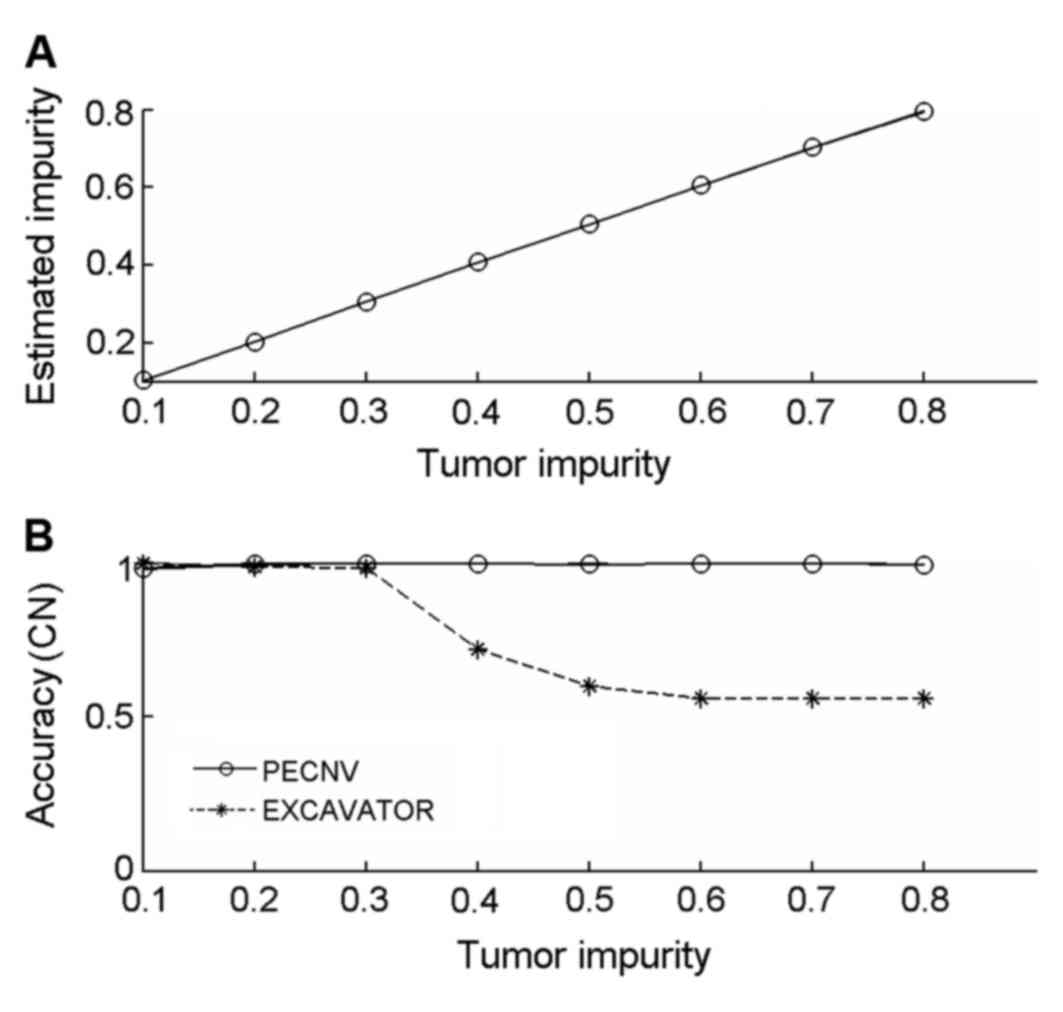
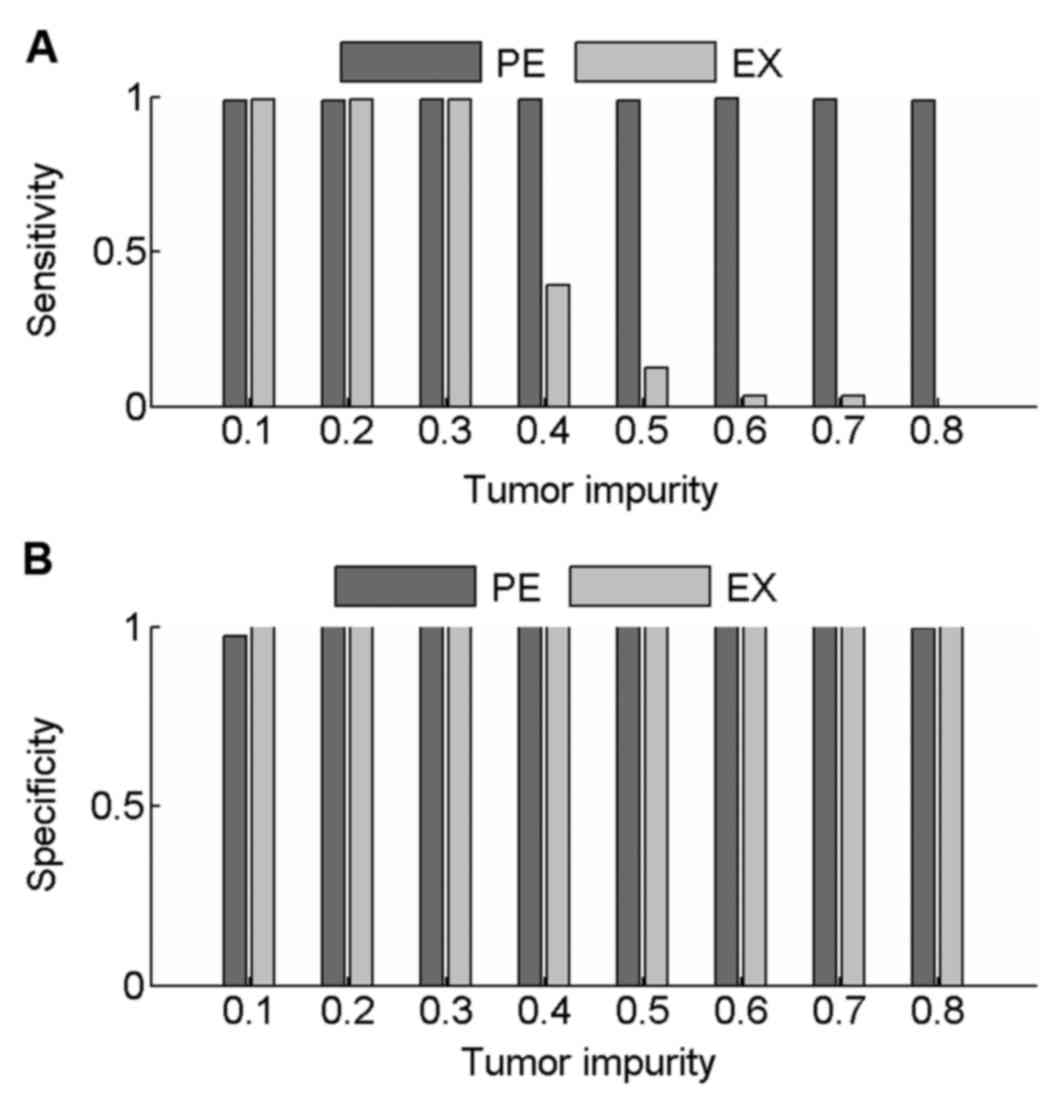
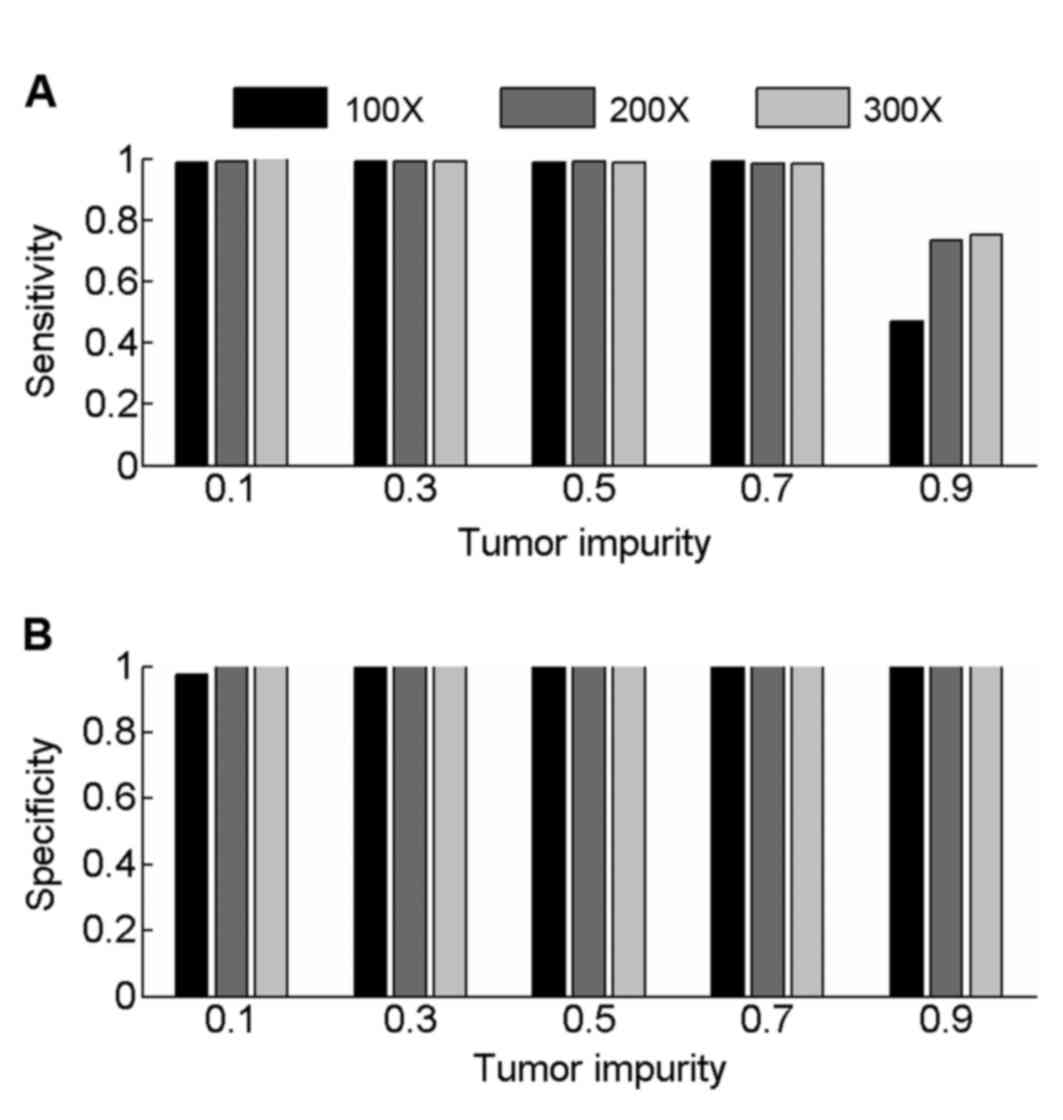
