Currently, cancer is a major threat human health
worldwide. Carcinogenesis is a multi-step process resulting mainly
from the activation of oncogenes and the deactivation of
tumor-suppressor genes. Etiologically, emerging evidences have
demonstrated that epigenetic mechanisms are equally vital to
carcinogenesis (1), including the
chemical modifications of DNA and histone proteins,
post-transcriptional regulation of microRNAs (miRNA)s and
associated signaling pathways (2).
Epigenetic modifications have been suggested to be a nearly event
in carcinogenesis, and maybe useful as potential targets for early
diagnosis, cancer treatment and prognosis evaluation (3). Based on the increasing number of
studies, the focus of investigations of carcinogenesis mechanisms
have also shifted from the genetic to epigenetic (4). Epidemiologically, epigenetic mechanisms
are stressed by foreign substances, including xenobiotics and
environmental conditions (5).
Identifying an association between environmental factors and
tumorigenesis may enable the development of personalized epigenetic
medicines. In the present review, the environmental factors
involved in epigenetic actions of carcinogenesis and the recent
advancements in epigenetic drugs for cancer treatment are
summarized.
Epigenetic modifications are defined as heritable
alterations of gene expression levels induced by environment-gene
interactions, including DNA methylation, DNA hydroxy methylation,
histone modifications, non-coding RNA and miRNA (1). The manifestations of epigenetic
alterations are various post-translational modifications (PTMs),
including acetylation, methylation, phosphorylation and
ADP-ribosylation (6). PTMs drive
local changes in chromatin structure and allow for selective access
of transcriptional machinery to the DNA. They can also induce
various types of signals, subsequently activating mechanisms that
induce specific cellular responses to the environment (7).
Epidemiologically, the majority of environmental
factors, including geographical regions, stress, nutrition and
toxicants, affect malignant diseases by inducing epigenetic
modifications (8). Additionally, the
environmental factors include race, climate, life style, diet,
nutritional factors (9), airborne
polycyclic aromatic hydrocarbons (10), toxicants (e.g., cocaine) (11), alcohol (5), fungicides or pesticides (e.g., dicofol
and vinclozolin) (12), aflatoxin
(13), bacteria (e.g.,
Helicobacter Pylori), viruses (e.g., hepatitis virus)
(14), heavy metal exposure (e.g.,
cadmium, arsenic) (15) and endocrine
disruptors (e.g., bisphenol-A) (16).
Previous studies have demonstrated that the majority
of environmental factors have the ability to interfere with DNA
methylation by altering the availability of the methyl donor or the
activity of DNA methyltransferases (DNMTs) (17). Compounds in the environment, including
the endocrine disruptors (e.g., diethylstilbestrol), tobacco and
ethanol, may induce epigenetic modification (18). Dysplasia and sudden exposure in the
critical stage (e.g., early development) to environmental factors
promotes disease occurrence in adults (19). Environmental factors may permanently
change the epigenetic genome and gene expression levels, and result
in alterations of phenotypes and susceptibility to disease
(19).
Evidence from liver cancer tissue samples revealed
that ethanol altered the methylation status of histone H3 at two
lysine residues (e.g., lys-4/9) and increased the phosphorylation
of histone H3 at two serine residues (e.g., ser-10/28) (5). Chronic ethanol uptake may result in
upregulation of certain miRNAs (miR-34a, miR-107 and miR-122),
which can also alter the methylation pattern of DNA in liver
tumors, thereby affecting gene expression levels (20). Taken together, histone modification,
DNA methylation and miRNA may produce a synergistic effect in
ethanol-associated tumors. It was reported that the hepatitis B
virus X protein may induce aberrant epigenetic modifications in
human hepatocellular carcinoma by inducing the DNA hypermethylation
of tumor suppression genes (21),
promotion-associated gene-specific DNA hypomethylation, histone
acetylation or deacetylationand alterations of miRNAs (22).
Epigenetic modifications serve an important role in
cancer development; the deregulation of this has been identified as
a feature of cancer initiation (3).
Investigating the underlying mechanisms may aid the development of
specific therapeutic targets and personalized epigenetic medicines
(23). Epigenetic drugs have emerged
as potential agents for cancer treatment (Tables I and II).
Evidence has demonstrated that histone modifications
together with DNA methylation constitute an ‘epigenetic code’,
which regulates transcriptional status and disruptscode writing or
interpretation (23). These aberrant
alterations to the code may activate the expression of oncogenes,
including c-Myc, which promotes the development of specific small
molecule modulators of histone binding proteins (24). A few of these compounds have been used
in clinical development for tumor therapy, Tables I and II summarized the current reported
epigenetic inhibitors.
