Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of
the most lethal solid tumors and is the fourth leading cause of
cancer-related death in the United States. (1) The poor prognosis of pancreatic cancer
mainly results from its rapid growth, invasion and early metastasis
(2,3). Considerable effort has been made to
treat pancreatic cancer, however, no satisfactory progress has been
made during the past several years. Therefore, increased
understanding of the cancer biology of pancreatic cancer and
identification of novel therapeutic strategies are both urgently
needed.
In addition to the extreme malignancy of the cancer
epithelial cells, another hallmark of PDAC is a dense stroma
surrounding the cancer cells. The stroma of PDAC is mainly composed
of collagen fibers, extracellular matrix proteins, fibroblasts and
inflammatory cells. An abundant extracellular matrix defends the
cancer cells and is a leading cause of the enhanced malignant
potential (4). It is now widely
believed that the tumor-microenvironment plays key roles during
both tumorigenesis and the anticancer drug response. To identify
novel therapeutic pancreatic cancer targets, it is essential to
investigate the interaction between the tumor and its
microenvironment during the tumorigenesis of pancreatic cancer.
Among the factors associated with
tumor-microenvironment interaction that are potential targets of
anticancer therapies, members of the chemokine superfamily are
promising candidates (5,6). Chemokines are a superfamily of
chemotactic cytokines with a crucial role in the immune and
inflammation responses. Chemokines are classified into four groups
based on the spacing of their first two cysteine residues: CXC, CC,
C and CX3C chemokines (7,8). The most important function of
chemokines is to regulate the chemotactic migration of leukocytes
(9,10). Recent studies have also revealed
the pivotal role of chemokines in tumor progression (11,12).
CCL18, a member of CC chemokine family, was recently
found to play a pivotal role in the progression of malignant
tumors. CCL18 is mainly expressed in monocytes, macrophages and
immature dendritic cells (13–15).
CCL18 protein attracts lymphocytes and immature dendritic cells and
induces collagen deposition by fibroblasts (16). CCL18 is one of the most abundant
factors that were expressed by tumor-associated macrophages (TAMs).
Stimulators, such as IL4, can polarize macrophages to a phenotype
of M2 and upregulate the expression of CCL18 (17). Enhanced CCL18 production has been
demonstrated in tumor tissue, peripheral blood and dropsy of the
serous cavity associated with several malignancies. Highly
expressed CCL18 protein can suppress the maturation and recruitment
of killer cells, including lymphocytes and dendritic cells, and
destroy their immunocompetence (16). CCL18 can also promote the migration
and invasion of cancer cells by binding them directly (6,18).
Recently, in pathway analysis of a genome-wide association study,
Li et al (19) reported
that CCL18 might be a susceptibility factor for the progression of
pancreatic cancer through the Th1/Th2 immune response. Therefore,
we hypothesized that CCL18 may play key roles during the
progression of pancreatic cancer.
In this study, we evaluated the expression of CCL18
in human PDAC tissues by immunohistochemistry and preoperative
serum by ELISA, and analyzed the correlation between CCL18
expression and the clinicopathological factors of 62 PDAC patients.
Furthermore, we assessed the effects of CCL18 on the proliferation,
migration and invasion of in vitro cultured pancreatic
cancer cells.
Materials and methods
Patients and samples
Sixty-two patients (46 males and 16 females, median
age 59 years, range 35–75), who underwent resection of PDAC at our
institution between January 2007 and December 2011, were included
in this study. Clinical data was collected from a pathography of
the patients. The pathological classification of these cases was
based on the UICC (Union for International Cancer Control)-TNM
classification of malignant tumors (20). Preoperative serum was collected
from 24 PDAC patients and the control group consisted of eight age-
and gender-matched healthy volunteers. The use of the clinical
samples was approved by the Ethics Committee of the First
Affiliated Hospital of China Medical University, and written
informed consent was obtained from each patient.
Cell lines and treatment
Human pancreatic cancer cell lines PANC-1, BxPC-3,
CAPAN-2 and SW1990, and human monocyte cell lines U937 and THP-1
were purchased from American Type Culture Collection (ATCC,
Rockville, MD, USA). Cells were grown in RPMI-1640 medium
containing 10% fetal bovine serum (FBS), 100 U/ml penicillin and
100 μg/ml streptomycin and were incubated at 37°C in a 5%
CO2 atmosphere. For the collection of pancreatic cancer
cells for quantitative real-time PCR (qRT-PCR) or cell culture
supernatants of these cells for enzyme-linked immunosorbent assay
(ELISA), the four pancreatic cancer cell lines were cultured at a
density of 5×105/ml. After incubation for one day, the
medium was replaced by fresh medium with or without IL-4. After
incubation for additional 72 h, the supernatants and cells were
collected. For the collection of macrophages for qRT-PCR or cell
culture supernatant for ELISA, U937 and THP-1 cells were incubated
at a density of 5×105/ml and stimulated for two days by
10 ng/ml (U937) or 100 ng/ml (THP-1) of phorbol 12-myristate
13-acetate (PMA, Sigma-Aldrich), respectively. Non-adherent cells
were removed by washing and new medium with or without 45 ng/ml IL4
was added. After incubation for three days, the cells and
supernatants were collected. The two monocyte cell lines were
incubated at a density of 5×105/ml for seven days to
collect cells and the supernatant.
Immunohistochemistry and
immunofluorescence
Patient PDAC tissues were formalin-fixed and
paraffin-embedded. The tissue sections (3–4 μm) were rehydrated and
treated with 3% hydrogen peroxide in methanol, followed by antigen
retrieval. After being blocked with Dako® Protein Block,
the sections were incubated with primary antibodies at 4°C
overnight. This was followed by incubation with a secondary
antibody for 30 min at room temperature. Slides were then treated
with streptavidin-peroxidase reagent at 37°C for 15 min. For
immunohistochemistry, Dako® DAB Chromogen was used for
the color-reaction followed by nucleus counterstaining with
hematoxylin. For immunofluorescence, the nucleus was counterstained
by DAPI and imaged using a Nikon Eclipse E600 microscope with DP
Manager Version 1.2.1.107 software. The following antibodies were
used: primary antibodies: rabbit anti-CCL18 (1:200, Abcam); mouse
anti-CD68 (1:100, Dako); mouse anti-CD163 (1:100, Novocastra);
mouse anti-Ki67 (1:50, Abcam); mouse anti-p53 (1:200, Abcam), mouse
anti-CEA (1:400, Abcam); mouse anti-CA19-9 (1:100, Abcam); mouse
anti-CD34 (1:100, Abcam); secondary antibodies: anti-rabbit or
anti-mouse IgG (1:200, Vector Laboratories); Alexa Fluor 594 donkey
anti-rabbit or 488 goat anti-mouse IgG (H+L) (1:200, Life
Technologies).
qRT-PCR
Total RNA was extracted from pancreatic cancer cells
or monocyte/macrophages using TRIzol reagent according to the
instructions of the manufacturer (Takara Bio). CDNA was synthesized
using GoScript™ Reverse Transcription system according to the
manufacturer’s instructions (Promega). The relative levels of
target gene mRNA to control GAPDH were determined by qRT-PCR in a
7900 HT Fast Real-Time PCR system (Applied Biosystems) using the
GoTaq® qPCR Master Mix (Promega). The data were analyzed
by the 2−Δ ΔCt method. The sequences are listed in
Table I.
 | Table IThe sequences of PCR primers used in
this study. |
Table I
The sequences of PCR primers used in
this study.
| Gene | Forward primer | Reverse primer | Amplicon (bp) |
|---|
| CCL18 |
5′-CTCTGCTGCCTCGTCTATACCT-3′ |
5′-CTTGGTTAGGAGGATGACACCT-3′ | 108 |
| PITPNM3 |
5′-GATGCCAGAGGAGAAGGGAC-3′ |
5′-TCGCTGTCTTCGTGGATCTC-3′ | 134 |
| CCR6 |
5′-GCTCAAGTGTTCACAACCTGGAAG-3′ |
5′-TCCTAATGGCCCACTACAACCTG-3′ | 118 |
| GPR3 |
5′-TCCTCTCTCTAGCCCTGCTC-3′ |
5′-CTCTCTGGGTACCTGGGTTG-3′ | 148 |
| SNAIL1 |
5′-CATCCTTCTCACTGCCATGGA-3′ |
5′-AGGCAGAGGACACAGAACCAGA-3′ | 107 |
| VEGF |
5′-ATGACGAGGGCCTGGAGTGTG-3′ |
5′-CCTATGTGCTGGCCTTGGTGAG-3′ | 91 |
| IL8 |
5′-AAACCACCGGAAGGAACCAT-3′ |
5′-CCTTCACACAGAGCTGCAGAAA-3′ | 101 |
| E-cadherin |
5′-AGTGCCAACTGGACCATTCA-3′ |
5′-TCTTTGACCACCGCTCTCCT-3′ | 314 |
| CD44 |
5′-TGCCGCTTTGCAGGTGTAT-3′ |
5′-GGCCTCCGTCCGAGAGA-3′ | 66 |
| CXCR4 |
5′-GCCTTATCCTGCCTGGTATTGTC-3′ |
5′-GCGAAGAAAGCCAGGATGAGGAT-3′ | 130 |
| GAPDH |
5′-TGCACCACCAACTGCTTAGC-3′ |
5′-GGCATGGACTGTGGTCATGAG-3′ | 87 |
ELISA
CCL18 levels in cell culture supernatants and the
serum of healthy volunteers or PDAC patients were determined
quantitatively using a human PARC (CCL18) ELISA kit (Raybiotech) as
described by the manufacturer.