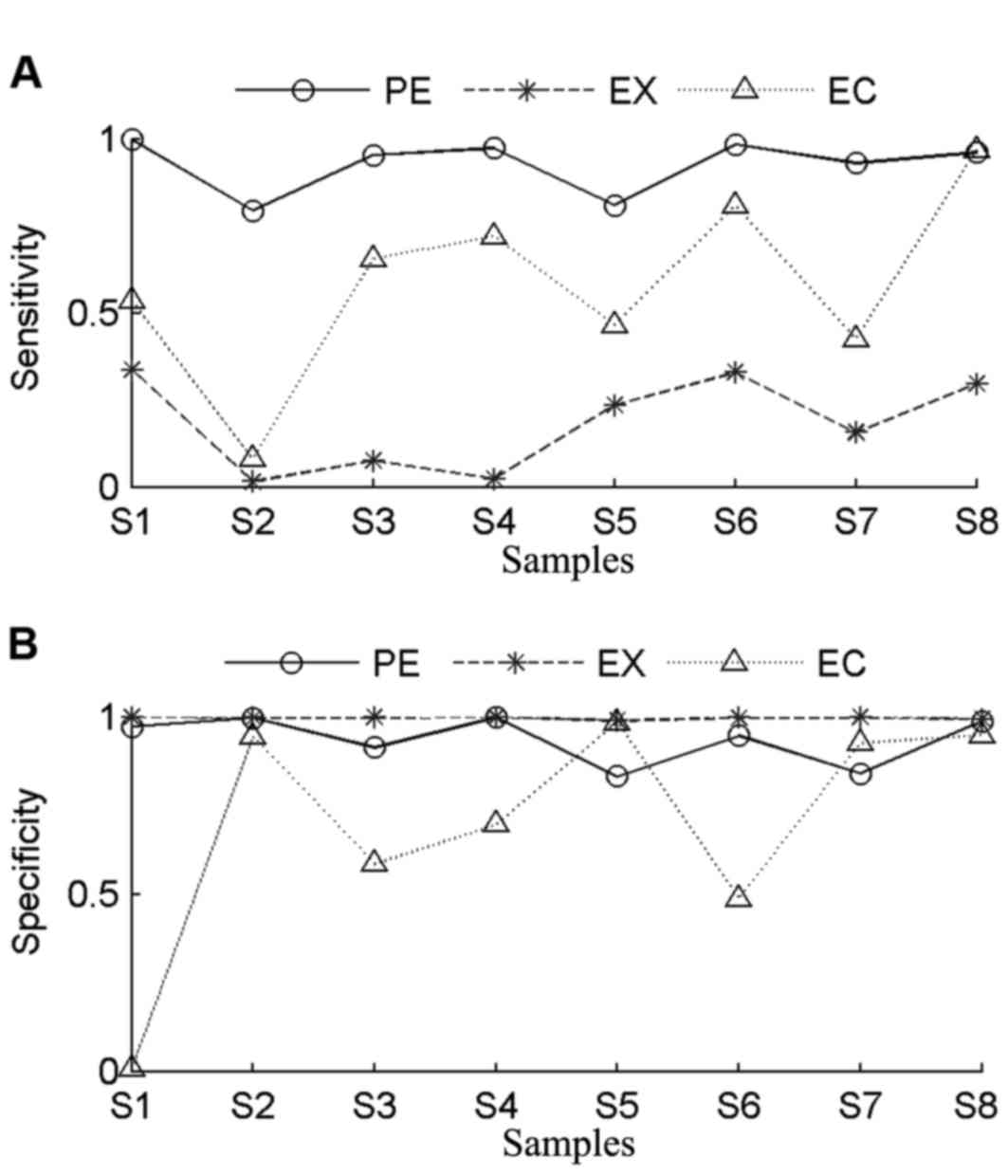
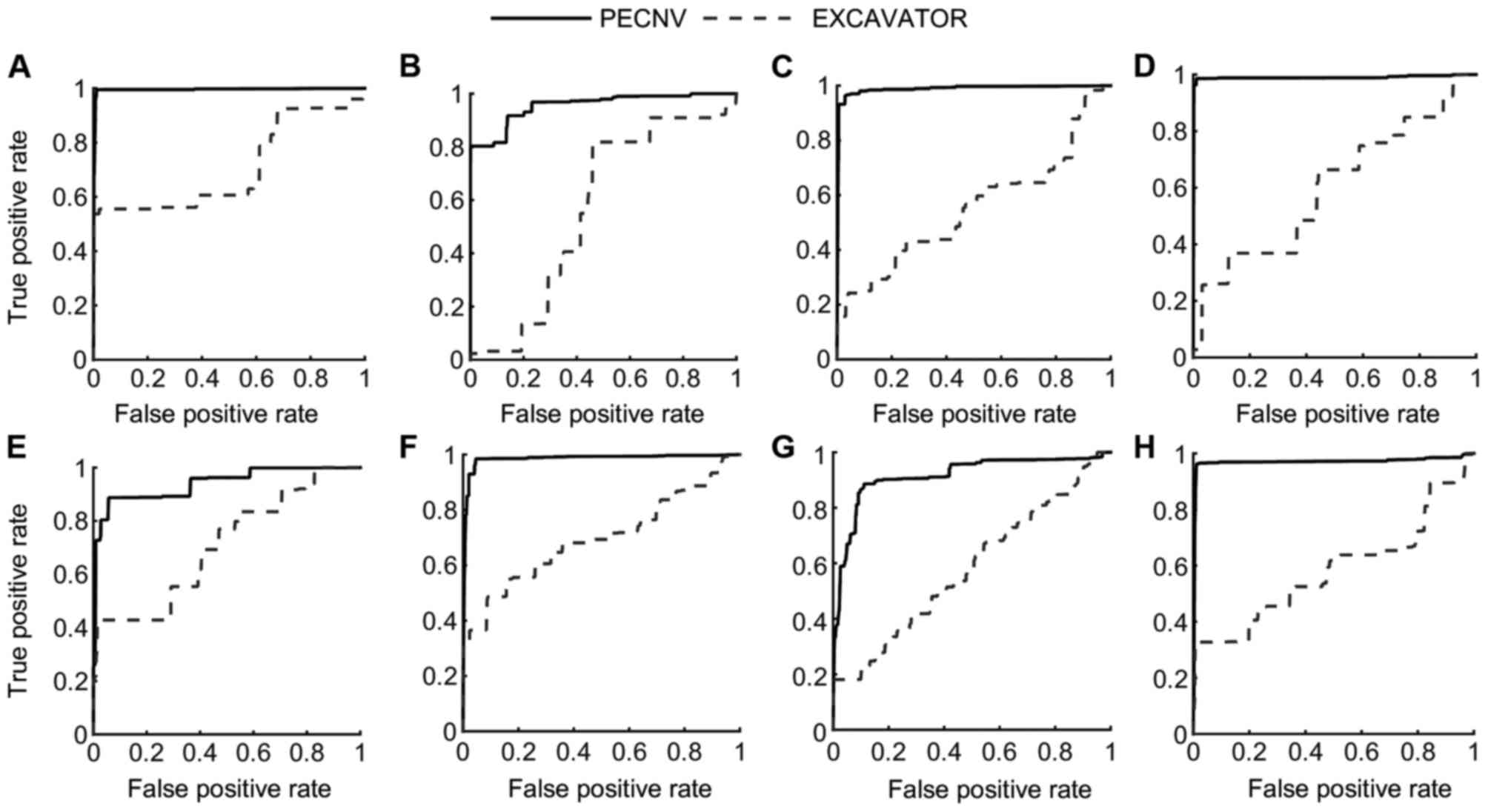
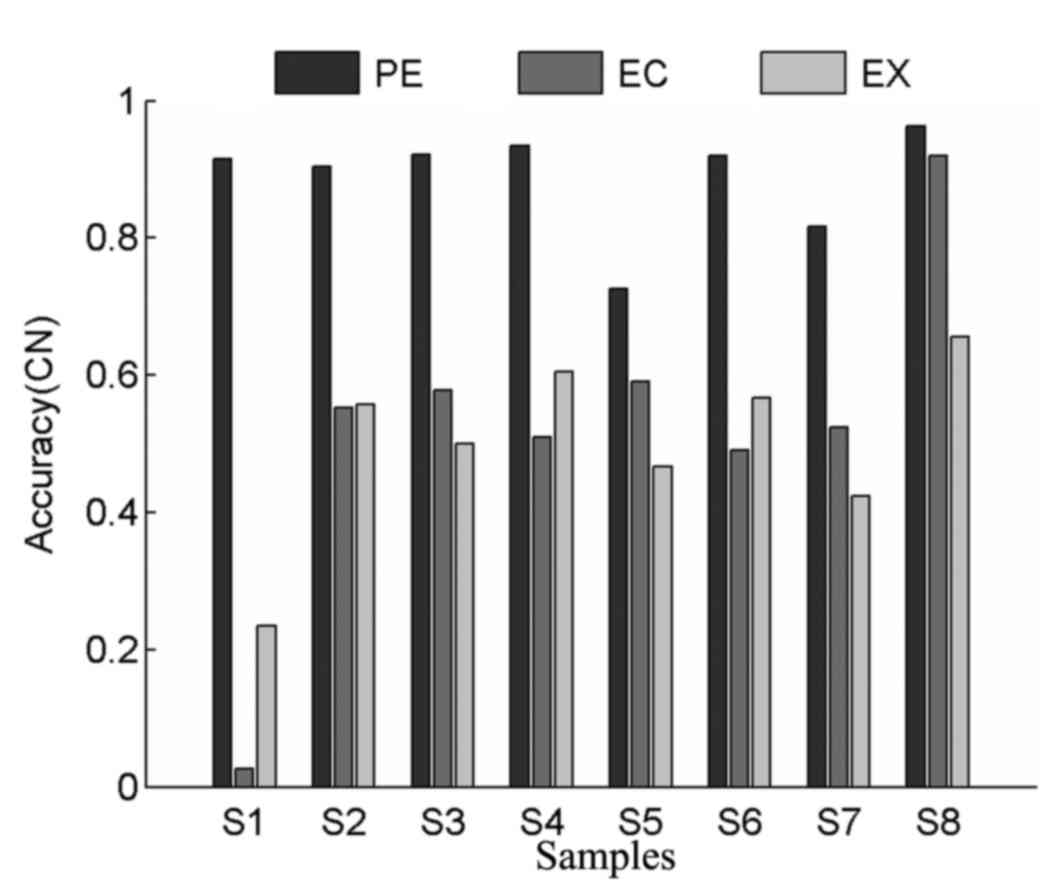
|
1
|
Albertson DG, Collins C, McCormick F and
Gray JW: Chromosome aberrations in solid tumors. Nat Genet.
34:369–376. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Weir BA, Woo MS, Getz G, Perner S, Ding L,
Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al:
Characterizing the cancer ge-nome in lung adenocarcinoma. Nature.
