Introduction
Multiple myeloma is defined as a plasma cell
malignancy characterized by the hyperplasia of plasma cells in the
bone marrow; this malignancy is usually associated with high levels
of monoclonal immunoglobulin in the blood, and leads to
pathological fracture, ostealgia, infection, hypercalcemia and
anemia (1). For over half a
century, multiple myeloma therapy has shown only limited success,
with approximately one third of patients not responding to
chemotherapy and the remainder eventually relapsing if they do not
succumb to other diseases (2).
Oridonin is a diterpenoid compound isolated from the
Chinese medicinal herb, Rabdosia rubescens and exhibits
marked antitumor activity. Accumulating evidence has suggested that
oridonin is able to inhibit the progression of tumors, thereby
alleviating the tumor burden and cancer syndrome (3–6).
Oridonin has been reported to induce apoptosis and autophagy
through the Fas/FasL-mediated signaling cascade (7). Additionally, oridonin has been shown
to be involved in the suppression of cell cycle progression and/or
the induction of cancer cell death in various in vitro
trials (8–10). Although oridonin is closely
associated with the induction of apoptosis, the definitive
systematic molecular mechanism underlying the action of oridonin in
multiple myeloma therapy remains to be elucidated and awaits
further investigation.
Currently, a proteomic approach is used in the
molecular analysis of various types of human cancer (11). However, there has been no
systematic identification of the global proteome of
oridonin-induced apoptosis in multiple myeloma LP-1 cell lines
until now. To further understand the molecular mechanism of
oridonin in multiple myeloma therapy, the present study performed
proteomic analysis using a two-dimensional gel electrophoresis
(2-DE)-based system and mass spectrometry (MS).
Materials and methods
Cell culture
The LP-1 multiple myeloma cell line was obtained
from the China Center for Type Culture Collection (Wuhan, China)
and was stored in Shanghai Institute of Hematology, Ruijin Hospital
(Shanghai, China). The 8226 human multiple myeloma cell line was
purchased from American Type Culture Collection (Manassas, VA,
USA). The cells were cultured in RPMI-1640 (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA USA) and supplemented with 10%
heat-inactivated fetal bovine serum (FBS; Hyclone; GE Healthcare
Life Sciences, Logan, UT, USA) at 37°C in an atmosphere containing
5% CO2. The cells were maintained at an optimal cell
density between 5×105 and 1×106/ml. Oridonin was purchased from
Xi'an Traditional Chinese Drug Company (Xi'an, China), the purity
of which was determined to be 97% using high-performance liquid
chromatography. A stock solution of oridonin was prepared in
dimethyl sulfoxide (DMSO; Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) and the aliquots were stored at −20°C.
Cell viability assay
Cytotoxicity was assessed using a
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT)
assay. The control and oridonin-treated LP-1 cells (0, 5, 10, 15,
20, 25 and 50 µM) in the logarithmic growth phase were cultured in
a sterile 96-well plate at an optimal cell density of 0.5–1×105/ml
per well, and were incubated at 37°C in a 5% CO2
incubator for 12, 24, 48 and 72 h. Subsequently, 20 µl of MTT
working solution was added to each of the cultured wells, and the
cells were incubated for 4 h at 37°C. The culture medium
supernatant was removed from the wells after each plate was
centrifuged (800 × g for 15 min) and replaced with 200 µl
DMSO at room temperature. Following solubilization, the absorbance
of each well was measured using a computer-controlled microplate
analyzer at 490 nm. Each treatment was performed in triplicate and
each experiment was repeated three times.
Detection of apoptosis using Annexin V
FITC/propidium iodide (PI) binding analysis
Flow cytometric analysis using Annexin V FITC
(Sigma-Aldrich; Merck KGaA) and PI (Sigma-Aldrich; Merck KGaA) was
performed to measure the ratio of apoptotic LP-1 cells. Oridonin
was added at a final concentration of 25 µM for the experimental
group, and the same volume of DMSO was added for the control group.
Overall, 106 cultured cells were incubated for 24 h prior to being
harvested; the cells were washed twice with cold PBS and
resuspended in 500 µl Annexin V binding buffer. The cell suspension
was then transferred to a centrifuge tube and incubated with 5 µl
of Annexin V-FITC at room temperature in the dark for 15 min. The
analysis was performed using FACSort flow cytometer (BD
Biosciences, Franklin Lakes, NJ, USA) and evaluated using the
CellQuest software system version 7.5.3 (BD Biosciences).
Observation using transmission
electron microscopy
The apoptotic morphology was monitored using
transmission electron microscopy. In the experimental group, the
culture medium was replaced with 25 µM of oridonin solution. After
24 h, LP-1 cells were collected and fixed with 2.5% glutaraldehyde
for 2 h at 4°C. Subsequently; the cells were rinsed three times
with PBS and fixed with 1% osmic acid for 2 h at 4°C. The cells
were subsequently dehydrated with 30, 50 and 70% ethanol for 10 min
in sequence. Finally, the cells were dipped in Epoxy resin
(Epon812) for at least 2 h, freeze-dried for 3 h and then loaded
onto the transmission electron microscope (JEM-100sx; JEOL, Ltd.,
Tokyo, Japan) for observation.
Proteomic sample preparation and
2-DE
The frozen cell samples were dissolved in lysis
buffer (100 µl per 107 cells), containing 40 mM Tris base, 8 M
urea, 2 M thiourea, 4% CHAPS, 1% dithiothreitol (DTT), 1 mM EDTA
and a 1X protease inhibitor cocktail (Roche Diagnostics GmbH,
Mannheim, Germany). The oridonin treated group and control group
cell precipitates were resuspended, oscillated by vortexing for 2
min and then freeze-thawed three times in liquid nitrogen;
solubilization was achieved using ultrasound in ice water.
Following centrifugation at 25,000 × g or 30 min at 4°C, the
supernatant was used as the 2-DE sample, and the protein
concentration was determined using the Bradford method (12) and a Bradford assay kit (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The protein samples were
stored in aliquots and were frozen at −80°C until further use.