Cell proliferation assay (MTT assay)
Cells (BxPC-3, PANC-1) were seeded in 96-well plates
at a density of 5×104/ml in 100 μl of complete medium
and grown for 24 h. Then the medium was replaced with serum-free
medium containing different concentrations of recombinant human
CCL18 (rh-CCL18, Peprotech). After incubation for 24–72 h, cell
Titer 96 AQueous One Solution Cell Proliferation assay (Promega)
was added (20 μl/well) and incubated for 90 min. Finally, the
optical densities (OD) were measured at 492 nm.
Transwell chamber assay
Transwell chamber migration assay was performed
using Nunc 24-well 8.0-μm-pore transwell plates (Thermo Fisher
Scientific) according to the manufacturer’s instructions.
Pancreatic cancer cells were plated into the upper chambers at
5×104/ml. The lower chambers contained 5–25 ng/ml
rh-CCL18 or macrophages prepared beforehand. After incubation for
24 h, non-invading cells were removed from the upper surface of the
membrane using a cotton-tipped swab. Then the invading cells were
fixed in methanol for 10 min and stained with 0.1% crystal violet
hydrate (Sigma) for 30 min. The invading cells were counted as
cells per field at 10× magnification. The invasion assay was
performed in a similar fashion except the 8.0-μm pore size membrane
inserts were coated with matrigel (BD Biosciences) that was diluted
at 1:6 with serum-free media.
Western blotting
Total proteins were extracted from cells with RIPA
cell lysis buffer (Cell Signaling) on ice for 30 min. Equal amounts
of proteins were separated by 10% SDS-PAGE and transferred to
polyvinylidene difluoride membranes (Millipore). The membranes were
blocked with 2% fat-free milk in PBS at room temperature for 1 h
and probed with primary anti-SNAIL1 antibody (1:500, Abcam),
E-cadherin (1:500, Abcam) or β-actin (1:1,000, Peproteck)
antibodies at 4°C overnight. Membranes were incubated with
peroxidase-conjugated anti-rabbit IgG secondary antibody (1:5,000,
Beyotime) at room temperature for 45 min. Immunoreactive protein
bands were visualized with an ECL detection kit (Thermol Biotech).
The experiment was repeated three times.
Statistical analysis
Statistical comparisons of means were performed by
Student’s t-test, whereas χ2 test was applied to analyze
the relationship between CCL18 expression status and
clinicopathological factors. Cutoff value of CCL18-positive
macrophage counts was calculated by performing non-parametric
receiver operating characteristics (ROC) using Dr. SPSS II for
windows. The cutoff value was defined according to the best
predictive values calculated by ROC analysis (cutoff value: 19.5
cells/40× magnification). Statistic significance was defined as
P<0.05.
Results
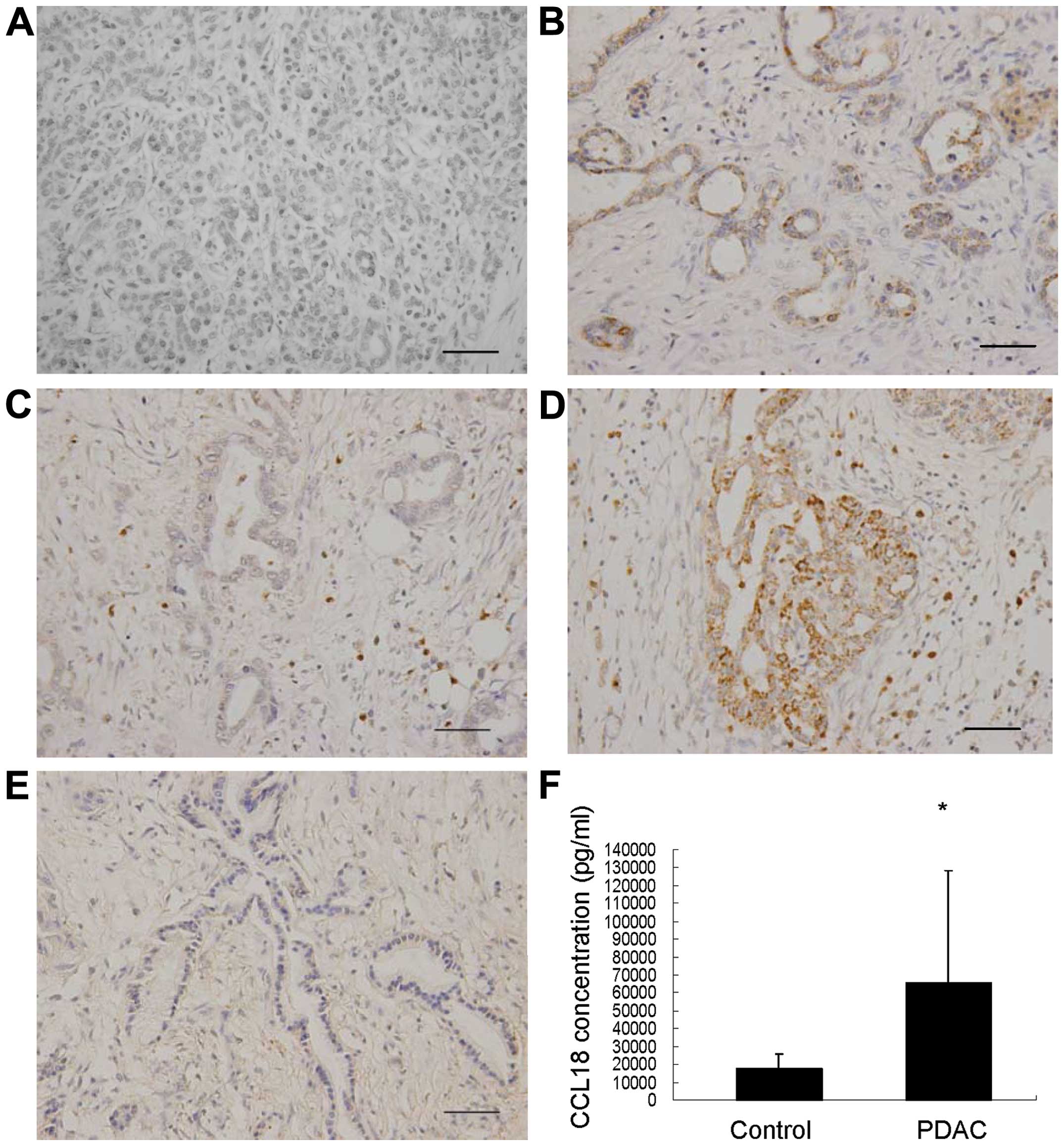
CCL18 is expressed in PDAC tissue
To evaluate the expression status of CCL18 in human
PDAC tissues, we performed immunohistochemical analysis of CCL18 in
PDAC tissues from 62 patients. Compared with normal pancreas, there
was dramatically increased expression of CCL18 in PDAC tissues
(Fig. 1A–E). Among all the cases
tested, 61.29% (38/62) stained positively for CCL18 in cancer cells
and 70.97% (44/62) were positive in mesenchymal cells, while 45.16%
(28/62) stained positively in both cancer and mesenchymal cells.
Thus, 85.48% (53/62) showed a positive expression of CCL18 in
cancer and/or mesenchymal cells. Notably, in cases with positive
staining in both cancer and mesenchymal cells, the staining of
CCL18 in cancer epithelial cells was weaker than that in
mesenchymal cells (Fig. 1D). These
results demonstrated that both the cancer epithelial and
mesenchymal cells of PDAC tissue positively express CCL18.
The concentration of CCL18 in the
peripheral blood serum of patients with PDAC is significantly
higher than that of healthy controls
Currently, several serum markers, such as CEA and
CA19-9, are commonly used to detect the origin, recurrence and
metastasis of pancreatic cancer (21,22).
