Introduction
Sirtuins (SIRT1-SIRT7) are NAD+-dependent
hydrolases (1,2). Derivatives of the yeast SIR2 histone
deacetylase have a common catalytic domain, which is highly con
served in multiple organisms ranging from bacteria to humans. The
sirtuin domain is composed of two distinct motifs binding
NAD+ and the acetyl-lysine substrate, respectively
(3,4). They catalyse the deacetylation of
acetylated lysine residues of histone substrates and act on
non-histone substrates, e.g., several transcription factors such as
the p53 tumor suppressor protein (5), the cytoskeletal protein α-tubulin
(6) or the acetyl-CoA synthetase
(7). Besides the deacetylation
reaction, some sirtuins show ADP ribosyltransferase activity
(8,9).
A dysregulation of the tightly regulated equilibrium
of acetylation and deacetylation plays an important role both, in
the pathogenesis and in the suppression of cancer (10). Histone acetylation modifiers are
therefore becoming more important as potential targets in the
treatment of cancer. Relaxation of the chromatin fiber facilitates
transcription and is controlled by two competing enzymatic
activities, histone acetyltransferases (HATs) and histone
deacetylases (HDACs), which modify the acetylation state of histone
proteins and other promoter-bound transcription factors. While
HATs, which are frequently part of multisubunit coactivator
complexes, induce the relaxation of chromatin structure and
transcriptional activation, HDACs tend to associate with
multisubunit corepressor complexes, which result in chromatin
condensation and transcriptional repression of specific target
genes. HAT and HDAC enzymatic activities are involved in the
generation and in the suppression of cancer. Some of the genes
encoding these enzymes have been shown to be rearranged in the
context of chromosomal translocations in human acute leukemias and
solid tumors, where fusions of regulatory and coding regions of a
variety of transcription factor genes result in completely new gene
products, which may interfere with regulatory cascades that control
cell growth and differentiation (10). On the other hand, some histone
acetylation modifying enzymes have been located within chromosomal
regions being particularly prone to chromosomal breaks. In such
cases, gains and losses of chromosomal material may affect the
availability of functionally active HATs and HDACs, which in turn
disturbs the tightly controlled equilibrium of histone acetylation
(11).
Sirtuins are important for various biological
processes, including cellular development, heterochromatin
formation, gene/transcriptional silencing (12–16),
DNA repair, genome stability and cellular processes such as the
response to stress, adipogenesis and metabolism and are an
important mediator of organismal longevity through a number of
different mechanisms such as the induction of cell cycle arrest,
resistance to oxidative stress and the inhibition of apoptosis
(17). Sirtuins connect aging,
cancer, and diet and thus are potential molecular targets for the
development of pharmaceuticals to treat human malignant, metabolic
and neurological diseases (18).
Based on structural and functional similarities,
mammalian histone deacetylases are classified into four categories,
of which three contain non-sirtuin HDACs comprising the yeast RPD3
homologs (class I HDACs), the HDA1 mammalian homologs (class II
HDACs) and HDAC11-related enzymes (class IV HDACs), while one
category consists of sirtuin histone deacetylases (class III
HDACs), being homologs of the yeast Sir2 protein (19). In contrast to SIRT1, the SIRT5
protein is still poorly investigated (6,7,19,20).
Materials and methods
Identification of the murine Sirt5
cDNA
Homology searches of the EST database at NCBI
(National Center for Biotechnology Information) with the yeast SIR2
protein sequence (GenPept P06700) yielded 4 mRNA sequences of
variable length, of which mRNA sequence NM_178848.3, which in the
meantime is referred to as the NCBI reference sequence for Sirt5,
contained the full length murine Sirt5 mRNA which was then used for
the identification of the murine Sirt5 genomic clone.
Identification of BAC genomic clone
RP24-62L21
The murine Sirt5 genomic clone was identified from a
murine BAC genomic library (RZPD, Berlin, Germany) after in
silico screening with the Sirt5 cDNA (GenBank clone
NM_178848.3), which was shown to contain the full-length murine
Sirt5 cDNA. BAC clone RP24-62L21 was identified to contain
an insert with a size of ∼120 kb in the vector pBACe3.6, which
included the murine Sirt5 genomic sequence. BAC genomic DNA was
prepared according to published protocols (21) and the murine Sirt5 insert was con
firmed by cycle sequencing (22).
Instrumental methods
Dye terminator cycle sequencing was performed using
the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit
with AmpliTaq DNA polymerase (Perkin-Elmer, Branchburg, NJ) and
analyzed with an ABI PRISM 310 Genetic Analyzer which utilizes the
four-color sequencing chemistry.
PCR methods
The sirt5 sequence was partially sequenced by
primer walking on both strands using a direct sequencing strategy
(22). Sequencing reactions were
performed using 0.6 μg cDNA and 20–30mer oligonucleotide
primers (Thermo Electon, Dreieich, Germany). Sequencing reactions
were set up in a volume of 20 μl containing 10 pmol of the
sequencing primer, 4 μl BigDye Terminator Cycle Sequencing
Ready Reaction Mix (Perkin-Elmer, Norwalk, CT), DNA as indicated
and ddH2O added up to a final volume of 20 μl.
The thermal cycling profile for the se quencing of the cDNA-clones
was as follows: denaturation at 95°C for 30 sec, annealing at 50°C
for 15 sec, extension at 60°C for 4 min (25 cycles) and storage at
4°C.
Chromosomal localization by fluorescence
in situ hybridization (FISH)
Cell culture and chromosome
preparation
Standard chromosome preparations were used from a
mouse embryonic fibroblast cell line.
Slide preparation
In order to remove excess of cytoplasm, slides were
treated with pepsin (0.5 mg/ml in 0.01 M HCl, pH 2.0) at 37°C for
40 min. Slides were then washed 10 min in 1X PBS at room
temperature followed by an ethanol series (70, 90 and 100%) and
air-dried. BAC genomic clone RP24-62L21, which was shown to
contain the murine Sirt5 gene, was used as a probe.