The 2-DE was performed as described by Görg et
al (13). The first-dimension
separation was performed using commercial immobilized pH gradient
dry strips (18 cm; pH 3–10; nonlinear; GE Healthcare Life Sciences,
Chalfont, UK). The samples were diluted in rehydration solution
containing 8 M urea, 2% CHAPS, 0.5% immobilized pH gradient (IPG)
buffer (GE Healthcare Life Sciences) and 20 mM DTT. The strips were
rehydrated at 20°C. The proteins were then focused using the
IPGphor system (GE Healthcare Life Sciences) according to the
manufacturer's protocol (14). The
IPG strips were rehydrated at 30 V for 12 h and focused at 500 V
for 30 min, 2,000 V for 30 min and 5,000 V for 30 min, following
which the voltage was gradually increased to 8,000 V and maintained
at 8,000 V for 160,000 Vh. The strips were then equilibrated twice
for 15 min in equilibration buffer, containing 6 M urea, 30%
glycerol and 2% SDS in 50 mM Tris-HCl buffer (pH 8.8), which was
supplemented with 65 mM DTT for the first treatment and 259 mM
iodoacetamide for the second treatment. 2-DE was performed using
the Protean II Cell system (Bio-Rad Laboratories, Inc.) with a 13%
SDS-polyacrylamide gel at a constant current of 20 mA/gel for the
initial 40 min, followed by a current of 30 mA/gel until the
bromophenol blue dye marker reached the bottom of the gel.
Protein spot visualization and
analysis of the 2-DE images
Following electrophoresis, the protein spots were
visualized using silver nitrate staining, according to the protocol
described by Pasquali et al (15), and Coomassie Brilliant Blue R-250
(0.05% Brilliant Blue) for the analytical and micropreparative
gels, respectively. The gels were scanned with an ImageScanner (GE
Healthcare Life Sciences) and the 2-DE images were analyzed using
ImageMaster™ 4.01 software (GE Healthcare Life Sciences). Only the
spots found to be significantly (Student's t-test; P<0.05)
upregulated or downregulated (>2-fold) were selected for in-gel
digestion and MS) analysis as the lower variations were not
reproducible.
In-gel digestion and Matrix-assisted
laser desorption/ionization time of flight mass spectrometry
(MALDI-TOF-MS/MS) analysis
For 2-DE, 1.2 mg of the protein sample was dyed with
Coomassie brilliant blue. The corresponding differential protein
spots were identified, cut, decolorized and in-gel digested, and
the peptides were extracted according to the Thermo Finnigan
operation process (16). A 1 µl
volume of sample solution and an equal volume of the saturated
matrix solution were mixed and applied to the target plate. All
mass spectra of MALDI-TOF-MS were obtained on a Bruker Reflex
MALDI-TOF-MS (Bruker-Franzen, Bremen, Germany) in positive ion mode
at an accelerating voltage of 20 kV. Monoisotopic peptide masses
were used to for database searching, allowing for a peptide mass
accuracy of 0.3 Da and one partial cleavage. The oxidation of
methionine and carbamidomethyl modification of cysteine was
considered. The protein identification was performed automatically
by searching the NCBInr database using the MASCOT search engine
(www.matrixscience.co.uk).
Western blot analysis
The oridonin-treated LP-1 and 8226 cells were
collected and the proteins were extracted using RIPA lysis buffer
containing a protease inhibitor cocktail (Roche Diagnostics GmbH).
The protein concentrations were determined using a Bio-Rad protein
assay (Bio-Rad Laboratories, Inc.). Protein samples (50 µg) were
separated by 12% SDS-PAGE and then transferred to polyvinylidene
fluoride membranes (Merck KGaA, Darmstadt, Germany) at a constant
voltage of 25 V for 50 min. Following this, membranes were blocked
with 5% fat-free milk solution at room temperature for 2 h,
incubated with primary antibodies overnight at 4°C and subsequently
washed 3 times with TBST. The following primary antibodies were
used: monoclonal rabbit anti-Stathmin (catalog no. ab52630; 1:500;
Abcam, Cambridge, MA, USA), monoclonal mouse anti-dihydrofolate
reductase (catalog no. sc74594; DHFR; 1:1,000; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) and monoclonal rabbit
anti-thioredoxin reductase (catalog no. sc365658; TrxR; 1:1,000;
Santa Cruz Biotechnology, Inc.). Monoclonal mouse anti-β-actin was
used as a loading control (catalog no. ab6276; 1:1,000; Abcam).
Following incubation with horseradish peroxidase-conjugated goat
anti-rabbit IgG or goat anti-mouse IgG secondary antibodies occured
(catalog no. 31460WB, 31430WB; 1:5,000; Pioneer Biotechnology,
Xi'an, China) for 2 h at room temperature. ECL detection reagents
(GE Healthcare Life Sciences) were used to detect signals. The
intensity of the stained bands was quantified using a densitometer
and normalized to the intensity of the β-actin bands, which was
used as an internal control.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated from cells using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. A total of 1 µg RNA and 1 µl
oligo(dT)18 primer was used for reverse transcription
using a Reverse Transcription kit (Fermentas; Thermo Fisher
Scientific, Inc.). The PCR primers were designed as follows:
Stathmin, sense 5′-AAGGCAAGATTGAAGACACAGGAG-3′ and antisense
5′-TGAGGACACCCAAGCAAGACC-3′; DHFR, sense 5′-GGATGCCTTTGTGGAACTGT-3′
and antisense 5′-CGTTTCATGGGACATCACTG-3′; and TrxR, sense
5′-GCCCTGCAAGACTCTCGAAATTA-3′ and antisense
5′-GCCCATAAGCATTCTCATAGACGA-3′. To normalize the quantities of RNA
in the samples, PCR analysis was also performed using primers of
GAPDH (sense 5′-GAAGGTGAAGGTCGGAGTC-3′ and antisense
5′-GAAGATGGTGATGGGATTTC-3′). The PCR analysis was performed in a
total volume of 50 µl, containing 1 µl cDNA, 1 µl forward primers,
1 µl reverse primers, 22 µl ddH2O, 25 µl 2X Taq PCR
Master mix (Takara Bio, Inc., Otsu, Japan) in an ABI PRISM7000
real-time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). All samples were run in triplicate. The RNA expression
levels were calculated using the 2ΔΔCq method (17).
Statistical analysis
The results are expressed as the mean ± standard
deviation of triplicate samples. Statistical analyses were
performed using Student's t-test or one-way analysis of
variance. P<0.05 was considered to indicate a statistically
significant difference.