Since CCL18 is a soluble protein and is upregulated in PDAC
tissues, we hypothesized that serum levels of CCL18 might serve as
a diagnostic or follow-up marker in PDAC. We measured the
concentration of soluble CCL18 in serum samples of patients with
PDAC (n=24) and in healthy donors (n=8) by ELISA. The serum level
of CCL18 in PDAC patients was 8,913.60 pg/ml to 270,117.30 pg/ml
(65,337.19±63,287.63 pg/ml) while that of healthy donors was
3,721.57 pg/ml to 25,046.21 pg/ml (17,510.83±8,717.47 pg/ml)
(P=0.039) (Fig. 1F). Moreover, 75%
(18/24) of the PDAC samples demonstrated higher CCL18 levels than
the highest value measured in the control group. Therefore, the
serum CCL18 levels were significantly higher in patients with PDAC
in comparison to healthy controls, suggesting that serum CCL18
level is a potentially useful biomarker for the diagnosis and
prognosis of PDAC.
CCL18 expression in both cancer and
mesenchymal cells correlates with PDAC tumor progression and a
worse survival rate for PDAC patients
We next analyzed the correlation between the
expression of CCL18 and the clinicopathological factors of 62 PDAC
patients. The clinical data are summarized in Tables II and III. The results revealed that CCL18
expression in both cancer and mesenchymal cells was significantly
correlated with lymph node metastasis (P=0.015) and UICC stage
(P=0.037) (Table II). Moreover,
the number of CCL18-expressing mesenchymal cells was significantly
associated with tissue CA19-9 expression level (P=0.030) (Table III).
 | Table IICorrelation of CCL18 expression with
clinical data from PDAC patients. |
Table II
Correlation of CCL18 expression with
clinical data from PDAC patients.
| | Cancer cells | | Mesenchymal | | Mesenchymal
positive cell counts | | Positive
mesenchymal and cancer cells | |
|---|
| |
| |
| |
| |
| |
|---|
| Parameters | No. of
patients | − | + | P-value | − | + | P-value | < 20 | ≥20 | P-value | No | Yes | P-value |
|---|
| Cases | 62 | 24 | 38 | | 18 | 44 | | 43 | 19 | | 34 | 28 | |
| Age | | | | | | | | | | | | | |
| ≤60 | 35 | 15 | 20 | 0.309 | 12 | 23 | 0.226 | 25 | 10 | 0.448 | 21 | 14 | 0.251 |
| >60 | 27 | 9 | 18 | | 6 | 21 | | 18 | 9 | | 13 | 14 | |
| Gender | | | | | | | | | | | | | |
| Male | 46 | 18 | 28 | 0.576 | 13 | 33 | 0.528 | 30 | 16 | 0.19 | 26 | 20 | 0.435 |
| Female | 16 | 6 | 10 | | 5 | 11 | | 13 | 3 | | 8 | 8 | |
| Tumor location | | | | | | | | | | | | | |
| Head | 56 | 22 | 34 | 0.572 | 16 | 40 | 0.567 | 38 | 18 | 0.397 | 3 | 26 | 0.434 |
| Body/tail | 6 | 2 | 4 | | 2 | 4 | | 5 | 1 | | 4 | 2 | |
| Tumor size | | | | | | | | | | | | | |
| ≤2.5 cm | 16 | 4 | 12 | 0.157 | 6 | 10 | 0.288 | 11 | 5 | 0.592 | 8 | 8 | 0.435 |
| >2.5 cm | 46 | 20 | 26 | | 12 | 34 | | 32 | 14 | | 26 | 20 | |
|
Differentiation | | | | | | | | | | | | | |
| Well | 20 | 7 | 13 | 0.449 | 7 | 13 | 0.335 | 16 | 4 | 0.169 | 11 | 9 | 0.602 |
| Moderate/poor | 42 | 17 | 25 | | 11 | 31 | | 27 | 15 | | 23 | 19 | |
| T stage | | | | | | | | | | | | | |
| T1+T2 | 22 | 12 | 10 | 0.052 | 9 | 13 | 0.109 | 16 | 6 | 0.449 | 15 | 7 | 0.096 |
| T3+T4 | 40 | 12 | 28 | | 9 | 31 | | 27 | 13 | | 19 | 21 | |
| Lymph node
metastasis | | | | | | | | | | | | | |
| Negative | 41 | 19 | 22 | 0.072 | 14 | 27 | 0.173 | 31 | 10 | 0.115 | 27 | 14 | 0.015 |
| Positive | 21 | 5 | 16 | | 4 | 17 | | 12 | 9 | | 7 | 14 | |
| TNM stage | | | | | | | | | | | | | |
| I+IIA | 31 | 15 | 16 | 0.096 | 11 | 20 | 0.201 | 24 | 7 | 0.135 | 21 | 10 | 0.037 |