DNMTs, including DNMT1, DNMT3A and DNMT3B, catalyze
a methyl group transformation from the methyl donor
S-adenosylmethionine to the C-5 of cytosine in DNA. In malignant
cells, hypermethylation at the CpG island induces suppression of
numerousvital tumor suppressor genes, including p16 (25). Thus, small molecules targeting DNMTs
may potentially reverse epigenetic silencing of cancer suppressor
genes in a number of different cancer types. The DNMT inhibitors
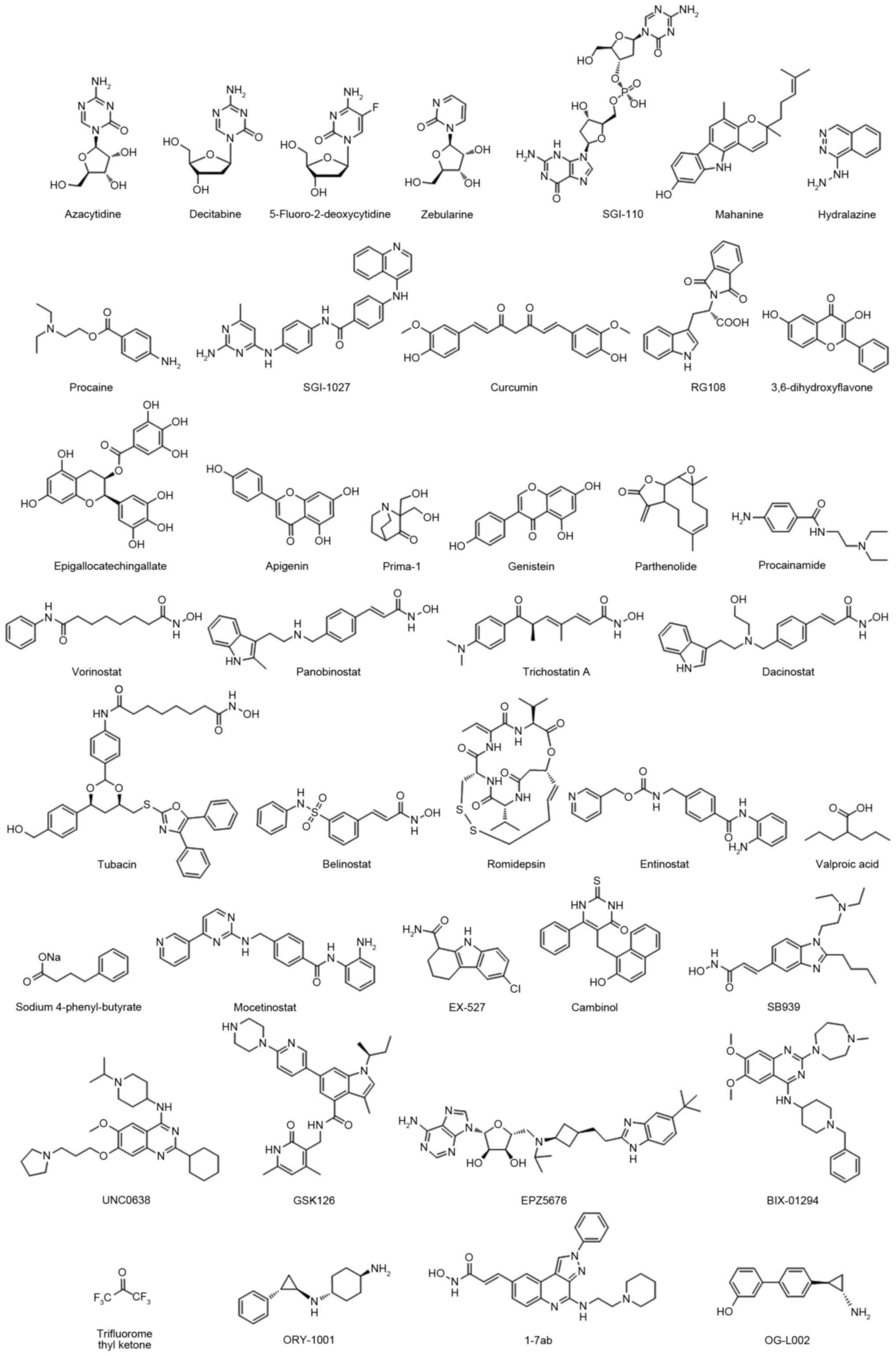
were used in tumor clinical treatments, including azacitidine,
decitabine and SGI-110 (others are presented in Table I and Fig.