Results
MTT assay
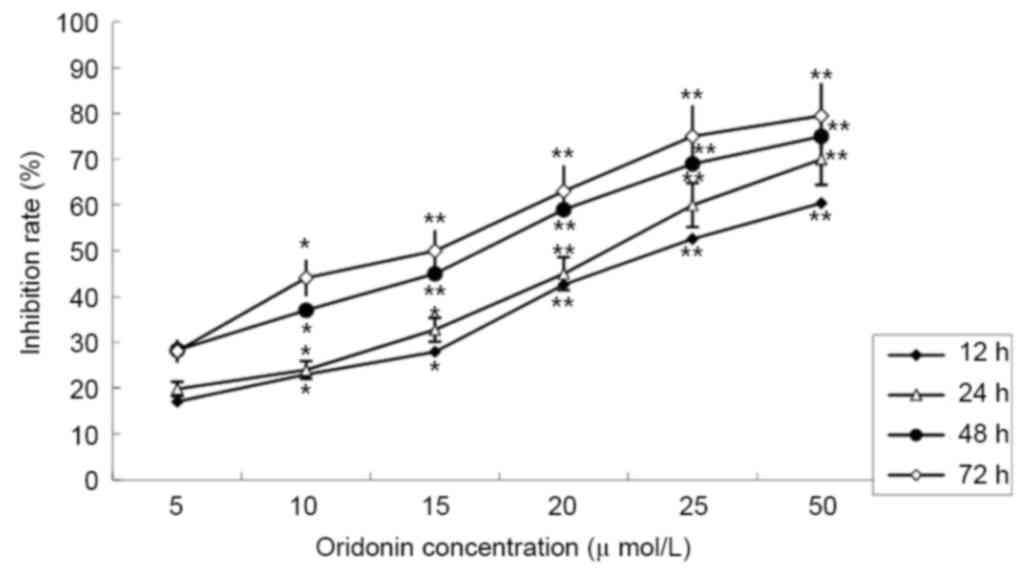
The MTT assay revealed that treatment with oridonin
caused a significant decrease in cell viability. The LP-1 cells
were treated with different concentrations of oridonin (0–50 µM),
and the inhibition rates ranged between 19.81±2.89% at 24 h and
79.52±2.37% at 72 h. The inhibition effect was dose- and
time-dependent. The half maximal inhibitory concentration
(IC50) value was determined to be 25.01 µmol/l at 24 h;
this time point was selected for subsequent assays (Fig. 1).
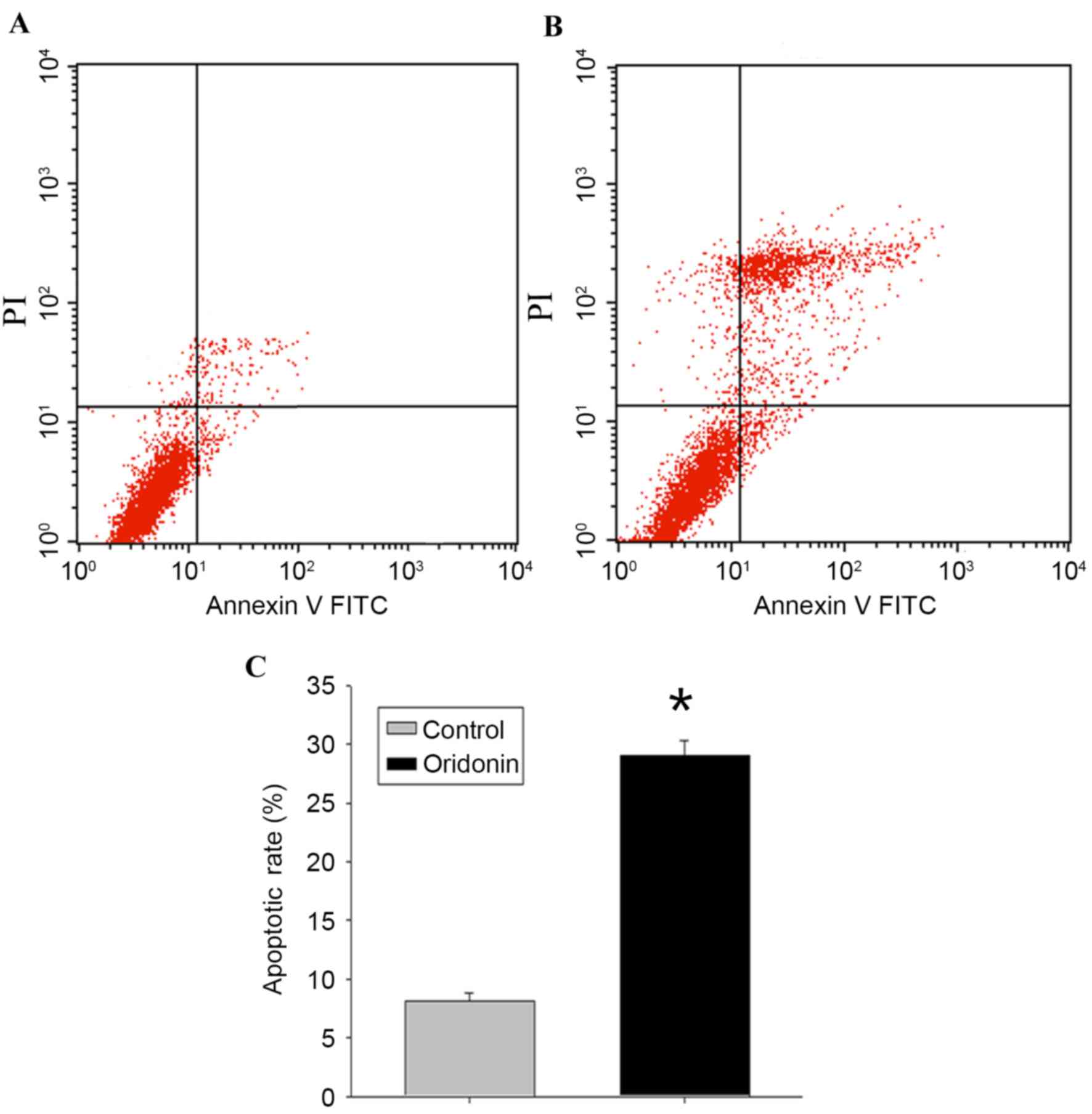
Detection of apoptosis
To determine whether the decrease in MTT activity
following exposure to oridonin was attributable to the induction of
cellular apoptosis, flow cytometric analysis and evaluation of the
ultrastructural characteristics of the cells were performed.