| IIB+III | 31 | 9 | 22 | | 7 | 24 | | 19 | 12 | | 13 | 18 | |
| Perineural
invasion | | | | | | | | | | | | | |
| Absent | 52 | 20 | 32 | 0.596 | 16 | 36 | 0.394 | 34 | 18 | 0.117 | 30 | 22 | 0.247 |
| Present | 10 | 4 | 6 | | 2 | 8 | | 9 | 1 | | 4 | 6 | |
| Vascular
permeation | | | | | | | | | | | | | |
| Absent | 43 | 17 | 26 | 0.536 | 12 | 31 | 0.497 | 32 | 11 | 0.158 | 23 | 20 | 0.484 |
| Present | 19 | 7 | 12 | | 6 | 13 | | 11 | 8 | | 11 | 8 | |
| Pre-therapeutic
CA19-9 level | | | | | | | | | | | | | |
| <37 U/ml | 19 | 7 | 12 | 0.536 | 8 | 11 | 0.115 | 13 | 6 | 0.57 | 11 | 8 | 0.484 |
| ≥37 U/ml | 43 | 17 | 26 | | 10 | 33 | | 30 | 13 | | 20 | 23 | |
 | Table IIICorrelation of CCL18 expression with
Ki67, P53, CEA, CA19-9 and CD34 expression in 62 cases of PDAC. |
Table III
Correlation of CCL18 expression with
Ki67, P53, CEA, CA19-9 and CD34 expression in 62 cases of PDAC.
| | Cancer cells | | Mesenchymal | | Mesenchymal
positive cell counts | | Positive
mesenchymal and cancer cells | |
|---|
| |
| |
| |
| |
| |
|---|
| No. of
patients | − | + | P-value | − | + | P-value | < 20 | ≥20 | P-value | No | Yes | P-value |
|---|
| Cases | 62 | 24 | 38 | | 18 | 44 | | 43 | 19 | | 34 | 28 | |
| Ki67 |
| Negative | 25 | 13 | 12 | 0.067 | 6 | 19 | 0.336 | 19 | 6 | 0.259 | 15 | 10 | 0.341 |
| Positive | 37 | 11 | 26 | | 12 | 25 | | 24 | 13 | | 19 | 18 | |
| P53 |
| Negative | 28 | 12 | 16 | 0.364 | 7 | 21 | 0.363 | 22 | 6 | 0.124 | 16 | 12 | 0.471 |
| Positive | 34 | 12 | 22 | | 11 | 23 | | 21 | 13 | | 18 | 16 | |
| CEA |
| Negative | 15 | 7 | 8 | 0.333 | 2 | 13 | 0.110 | 8 | 7 | 0.112 | 8 | 7 | 0.563 |
| Positive | 47 | 17 | 30 | | 16 | 31 | | 35 | 12 | | 26 | 21 | |
| CA19-9 |
| Negative | 29 | 11 | 18 | 0.557 | 8 | 21 | 0.519 | 24 | 5 | 0.030 | 14 | 15 | 0.237 |
| Positive | 33 | 13 | 20 | | 10 | 23 | | 19 | 14 | | 20 | 13 | |
| CD34 |
| Negative | 48 | 19 | 29 | 0.525 | 13 | 35 | 0.377 | 31 | 17 | 0.117 | 27 | 21 | 0.455 |
| Positive | 14 | 5 | 9 | | 5 | 9 | | 12 | 2 | | 7 | 7 | |
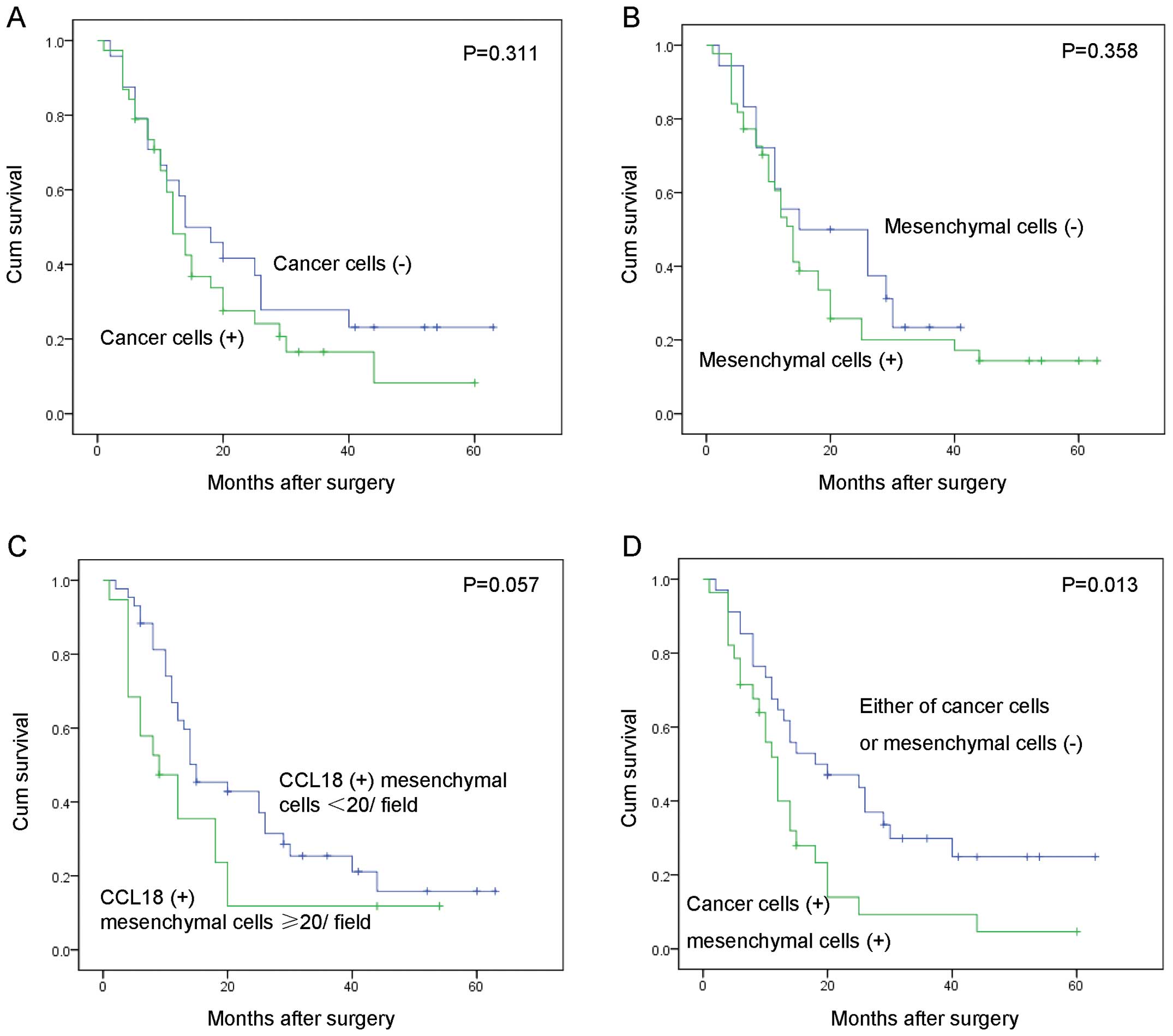
We further examined the correlation between the
expression of CCL18 and the overall survival of these 62 PDAC
patients by Kaplan-Meier analysis. CCL18 expression in cancer cells
(Fig. 2A, P=0.311), mesenchymal
cells (Fig. 2B, P=0.358), and the
count of mesenchymal positive cells (Fig. 2C, P=0.057) was not statistically
associated with survival of PDAC patients. Interestingly, PDAC
patients with CCL18 expression in both cancer and mesenchymal cells
had a significantly worse overall survival rate than patients
without CCL18 expression in either cell type (Fig. 2D, χ2 = 6.165,
P=0.013).
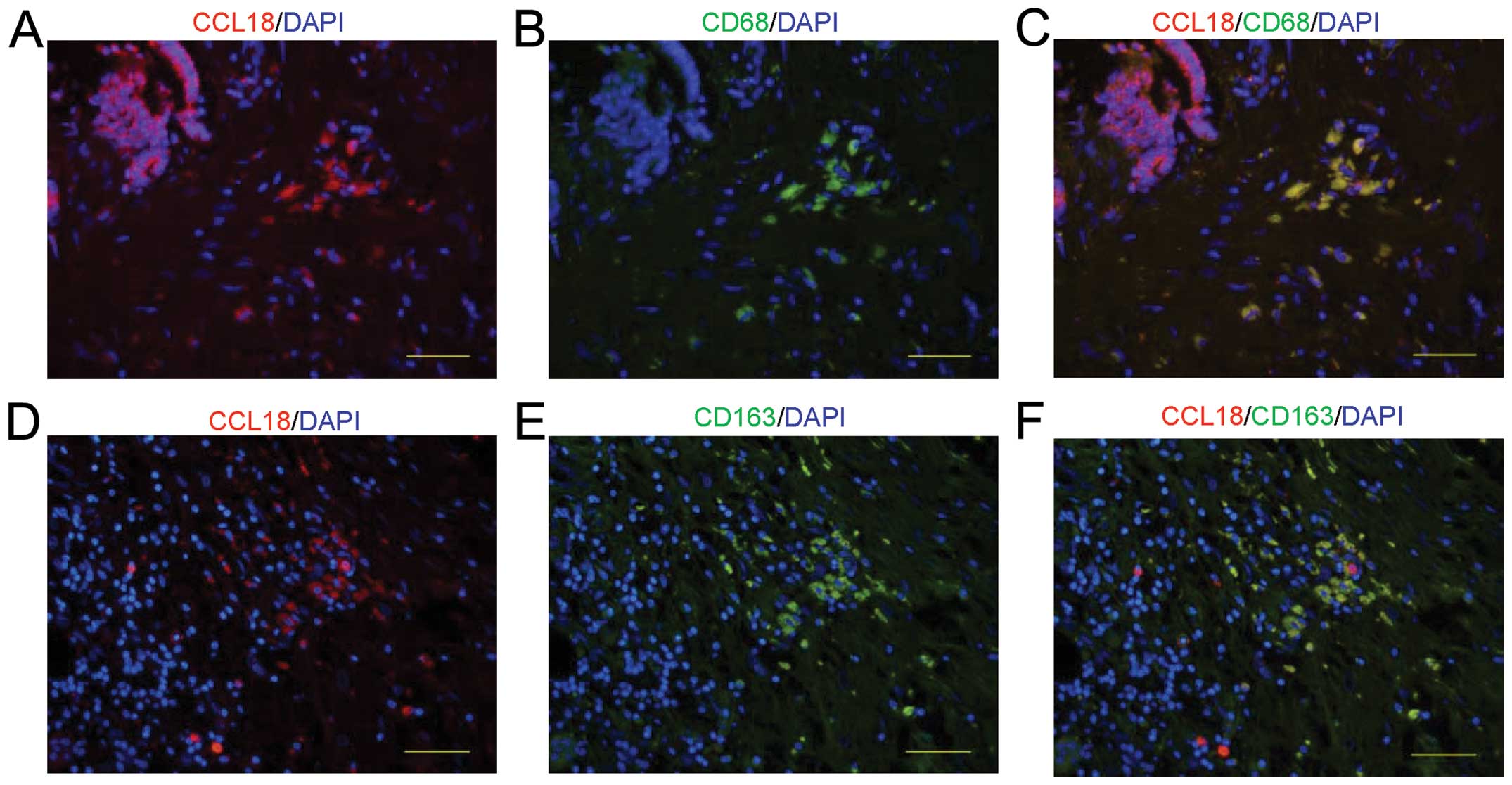
CCL18-positive cells in the mesenchyme of
PDAC tissues are M2-polarized macrophages
Previous reports showed that CCL18 is mainly
expressed in the monocyte-macrophage system and is highly expressed
in tumor-associated macrophages (TAMs) (17). It is thus possible that the
CCL18-positive mesenchymal cells in our tested PDAC tissues are
macrophages. To test this hypothesis, we performed
immunofluorescence staining of both CCL18 and the macrophage marker
CD68. CCL18-expressing cells co-localized with CD68 positive
staining (Fig. 3A–C), indicating
that CCL18-expressing cells are macrophages. Moreover, in agreement
with a previous report (6),
additional double staining of CD163 and CCL18 indicated that
CCL18-expressing macrophages were a subset of CD163-positive
M2-polarized macrophages (Fig.
3D–F).