Probe labeling
The BAC DNA was labelled by a standard nick
translation procedure. Digoxigenin (Roche Diagnostics) was used as
labelled dUTP at the concentration of 40 μM. Probe length
was analyzed on a 1% agarose gel. The probe showed the optimal
average length of ∼300 bp after nick translation.
Hybridization and probe detection
DNA (∼50 ng) was pooled together with 2 μg
cot-1 in 10 μl hybridization buffer (50% formamide, 2X SSC,
10% dextran sulfate). The DNA was applied to chromosomes fixed on a
slide, mounted with a cover slip and sealed with rubber cement.
Probe DNA and chromosomes were denatured together at 72°C for 3
min. Hybridization was overnight at 37°C in a wet chamber. After
hybridization the cover slip was carefully removed and the slide
was washed in 2X SSC for 8 min. Slides were then incubated at 70°C
in 0.4X SSC/0.1% Tween for 1 min. After equilibration in 4X
SSC/0.1% Tween for 5 min the rhodamine coupled antibody was applied
(dilution of 1:400). Incubation was for 45 min at 37°C. The slide
was then washed twice in 4X SSC/0.1% Tween for 10 min at 45°C
followed by staining in DAPI (4’,6-diamidino-2-phenylindole) for 10
min. For microscopy the slide was mounted in antifade solution
(Vectashield).
Microscopy
In situ hybridization signals were analyzed
on a Zeiss Axioplan II microscope. Each image plain (blue and
orange) was recorded separately with a b/w CCD camera. Chromosomes
and FISH signals were then displayed in false colors and images
merged on the computer. Camera control, image capture and merging
were done with SmartCapture X software (Digital Scientific,
Cambridge, UK).
Sequence analysis and computer
database searches
DNA sequence analysis was performed using the HUSAR
(Heidelberg Unix Sequence Analysis Resources) server hosted by the
Biocomputing Service Group at the German Cancer Research Center
(Heidelberg, Germany) and the UniGene and LocusLink programs at the
National Center for Biotechnology Information (NCBI). Sequence
comparisons were performed with the BLAST algorithm of the GenBank
and EMBL databases (23). Protein
similarity scores were calculated from fast alignments generated by
the method of Wilbur and Lipman with the Clustal W Multiple
Alignment Program Version 1.7 and with the BLAST algorithm at NCBI
(Table I) (25). Protein motifs were identified
online at the ExPASy (Expert Protein Analysis System) proteomics
server of the Swiss Institute of Bioinformatics (SIB) with the
program PROSITE and double-checked using the MotifFinder program
hosted by the GenomeNet WWW server at Institute for Chemical
Research, Kyoto University (Japan), but still remain to be
experimentally confirmed. Potential transcription factor binding
sites were identified with the TRANSFAC program, which is part of
the GenomeNet Computation Service, which is hosted by the
Bioinformatics Center at the Institute for Chemical Research at the
Kyoto University. Sequence similarities were calculated with the
GAP software, which considers all possible alignments and gap
positions between two sequences and creates a global alignment that
maximizes the number of matched residues and minimizes the number
and size of gaps on the HUSAR server (26). Repetitive elements were identified
on the Repeat Masker Server at the University of Washing ton and
CpG elements were found with the CPG software hosted by the
European Bioinformatics Institute (EMBL outstation) (Figs. 1 and 2).
 | Table I.Sequence identity and similarity
among class III sirtuin-HDACs.a |
Table I.
Sequence identity and similarity
among class III sirtuin-HDACs.a
| Identity
similarity | Mouse
SIRT1 | Mouse
SIRT2 | Mouse
SIRT3 | Mouse
SIRT4 | Mouse
SIRT5 | Mouse
SIRT6 | Mouse
SIRT7 | Yeast
SIR2 |
|---|
Mouse
SIRT1 | | 41 | 42 | 31 | 27 | 23 | 23 | 42 |
Mouse
SIRT2 | 59 | | 51 | 29 | 26 | 27 | 27 | 30 |
Mouse
SIRT3 | 64 | 66 | | 29 | 30 | 32 | 28 | 38 |
Mouse
SIRT4 | 48 | 46 | 43 | | 29 | 27 | 27 | 28 |
Mouse
SIRT5 | 41 | 44 | 45 | 48 | | 23 | 23 | 28 |
Mouse
SIRT6 | 40 | 42 | 45 | 42 | 38 | | 41 | 24 |
Mouse
SIRT7 | 41 | 45 | 43 | 43 | 37 | 55 | | 23 |
Yeast
SIR2 | 59 | 48 | 53 | 46 | 42 | 40 | 40 | |
Results
Identification of cDNAs encoding
murine Sirt5
Homology searches of the EST database at NCBI
(National Center for Biotechnology Information) with the yeast SIR2
protein sequence (GenPept P06700) yielded 4 mRNA sequences of
variable length: AK002609.1 (1,397 bp), AK005346.1 (1,381 bp),
BC031770.1 (1,403 bp), BC087898.1 (1,547 bp) and NM_178848.3 (1,369
bp). NM_178848.3, which is the sirt5 reference sequence contained
the full length murine Sirt5 mRNA sequence, which was then used for
the identification of the murine Sirt5 genomic clone. The
authenticity of its insert was confirmed by DNA cycle sequencing.