Following treatment of the LP-1 cells with oridonin (25 µM) for 24
h, the flow cytometry data (Fig. 2A
and B) revealed that the apoptotic rate of the cells was
29.03±1.27%, compared with the rate of 8.11±0.73% in the control
cells (P<0.05; Fig. 2C).
To confirm the occurrence of apoptosis, the cells
were observed using transmission electron microscopy. The
transmission images of the untreated control cells showed intact
nuclei and membranes (Fig. 3A-B).
By contrast, the LP-1 cells treated with oridonin for 24 h
exhibited chromatin and cytoplasmic condensation, vacuolization,
prominent nuclear fragmentation and marginalization (Fig. 3C-F), in addition to mitochondrial
swelling.
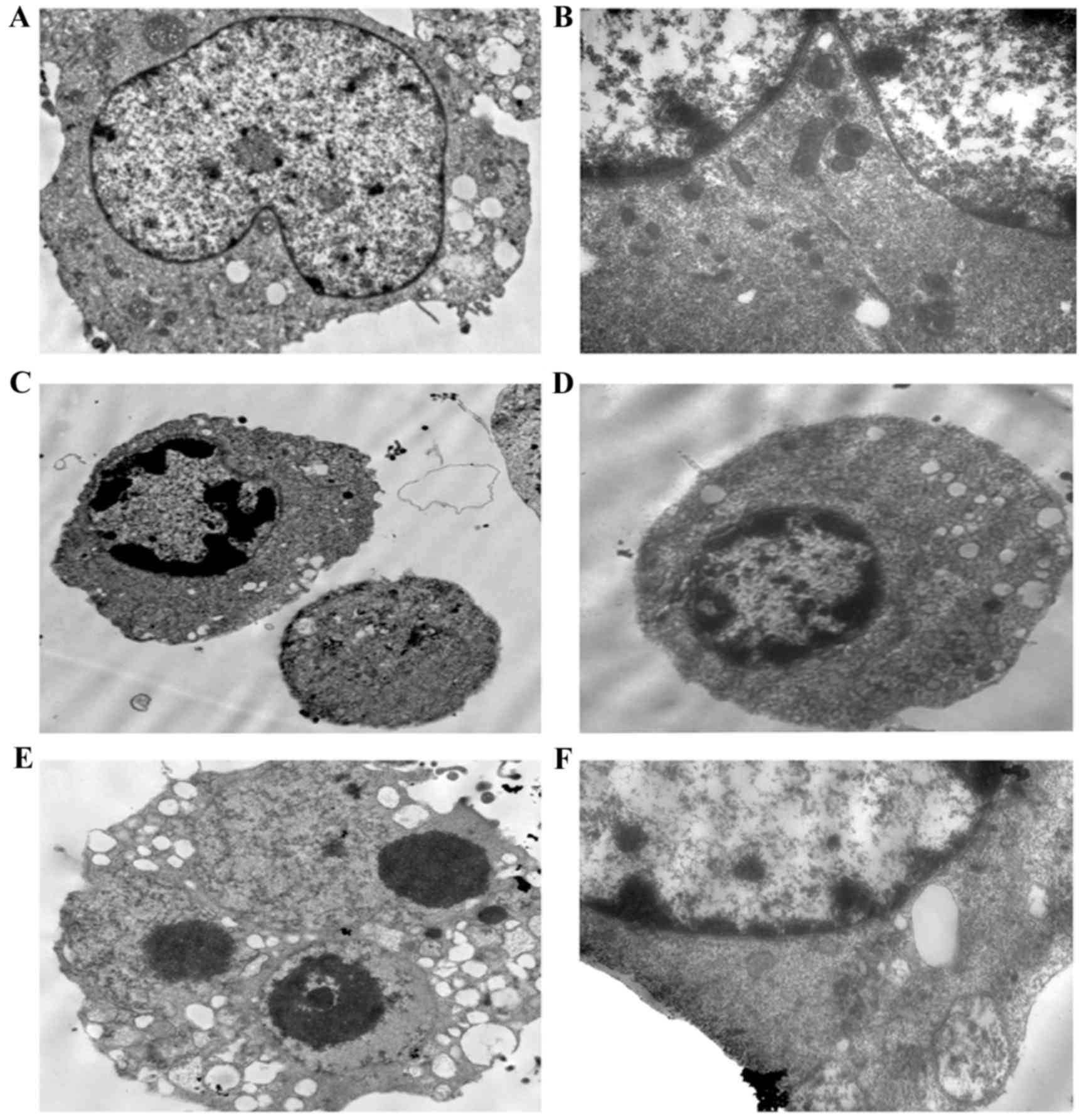 | Figure 3.Ultrastructural alterations in LP-1
cells following exposure to 25 µM oridonin for 24 h. (A) Control
LP-1 cells (magnification, ×4,000). (B) Control LP-1 cells
(magnification, ×8,000). (C and D) Oridonin treatment for 24 h
revealed chromatin condensation and cytoplasmic vacuolization
(magnification, ×4,000). (E) Oridonin-treated cells exhibited
cytoplasmic vacuolization and apoptotic bodies (magnification,
×8,000). (F) At high magnification, mitochondrial swelling was
observed (magnification, ×20,000). |
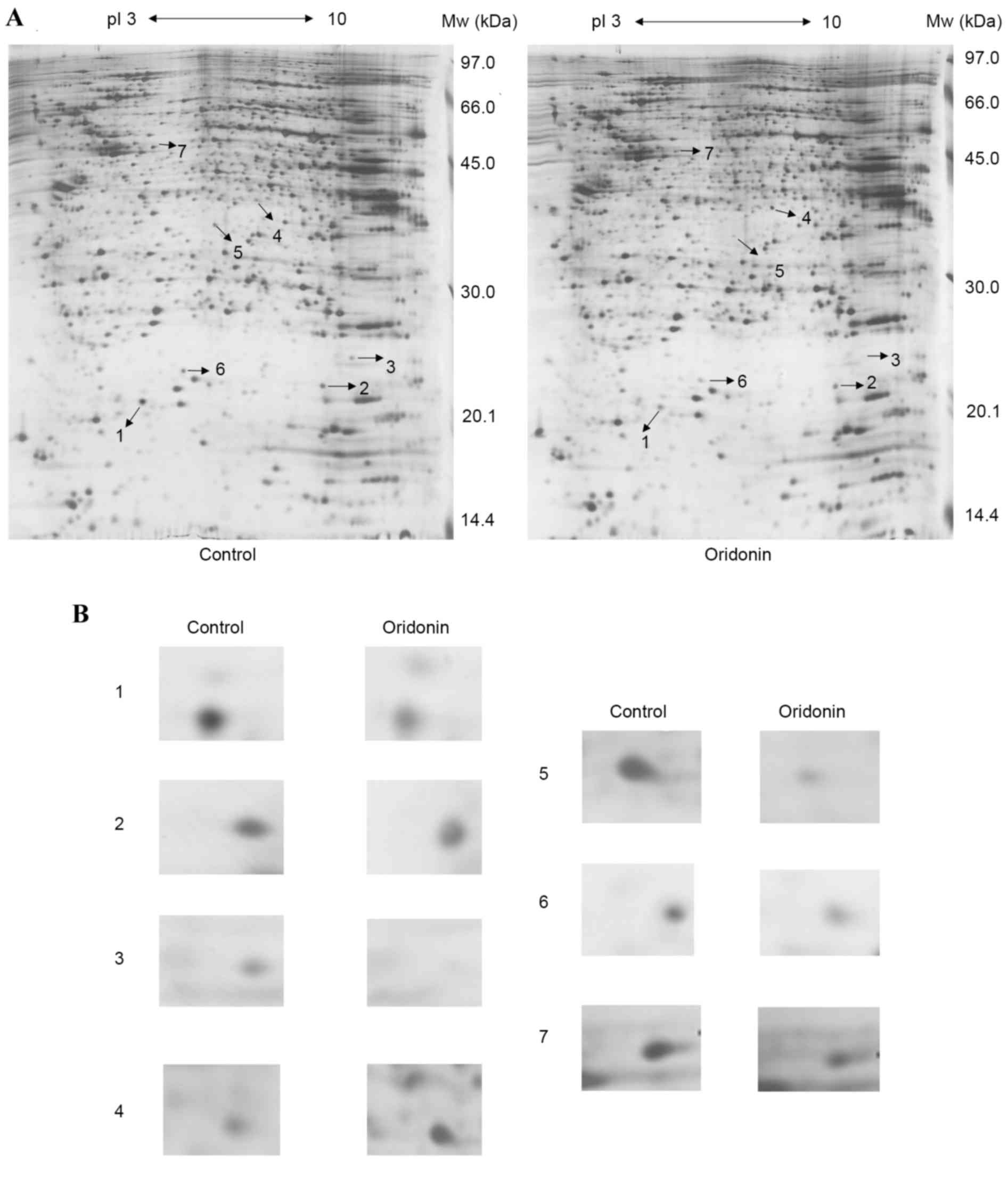
2-DE and MS analysis
The proteome expression of the cells prior to and
following oridonin treatment were monitored using 2-DE-based
proteomics. Representative gel images are shown in Fig. 4A. Proteins in the range of 14.4–97
kDa and with isoelectric points of 3–10 were well separated. In
total, >1,000 spots were detected on the silver staining gel
using ImageMaster™ 2D Platinum software and manual clear-up. By
combining the artificial comparison and the published 2-DE gels, a
total of seven significantly and consistently upregulated or
downregulated protein spots (Student's t-test; P<0.05)
with fold-changes >2 in volume intensity were selected for MS
(Fig. 4B).
Overall, six downregulated protein spots and one
upregulated protein spot were successfully identified by the
MALDI-TOF-MS spectra. The results of the MALDI-TOF MS/MS analysis
and NCBInr database search for the proteins are listed in Table I, including the experimental
molecular weight, PI, protein score of each protein spot, NCBI
accession number and the coverage of peptides. The proteins were
classified into the following major functional groups based on
their functions: Oxidative stress-associated proteins, energy
metabolism-associated enzymes, apoptosis induction-associated