1) (26–42). These compounds demonstrated good
anti-proliferative effects in various cancer cell lines, including
breast, prostate, lung, pancreas, liver and leukemia (23). However, the practical utility in
clinics has been limited by systemic toxicity and off-target
effects, including in certain heme malignancies.
The other major category is the histone deacetylase
(HDACs) inhibitor, which enables the catalysis of N-acetyl residues
hydrolysis in histones and activation of histone acetyl
transferases. A previous study revealed that HDACs serve roles as
crucial mediators in tumor survival and progression (43). A total of four HDAC inhibitors were
approved by the Food and Drug Administration (FDA): Vorinostat,
belinostat, panobinostat and romidepsin (details are presented in
Table II and Fig. 1) (44–58).
Following the development of epigenetic drugs,
second-generation epigenetic inhibitors emerged, including histone
methyltransferase inhibitors, euchromatic histone lysine
methyltransferase 2 (G9a) inhibitors, enhancer of zeste 2 polycomb
repressive complex 2 subunit inhibitors, DOT1 like histone lysine
methyltransferase inhibitors, histone demethylases and Jumonji C
inhibitors (Table III and Fig. 1) (59–64). These
epigenetic clinical agents have intrinsically greater binding
specificity to their molecular targets and may be developed as
drugs for malignant disease.
Valproic acid (VPA; valproate), an acidic chemical
compound, was mainly used in the treatment of epilepsy, bipolar
mania and migraine prophylaxis previously (65). In 1996, Cinatl et al (66), reported the inhibiting effect of VPA
on N-myconco protein expression in human neuroblastoma cells,
suggesting that VPA may have anticancer properties. In the past few
decades, great effort has been made to study its epigenetic
mechanism in various types of cancer, the majority of which focused
on transcriptionally activating chromatin structures (67). Recently a phase I/II clinical trial
headed by Iwahashi et al (68)
demonstrated that S-1 (an oral fluoropyrimidine derivative
consisting of the 5-fluorouracil prodrug tegafur combined with VPA
for patients with pancreatobiliary tract cancer, had a manageable
safety profile and preliminary antitumor activity. Sugimoto et
al (69), reported that combined
VPA with PEG-interferon (IFN)-α increased caspase-3/7 activity,
induced IFN-α and -β receptor subunit (IFNAR)1 and IFNAR2
expression and increased the expression levels of IFN-α receptor
and IFN regulatory factor 8 in pancreatic cancer, which revealed
that VPA may be useful for the treatment of pancreatic cancer via
enhancing the function of IFN-α.
Increasing interest has been drawn to the idea of
‘drug repositioning’. Although it is a costly approach to novel
drug development, the clinical value is low as the majority of the
drugs have not passed the phase I trial. Therefore, certain
existing drugs have been re-examined (70). A typical and successful example is
Viagra, which had high expectations for use in the treatment of
cardiovascular disease, but serves a role in the treatment of male
sexual dysfunction. Another example is vorinostat, which was
initially designed for cutaneous T-cell lymphoma but facilitated a
breakthrough in HIV treatment by disturbing HIV's latency in
stationary phase patients (71). Due
to the potential effects and characteristics of targeted treatment
for epigenetic-associated disease, epigenetic drugs are making
progress and attracting attention for cancer therapy (72). The FDA approved the aforementioned
epigenetic drugs, including the DNMT inhibitors azacitidine and
decitabine, which were revealed to be effective in myelody splastic
syndrome therapy (73). The HDAC
inhibitors, vorinostat, romidepsin and belinostat, also acquired
recognition in the treatment of cutaneous and peripheral T cell
lymphoma (74). Emerging evidence
demonstrated that azacitidine and decitabine also possessed
anticancer effects on liver cancer, pancreatic cancer and breast
cancer cells (75). It is reasonable
to speculate that combining azacitidine and decitabine with other
anticancer drugs, including platinum compounds and monoclonal
antibodies may produce a stronger anticancer effect (76). Furthermore, vorinostat and romidepsin
were also popular for gastric and lung cancer therapy (77,78). Novel
drug development also requires investigation using cutting-edge
technology, including gene sequencing.