Sequences flanking the 5′- and 3′-ends of the Sirt5 open
reading frame were identified from the Sirt5 murine genomic
clone BAC RP24-62L21. Characterization of the 5′-flanking
genomic region, which precedes the Sirt5 open reading frame,
revealed a number of putative optimal transcription factor binding
sites for GATA, Evi-1, C/EBPb, AP-1, SRY, MZF1 and CdxA (Fig. 1). However, their biological
relevance still awaits to be investigated ex perimentally. Islands
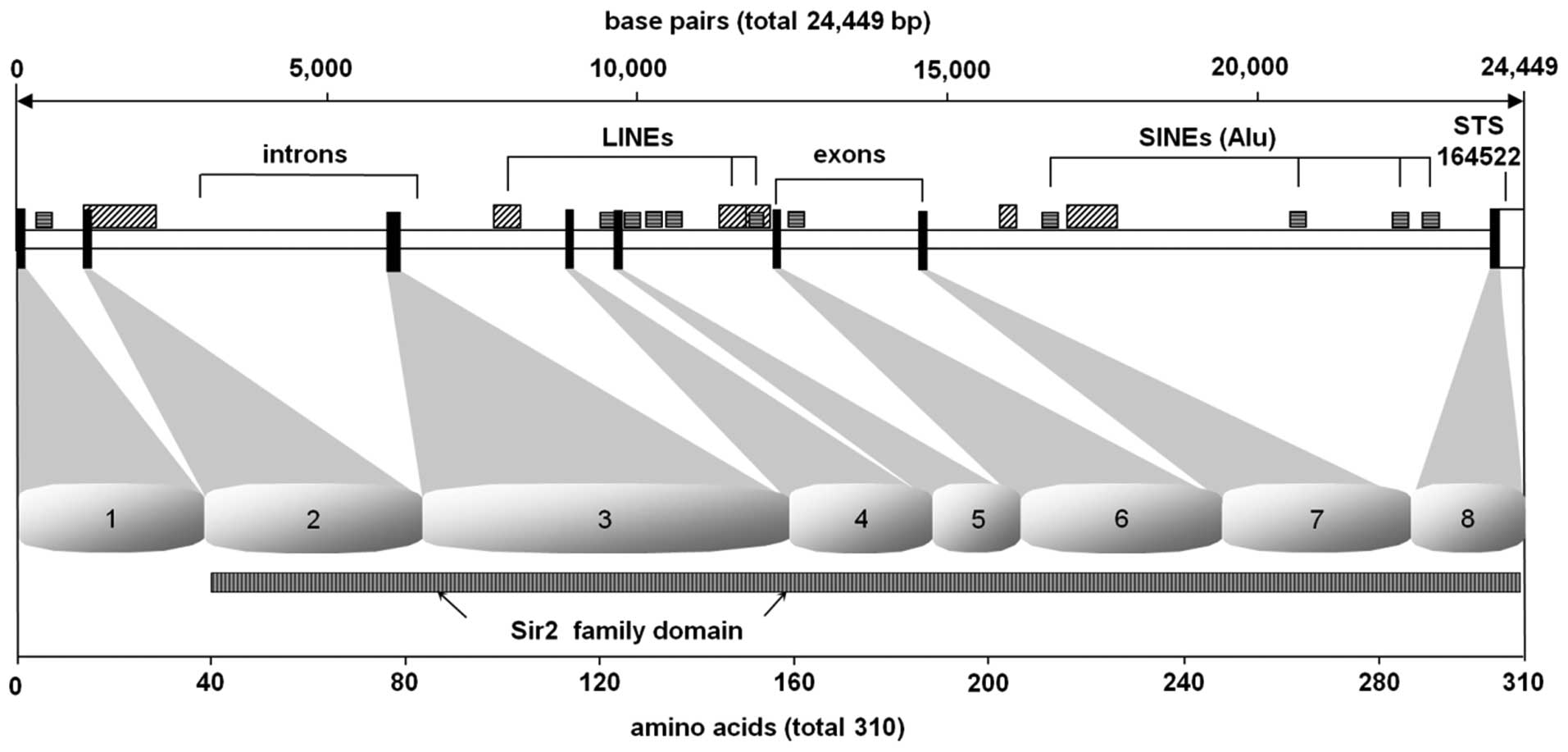
of unusual CG composition were not observed. The 24,449-bp murine
Sirt5 gene encodes a 310-aa protein (Fig. 2) with a predictive mo lecular
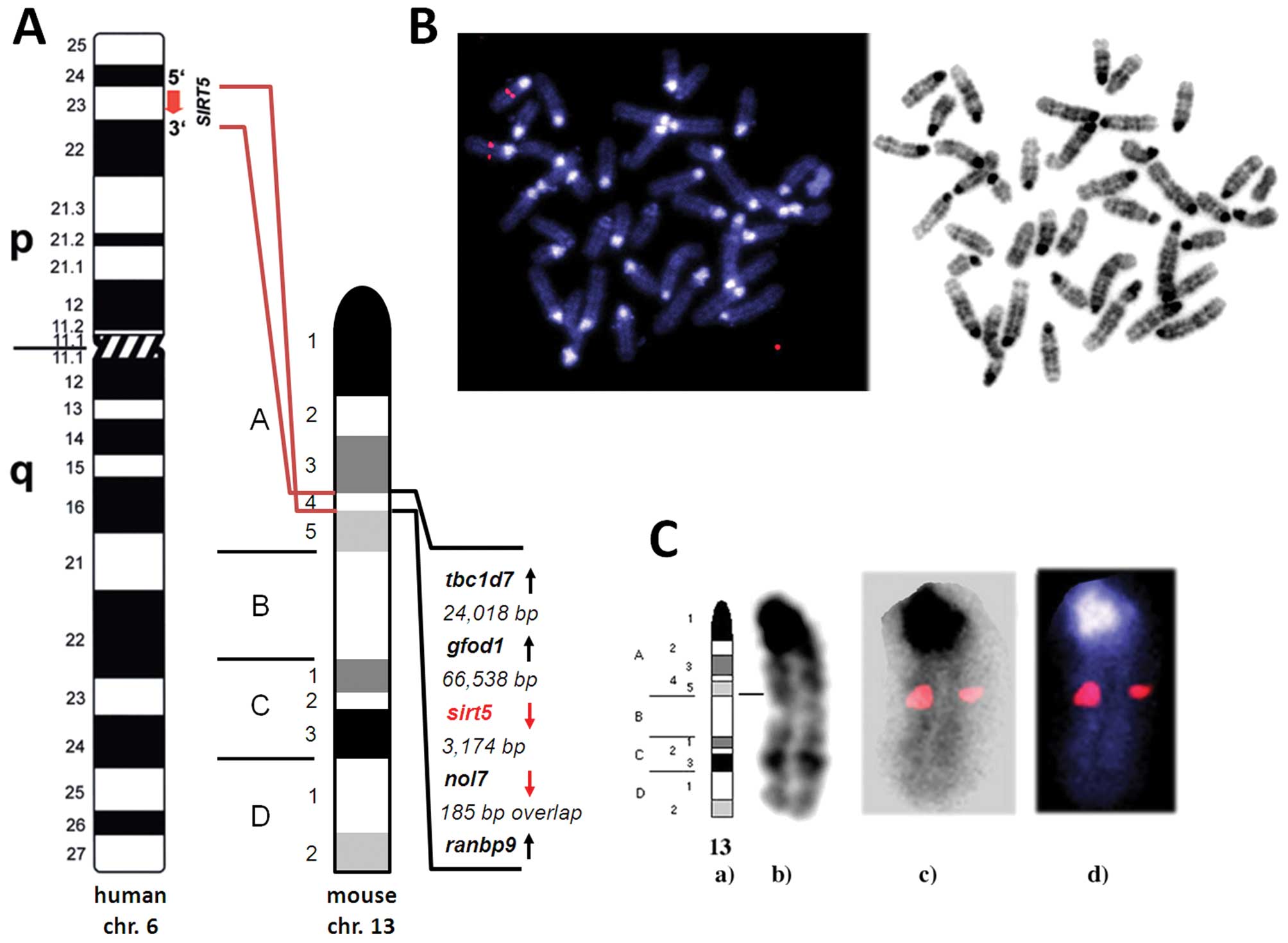
weight of 34.1 kDa and an isoelectric point of 8.90. Fluorescence
in situ hybridization analysis localized the murine
Sirt5 gene to mouse chromosome 13A4 (Fig. 3). Translational stop codons in all
reading frames precede the human sirt5 open reading frame.
The 3′-flanking region was shown to contain the eukaryotic
polyadenylation consensus signal ATTAAA 295 bp downstream of the
termination of translation signal TAA (Fig. 1) (27).
Identification and characterization of
the murine Sirt5 genomic locus
The murine Sirt5 genomic clone was obtained
from an arrayed murine BAC genomic library from the RZPD German
Resource Center for Genome Research (Berlin, Germany) after in
silico screening with the murine Sirt5 cDNA (GenBank
clone NM_178848.3), which was shown to contain the full-length
murine Sirt5 cDNA sequence. BAC clone RP24-62L21 was
identified to contain inserts with an average size of ∼120 kb in
the 11.6-kb vector pBACe3.6, which included the murine Sirt5
genomic sequence. BAC genomic DNA was prepared according to
published protocols (22) and the
Sirt5 insert was confirmed by cycle sequencing (23). Genomic sequence comparison analyses
with the BLAST algorithm helped us with the identification of mouse
chromosome 13 genomic contig GenBank NC_000079. This sequence is
part of the largely finished reference sequence (C57BL/6J) that
contains small amounts of WGS and HTGS draft sequence and was
assembled by NCBI in consultation with the Mouse Genome Sequencing
Consortium. We have used this sequence for the determination of