proteins, and cytoskeletal proteins (Table I).
 | Table I.Matrix-assisted laser
desorption/ionization time of flight mass spectrometry
identification results of differentially expressed protein spots in
oridonin-treated LP-1 cells. |
Table I.
Matrix-assisted laser
desorption/ionization time of flight mass spectrometry
identification results of differentially expressed protein spots in
oridonin-treated LP-1 cells.
| Spot | NCBInr ID | Mr (Da) | pI | Protein | Expression | Sequence coverage
(%) | Score | Function |
|---|
| 1 | gi|197692597 | 17,320 | 5.76 | Stathmin | Decrease | 46 | 102 | Proliferation
inhibition |
| 2 | gi|170696334 | 18,438 | 6.43 | DHFR | Decrease | 46 | 93 | Oxidation
reduction, energy metabolism |
| 3 | gi|37496526 | 18,491 | 8.22 | Cofilin1 | Decrease | 51 | 101 | Cytoskeleton |
| 4 | gi|304373302 | 35,707 | 6.20 | PDHB | Increase | 24 | 117 | Oxidation
reduction |
| 5 | gi|304373227 | 33,180 | 5.85 | TrxR | Decrease | 36 | 127 | Oxidation
reduction |
| 6 | gi|62901936 | 20,210 | 6.03 | LAP p18 | Decrease | 39 | 109 | Proliferation
inhibition |
| 7 | gi|52783267 | 70,854 | 5.37 | HSP 70 | Decrease | 36 | 107 | Stress
resistance |
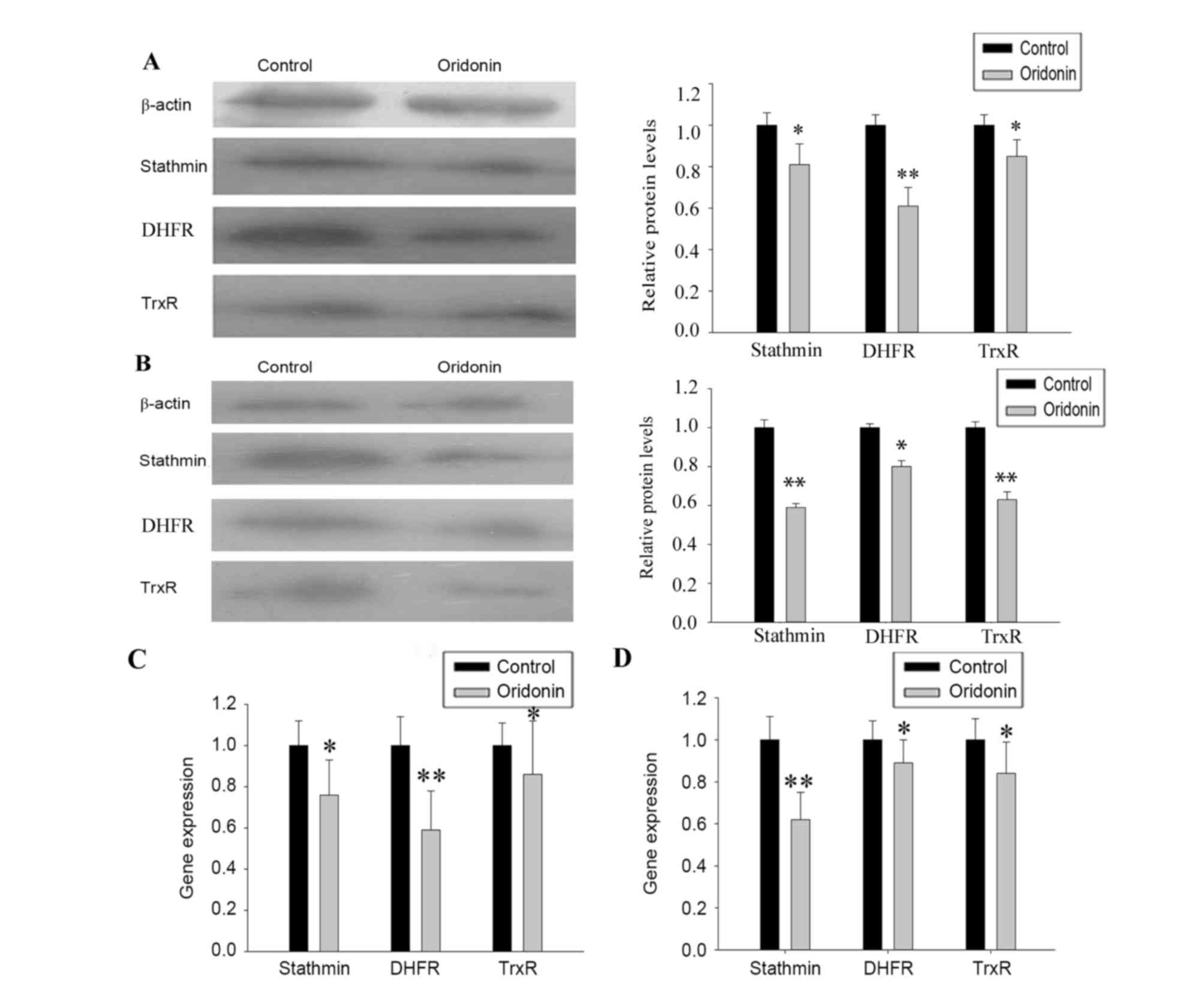
Verification of 2-DE and MS
analysis
The protein expression levels of stathmin, DHFR and
TrxR were confirmed using western blot analysis (Fig. 5A and B) and the mRNA expression
levels were assessed using RT-qPCR analysis (Fig. 5C and D). The changes in the protein
and mRNA expression levels were comparable to the changes in
protein expression levels found using 2-DE.
Discussion
Oridonin has exhibited antitumor properties in
various cancer cell lines via the regulation of cell cycle,
apoptosis and autophagy (4,18).
Investigations on the antitumor mechanism of oridonin have been
limited to date. In the present study, it was found that oridonin
inhibited the growth of LP-1 multiple myeloma cells in a time- and
dose-dependent manner; an IC50 value of 25.1 µmol/l for
24 h was determined using an MTT assay. Flow cytometry and
transmission electron microscopy analyses confirmed that oridonin
inhibited the growth of the LP-1 cells by inducing apoptosis. To
further investigate the molecular mechanisms underlying the
anticancer effects of oridonin, proteomic analysis was performed to
identify the differentially expressed proteins in the
oridonin-treated LP-1 cells. In total, seven proteins were
successfully identified. Among these were proteins associated with
cell proliferation and apoptosis, and proteins involved in
