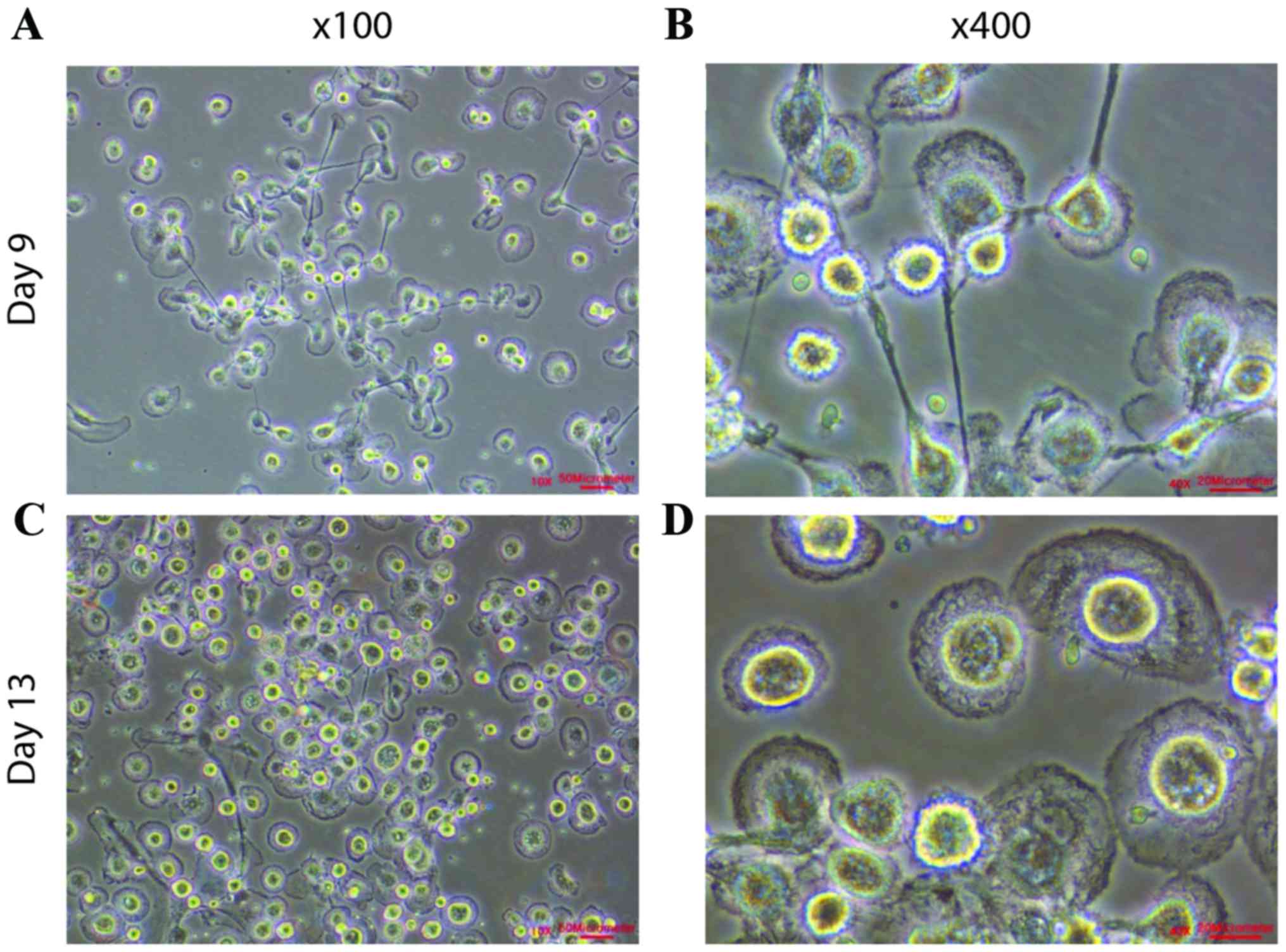
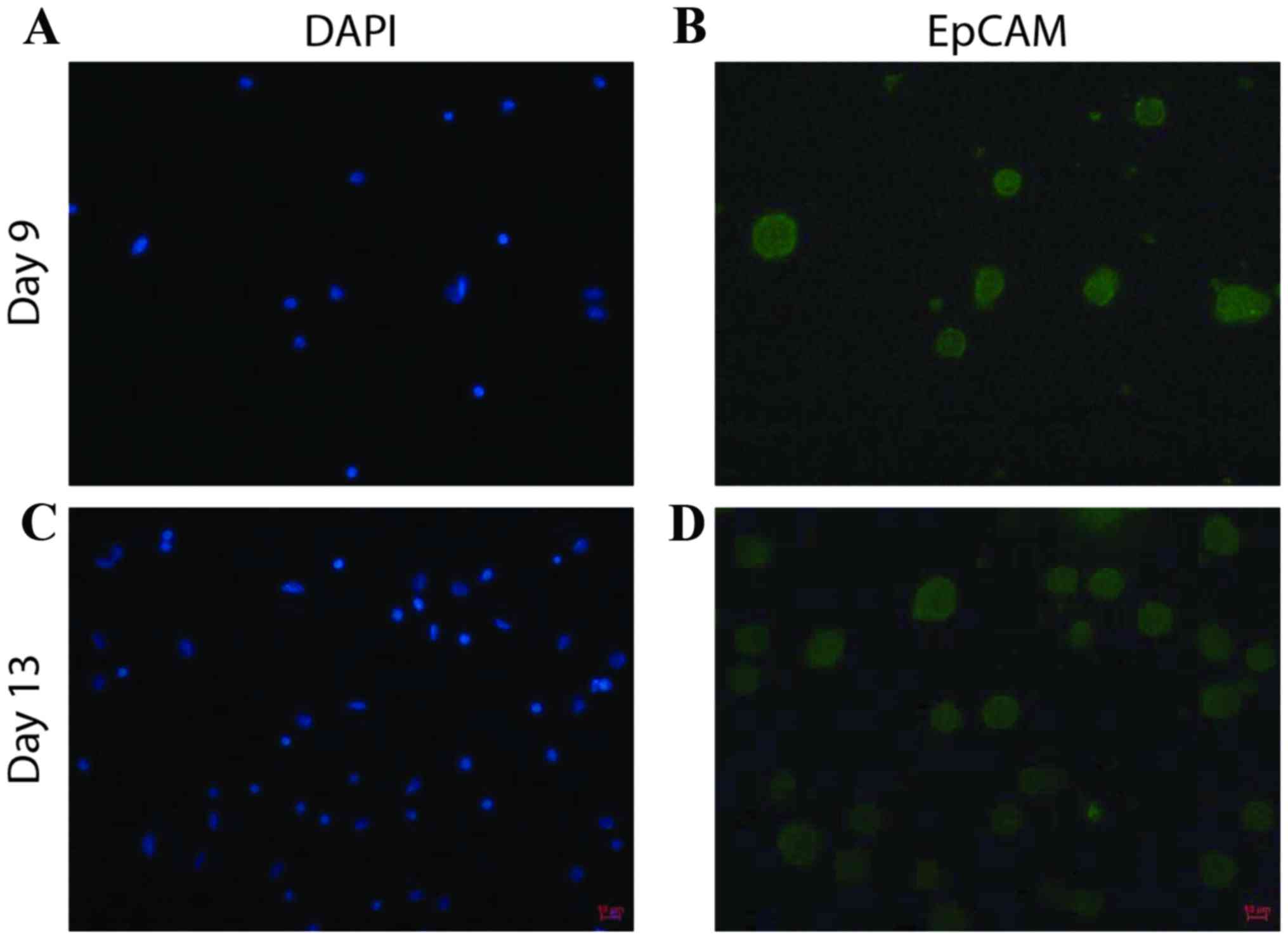
|
1
|
Ashworth TR: A case of cancer in which
cells similar to those in the tumors were seen the blood after
death. Aust Med J. 14:146–149. 1869.
|
|
2
|
Aceto N, Bardia A, Miyamoto DT, Donaldson
MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H, et al:
Circulating tumor cell clusters are oligoclonal precursors of
breast cancer metastasis. Cell. 158:1110–1122. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cristofanilli M, Hayes DF, Budd GT, Ellis
MJ, Stopeck A, Reuben JM, Doyle GV, Matera J, Allard WJ, Miller MC,
et al: Circulating tumor cells: A novel prognostic factor for newly
diagnosed metastatic breast cancer. J Clin Oncol. 23:1420–1430.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Markiewicz A, Książkiewicz M,
Wełnicka-Jaśkiewicz M, Seroczyńska B, Skokowski J, Szade J and
Żaczek AJ: Mesenchymal phenotype of CTC-enriched blood fraction and
lymph node metastasis formation potential. PLoS One. 9:e939012014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marchetti A, Del Grammastro M, Felicioni
L, Malatesta S, Filice G, Centi I, de Pas T, Santoro A, Chella A,
Brandes AA, et al: Assessment of EGFR mutations in circulating
tumor cell preparations from NSCLC patients by next generation
sequencing: Toward a real-time liquid biopsy for treatment. PLoS
One. 9:e1038832014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Marrinucci D, Bethel K, Luttgen M, Bruce
RH, Nieva J and Kuhn P: Circulating tumor cells from
well-differentiated lung adenocarcinoma retain cytomorphologic
features of primary tumor type. Arch Pathol Lab Med. 133:1468–1471.
2009.PubMed/NCBI
|
|
7
|
van de Stolpe A, Pantel K, Sleijfer S,
Terstappen LW and den Toonder JM: Circulating tumor cell isolation
and diagnostics: Toward routine clinical use. Cancer Res.