Sanger sequencing, first-generation sequencing that
markedly impacted gene research has now evolved into next
generation sequencing (NGS), which has a lower cost, higher speed
and improved throughput. Recently, an epigenetic study used NGS and
achieved a novel understanding of ependymoma in children. The
previous study investigated DNA methylation patterns and defined a
tumor CpG island methylator phenotype for infant nervous system
malignancy, using whole genome sequencing and whole-exome
sequencing (79). They revealed that
the development of posterior fossa ependymomas group A (PFA), which
had a poor prognosis, occurred primarily in infants and was
associated with epigenetic modifications. The PFA exhibits an
increased number of methylated CpG sites, an increased number of
genes with CpG methylation and an increased number of genes that
are transcriptionally silenced by CpG hypermethylation in tumor
development and maintenance. The in vivo data demonstrated
that treatment with decitabine and Gsk343 is able to attenuate the
proliferation of PFA cells. This may further support the concept of
‘drug repositioning’. Widely applicable in modern cancer clinical
research (80), NGS has begun to
elucidate the underlying epigenetic mechanisms; however, there is a
large amount of data. Methodological improvement is required for
convenient clinical application.
Epigenetics provides a molecular and etiological
mechanism for the incidence of malignant cancer. Early ectogenic
exposure can program later life physiology and adult onset disease
due to the replication of the epigenome during somatic cell
mitosis, during which ‘epigenetic transgenerational inheritance’
initiates. Although an increasing number of approved antitumor
drugs have emerged, the outcomes of clinical trials have been
unsatisfactory. This may be due to the lack of specificity and the
combination with environmental exposure. In view of the critical
roles of ectogenic cues on tumorigenesis, comprehensive analysis
and treatment is required for early diagnosis, standardized and
personalized treatment. The presence of epigenetic factors is
associated with gene abnormality in premalignant cancer, and its
potential reversibility indicated that epigenetic alterations may
be promising biomarkers and potential novel mechanism-based
strategies for tumor early diagnosis and treatment.
Previous clinical trials revealed that first
generation inhibitors, including DNMTs and HDACs, have been
observed to have limited utility due to toxicity and off-target
effects. However, second generation compounds have been suggested
to have more promise. These clinical agents have greater
selectivity for their molecular target and may be a robust driver
or key mediator at safe doses in malignancies. Additionally, drug
repositioning still requires further enhancement and study. The
improvement of epigenetic therapeutic strategies needs to be
combined with cytotoxic factors, immunotherapy, targeted kinase
inhibitors, NSG and the possible environmental cues.
The present study was supported by the National
Natural Science Fund of China (grant nos. 31570509 and 81702326),
the Gansu Province Science Foundation for Distinguished Young
Scholars (grant no. 1606RJDA317) and Science and Technology Program
of Lanzhou City (grant no. 2015-3-93).
|
1
|
Skinner MK, Manikkam M and
Guerrero-Bosagna C: Epigenetic transgenerational actions of
environmental factors in disease etiology. Trends Endocrinol Metab.
21:214–222. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lopez-Ramirez MA and Nicoli S: Role of
miRNAs and epigenetics in neural stem cell fate determination.
Epigenetics. 9:90–100. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Graça I, Pereira-Silva E, Henrique R,
Packham G, Crabb SJ and Jerónimo C: Epigenetic modulators as
therapeutic targets in prostate cancer. Clin Epigenetics. 8:982016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Do C, Shearer A, Suzuki M, Terry MB,
Gelernter J, Greally JM and Tycko B: Genetic-epigenetic
interactions in cis: A major focus in the post-GWAS era. Genome
Biol. 18:1202017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shukla SD and Lim RW: Epigenetic effects
of ethanol on the liver and gastrointestinal system. Alcohol Res.