tumorigenesis and progression, including stathmin and DHFR. A
number of the differentially expressed proteins were found to be
involved in energy metabolism, including TrxR and pyruvate
dehydrogenase E1β (PDHB). Heat shock protein 70 is considered to be
an oxidative stress-inducible protein involved in oridonin-induced
apoptosis (19).
Stathmin is a microtubule destabilizing protein,
which is involved in the assembly of microtubules and spindles by
binding to the tubulin protein (20). It is important in cell
proliferation, differentiation, regeneration and migration, and has
regulatory effects on signal transduction. In addition, stathmin
has been reported to be overexpressed in a variety of human
malignancies (21–23) and has been shown to induce tumor
cell migration and invasion by regulating microtubule
depolymerization. The post-translational modification of stathmin
affects the interaction with the p53 protein, and is involved in
the initiation and progression of malignant tumors (24). In the present study, it was
demonstrated that the downregulation of stathmin was correlated
with an inhibitory effect on the growth of the LP-1 cells following
oridonin treatment. These results suggested that oridonin may
inhibit proliferation of the LP-1 cells partially by downregulating
stathmin. Stathmin may also have pro-apoptotic functions. As
stathmin has been used, either alone or in combination with
chemotherapeutics, for tumor therapy (25,26),
stathmin may be a potential target for anticancer drugs.
DHFR is a member of the reductase enzyme family,
which is important in the carbon transfer process. DHFR catalyzes
the NADPH-dependent reduction of dihydrofolate (DHF) to
tetrahydrofolate (THF), which is necessary for several one-carbon
transfer reactions in purine and pyrimidine synthesis (27). The reduction of DHFR enzymatic
activity reduces the THF pool inside the cell, which affects the
level of folate coenzymes, and thus reduces purine and pyrimidine
synthesis (28). Due to the
crucial role of DHFR in the conversion of DHF to THF, the
inhibition of DHFR inhibits the key enzymes involved in folate
metabolism; this inhibition can result in the disruption of purine
and thymidylate biosynthesis, thereby inhibiting DNA replication
and inducing tumor cell apoptosis. In the present study, the
downregulation of DHFR may have contributed to the apoptotic and
cell cycle-arresting effects of oridonin. The inhibition of DHFR is
essential to the action of antifolate medications used to treat
cancer and certain inflammatory diseases (29,30).