Sirt5 introns and exon/intron boundaries (Table II). The murine Sirt5 gene
spans a region of 24,449 bp. Determination of the exon-intron
splice junctions established that the gene Sirt5 is encoded
by 8 exons ranging in size from 54 bp (exon 5) to 466 bp (exon 8).
Within introns 5, 6 and 7 in particular, we identified an
accumulation of interspersed repetitive elements, SINEs (short
inter spersed nuclear elements) and LINEs (long interspersed
nuclear elements). Additionally, we have identified STS-marker
164522 (synonymous WI MRC-RH: 506859) within the untranslated
proportion of murine Sirt5 exon 8 between the Sirt5
translational termination signal (TAA) and the polyadenylation
consensus signal ATTAAA. The sirtuin catalytic domain, which is
highly con served in all members of mammalian sirtuins that have
been described so far as well as in their Sir2 yeast ancestor
protein, is found between amino acid residues 41 and 309, i.e.,
within exons 1 and 8 of the protein (Fig. 2).
 | Table II.Exon/intron splice-junctions of the
murine Sirt5 gene. |
Table II.
Exon/intron splice-junctions of the
murine Sirt5 gene.
| Exon no. | Exon size | 5′-Splice
donor | Intron no. | Intron size | 3′-Splice
acceptor |
|---|
| 1 | 115 |
CATGGCTCGTgtaagtcatctg | 1 | 963 |
ttttctgtttagATATGGCAGA |
| 2 | 134 |
GCAGGCTCAGgttagtaacgct | 2 | 4729 |
ctctcccctcagGACCTGGCAA |
| 3 | 226 |
GAAATCCACGgtgaggagaacg | 3 | 2752 |
cttttttttcagGAACCTTATT |
| 4 | 88 |
CAGGAAAAGGgtaagtatagca | 4 | 739 |
tatttgctccagGGCCCCAGAG |
| 5 | 54 |
AACTTCCCCGgtaggtaaaaca | 5 | 2555 |
atgctcttccagGTGCGAGGAG |
| 6 | 124 |
GTGTCTAGTGgtaagtcacatg | 6 | 2265 |
cttgctttgtagGTGGGAACAT |
| 7 | 116 |
ACAGATTCAGgtacagggacaa | 7 | 9123 |
tcttgtgtttagGTTTCATTTT |
| 8 | 466 | | | | |
Murine Sirt5 is a single copy
gene
Both sequencing and results obtained by electronic
PCR of BAC clone RP24-62L21 identified STS-marker 164522
(synonymous WI MRC-RH: 506859) within the untranslated proportion
of murine sirt5 exon 8 genomic sequence (Fig. 2). Our fluorescence in situ
hybridization studies localized BAC clone RP24-62L21, which
contains the murine sirt5 genomic sequence to the chromosome
13A4/5 border region. However, the more precise high-resolution
analyses assigned murine sirt5 to mouse chromosome 13A4.