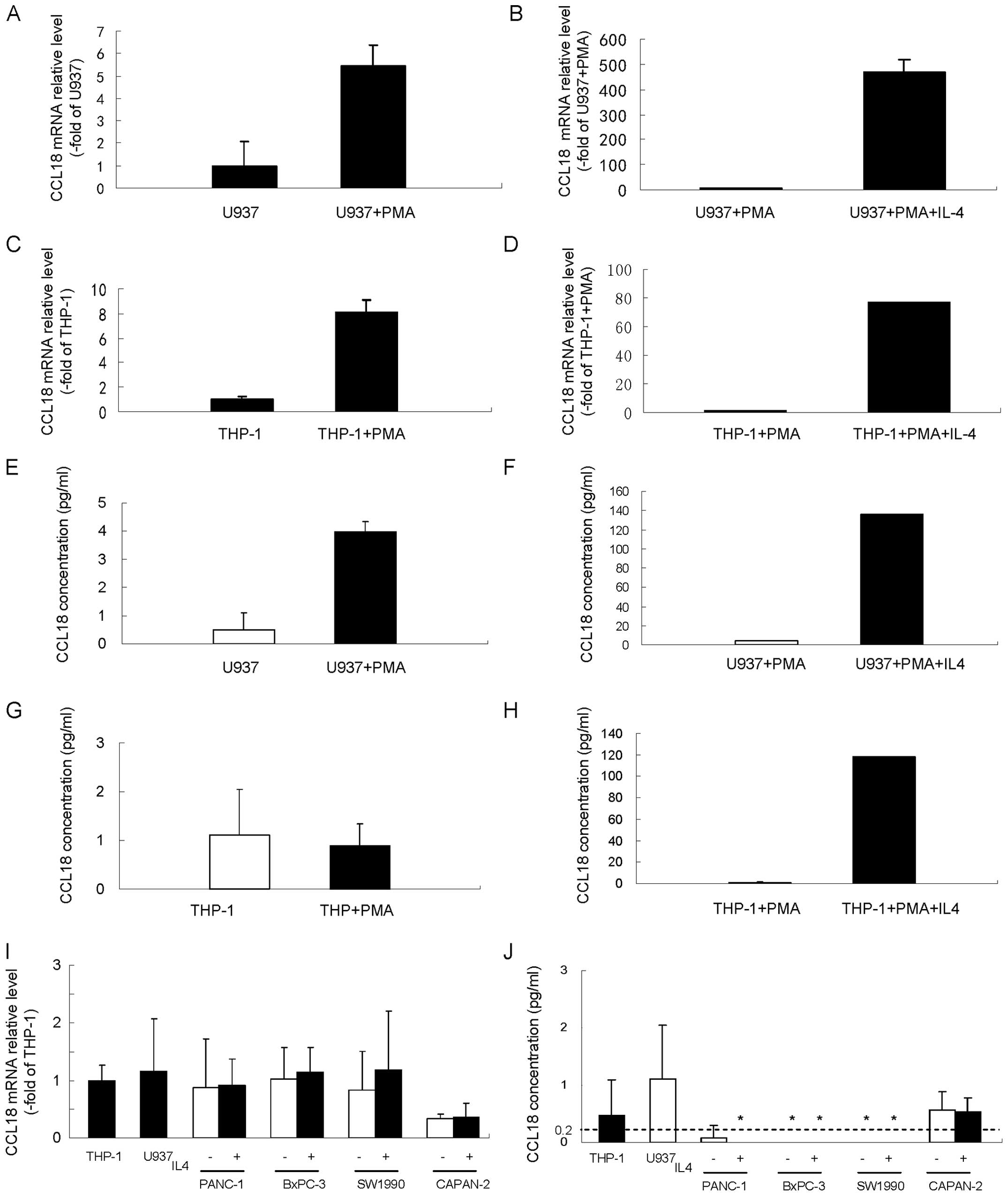
U937 and THP-1 cell derived macrophages
secrete high levels of CCL18 while cultured pancreatic cancer cells
express limited levels of CCL18
U937 and THP-1 cell lines are representative
monocyte/macrophage cells and widely used as human monocyte cells.
Macrophage differentiation can be induced in both cell lines by
stimulation with PMA (23,24). Previous reports showed that IL4
could stimulate the polarization of peripheral blood monocytes,
causing the monocytes to adopt an M2 phenotype and release of high
levels of CCL18 (6). We used these
two cell lines as monocyte models to verify the expression of CCL18
in monocytes and macrophages. In agreement with previous reports,
our results showed that CCL18 mRNA level (Fig. 4A) and secreted CCL18 protein
(Fig. 4E) were low in untreated
U937 cells, while both were significantly upregulated after
conversion of U937 cells to macrophages by PMA. Impressively, IL4
further dramatically enhanced the CCL18 mRNA level (Fig. 4B) and secretion of CCL18 protein
(Fig. 4F) in PMA stimulated U937
cells. We observed no stimulation of CCL18 protein expression in
THP-1 cells after PMA treatment (Fig.
4G), but PMA treatment significantly increased the levels of
CCL18 mRNA (Fig. 4C). Similar to
U937 cells, IL4 significantly upregulated the mRNA level (Fig. 4D) and secretion of CCL18 protein
(Fig. 4H) in THP-1 cells
stimulated with PMA.
To determine gene and protein expression of CCL18 in
pancreatic cancer cells, various pancreatic cancer cell lines
(PANC-1, BxPC-3, SW1990 and CAPAN-2) were subjected to qRT-PCR and
ELISA analysis of CCL18. All four pancreatic cancer cell lines
showed faint CCL18 mRNA expression levels that were similar to
those of THP-1 and U937 cells (Fig.
4I). With the exception of the CAPAN-2 cells, which displayed a
faintly detectable level of CCL18, the soluble CCL18 protein level
in the cell culture supernatant was less than the detection limit
of CCL18 ELISA kit (0.2 pg/ml) (Fig.
4J). IL4, which can upregulate the expression of CCL18 in
monocyte and macrophage U937 and THP-1 cells, did not induce CCL18
mRNA expression (Fig. 4I) or the
soluble CCL18 protein level in cell culture supernatant (Fig. 4J). These results showed that in
cultured pancreatic cancer cells, CCL18 was either not expressed or
was only expressed at a very low level.
Pancreatic cancer cell lines express the
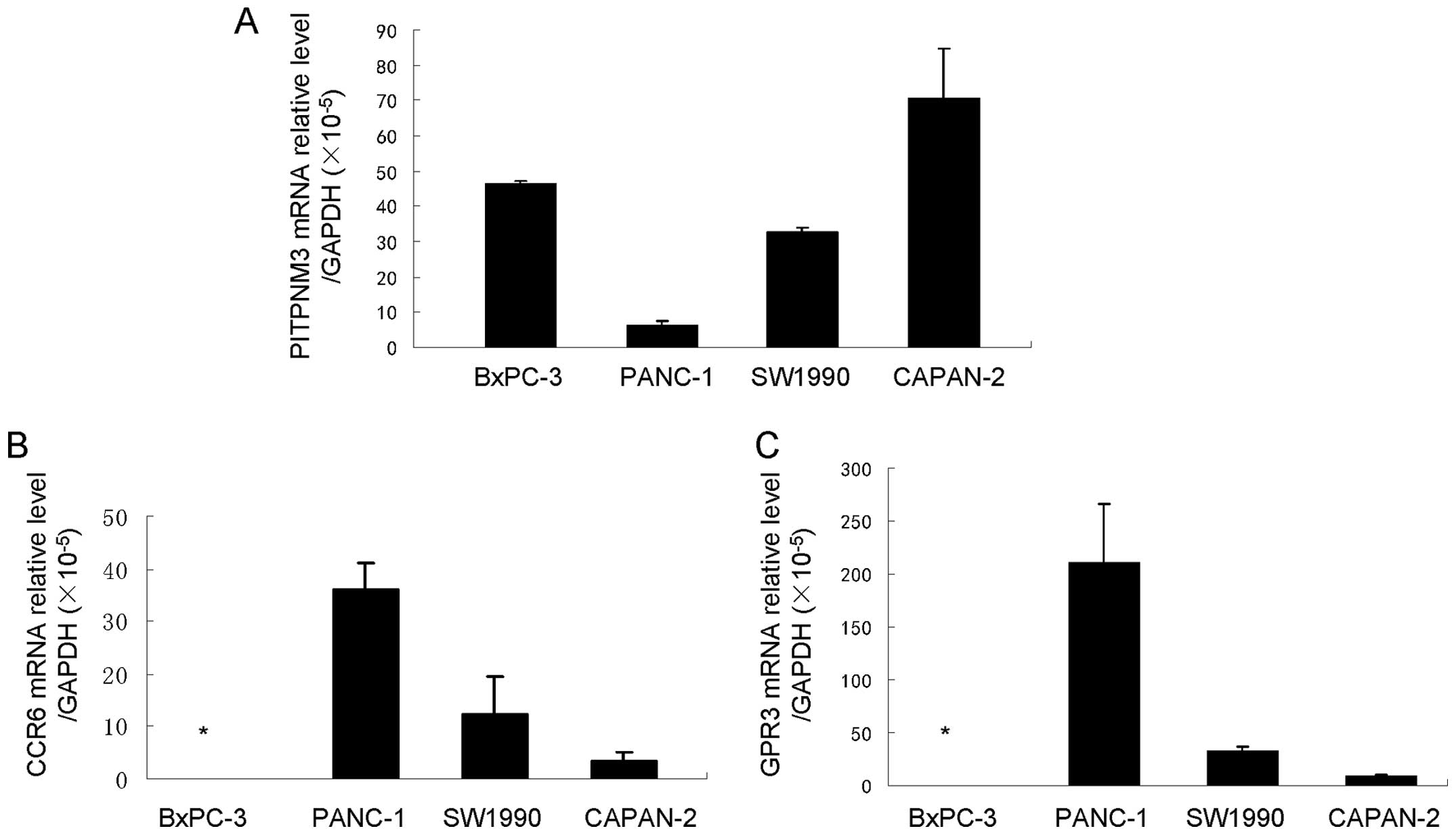
potential CCL18 receptors PITPNM3, CCR6 and GPR3
CCL18 is considered an ‘orphan ligand’, with its
cognate receptor and the underlying pathways unidentified. Recent
studies have suggested some potential receptors of CCL18. The study
by Catusse et al revealed that CCL18 acts agonistically to
and diminishes the CXCR4-mediated effects of CXCL12 via GPR30
(25). Chen et al
demonstrated that CCL18 could promote the invasion and migration of
breast cancer cells by binding PITPNM3, a membrane-associated
phosphatidylinositol transfer domain-containing protein (6). Zissel et al (26) proposed that the chemokine receptor
CCR6 is a CCL18 receptor with the ability to initiate fibroblast
activity. We examined the gene expression of these three potential
receptors of CCL18 in pancreatic cell lines by qRT-PCR and found
that they were expressed at different levels in different cell
lines. BxPC-3 cells expressed only PITPNM3, while PANC-1, CAPAN-2
and SW1990 cells expressed various levels of PITPNM3, CCR6 and GPR3
(Fig. 5). These results suggest
the possibility of the existence of CCL18 receptors in pancreatic
cancer cells and that CCL18 may have effects on the biological
behavior of pancreatic cancer cells.