71:5955–5960. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Giuliano M, Giordano A, Jackson S, de
Giorgi U, Mego M, Cohen EN, Gao H, Anfossi S, Handy BC, Ueno NT, et
al: Circulating tumor cells as early predictors of metastatic
spread in breast cancer patients with limited metastatic
dissemination. Brest Cancer Res. 16:4402014. View Article : Google Scholar
|
|
9
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lorusso G and Rüegg C: New insights into
the mechanisms of organ-specific breast cancer metastasis. Semin
Cancer Biol. 22:226–233. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cardoso F, Harbeck N, Fallowfield L,
Kyriakides S and Senkus E; ESMO Guidelines Working Group, : Locally
recurrent or metastatic breast cancer: ESMO clinical practice
guidelines for diagnosis, treatment and follow-up. Ann Oncol. 23
Suppl 7:vii11–vii19. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Suzuki M and Tarin D: Gene expression
profiling of human lymph node metastases and matched primary breast
carcinomas: Clinical implications. Mol Oncol. 1:172–180. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Giuliano M, Giordano A, Jackson S, de
Giorgi U, Mego M, Cohen EN, Gao H, Anfossi S, Handy BC, Ueno NT, et
al: Circulating tumor cells as early predictors of metastatic
spread in breast cancer patients with limited metastatic
dissemination. Breast Cancer Res. 16:4402014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rack B, Schindlbeck C, Jückstock J,
Andergassen U, Hepp P, Zwingers T, Friedl TW, Lorenz R, Tesch H, et
al: Circulating tumor cells predict survival in early
average-to-high risk breast cancer patients. J Natl Cancer Inst.
106:pii: dju066. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bidard FC, Fehm T, Ignatiadis M, Smerage
JB, Alix-Panabières C, Janni W, Messina C, Paoletti C, Müller V,
Hayes DF, et al: Clinical application of circulating tumor cells in
breast cancer: Overview of the current interventional trials.
Cancer Metastasis Rev. 32:179–188. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nadal R, Lorente JA, Rosell R and Serrano
MJ: Relevance of molecular characterization of circulating tumor
cells in breast cancer in the era of targeted therapies. Expert Rev
Mol Diagn. 13:295–307. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Franken B, de Groot MR, Mastboom WJ,
Vermes I, van der Palen J, Tibbe AG and Terstappen LW: Circulating
tumor cells, disease recurrence and survival in newly diagnosed
breast cancer. Breast Cancer Res. 14:R1332012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cristofanilli M, Budd GT, Ellis MJ,
Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ,
Terstappen LW and Hayes DF: Circulating tumor cells, disease
progression, and survival in metastatic breast cancer. N Engl J
Med. 351:781–791. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fernandez SV, Bingham C, Fittipaldi P,
Austin L, Palazzo J, Palmer G, Alpaugh K and Cristofanilli M: TP53
mutations detected in circulating tumor cells present in the blood
of metastatic triple negative breast cancer patients. Breast Cancer
Res. 16:4452014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu M, Stott S, Toner M, Maheswaran S and
Haber DA: Circulating tumor cells: Approaches to isolation and
characterization. J Cell Biol. 192:373–382. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th Edition of the AJCC Cancer Staging
Manual and the Future of TNM. Ann Surg Oncol. 17:14712010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim EH, Lee JK, Kim BC, Rhim SH, Kim JW,
Kim KH, Jung SM, Park PS, Park HC, Lee J and Jeon BH: Enrichment of
cancer cells from whole blood using a microfabricated porous
filter. Anal Biochem. 440:114–116. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gorges TM, Tinhofer I, Drosch M, Röse L,
Zollner TM, Krahn T and von Ahsen O: Circulating tumour cells
escape from EpCAM based detection due to epithelial-to mesenchymal
transition. BMC Cancer. 12:1782012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tung N, Battelli C, Allen B, Kaldate R,
Bhatnagar S, Bowles K, Timms K, Garber JE, Herold C, Ellisen L, et
al: Frequency of mutations in individuals with breast cancer
referred for BRCA1 and BRCA2 testing using next-generation
sequencing with a 25-gene panel. Cancer. 121:25–33. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Inês C, Fernanda M, Albino M, Rui MR and
Fernando S: Overexpression of platelet-derived growth factor
receptor alpha in breast cancer is associated with tumour
progression. Breast Cancer Res. 7:R788–R795. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fernández-Medarde A and Santos E: Ras in
cancer and developmental diseases. Genes Cancer. 2:344–358. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yong HY, Hwang JS, Son H, Park HI, Oh ES,
Kim HH, Kim DK, Choi WS, Lee BJ, Kim HR and Moon A: Identification
of H-Ras-specific motif for the activation of invasive signaling
program in human breast epithelial cells. Neoplasia. 13:98–107.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Watson DM, Elton RA, Jack WJ, Dixon JM,
Chetty U and Miller WR: The H-ras oncogene product p21 and
prognosis in human breast cancer. Breast Cancer Res Treat.
17:161–169. 1991. View Article : Google Scholar : PubMed/NCBI
|