Taken together, the results obtained by FISH, electronic PCR and
the already known location of the STS marker listed above,
indicated one single site of hybridization of sirt5 on mouse
metaphase chromosomes and its specific localization on chromosome
13A4 (Fig. 3).
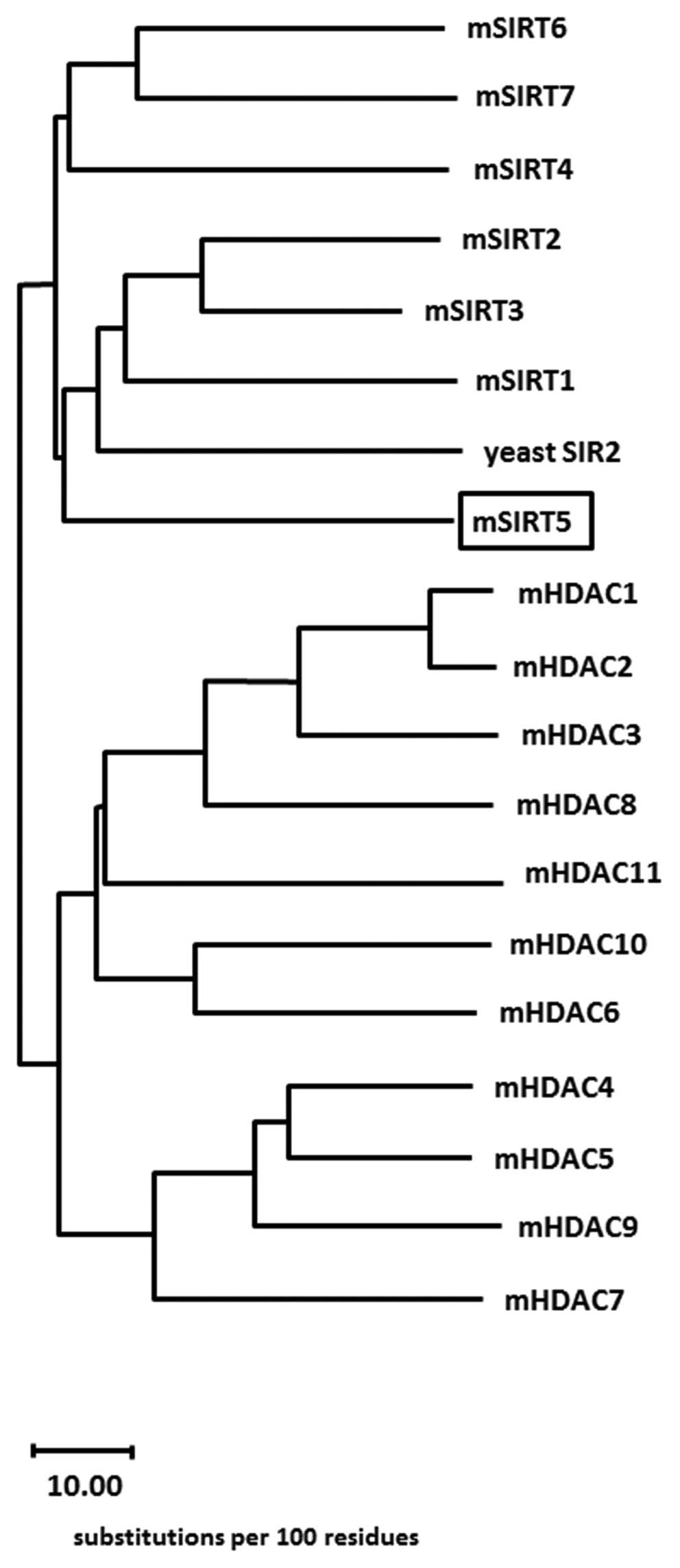
Phylogenetic analyses and pairwise
sequence comparisons
Using the consensus murine sirtuin (class III
deacetylase) protein sequences together with the class I, II and IV
murine histone deacetylase protein sequences, a consensus
evolutionary tree was calculated (Fig.
4), which reveals the evolutionary position of the murine SIRT5
protein. The accession numbers of the sequences that have been used
in this phylogenetic analysis were as follows: Yeast Sir2 (GenPept
P06700), Mus musculus SIRT3 (GenPept CAJ18608.1), Mus musculus
SIRT2 (GenPept AAH21439.1), Mus musculus SIRT1 (GenPept Q923E4),
Mus musculus SIRT4 (GenPept XP_993153), Mus musculus SIRT7 (GenPept
NP_694696.2), Mus musculus SIRT6 (GenPept AAH52763.1), Mus musculus
SIRT5 (GenPept AAH31770.1), Mus musculus HDAC1 (GenPept CAQ
51569.1), Mus musculus HDAC2 (GenPept AAI38518.1), Mus musculus
HDAC3 (GenPept AAF36425.1), Mus Musculus HDAC4 (GenPept
AAH66052.1), Mus musculus HDAC5 (GenPept CAX15947.1), Mus musculus
HDAC6 (GenPept AAH41105.1), Mus musculus HDAC7 (GenPept
AAH57332.1), Mus musculus HDAC8 (GenPept AAH 61257.1), Mus musculus
HDAC9 (GenPept AAH98187.1), Mus musculus HDAC10 (GenPept
AAH64018.1) and Mus musculus HDAC11 (GenPept AAH16208.1). In
Table I the sequence identity and
similarity among class III sirtuin-HDACs is demonstrated. The
indicated numbers represent the percentage of sequence identity and
similarity from pairwise sequence comparisons.