35:47–55. 2013.PubMed/NCBI
|
|
6
|
Rothbart SB and Strahl BD: Interpreting
the language of histone and DNA modifications. Biochim Biophys
Acta. 1839:627–643. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wilkins BJ, Rall NA, Ostwal Y, Kruitwagen
T, Hiragami-Hamada K, Winkler M, Barral Y, Fischle W and Neumann H:
A cascade of histone modifications induces chromatin condensation
in mitosis. Science. 343:77–80. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tracey R, Manikkam M, Guerrero-Bosagna C
and Skinner MK: Hydrocarbons (jet fuel JP-8) induce epigenetic
transgenerational inheritance of obesity, reproductive disease and
sperm epimutations. Reprod Toxicol. 36:104–116. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Heijmans BT, Tobi EW, Stein AD, Putter H,
Blauw GJ, Susser ES, Slagboom PE and Lumey LH: Persistent
epigenetic differences associated with prenatal exposure to famine
in humans. Proc Natl Acad Sci USA. 105:17046–17049. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Perera F, Tang WY, Herbstman J, Tang D,
Levin L, Miller R and Ho SM: Relation of DNA methylation of 5′-CpG
island of ACSL3 to transplacental exposure to airborne polycyclic
aromatic hydrocarbons and childhood asthma. PLoS One. 4:e44882009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Novikova SI, He F, Bai J, Cutrufello NJ,
Lidow MS and Undieh AS: Maternal cocaine administration in mice
alters DNA methylation and gene expression in hippocampal neurons
of neonatal and prepubertal offspring. PLoS One. 3:e19192008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiménez-Chillarón JC, Nijland MJ, Ascensão
AA, Sardão VA, Magalhães J, Hitchler MJ, Domann FE and Oliveira PJ:
Back to the future: Transgenerational transmission of
xenobiotic-induced epigenetic remodeling. Epigenetics. 10:259–273.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y: Detection of epigenetic
aberrations in the development of hepatocellular carcinoma. Methods
Mol Biol. 1238:709–731. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tian Y, Yang W, Song J, Wu Y and Ni B:
Hepatitis B virus X protein-induced aberrant epigenetic
modifications contributing to human hepatocellular carcinoma
pathogenesis. Mol Cell Biol. 33:2810–2816. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ryu HW, Lee DH, Won HR, Kim KH, Seong YJ
and Kwon SH: Influence of toxicologically relevant metals on human
epigenetic regulation. Toxicol Res. 31:1–9. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Khan D and Ahmed SA: Epigenetic regulation
of non-lymphoid cells by Bisphenol A, a model endocrine disrupter:
Potential implications for immunoregulation. Front Endocrinol
(Lausanne). 6:912015.PubMed/NCBI
|
|
17
|
Guerrero-Bosagna C and Skinner MK:
Environmentally induced epigenetic transgenerational inheritance of
phenotype and disease. Mol Cell Endocrinol. 354:3–8. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Godfrey KM, Costello PM and Lillycrop KA:
The developmental environment, epigenetic biomarkers and long-term
health. J Dev Orig Health Dis. 6:399–406. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jirtle RL and Skinner MK: Environmental
epigenomics and disease susceptibility. Nat Rev Genet. 8:253–262.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dippold RP, Vadigepalli R, Gonye GE, Patra
B and Hoek JB: Chronic ethanol feeding alters miRNA expression
dymanics during liver regeneration. Alcohol Clin Exp Res.
37:E59–E69. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pogribny IP and Rusyn I: Role of
epigenetic aberrations in the development and progression of human
hepatocellular carcinoma. Cancer Lett. 342:223–230. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wei X, Xiang T, Ren G, Tan C, Liu R, Xu X
and Wu Z: miR-101 is down-regulated by the hepatitis B virus X
protein and induces aberrant DNA methylation by targeting DNA
methyltransferase 3A. Cell Signal. 25:439–446. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dhanak D and Jackson P: Development and
classes of epigenetic drugs for cancer. Biochem Biophys Res Commun.
455:58–69. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Z and Patel DJ: Small molecule
epigenetic inhibitors targeted to histone lysine methyltransferases
and demethylases. Q Rev Biophys. 46:349–373. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Enjuanes A, Albero R, Clot G, Navarro A,
Beà S, Pinyol M, Martín-Subero JI, Klapper W, Staudt LM, Jaffe ES,
et al: Genomewide methylation analyses identify a subset of mantle
cell lymphoma with a high number of methylated CpGs and aggressive
clinicopathological features. Int J Cancer. 133:2852–2863.
2013.PubMed/NCBI
|
|
26
|
Müller AM and Florek M:
5-Azacytidine/5-Azacitidine. Recent Results Cancer Res.
201:299–324. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Momparler RL: Epigenetic therapy of cancer
with 5-aza-2′-deoxycytidine (decitabine). Semin Oncol. 32:443–451.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao Q, Fan J, Hong W, Li L and Wu M:
Inhibition of cancer cell proliferation by
5-fluoro-2′-deoxycytidine, a DNA methylation inhibitor, through
activation of DNA damage response pathway. Springerplus. 1:652012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Marquez VE, Kelley JA, Agbaria R,
Ben-Kasus T, Cheng JC, Yoo CB and Jones PA: Zebularine: A unique
molecule for an epigenetically based strategy in cancer
chemotherapy. Ann N Y Acad Sci. 1058:246–254. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Srivastava P, Paluch BE, Matsuzaki J,
James SR, Collamat-Lai G, Taverna P, Karpf AR and Griffiths EA:
Immunomodulatory action of the DNA methyltransferase inhibitor
SGI-110 in epithelial ovarian cancer cells and xenografts.