Therefore, oridonin may be offer potential as an antifolate drug in
myeloma therapy.
TrxR is a homodimeric selenoprotein, which catalyzes
the NADPH-dependent reduction of thioredoxin. Thioredoxin is a
cofactor in protein disulfide reduction and DNA synthesis;
independently, thioredoxin inhibits apoptosis, stimulates cell
proliferation and increases transcription factor activity (31). TrxR is a potential molecular target
of anticancer agents as it is overexpressed in several types of
tumor cells, exhibits a prosurvival effect, and enhances tumor
proliferation and resistance to therapeutic treatments (32). It has been shown that decreasing
the expression of TrxR using a small interfering RNA construct
reverses the tumor morphology and the tumorigenic properties of
lung cancer cells (33). In
addition, targeting TrxR is a basis for cancer therapy by arsenic
trioxide and cyclophosphamide (34,35).
These findings, together with the downregulation of TrxR observed
the LP-1 cells treated with oridonin, indicate that the
downregulation of TrxR may be one mechanism by which oridonin
mediates its antitumor effects. Oridonin may induce the apoptosis
of LP-1 cells via a reduction of the prosurvival effect and cell
proliferation, contributing to antiproliferation and apoptosis of
the LP-1 cells.
The PDH complex (PDC) is a mitochondrial
multi-enzyme complex, which catalyzes the overall conversion of
pyruvate to acetyl-CoA. The E1 component of PDC is a heterotetramer
of two α and two β subunits (PDHB), which are key in the
decarboxylation of pyruvate. The majority of types of cancer rely
disproportionately on glycolysis for energy, even in the presence
of an adequate supply of oxygen, which is a condition known as the
Warburg effect (36). Reversal of
the Warburg effect has been shown to cause the selective apoptosis
of tumor cells by stimulating mitochondrial respiratory chain
activity (37). The present study
hypothesized that oridonin may act as an antitumor agent in a
similar manner by upregulating the expression of PDHB and the
activity of PDC, which subsequently leads to the upregulation of
glucose oxidation and downregulation of glycolysis in the cytosol.
Additionally, the increased carbon flux through the tricarboxylic
cycle and respiratory chain activity catalyze the production of
reactive oxygen species, which consequently induces depolarization
of the mitochondrial membrane potential, resulting in the release
of cytochrome c and the stimulation of caspase-mediated cell
death (38).
In conclusion, the present study is, to the best of
our knowledge, the first to systematically identify and
characterize the global proteome of apoptosis induced by oridonin
in LP-1 cells. The proteomic profiling technique provided an
effective approach to elucidate the antitumor mechanism of
oridonin.
The present study demonstrated that treatment of the
LP-1 cells with oridonin induced significant changes in the
expression of multiple proteins. The identification of functionally
modulated proteins involved in the oridonin-treated LP-1 cells
improves current understanding of the antitumor effect of oridonin
at the molecular level. The expression and functional regulation of
target proteins, stathmin, DHFR and PDHB, may represent novel
effective therapeutic strategies for multiple myeloma. These
observations improves understanding of the molecular mechanism
underlying the oridonin-induced apoptosis of LP-1 cells in
vitro and assists in the identification of possible targets for
cancer intervention.
Acknowledgements
This study was supported by the Natural Science
Foundation for Young Scholars of China (grant no. 81000218). The
authors would like to thank the Proteome Laboratory of the
Institute of Basic Medical Sciences, National Center of Biomedical
Analysis (Beijing, China), for the proteomic analyses.
References
|
1
|
Sirohi B and Powles R: Multiple myeloma.
Lancet. 363:875–887. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mahindra A, Laubach J, Raje N, Munshi N,
Richardson PG and Anderson K: Latest advances and current
challenges in the treatment of multiple myeloma. Nat Rev Clin
Oncol. 9:135–143. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cheng Y, Qiu F, Ye YC, Tashiro S, Onodera
S and Ikejima T: Oridonin induces G2/M arrest and apoptosis via
activating ERK-p53 apoptotic pathway and inhibiting PTK-Ras-Raf-JNK
survival pathway in murine fibrosarcoma L929 cells. Arch Biochem
Biophys. 490:70–75. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang J, Jiang H, Wang C, Yang B, Zhao L,
Hu D, Qiu G, Dong X and Xiao B: Oridonin triggers apoptosis in
colorectal carcinoma cells and suppression of microRNA-32
expression augments oridonin-mediated apoptotic effects. Biomed
Pharmacother. 72:125–134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kang N, Zhang JH, Qiu F, Tashiro S,
Onodera S and Ikejima T: Inhibition of EGFR signaling augments
oridonin-induced apoptosis in human laryngeal cancer cells via
enhancing oxidative stress coincident with activation of both the
intrinsic and extrinsic apoptotic pathways. Cancer Lett.
294:147–158. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lou H, Zhang X, Gao L, Feng F, Wang J, Wei
X, Yu Z, Zhang D and Zhang Q: In vitro and in vivo antitumor
activity of oridonin nanosuspension. Int J Pharm. 379:181–186.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu YQ, Mu ZQ, You S, Tashiro S, Onodera S
and Ikejima T: Fas/FasL Signaling allows extracelluar-signal
regulated kinase to regulate cytochrome c release in
oridonin-induced apoptotic u937 cells. Biol Pharm Bull.
29:1873–1879. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ren KK, Wang HZ, Xie LP, Chen DW, Liu X,
Sun J, Nie YC and Zhang RQ: The effects of oridonin on cell growth,
cell cycle, cell migration and differentiation in melanoma cells. J
Ethnopharmacol. 103:176–180. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang Y, Wu Y, Tashiro S, Onodera S and
Ikejima T: Involvement of PKC signal pathways in oridonin-induced
autophagy in HeLa cells: A protective mechanism against apoptosis.