Discussion
Sirtuins show different subcellular distribution,
different substrate specificity and cellular function. In the
nucleus, SIRT1 functions (31) as
a transcriptional repressor via histone deacetylation. SIRT1 also
regulates transcription by modifying the acetylation levels of
transcription factors, e.g., MyoD, FOXO, p53, and NF-κB (5,32–36).
In the cytoplasm the SIRT2 protein associates with microtubules and
deacetylates α-tubulin lysine 40 (6). SIRT6 regulates telomeric chromatin
and is a histone H3K9 deacetylase (37). In the nucleolus, SIRT7 functions as
a positive regulator of RNA polymerase I transcription (38).
Three mammalian sirtuins (Sirt3, 4, and 5) are
localized in the mitochondria, the center of energy metabolism and
apoptosis initiation (39).
Electrons pass through electron transport complexes (I–IV),
generating a proton gradient that is used to drive ATP synthase to
generate ATP. SIRT3 binds to complex I, binds and deacetylates
acetyl-CoA synthetase 2 (AceCS2) and glutamate dehydrogenase (GDH),
activating their enzymatic activities. SIRT4 binds and represses
GDH activity via ADP-ribosylation (40).
The exact localization of Sirt5 within the
mitochondria differs. In transfected COS7 cells, murine FLAG-tagged
Sirt5 was solely found in the mitochondrial intermembrane space
(41), whereas endogenous murine
Sirt5 protein was exclusively found in the mitochondrial matrix
(42) in liver cells (31,45).
SIRT5 may localize to the mitochondrial intermembrane space after
overexpression or after mitochondrial import in vitro
(31,41,43).
It is therefore discussed that the localization of SIRT5 may in
fact depend on the type of underlying stimulus, whether SIRT5 is
translocated predominantly into the mitochondrial intermembrane
space or to the mitochondrial matrix (40).
Because of its localization in mitochondria, SIRT5
deacetylates and activates carbamoyl phosphate synthetase 1 (CPS1),
the rate-limiting step of the urea cycle (40) and urate oxidase in murine liver
mitochondria (46). SIRT5 exhibits
deacetylase activity against acetylated cytochrome C, a conserved
mitochondrial intermembrane space protein (31,43)
and regulates various mitochondrial metabolic pathways together
with SIRT3 and SIRT4 (31). In
contrast to Sirt3, Sirt5 does not deacetylate any of the
mitochondrial matrix proteins tested (43). SIRT5 exhibits weak but detectable
deacetylase activity against acetylated histone H4 (31,47),
as well as chemically acetylated histones or acetylated BSA
(2,31). Whether Sirt5 has an additional
function in the regulation of apoptosis, remains speculative
(44), but there is a
physiological role of Sirt5 in the regulation of cellular
metabolism and cellular senescence (2,39).
The role of Sirt5 in the pathogenesis of human
diseases is currently being discussed since repetitive elements in
the gene structure of Sirt5 may partly induce genomic instability
and thus malignant transformation (44). Besides, Sirt5 could contribute to
liver damage as a consequence of chronic alcohol consumption
(48), since alcohol exposure may
induce posttranslational modification such as hyperacetylation of
numerous proteins including p53, ACS2 and tubulin (44,49).
Since hepatic expression levels of Sirt5, but not Sirt3, are
significantly diminished upon exposure to ethanol (48); this suggests that Sirt5 may
contribute to the hyperacetylation of ethanol-dependent proteins
(44). Also, the depletion of the
Sirtuin cosubstrate NAD+ is known to accompany alcohol
exposure and the highly reactive intermediates that are associated
with alcohol metabolism indicate an additional, more direct
mechanism how Sirt5 activity may be affected. The emerging role of
Sirt5 in energy and amino acid metabolism in liver mitochondria
suggests that changes of its activity may indeed contribute to the
pathology of alcohol associated organ disease, but details will
have to await further studies (44).
In the study presented herein, we report the
cloning, characterization and mapping of murine sirt5 on the
genomic level. Murine Sirt5 is a single-copy gene that spans
a region of 24,449 bp. It is composed of 8 exons ranging in size
from 54 bp (exon 5) to 466 bp (exon 8) (Table II). Particularly within introns 5,
6 and 7 we identified an accumulation of interspersed repetitive
elements, which consist of Alu or KpnI and
BamH1 repeats as representative examples of short and long
interspersed nuclear elements, known as SINEs (Alu repeats)
and LINEs (KpnI and BamH1 repeats) (50). Additionally, we identified an
internal STS-marker 164522 (synonymous WI MRC-RH: 506859), within
the untranslated proportion of sirt5 exon 8 between the
Sirt5 translational termination signal (TAA) and the
polyadenylation consensus signal ATTAAA. The sirtuin catalytic
domain, which is highly conserved in all members of mammalian
sirtuins that have been described so far as well as in their Sir2
yeast ancestor protein, is found between amino acid residues 41 and
309, i.e., within exons 1 and 8 of the protein. The 1,369-bp human
Sirt5 mRNA (GenBank NM_178848) has an open reading frame of
930 bp that encodes 310-aa protein with a predictive molecular
weight of 34.1 kDa and an isoelectric point of 8.90.
Characterization of the 5′-flanking genomic region, which precedes
the Sirt5 open reading frame, revealed a TATA- and CCAAT-box
less promoter that contained a number of putative optimal
transcription factor binding sites for GATA, Evi-1, C/EBPb, AP-1,
SRY, MZF1 and CdxA. However, their biological relevance awaits
investigation ex perimentally. Islands of unusual CG composition
were not observed. The sirtuin deacetylase catalytic domain is
highly conserved within all members of mammalian sirtuins described
so far and located within exons 1–8 (Fig. 2).
Fluorescence in situ hybridization analysis
in conjunction with electronic PCR localized the murine
Sirt5 gene to chromosome 13A4 (Fig. 3); a genomic area, which shows
syntenic conservation with human chromosome 6p23, the human Sirt5
genomic locus, a region which has been found to be involved in
numerous chromosomal abnormalities associated with malignant
disease, especially as part of both balanced and unbalanced
chromosomal abnormalities in acute myeloid leukemia in accordance
with data that have been retrieved from the Cancer Genome Anatomy
Project (CGAP) database at the National Cancer Institute (19,51).
