at both mRNA and
protein level in BGC823 cells, which indicated that S100A4
might be a downstream target of RPS12. Our further study on
luciferase reporter assay showed that RPS12 suppression
significantly reduced the activity of S100A4 promoter,
suggesting the involvement of RPS12 in the transcriptional
regulation of S100A4 gene. How RPS12 regulates the
transcription of the S100A4 needs to be clarified. Other
reports have proved that some ribosomal proteins are gene
transcription regulators. An example of RPs as a gene transcription
regulator is the mammalian RPL11. It binds to the Myc
oncoprotein and inhibits Myc-mediated transcriptional activation of
target genes (26). L5, L23 and S7
were shown to bind to MDM2 and inhibit MDM2-mediated p53
ubiquitination and degradation, leading to p53 activation (27–29).
Based on these studies, we hypothesize that RPS12 may
interact with or affect the stability of certain transcription
factors regulating S100A4 expression. In what way
RPS12 control S100A4 gene transcription is under
investigation by our group.
In order to further investigate whether
S100A4 mediates the biological effects of RPS12 in
gastric cancer cells, we carried out rescue assays. The results
showed that ectopic expression of S100A4 reversed the
reduced migration and proliferation ability, which suggested that
as a downstream effector, S100A4 could partly mediate the
roles of RPS12 in BGC823 cells.
In summary, the current study investigated the
effects of RPS12 on gastric cancer cells and the underlying
mechanisms. We demonstrated for the first time that RPS12
suppression was able to inhibit the proliferation and migration of
gastric cancer cells. RPS12 inhibition reduces the activity
of S100A4 promoter and leads to the downregualtion of
S100A4 expression. Ectopic expression of S100A4 is
able to reverse the reduced proliferation, and migration by
RPS12 inhibition. Collectively, the evidence suggests that
RPS12 may take part in the development and progression of
gastric cancer by affecting the proliferation and migration, and
the effects may be at least partly mediated by S100A4. RPS12
might be a new potential target for diagnosis and therapy of
gastric cancer.
Acknowledgments
This study was supported by grants from the
National Natural Science Foundation of China (no. 30570848) and
Liaoning Natural Science Foundation (no. 20102289).
References
|
1.
|
Sun XJ, Sun KL, Zheng ZH, Hao DM, Fu WN,
Xu HM, Chen JQ and Li XM: Analysis of gene expression profiles for
distinct stages of intestinal-type gastric cancer using suppression
subtractive hybridization and cDNA microarray. Scand J
Gastroenterol. 40:1244–1245. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Sun XJ, Hao DM, Zheng ZH, Fu H, Xu HM,
Wang MX and Sun KL: Screening and analysis of associated genes in
the carcinogenesis and progression of gastric cancer. Zhonghua Yi
Xue Yi Chuan Xue Za Zhi. 22:31–34. 2005.(In Chinese).
|
|
3.
|
Yan Y, Fu H, Hua J, Zhang RX, Chen DQ, Sun
KL and Sun XJ: The effects of silencing the RPS12 gene by RNAi on
apoptosis of gastric cancer cells and the possible molecular
mechanism. Chin J Clin Oncol. 35:1415–1418. 2008.(In Chinese).
|
|
4.
|
Hua J, Chen DQ, Fu H, Zhang RX, Shen W,
Liu SS, Sun KL and Sun XJ: Short haripin RNA-mediated inhibition of
S100A4 promotes apoptosis and suppresses proliferation of BGC823
gastric cancer cells in vitro and in vivo. Cancer Lett. 292:41–47.
2010. View Article : Google Scholar
|
|
5.
|
Zhang RX, Fu H, Chen DQ, Hua J, Hu YP, Sun
KL and Sun XJ: Subcellular distribution of S100A4 and its
transcriptional regulation under hypoxic conditions in gastric
cancer cell line BGC823. Cancer Sci. 101:1141–1146. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Shen W, Chen DQ, Fu H, Liu SS, Sun KL and
Sun XJ: S100A4 protects gastric cancer cells from anoikis through
regulation of αv and α5 integrin. Cancer Sci. 102:1014–1018.
2011.PubMed/NCBI
|
|
7.
|
Frank J: The ribosome - a macromolecular
machine par excellence. Chem Biol. 7:R133–R141. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Lai MD and Xu J: Ribosomal proteins and
colorectal cancer. Curr Genomics. 8:43–49. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Kobayashi T, Sasaki Y, Oshima Y, Yamamoto
H, Mita H, Suzuki H, Toyota M, Tokino T, Itoh F, Imai K and
Shinomura Y: Activation of the ribosomal protein L13 gene in human
gastrointestinal cancer. Int J Mol Med. 18:161–170. 2006.PubMed/NCBI
|
|
10.
|
Huang CJ, Chen CC, Yang SH, Chang CC, Sun
HL, Cheng YC, Liu CC, Lin SC and Lin CM: Faecal ribosomal protein
L19 is a genetic prognostic factor for survival in colorectal
cancer. J Cell Mol Med. 12:1936–1943. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Cheng Q, Lau WM, Chew SH, Ho TH, Tay SK
and Hui KM: Identification of molecular markers for the early
detection of human squamous cell carcinoma of the uterine cervix.