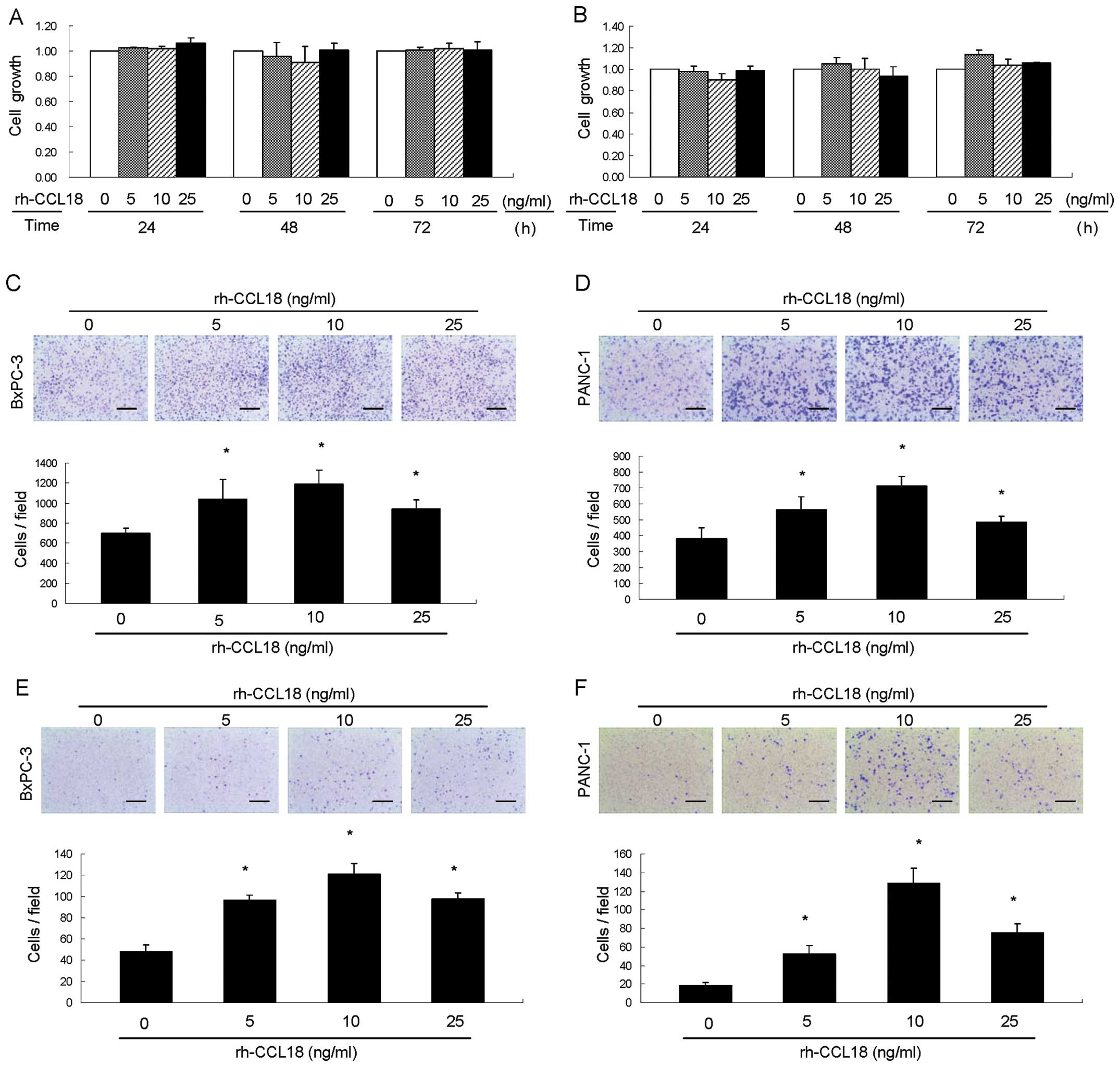
CCL18 promotes migration and invasion of
pancreatic cancer cells in vitro, but has no effect on cell
proliferation
To investigate whether CCL18 could promote the
progression of PDAC, we assessed the cell proliferation, migration
and invasion of pancreatic cancer cells by MTT assay, transwell
migration assay and transwell invasion assay, respectively. MTT
assay showed that treatment of pancreatic cancer BxPC-3 (Fig. 6A) and PANC-1 (Fig. 6B) cells with 5 ng/ml to 25 ng/ml
rh-CCL18 for 24, 48 and 72 h did not noticeably alter cell
proliferation. In contrast, cell migration after treatment with
rh-CCL18 was significantly increased in both BxPC-3 and PANC-1
cells. Migratory capability of BxPC-3 cells was significantly
higher after treatment with 10 ng/ml rh-CCL18 in both BxPC-3 cells
(1,191±141 versus control 699±54 migratory cells/field, P=0.0006,
Fig. 6C) and PANC-1 cells (715±59
versus control 383±66 migratory cells/field, P<0.0001, Fig. 6D). Transwell invasion assay
revealed that rh-CCL18 significantly increased the number of
invading cancer cells compared to serum-free medium in both BxPC-3
(Fig. 6E, P<0.0001) and PANC-1
(Fig. 6F, P<0.0001) cells.
Invading cancer cells reached their maximum at a dose of 10 ng/ml
rh-CCL18. These results indicated that CCL18 was involved in
migration and invasion but not proliferation of pancreatic cancer
cells.