Epigenetics. 10:237–246. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Agarwal S, Amin KS, Jagadeesh S, Baishay
G, Rao PG, Barua NC, Bhattacharya S and Banerjee PP: Mahanine
restores RASSF1A expression by down-regulating DNMT1 and DNMT3B in
prostate cancer cells. Mol Cancer. 12:992013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dueñas-Gonzalez A, Coronel J, Cetina L,
González-Fierro A, Chavez-Blanco A and Taja-Chayeb L:
Hydralazine-valproate: A repositioned drug combination for the
epigenetic therapy of cancer. Expert Opin Drug Metab Toxicol.
10:1433–1444. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gao Z, Xu Z, Hung MS, Lin YC, Wang T, Gong
M, Zhi X, Jablons DM and You L: Procaine and procainamide inhibit
the Wnt canonical pathway by promoter demethylation of WIF-1 in
lung cancer cells. Oncol Rep. 22:1479–1484. 2009.PubMed/NCBI
|
|
34
|
Rilova E, Erdmann A, Gros C, Masson V,
Aussagues Y, Poughon-Cassabois V, Rajavelu A, Jeltsch A, Menon Y,
Novosad N, et al: Design, synthesis and biological evaluation of
4-amino-N-(4-aminophenyl) benzamide analogues of quinoline-based
SGI-1027 as inhibitors of DNA methylation. ChemMedChem. 9:590–601.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ramasamy TS, Ayob AZ, Myint HH,
Thiagarajah S and Amini F: Targeting colorectal cancer stem cells
using curcumin and curcumin analogues: Insights into the mechanism
of the therapeutic efficacy. Cancer Cell Int. 15:962015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Graça I, Sousa EJ, Baptista T, Almeida M,
Ramalho-Carvalho J, Palmeira C, Henrique R and Jerónimo C:
Anti-tumoral effect of the non-nucleoside DNMT inhibitor RG108 in
human prostate cancer cells. Curr Pharm Des. 20:1803–1811. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee E, Jeong KW, Jnawali HN, Shin A, Heo
YS and Kim Y: Cytotoxic activity of 3,6-dihydroxyflavone in human
cervical cancer cells and its therapeutic effect on c-Jun
N-terminal kinase inhibition. Molecules. 19:13200–13211. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chakrabarty S, Ganguli A, Das A, Nag D and
Chakrabarti G: Epigallocatechin-3-gallate shows anti-proliferative
activity in HeLa Cells targeting tubulin-microtubule equilibrium.
Chem Biol Interact. 242:380–389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shukla S and Gupta S: Apigenin: A
promising molecule for cancer prevention. Pharm Res. 27:962–978.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Qiang W, Jin T, Yang Q, Liu W, Liu S, Ji
M, He N, Chen C, Shi B and Hou P: PRIMA-1 selectively induces
global DNA demethylation in p53 mutant-type thyroid cancer cells. J
Biomed Nanotechnol. 10:1249–1258. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Spagnuolo C, Russo GL, Orhan IE,
Habtemariam S, Daglia M, Sureda A, Nabavi SF, Devi KP, Loizzo MR,
Tundis R and Nabavi SM: Genistein and cancer: Current status,
challenges, and future directions. Adv Nutr. 6:408–419. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wyrębska A, Gach K and Janecka A: Combined
effect of parthenolide and various anti-cancer drugs or anticancer
candidate substances on malignant cells in vitro and in vivo. Mini
Rev Med Chem. 14:222–228. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Minami J, Suzuki R, Mazitschek R, Gorgun
G, Ghosh B, Cirstea D, Hu Y, Mimura N, Ohguchi H, Cottini F, et al:
Histone deacetylase 3 as a novel therapeutic target in multiple
myeloma. Leukemia. 28:680–689. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Richon VM: Targeting histone deacetylases:
Development of vorinostat for the treatment of cancer. Epigenomics.
2:457–465. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li X, Zhang J, Xie Y, Jiang Y, Yingjie Z
and Xu W: Progress of HDAC inhibitor panobinostat in the treatment
of cancer. Curr Drug Targets. 15:622–634. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang X, Xu J, Wang H, Wu L, Yuan W, Du J
and Cai S: Trichostatin A, a histone deacetylase inhibitor,
reverses epithelial-mesenchymal transition in colorectal cancer
SW480 and prostate cancer PC3 cells. Biochem Biophys Res Commun.