The sirt5 gene is being transcribed towards the downstream distal
end of chromosome 13 and is closely neighboured by the nol7
(nucleolar protein 7) gene (3,174 bp downstream) and the gfod1
(glucosefructose oxidoreductase domain containing 1) gene (66,538
bp upstream).
It is currently not clear whether and to what extent
chromosomal abnormalities involving the human chromosomal locus
6p23 or the murine chromosomal locus 13A4 affect SIRT5-mediated
functional effects. It is, however, obvious that several genes
encoding sirtuin proteins are located within chromosomal regions
that are particularly prone to chromosomal alterations. In such
cases, gains and losses of chromosomal material may influence the
availability of functionally active sirtuin proteins and thus the
tightly controlled intracellular equilibrium of protein acetylation
and/or ADP ribosylation, respectively (11). The murine SIRT5 gene is localized
at mouse chromosome 13A4, a chromosomal region that has been
reported to be associated with malignant disease such as pancreatic
cancer in humans (19,31,40).
SIRT5 is ubiquitously expressed in various tissues, with relatively
high Sirt5 expression level in the heart, skeletal muscle, brain,
liver, testis and kidney (7,19,28,29,31,39,40,42).
Sirtuin 5 (SIRT5) is distributed widely in all prokaryotes either
bacteria or archaea (52). The
wide tissue distribution for Sirt5 expression, with significant
levels in all tissues tested, might indicate that Sirt5 fulfills
general functions needed in all tissues (44). In humans, analyses of expressed
sequence tag databases indicated that Sirt5 is predominantly
expressed in lymphoblasts, beside heart muscle cells and thymus,
leading to the suggestion that chromosomal breaks in SIRT5 might
contribute to myeloid leukemia (19). However, a direct involvement of
SIRT5 and its repetitive elements in malignant diseases remains to
be shown. The further functional characterization of murine SIRT5
may help to elucidate its potential role, and possibly becoming an
exciting en deavor.
References
|
1.
|
Imai S, Armstrong CM, Kaeberlein M and
Guarente L: Transcriptional silencing and longevity protein Sir2 is
an NAD-dependent histone deacetylase. Nature. 403:795–800. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Schuetz A, Min J, Antoshenko T, et al:
Structural basis of inhibition of the human
NAD+-dependent deacetylase SIRT5 by suramin. Stucture.
15:377–389. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Brachmann CB, Sherman JM, Devine SE,
Cameron EE, Pillus L and Boeke JD: The SIR2 gene family, conserved
from bacteria to humans, functions in silencing, cell cycle
progression, and chromosome stability. Genes Dev. 9:2888–2902.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Voelter-Mahlknecht S and Mahlknecht U:
Cloning, chromosomal characterization and mapping of the
NAD-dependent histone deacetylases gene sirtuin 1. Int J Mol Med.
17:59–67. 2006.PubMed/NCBI
|
|
5.
|
Vaziri H, Dessain SK, Ng Eaton E, et al:
hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell.
107:149–159. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
North BJ, Marshall BL, Borra MT, Denu JM
and Verdin E: The human Sir2 ortholog, SIRT2, is an
NAD+-dependent tubulin deacetylase. Mol Cell.
11:437–444. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Starai VJ, Celic I, Cole RN, Boeke JD and
Escalante-Semerena JC: Sir2-dependent activation of acetyl-CoA
synthetase by deacetylation of active lysine. Science.
298:2390–2392. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Sauve AA and Schramm VL: SIR2: the
biochemical mechanism of NAD(+)-dependent protein deacetylation and
ADP-ribosyl enzyme intermediates. Curr Med Chem. 11:807–826.
2004.
|
|
9.
|
Liszt G, Ford E, Kurtev M and Guarente L:
Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J
Biol Chem. 280:21313–21320. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Mahlknecht U and Hoelzer D: Histone
acetylation modifiers in the pathogenesis of malignant disease. Mol
Med. 6:623–644. 2000.PubMed/NCBI
|
|
11.
|
Mahlknecht U, Ottmann OG and Hoelzer D:
When the band begins to play: histone acetylation caught in the
crossfire of gene control. Mol Carcinog. 27:268–271. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Vaquero A, Scher M, Lee D,
Erdjument-Bromage H, Tempst P and Reinberg D: Human SirT1 interacts
with histone H1 and promotes formation of facultative
heterochromatin. Mol Cell. 16:93–105. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Blander G, Olejnik J, Krzymanska-Olejnik
E, et al: SIRT1 shows no substrate specificity in vitro. J Biol
Chem. 280:9780–9785. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Blander G and Guarente L: The Sir2 family
of protein deacetylases. Annu Rev Biochem. 73:417–435. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Straight AF, Shou W, Dowd GJ, et al: Net1,
a Sir2-associated nucleolar protein required for rDNA silencing and
nucleolar integrity. Cell. 97:245–256. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Fritze CE, Verschueren K, Strich R and
Easton Esposito R: Direct evidence for SIR2 modulation of chromatin
structure in yeast rDNA. EMBO J. 16:6495–6509. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Haigis MC, Mostoslavsky R, Haigis KM, et
al: SIRT4 inhibits glutamate dehydrogenase and opposes the effects
of calorie restriction in pancreatic beta cells. Cell. 126:941–954.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Hede K: Histone deacetylase inhibitors sit
at crossroads of diet, aging, cancer. J Natl Cancer Inst.