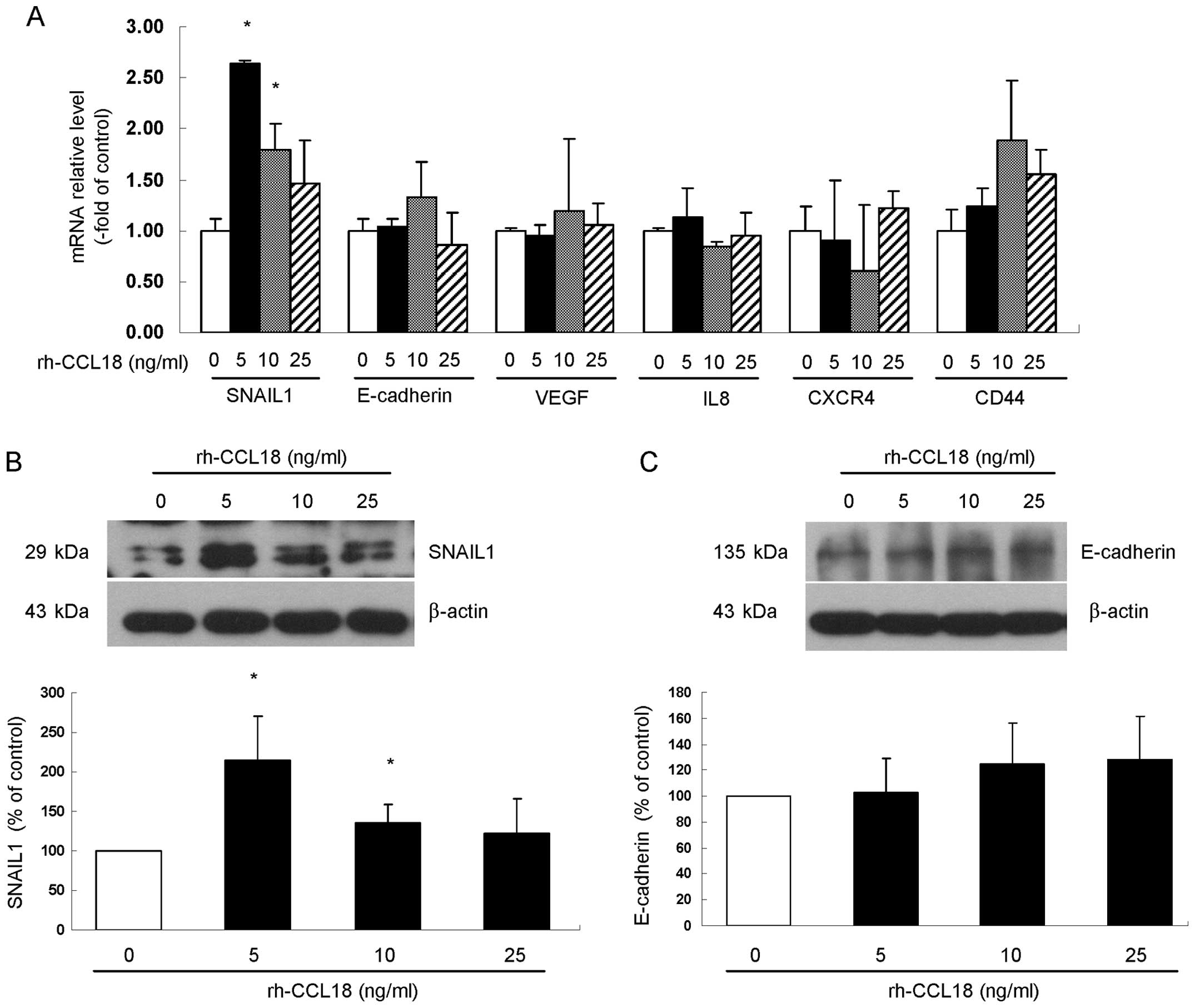
CXCL16 increased the expression level of
EMT-associated factor SNAIL1 in pancreatic cancer cells
To investigate the molecular mechanisms by which
CCL18 enhances cell migration and invasion, we examined BxPC-3
cells after stimulation with rh-CCL18 and measured the mRNA
expression of EMT-associated factors SNAIL1 and E-cadherin,
angiogenesis-associated factors VEGF and IL8 and pancreatic cancer
stem cell specific markers CD44 and CXCR4. After 24 h rh-CCL18
stimulation, gene expression of these factors was examined by
qRT-PCR. The results showed that, among all six factors examined,
only stimulation with rh-CCL18 significantly upregulated the gene
expression of EMT-related factor SNAIL1 at the concentrations of 5
ng/ml (2.64±0.03-fold of control, P=0.020) and 10 ng/ml
(1.79±0.26-fold of control, P=0.035) (Fig. 7A). Consistent with the increase in
SNAIL1 mRNA after stimulation by rh-CCL18 for 24 h, the protein
expression of SNAIL1 was upregulated at the concentrations of 5
ng/ml (2.14±0.56-fold of control, P=0.011) and 10 ng/ml
(1.35±0.23-fold of control, P=0.046) (Fig. 7B), while different concentrations
of rh-CCL18 treatment for 24 h did not change the protein level of
E-cadherin (Fig. 7C).
Discussion
In this study, we found that CCL18 was positively
expressed in both the epithelial and mesenchymal cells of human
PDAC tissues. Moreover, serum CCL18 levels were significantly
higher in patients with PDAC in comparison to those of healthy
controls. Furthermore, CCL18 expression in cancer epithelial and
mesenchymal cells correlated with malignant progression and shorter
overall survival of the 62 PDAC patients examined. Most
importantly, we observed that treatment with recombinant human
CCL18 promoted the migration and invasion of in vitro
cultured pancreatic cancer cells.
Chronic inflammation is implicated in a variety of
human cancers, including pancreatic cancer (27,28).
It is now becoming clear that the tumor microenvironment, which is
largely coordinated by inflammatory cells including
tumor-associated macrophages (TAM), tumor-associated dendritic
cells (TADC) and tumor-infiltrating T cells (TIL), is an
indispensable participant in the neoplastic process, proliferation,
survival and migration of cancer cells (29,30).
As key players in the creation of the tumor-microenvironment,
chemokines, which can be produced by the tumor cells and
tumor-associated inflammatory cells, may contribute directly to
malignant progression (6,31).
CCL18, a vital chemokine in Th-2 immune response,
was recently demonstrated to be associated with progression of
various malignant tumors, including pancreatic cancer (19,32–37).
Our immunohistochemistry results revealed that CCL18 was highly
expressed in human pancreatic cancer tissues. High CCL18 expression
was not restricted to mesenchymal cells, 61.29% of the cancer
tissues examined showed positive CCL18 staining in cancer
epithelial cells, although the expression level was relatively weak
in contrast to mesenchymal cells (Fig.
1). Furthermore, we demonstrated that CCL18 expression level
correlated with the stage of progression and overall survival of
PDAC patients (Tables II and
III and Fig. 2). These data suggest that CCL18
might contribute to the tumor progression of PDAC in both cancer
epithelial and mesenchymal cells.
Using immunofluorescence staining, we found that
CCL18-positive mesenchymal cells were CD68 or CD163-positive
macrophages (Fig. 3). In in
vitro cell cultures, we observed that CCL18 was highly
expressed in U937 and THP-1 monocytes and macrophages but very low
or undetectable in pancreatic cancer PANC-1, BxPC-3, SW1990 and
CAPAN-2 cells. In agreement with previous reports, (6) IL4, a stimulator of CCL18 expression,
significantly upregulated the expression level of CCL18 in both PMA
activated U937 and THP-1 macrophages. In sharp contrast, little to
no CCL18 was detected in pancreatic cancer cells, even after
stimulation with IL4 (Fig. 4).
This result agrees with previous reports showing CCL18 expression
under the detection limit and no CCL18 response to conventional
stimulators in various carcinoma cells (32). However, the results observed in
cultured pancreatic cancer cells were quite different from the
immunohistochemistry results we observed using clinical PDAC
samples. The discrepancy between the cell culture results and the
results observed in the clinical samples is probably due to the
distinct differences between the in vivo and in vitro
conditions. No CCL18 homologue has yet been found in rodents
(16), and the lack of an animal
model makes it difficult to confirm the expression of CCL18 in
vivo. Nevertheless, the high expression of CCL18 was mainly
found in macrophages in the microenvironment of PDAC, implying that
macrophages around cancer cells might promote the migration and
invasion of pancreatic cancer cells and that CCL18 expression might
be essential to this process.
Previous reports showed that overexpression of CCL18
by tumor tissues could promote the invasiveness of cancer cells by
interacting with other tumorigenic factors (6,18).
Our qRT-PCR results revealed expression of the potential CCL18
receptors PITPNM3, CCR6 and GPR3 in all four of the pancreatic
cancer cell lines tested (Fig. 5).
Consistently, our in vitro experiments demonstrated that
rh-CCL18 promoted the migration and invasion activity of the
pancreatic cancer cells BxPC-3 and PANC-1 (Fig. 6). These data suggest that CCL18 may
affect the biological behavior of pancreatic cancer cells by
binding cell surface receptors.
Finally, we investigated the molecular mechanism by
which CCL18 enhanced the progression of pancreatic cancer. Of the
six factors we examined, rh-CCL18 upregulated the gene expression
of SNAIL1, a marker of EMT, in BxPC-3 cells (Fig. 7A). Increased SNAIL1 expression was
confirmed by western blotting (Fig.
7B). Our results were consistent with previous observations
that CCL18 or other chemokines are associated with EMT in various
tumors (33,38–40).
Taken together, these facts suggest that, at least in part, CCL18
promotes the migration and invasion of pancreatic cancer cells
through SNAIL1 signaling.
In conclusion, we found that cancer epithelial cells
and mesenchymal macrophages from human PDAC tissues positively
expressed CCL18. The expression level of CCL18 correlated with
tumor progression and the overall survival of PDAC patients. Our
findings suggest that serum CCL18 level is a potential biomarker
for the diagnosis and prognosis of PDAC, and that CCL18 plays an
important role during the tumorigenesis of PDAC.
Acknowledgements
This study was supported by the Project for
Construction of Major Discipline Platform in Universities of
Liaoning province, the National Natural Science Foundation of China
(no. 81170423), the Science and Technology Program Foundation of
Shenyang city, China (no. F13-220-9-01) and the Science and
Technology Program Foundation of Liaoning province of China (no.
2011225019).