Biochem Biophys Res Commun. 378:273–278. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hsieh TC, Wijeratne EK, Liang JY,
Gunatilaka AL and Wu JM: Differential control of growth, cell cycle
progression and expression of NF-kappaB in human breast cancer
cells MCF-7, MCF-10A, and MDA-MB-231 by ponicidin and oridonin,
diterpenoids from the chinese herb Rabdosia rubescens. Biochem
Biophys Res Commun. 337:224–231. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hanash SM, Madoz-Gurpide J and Misek DE:
Identification of novel targets for cancer therapy using expression
proteomics. Leukemia. 16:478–485. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ramagli L: Quantifying protein in 2-D PAGE
solubilization buffers2-D Proteome Analysis Protocols. Link AJ:
112. Humana Press; Totowa, NJ: pp. 99–103. 1999, View Article : Google Scholar
|
|
13
|
Görg A, Postel W and Günther S: The
current state of two-dimensional electrophoresis with immobilized
pH gradients. Electrophoresis. 9:531–546. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
B Tom ST: 2-D electrophoresis using
immobilized pH gradients, pricinples and methods. Amersham
Pharmacia Biotech. 1998.
|
|
15
|
Pasquali C, Fialka I and Huber LA:
Preparative two-dimensional gel electrophoresis of membrane
proteins. Electrophosis. 18:2573–2781. 1997. View Article : Google Scholar
|
|
16
|
Iversen LF, Kastrup JS, Bjørn SE, Wiberg
FC, Larsen IK, Flodgaard HJ and Rasmussen PB: Structure and
function of the N-linked glycans of HBP/CAP37/azurocidin: Crystal
structure determination and biological characterization of
nonglycosylated HBP. Protein Sci. 8:2019–2026. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhu Y, Xie L, Chen G, Chen G, Wang H and
Zhang R: Effects of oridonin on proliferation of HT29 human colon
carcinoma cell lines both in vitro and in vivo in mice. Pharmazie.
62:439–444. 2007.PubMed/NCBI
|
|
19
|
Dal Piaz F, Cotugno R, Lepore L, Vassallo
A, Malafronte N, Lauro G, Bifulco G, Belisario MA and De Tommasi N:
Chemical proteomics reveals HSP70 1A as a target for the anticancer
diterpene oridonin in Jurkat cells. J Proteomics. 82:14–26. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Charbaut E, Curmi PA, Ozon S, Lachkar S,
Redeker V and Sobel A: Stathmin family proteins display specific
molecular and tubulin binding properties. J Biol Chem.
276:16146–16154. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Alli E, Yang JM and Hait WN: Silencing of
stathmin induces tumor-suppressor function in breast cancer cell
lines harboring mutant p53. Oncogene. 26:1003–1012. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mistry SJ, Bank A and Atweh GF: Targeting
stathmin in prostate cancer. Mol Cancer Ther. 4:1821–1829. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mistry SJ and Atweh GF: Therapeutic
interactions between stathmin inhibition and chemotherapeutic
agents in prostate cancer. Mol Cancer Ther. 5:3248–3257. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yuan RH, Jeng YM, Chen HL, Lai PL, Pan HW,
Hsieh FJ, Lin CY, Lee PH and Hsu HC: Stathmin overexpression
cooperates with p53 mutation and osteopontin overexpression, and is
associated with tumour progression, early recurrence, and poor
prognosis in hepatocellular carcinoma. J Pathol. 209:549–558. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang HZ, Wang Y, Gao P, Lin F, Liu L, Yu
B, Ren JH, Zhao H and Wang R: Silencing stathmin gene expression by
survivin promoter-driven siRNA vector to reverse malignant
phenotype of tumor cells. Cancer Biol Ther. 5:1457–1461. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Iancu C, Mistry SJ, Arkin S and Atweh GF:
Taxol and anti-stathmin therapy: A synergistic combination that
targets the mitotic spindle. Cancer Res. 60:3537–3541.
2000.PubMed/NCBI
|
|
27
|
Jensen DE, Black AR, Swick AG and Azizkhan
JC: Distinct roles for Sp1 and E2F sites in the growth/cell cycle
regulation of the DHFR promoter. J Cell Biochem. 67:24–31. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen MJ, Shimada T, Moulton AD, Cline A,
Humphries RK, Maizel J and Nienhuis AW: The functional human
dihydrofolate reductase gene. J Biol Chem. 259:3933–3943.
1984.PubMed/NCBI
|
|
29
|
Assaraf YG: Molecular basis of antifolate
resistance. Cancer Metastasis Rev. 26:153–181. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Morales C, García MJ, Ribas M, Miró R,
Muñoz M, Caldas C and Peinado MA: Dihydrofolate reductase
amplification and sensitization to methotrexate of
methotrexate-resistant colon cancer cells. Mol Cancer Ther.
8:424–432. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Biaglow JE and Miller RA: The thioredoxin
reductase/thioredoxin system: Novel redox targets for cancer
therapy. Cancer Biol Ther. 4:6–13. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nguyen P, Awwad RT, Smart DD, Spitz DR and
Gius D: Thioredoxin reductase as a novel molecular target for
cancer therapy. Cancer Lett. 236:164–174. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yoo MH, Xu XM, Carlson BA, Gladyshev VN
and Hatfield DL: Thioredoxin reductase 1 deficiency reverses tumor
phenotype and tumorigenicity of lung carcinoma cells. J Biol Chem.
281:13005–13008. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lu J, Chew EH and Holmgren A: Targeting
thioredoxin reductase is a basis for cancer therapy by arsenic
trioxide. Proc Natl Acad Sci USA. 104:12288–12293. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang X, Zhang J and Xu T: Cyclophosphamide
as a potent inhibitor of tumor thioredoxin reductase in vivo.
Toxicol Appl Pharmacol. 218:88–95. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Glushakova LG, Lisankie MJ, Eruslanov EB,
Ojano-Dirain C, Zolotukhin I, Liu C, Srivastava A and Stacpoole PW:
AAV3-mediated transfer and expression of the pyruvate dehydrogenase
E1 alpha subunit gene causes metabolic remodeling and apoptosis of
human liver cancer cells. Mol Genet Metab. 98:289–299. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu J, Tan M and Cai Q: The Warburg effect
in tumor progression: Mitochondrial oxidative metabolism as an
anti-metastasis mechanism. Cancer Lett. 356:156–164. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Michelakis ED, Webster L and Mackey JR:
Dichloroacetate (DCA) as a potential metabolic-targeting therapy
for cancer. Br J Cancer. 99:989–994. 2008. View Article : Google Scholar : PubMed/NCBI
|