98:377–379. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Mahlknecht U, Ho AD, Letzel S and
Voelter-Mahlknecht S: Assignment of the NAD-dependent deacetylase
sirtuin 5 gene (SIRT5) to human chromosome band 6p23 by in situ
hybridization. Cytogenet Genome Res. 112:208–212. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Frye RA: Phylogenetic classification of
prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res
Commun. 273:793–798. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Birnboim HC and Doly J: A rapid alkaline
extraction procedure for screening recombinant plasmid DNA. Nucleic
Acids Res. 7:1513–1523. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Mahlknecht U, Hoelzer D and Bucala R:
Sequencing of genomic DNA. Biotechniques. 27:406–408. 1999.
|
|
23.
|
Altschul SF, Madden TL, Schaffer AA, et
al: Gapped BLAST and PSI-BLAST: a new generation of protein
database search programs. Nucleic Acids Res. 25:3389–3402. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Mahlknecht U and Voelter-Mahlknecht S:
Genomic organization and localization of the NAD-dependent histone
deacetylase gene sirtuin 3 (Sirt3) in the mouse. Int J Oncol.
38:813–822. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Wilbur WJ and Lipman DJ: Rapid similarity
searches of nucleic acid and protein data banks. Proc Natl Acad Sci
USA. 80:726–730. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Needleman SB and Wunsch CD: A general
method applicable to the search for similarities in the amino acid
sequence of two proteins. J Mol Biol. 48:443–453. 1970. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Fitzgerald M and Shenk T: The sequence
5′-AAUAAA-3′forms parts of the recognition site for polyadenylation
of late SV40 mRNAs. Cell. 24:251–260. 1981.
|
|
28.
|
Su AI, Wiltshire T, Batalov S, et al: A
gene atlas of the mouse and human protein-encoding transcriptomes.
Proc Natl Acad Sci USA. 101:6062–6067. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Su AI, Cooke MP, Ching KA, et al:
Large-scale analysis of the human and mouse transcriptomes. Proc
Natl Acad Sci USA. 99:4465–4470. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Walker JR, Su AI, Self DW, et al:
Applications of a rat multiple tissue gene expression data set.
Genome Res. 14:742–749. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Huang JY, Hirschey MD, Shimazu T, Ho L and
Verdin E: Mitochondrial sirtuins. Biochim Biophys Acta.
1804:1645–1651. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Fulco M, Schiltz RL, Iezzi S, et al: Sir2
regulates skeletal muscle differentiation as a potential sensor of
the redox state. Mol Cell. 12:51–62. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Brunet A, Sweeney LB, Sturgill JF, et al:
Stress-dependent regulation of FOXO transcription factors by the
SIRT1 deacetylase. Science. 303:2011–2015. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Luo J, Nikolaev AY, Imai S, et al:
Negative control of p53 by Sir2alpha promotes cell survival under
stress. Cell. 107:137–148. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Yeung F, Hoberg JE, Ramsey CS, et al:
Modulation of NF-kappaB-dependent transcription and cell survival
by the SIRT1 deacetylase. EMBO J. 23:2369–2380. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Motta MC, Divecha N, Lemieux M, et al:
Mammalian SIRT1 represses forkhead transcription factors. Cell.
116:551–563. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Michishita E, McCord RA, Berber E, et al:
SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric
chromatin. Nature. 452:492–496. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Ford E, Voit R, Liszt G, Magin C, Grummt I
and Guarente L: Mammalian Sir2 homolog SIRT7 is an activator of RNA
polymerase I transcription. Genes Dev. 20:1075–1080. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Michishita E, Park JY, Burneskis JM,
Barrett JC and Horikawa I: Evolutionarily conserved and
nonconserved cellular localizations and functions of human SIRT
proteins. Mol Biol Cel. l6:4623–4635. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Haigis MC and Sinclair DA: Mammalian
sirtuins: biological insights and disease relevance. Annu Rev
Pathol. 5:253–295. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Nakamura Y, Ogura M, Tanaka D and Inagaki
N: Localization of mouse mitochondrial SIRT proteins: shift of
SIRT3 to nucleus by co-expression with SIRT5. Biochem Biophys Res
Commun. 366:174–179. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Nakagawa T, Lomb DJ, Haigis MC and
Guarente L: SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and
regulates the urea cycle. Cell. 137:560–570. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Schlicker C, Gertz M, Papatheodorou P,
Kachholz B, Becker CF and Steegborn C: Substrates and regulation
mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J
Mol Biol. 382:790–801. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Gertz M and Steegborn C: Function and
regulation of the mitochondrial sirtuin isoform Sirt5 in Mammalia.
Biochim Biophys Acta. 1804:1658–1665. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Nakahata Y, Kaluzova M, Grimaldi B, et al:
The NAD+-dependent deacetylase SIRT1 modulates
CLOCK-mediated chromatin remodeling and circadian control. Cell.
134:329–340. 2008.PubMed/NCBI
|
|
46.
|
Nakamura Y, Ogura M, Ogura K, Tanaka D and
Inagaki N: SIRT5 deacetylates and activates urate oxidase in liver
mitochondria of mice. FEBS Lett. 586:4076–4081. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
North BJ, Schwer B, Ahuja N, Marshall B
and Verdin E: Preparation of enzymatically active recombinant class
III protein deacetylases. Methods. 36:338–345. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Lieber CS, Leo MA, Wang X and Decarli LM:
Alcohol alters hepatic FoxO1, p53, and mitochondrial SIRT5
deacetylation function. Biochem Biophys Res Commun. 373:246–252.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
49.
|
Shepard BD and Tuma PL: Alcohol-induced
protein hyperacetylation: mechanisms and consequences. World J
Gastroenterol. 15:1219–1230. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50.
|
Singer MF, Thayer RE, Grimaldi G, Lerman
MI and Fanning TG: Homology between the KpnI primate and BamH1
(M1F-1) rodent families of long interspersed repeated sequences.
Nucleic Acids Res. 11:5739–5745. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
51.
|
Mitelman F, Mertens F and Johansson B: A
breakpoint map of recurrent chromosomal rearrangements in human
neoplasia. Nat Genet. 15:417–474. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
52.
|
Michan S and Sinclair D: Sirtuins in
mammals: insights into their biological function. Biochem J.
404:1–13. 2007. View Article : Google Scholar : PubMed/NCBI
|