References
|
1
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics. CA Cancer J Clin. 59:225–249. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Moore MJ: The treatment of advanced
pancreatic cancer: current evidence and future challenges. Ann
Oncol. 19(Suppl 7): vii304–vii308. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stathis A and Moore MJ: Advanced
pancreatic carcinoma: current reatment and future challenges. Nat
Rev Clin Oncol. 7:163–172. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hamada S, Masamune A and Shimosegawa T:
Novel therapeutic strategies targeting tumor-stromal interactions
in pancreatic cancer. Front Physiol. 4:3312013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hao NB, Lü MH, Fan YH, Cao YL, Zhang ZR
and Yang SM: Macrophages in tumor microenvironments and the
progression of tumors. Clin Dev Immunol. 2012:9480982012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen J, Yao Y, Gong C, et al: CCL18 from
tumor-associated macrophages promotes breast cancer metastasis via
PITPNM3. Cancer Cell. 19:541–555. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mantovani A: The chemokine system:
redundancy for robust outputs. Immunol Today. 20:254–257. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zlotnik A and Yoshie O: Chemokines: a new
classification system and their role in immunity. Immunity.
12:121–127. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sallusto F and Mackay CR: Chemoattractants
and their receptors in homeostasis and inflammation. Curr Opin
Immunol. 16:724–731. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bromley SK, Mempel TR and Luster AD:
Orchestrating the orchestrators: chemokines in control of T cell
traffic. Nat Immunol. 9:970–980. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Strieter RM, Burdick MD, Mestas J,
Gomperts B, Keane MP and Belperio JA: Cancer CXC chemokine networks
and tumour angiogenesis. Eur J Cancer. 42:768–778. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ijichi H, Chytil A, Gorska AE, et al:
Inhibiting Cxcr2 disrupts tumor-stromal interactions and improves
survival in a mouse model of pancreatic ductal adenocarcinoma. J
Clin Invest. 121:4106–4117. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schraufstatter I, Takamori H, Sikora L,
Sriramarao P and DiScipio RG: Eosinophils and monocytes produce
pulmonary and activation-regulated chemokine, which activates
cultured monocytes/macrophages. Am J Physiol Lung Cell Mol Physiol.
286:L494–L501. 2004. View Article : Google Scholar
|
|
14
|
Sallusto F, Palermo B, Lenig D, Miettinen
M, Matikainen S, Julkunen I, Forster R, Burgstahler R, Lipp M and
Lanzavecchia A: Distinct patterns and kinetics of chemokine
production regulate dendritic cell function. Eur J Immunol.
29:1617–1625. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pivarcsi A, Gombert M, Dieu-Nosjean MC, et
al: CC chemokine ligand 18, an atopic dermatitis-associated and
dendritic cell-derived chemokine, is regulated by staphylococcal
products and allergen exposure. J Immunol. 173:5810–5817. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schutyser E, Richmond A and Van Damme J:
Involvement of CC chemokine ligand 18 (CCL18) in normal and
pathological processes. J Leukoc Biol. 78:14–26. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schraufstatter IU, Zhao M, Khaldoyanidi SK
and Discipio RG: The chemokine CCL18 causes maturation of cultured
monocytes to macrophages in the M2 spectrum. Immunology.
135:287–298. 2012. View Article : Google Scholar :
|
|
18
|
Zhang B, Yin C, Li H, et al: Nir1 promotes
invasion of breast cancer cells by binding to chemokine (C-C motif)
ligand 18 through the PI3K/Akt/GSK3β/Snail signalling pathway. Eur
J Cancer. 49:3900–3913. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li D, Duell EJ, Yu K, et al: Pathway
analysis of genome-wide association study data highlights
pancreatic development genes as susceptibility factors for
pancreatic cancer. Carcinogenesis. 33:1384–1390. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sobin LH, Gospodarowicz MK and Wittekind
CH: International Union Against Cancer (UICC) TNM Classification of
Malignant Tumors. 7th edition. Wiley-Blackwell; Oxford: 2009
|
|
21
|
Ni XG, Bai XF, Mao YL, et al: The clinical
value of serum CEA, CA19-9, and CA242 in the diagnosis and
prognosis of pancreatic cancer. Eur J Surg Oncol. 31:164–169. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yasue M, Sakamoto J, Teramukai S, Morimoto
T, Yasui K, Kuno N, Kurimoto K and Ohashi Y: Prognostic values of
preoperative and postoperative CEA and CA19.9 levels in pancreatic
cancer. Pancreas. 9:735–740. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Harris P and Ralph P: Human leukemic
models of myelomonocytic development: a review of the HL-60 and
U937 cell lines. J Leukoc Biol. 37:407–422. 1985.PubMed/NCBI
|
|
24
|
Daigneault M, Preston JA, Marriott HM,
Whyte MK and Dockrell DH: The identification of markers of
macrophage differentiation in PMA-stimulated THP-1 cells and
monocyte-derived macrophages. PLoS One. 5:e86682010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Catusse J, Wollner S, Leick M, Schröttner
P, Schraufstätter I and Burger M: Attenuation of CXCR4 responses by
CCL18 in acute lymphocytic leukemia B cells. J Cell Physiol.
225:792–800. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zissel Gl, Höhne K, Kilic A, Maier C,
Goldmann T, Prasse A, Ploenes T, Trepel M, Eibel H and
Müller-Quernheim J: Identification of the CCL18 receptor - effects
of CCL18 on human lung fibroblasts in pulmonary fibrosis are
mediated via CCR6. Pneumologie. 66:P3_0122012. View Article : Google Scholar
|
|
27
|
Balkwill F, Charles KA and Mantovani A:
Smoldering and polarized inflammation in the initiation and
promotion of malignant disease. Cancer Cell. 7:211–217. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guerra C, Collado M, Navas C, Schuhmacher
AJ, Hernández-Porras I, Cañamero M, Rodriguez-Justo M, Serrano M
and Barbacid M: Pancreatitis-induced inflammation contributes to
pancreatic cancer by inhibiting oncogene-induced senescence. Cancer
Cell. 19:728–739. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Coussens LM and Werb Z: Inflammation and
cancer. Nature. 420:860–867. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Balkwill F and Mantovani A: Inflammation
and cancer: back to Virchow? Lancet. 357:539–545. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Darash-Yahana M, Gillespie JW, Hewitt SM,
et al: The chemokine CXCL16 and its receptor, CXCR6, as markers and
promoters of inflammation-associated cancers. PLoS One.
4:e66952009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schutyser E, Struyf S, Proost P, et al:
Identification of biologically active chemokine isoforms from
ascitic fluid and elevated levels of CCL18/pulmonary and
activation-regulated chemokine in ovarian carcinoma. J Biol Chem.
277:24584–24593. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ploenes T, Scholtes B, Krohn A, Burger M,
Passlick B, Müller-Quernheim J and Zissel G: CC-chemokine ligand 18
induces epithelial to mesenchymal transition in lung cancer A549
cells and elevates the invasive potential. PLoS One. 8:e530682013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Günther C, Zimmermann N, Berndt N, Grosser
M, Stein A, Koch A and Meurer M: Up-regulation of the chemokine
CCL18 by macrophages is a potential immunomodulatory pathway in
cutaneous T-cell lymphoma. Am J Pathol. 179:1434–1442. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Plönes T, Krohn A, Burger M, Veelken H,
Passlick B, Müller-Quernheim J and Zissel G: Serum level of
CC-chemokine ligand 18 is increased in patients with non-small-cell
lung cancer and correlates with survival time in adenocarcinomas.
PLoS One. 7:e417462012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Urquidi V, Kim J, Chang M, Dai Y, Rosser
CJ and Goodison S: CCL18 in a multiplex urine-based assay for the
detection of bladder cancer. PLoS One. 7:e377972012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Leung SY, Yuen ST, Chu KM, Mathy JA, Li R,
Chan AS, Law S, Wong J, Chen X and So S: Expression profiling
identifies chemokine (C-C motif) ligand 18 as an independent
prognostic indicator in gastric cancer. Gastroenterology.
127:457–469. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fanelli MF, Chinen LT, Begnami MD, Costa
WL Jr, Fregnami JH, Soares FA and Montagnini AL: The influence of
transforming growth factor-α, cyclooxygenase-2, matrix
metalloproteinase (MMP)-7, MMP-9 and CXCR4 proteins involved in
epithelial-mesenchymal transition on overall survival of patients
with gastric cancer. Histopathology. 61:153–161. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hao M, Zheng J, Hou K and Wang J, Chen X,
Lu X, Bo J, Xu C, Shen K and Wang J: Role of chemokine receptor
CXCR7 in bladder cancer progression. Biochem Pharmacol. 84:204–214.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bertran E, Caja L, Navarro E, Sancho P,
Mainez J, Murillo MM, Vinyals A, Fabra A and Fabregat I: Role of
CXCR4/SDF-1 alpha in the migratory phenotype of hepatoma cells that
have undergone epithelial-mesenchymal transition in response to the
transforming growth factor-beta. Cell Signal. 21:1595–606. 2009.
View Article : Google Scholar : PubMed/NCBI
|