456:320–326. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ganai SA: Strategy for enhancing the
therapeutic efficacy of histone deacetylase inhibitor dacinostat:
The novel paradigm to tackle monotonous cancer chemoresistance.
Arch Pharm Res. 2015.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xing LF, Wang DT, Yang Y and Pan SY:
Effect of HDAC-6 on PD cell induced by lactacystin. Asian Pac J
Trop Med. 8:855–859. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kirschbaum MH, Foon KA, Frankel P, Ruel C,
Pulone B, Tuscano JM and Newman EM: A phase 2 study of belinostat
(PXD101) in patients with relapsed or refractory acute myeloid
leukemia or patients over the age of 60 with newly diagnosed acute
myeloid leukemia: A California Cancer Consortium study. Leuk
Lymphoma. 55:2301–2304. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Bertino EM and Otterson GA: Romidepsin: A
novel histone deacetylase inhibitor for cancer. Expert Opin
Investig Drugs. 20:1151–1158. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ruiz R, Raez LE and Rolfo C: Entinostat
(SNDX-275) for the treatment of non-small cell lung cancer. Expert
Opin Investig Drugs. 24:1101–1109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Duenas-Gonzalez A, Candelaria M,
Perez-Plascencia C, Perez-Cardenas E, de la Cruz-Hernandez E and
Herrera LA: Valproic acid as epigenetic cancer drug: Preclinical,
clinical and transcriptional effects on solid tumors. Cancer Treat
Rev. 34:206–222. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tsunedomi R, Iizuka N, Harada S and Oka M:
Susceptibility of hepatoma-derived cells to histone deacetylase
inhibitors is associated with ID2 expression. Int J Oncol.
42:1159–1166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Nielsen TK, Hildmann C, Riester D, Wegener
D, Schwienhorst A and Ficner R: Complex structure of a bacterial
class 2 histone deacetylase homologue with a trifluoromethylketone
inhibitor. Acta Crystallogr Sect F Struct Biol Cryst Commun.
63:270–273. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tao YF, Lin F, Yan XY, Gao XG, Teng F, Fu
ZR and Wang ZX: Galectin-9 in combination with EX-527 prolongs the
survival of cardiac allografts in mice after cardiac
transplantation. Transplant Proc. 47:2003–2009. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Mahajan SS, Scian M, Sripathy S, Posakony
J, Lao U, Loe TK, Leko V, Thalhofer A, Schuler AD, Bedalov A and
Simon JA: Development of pyrazolone and isoxazol-5-one cambinol
analogues as sirtuin inhibitors. J Med Chem. 57:3283–3294. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yang L, Liang Q, Shen K, Ma L, An N, Deng
W, Fei Z and Liu J: A novel class I histone deacetylase inhibitor,
I-7ab, induces apoptosis and arrests cell cycle progression in
human colorectal cancer cells. Biomed Pharmacother. 71:70–78. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Eigl BJ, North S, Winquist E, Finch D,
Wood L, Sridhar SS, Powers J, Good J, Sharma M, Squire JA, et al: A
phase II study of the HDAC inhibitor SB939 in patients with
castration resistant prostate cancer: NCIC clinical trials group
study IND195. Invest New Drugs. 33:969–976. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Cui J, Sun W, Hao X, Wei M, Su X, Zhang Y,
Su L and Liu X: EHMT2 inhibitor BIX-01294 induces apoptosis through
PMAIP1-USP9X-MCL1 axis in human bladder cancer cells. Cancer Cell
Int. 15:42015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Fu L, Yan FX, An XR and Hou J: Effects of
the histone methyltransferase inhibitor UNC0638 on histone H3K9
dimethylation of cultured ovine somatic cells and development of
resulting early cloned embryos. Reprod Domest Anim. 49:e21–e25.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Tiffen JC, Gunatilake D, Gallagher SJ,
Gowrishankar K, Heinemann A, Cullinane C, Dutton-Regester K, Pupo
GM, Strbenac D, Yang JY, et al: Targeting activating mutations of
EZH2 leads to potent cell growth inhibition in human melanoma by
derepression of tumor suppressor genes. Oncotarget. 6:27023–27036.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Horiuchi KY, Eason MM, Ferry JJ, Planck
JL, Walsh CP, Smith RF, Howitz KT and Ma H: Assay development for
histone methyltransferases. Assay Drug Dev Technol. 11:227–236.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Maes T, Mascaró C, Ortega A, Lunardi S,
Ciceri F, Somervaille TC and Buesa C: KDM1 histone lysine
demethylases as targets for treatments of oncological and
neurodegenerative disease. Epigenomics. 7:609–626. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Qian S, Wang Y, Ma H and Zhang L:
Expansion and functional divergence of Jumonji C-containing histone
demethylases: Significance of duplications in ancestral angiosperms
and vertebrates. Plant Physiol. 168:1321–1337. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Verrotti A, Carelli A, di Genova L and
Striano P: Epilepsy and chromosome 18 abnormalities: A review.