Br J Cancer. 86:274–281. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Wang M, Hu Y, Amatangelo MD and Stearns
ME: Role of ribosomal protein RPS2 in controlling let-7a expression
in human prostate cancer. Mol Cancer Res. 9:36–50. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Kim JH, You KR, Kim IH, Cho BH, Kim CY and
Kim DG: Over-expression of the ribosomal protein L36a gene is
associated with cellular proliferation in hepatocellular carcinoma.
Hepatology. 39:129–138. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Sridharan S and Basu A: S6 kinase 2
promotes breast cancer cell survival via AKT. Cancer Res.
71:2590–2599. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Kasai H, Nadano D, Hidaka E, Higuchi K,
Kawakubo M, Sato TA and Nakayama J: Differential expression of
ribosomal proteins in human normal and neoplastic colorectum. J
Histochem Cytochem. 51:567–574. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Kondoh N, Shuda M, Tanaka K, Wakatsuki T,
Hada A and Yamamoto M: Enhanced expression of S8, L12, L23a, L27
and L30 ribosomal protein mRNAs in human hepatocellular carcinoma.
Anticancer Res. 21:2429–2433. 2003.PubMed/NCBI
|
|
17.
|
Shriver SP, Shriver MD, Tirpak DL, Bloch
LM, Hunt JD, Ferrell RE and Siegfried JM: Trinucleotide repeat
length variation in the human ribosomal protein L14 gene (RPL14):
localization to 3p21.3 and loss of heterozygosity in lung and oral
cancers. Mutat Res. 406:9–23. 1998.PubMed/NCBI
|
|
18.
|
Pogue-Geile K, Geiser JR, Shu M, Miller C,
Wool IG, Meisler AI and Pipas JM: Ribosomal protein genes are
over-expressed in colorectal cancer: isolation of a cDNA clone
encoding the human S3 ribosomal protein. Mol Cell Biol.
11:3842–3849. 1998.PubMed/NCBI
|
|
19.
|
O’Donohue MF, Choesmel V, Faubladier M,
Fichant G and Gleizes PE: Functional dichotomy of ribosomal
proteins during the synthesis of mammalian 40S ribosomal subunits.
J Cell Biology. 190:853–866. 2010.PubMed/NCBI
|
|
20.
|
Wang H, Zhao LN, Li KZ, Ling R, Li XJ and
Wang L: Overexpression of ribosomal protein L15 is associated with
cell proliferation in gastric cancer. BMC Cancer. 6:912006.
View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Khanna N, Sen S, Sharma H and Singh N: S29
ribosomal protein induces apoptosis in H520 cells and sensitizes
them to chemotherapy. Biochem Biophys Res Commun. 304:26–35. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Wu Q, Gou Y, Wang Q, Jin H, Cui L, Zhang
Y, He L, Wang J, Nie Y, Shi Y and Fan D: Downregulation of RPL6 by
siRNA inhibits proliferation and cell cycle progression of human
gastric cancer cell lines. PLoS One. 6:e264012011. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Forst B, Hansen MT, Klingelhöfer J, Møller
HD, Nielsen GH, Grum-Schwensen B, Ambartsumian N, Lukanidin E and
Grigorian M: Metastasis-inducing S100A4 and RANTES cooperate in
promoting tumor progression in mice. PLoS One. 5:e103742010.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Kwon YW, Chang IH, Kim KD, Kim YS, Myung
SC, Kim MK and Kim TH: Significance of S100A2 and S100A4 expression
in the progression of prostate adenocarcinoma. Korean J Urol.
51:456–462. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Wang YY, Ye ZY, Zhao ZS, Tao HQ and Chu
YQ: High-level expression of S100A4 correlates with lymph node
metastasis and poor prognosis in patients with gastric cancer. Ann
Surg Oncol. 17:89–97. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Dai MS, Sears R and Lu H: Feedback
regulation of c-Myc by ribosomal protein L11. Cell Cycle.
6:2735–2741. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Chen D, Zhang Z, Li M, Wang W, Li Y,
Rayburn ER, Hill DL, Wang H and Zhang R: Ribosomal protein S7 as a
novel modulator of p53-MDM2 interaction: binding to MDM2,
stabilization of p53 protein, and activation of p53 function.
Oncogene. 26:5029–5037. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Dai MS and Lu H: Inhibition of
MDM2-mediated p53 ubiquitination and degradation by ribosomal
protein L5. J Biol Chem. 279:44475–44482. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Dai MS, Zeng SX, Jin Y, Sun XX, David L
and Lu H: Ribosomal protein L23 activates p53 by inhibiting MDM2
function in response to ribosomal perturbation but not to
translation inhibition. Mol Cell Biol. 24:7654–7668. 2004.
View Article : Google Scholar : PubMed/NCBI
|