Seizure. 32:78–83. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Cinatl J Jr, Cinatl J, Scholz M, Driever
PH, Henrich D, Kabickova H, Vogel JU, Doerr HW and Kornhuber B:
Antitumor activity of sodium valproate in cultures of human
neuroblastoma cells. Anticancer Drugs. 7:766–773. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Woodworth AM and Holloway AF: The role of
epigenetic regulation in transcriptional memory in the immune
system. Adv Protein Chem Struct Biol. 106:43–69. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Iwahashi S, Utsunomiya T, Imura S, Morine
Y, Ikemoto T, Arakawa Y, Saito Y, Ishikawa D and Shimada M: Effects
of valproic acid in combination with S-1 on advanced
pancreatobiliary tract cancers: Clinical study phases I/II.
Anticancer Res. 34:5187–5191. 2014.PubMed/NCBI
|
|
69
|
Sugimoto K, Shimada M, Utsunomiya T,
Morine Y, Imura S, Ikemoto T and Iwahashi S: Valproic acid enhances
the anti-tumor effect of pegylated interferon-α towards pancreatic
cancer cell lines. Anticancer Res. 34:3403–3409. 2014.PubMed/NCBI
|
|
70
|
Bastos LF and Coelho MM: Drug
repositioning: Playing dirty to kill pain. CNS Drugs. 28:45–61.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Archin NM, Liberty AL, Kashuba AD,
Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney
MF, Strain MC, et al: Administration of vorinostat disrupts HIV-1
latency in patients on antiretroviral therapy. Nature. 487:482–485.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Di Costanzo A, Del Gaudio N, Migliaccio A
and Altucci L: Epigenetic drugs against cancer: An evolving
landscape. Arch Toxicol. 88:1651–1668. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Mummaneni P and Shord SS: Epigenetics and
oncology. Pharmacotherapy. 34:495–505. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Glaser KB: HDAC inhibitors: Clinical
update and mechanism-based potential. Biochem Pharmacol.
74:659–671. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Li X, Mei Q, Nie J, Fu X and Han W:
Decitabine: A promising epi-immunotherapeutic agent in solid
tumors. Expert Rev Clin Immunol. 11:363–375. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Garrido-Laguna I, McGregor KA, Wade M,
Weis J, Gilcrease W, Burr L, Soldi R, Jakubowski L, Davidson C,
Morrell G, et al: A phase I/II study of decitabine in combination
with panitumumab in patients with wild-type (wt) KRAS metastatic
colorectal cancer. Invest New Drugs. 31:1257–1264. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zhou C, Ji J, Shi M, Yang L, Yu Y, Liu B,
Zhu Z and Zhang J: Suberoylanilide hydroxamic acid enhances the
antitumor activity of oxaliplatin by reversing the
oxaliplatin–induced Src activation in gastric cancer cells. Mol Med
Rep. 10:2729–2735. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Karthik S, Sankar R, Varunkumar K and
Ravikumar V: Romidepsin induces cell cycle arrest, apoptosis,
histone hyperacetylation and reduces matrix metalloproteinases 2
and 9 expression in bortezomib sensitized non-small cell lung
cancer cells. Biomed Pharmacother. 68:327–334. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Mack SC, Witt H, Piro RM, Gu L, Zuyderduyn
S, Stütz AM, Wang X, Gallo M, Garzia L, Zayne K, et al: Epigenomic
alterations define lethal CIMP-positive ependymomas of infancy.
Nature. 506:445–450. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Tabatabaeifar S, Kruse TA, Thomassen M,
Larsen MJ and Sørensen JA: Use of next generation sequencing in
head and neck squamous cell carcinomas: A review. Oral Oncol.
50:1035–1040. 2014. View Article : Google Scholar : PubMed/NCBI
|