450:893–898. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Carén H, Kryh H, Nethander M, Sjöberg RM,
Träger C, Nilsson S, Abrahamsson J, Kogner P and Martinsson T:
High-risk neuroblastoma tumors with 11q-deletion display a poor
prognostic, chromosome instabil-ity phenotype with later onset.
Proc Natl Acad Sci USA. 107:4323–4328. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Solinas-Toldo S, Lampel S, Stilgenbauer S,
Nickolenko J, Benner A, Döhner H, Cremer T and Lichter P:
Matrix-based comparative genomic hybridization: Biochips to screen
for genomic imbalances. Genes Chromosomes Cancer. 20:399–407. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Park PJ: Experimental design and data
analysis for array comparative genomic hybridization. Cancer
Invest. 26:923–928. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
McCarroll SA, Kuruvilla FG, Korn JM,
Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PI, Maller JB,
Kirby A, et al: Integrated detection and population-genetic
analysis of SNPs and copy number variation. Nat Genet.
40:1166–1174. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peiffer DA, Le JM, Steemers FJ, Chang W,
Jenniges T, Garcia F, Haden K, Li J, Shaw CA, Belmont J, et al:
High-resolution genomic profiling of chromosomal aberrations using
Infinium whole-genome genotyping. Genome Res. 16:1136–1148. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Metzker ML: Sequencing technologies-the
next generation. Nat Rev Genet. 11:31–46. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Morozova O and Marra MA: Applications of
next-generation sequencing technologies in functional genomics.
Genomics. 92:255–264. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wheeler DA, Srinivasan M, Egholm M, Shen
Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, et al:
The complete genome of an individual by massively parallel DNA
sequencing. Nature. 452:872–876. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Teer JK and Mullikin JC: Exome sequencing:
The sweet spot before whole genomes. Hum Mol Genet. 19:R145–R151.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Clark MJ, Chen R, Lam HY, Karczewski KJ,
Chen R, Euskirchen G, Butte AJ and Snyder M: Performance comparison
of exome DNA sequencing technologies. Nat Biotechnol. 29:908–914.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ng SB, Turner EH, Robertson PD, Flygare
SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler
EE, et al: Targeted capture and massively parallel sequencing of 12
human exomes. Nature. 461:272–276. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Reese SE, Archer KJ, Therneau TM, Atkinson
EJ, Vachon CM, de Andrade M, Kocher JP and Eckel-Passow JE: A new
statistic for identifying batch effects in high-throughput genomic
data that uses guided principal component analysis. Bioinformatics.
29:2877–2883. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Karakoc E, Alkan C, O'Roak BJ, Dennis MY,
Vives L, Mark K, Rieder MJ, Nickerson DA and Eichler EE: Detection
of structural variants and indels within exome data. Nat Methods.
9:176–178. 2012. View Article : Google Scholar
|
|
16
|
Sathirapongsasuti JF, Lee H, Horst BA,
Brunner G, Cochran AJ, Binder S, Quackenbush J and Nelson SF: Exome
sequencing-based copy-number variation and loss of heterozygosity
detection: ExomeCNV. Bioinformatics. 27:2648–2654. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Krumm N, Sudman PH, Ko A, O'Roak BJ, Malig
M, Coe BP; NHLBI Exome Sequencing Project, ; Quinlan AR, Nickerson
DA and Eichler EE: Copy number variation detection and genotyping
from exome sequence data. Genome Res. 22:1525–1532. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fromer M, Moran JL, Chambert K, Banks E,
Bergen SE, Ruderfer DM, Handsaker RE, McCarroll SA, O'Donovan MC,
Owen MJ, et al: Discovery and statistical genotyping of copy-number
variation from whole-exome sequencing depth. Am J Hum Genet.
91:597–607. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Backenroth D, Homsy J, Murillo LR,
Glessner J, Lin E, Brueckner M, Lifton R, Goldmuntz E, Chung WK and
Shen Y: CANOES: Detecting rare copy number variants from whole
exome sequencing data. Nucleic Acids Res. 42:e972014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Magi A, Tattini L, Cifola I, D'Aurizio R,
Benelli M, Mangano E, Battaglia C, Bonora E, Kurg A, Seri M, et al:
EXCAVATOR: Detecting copy number variants from whole-exome
sequencing data. Genome Biol. 14:R1202013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li A, Liu Z, Lezon-Geyda K, Sarkar S,
Lannin D, Schulz V, Krop I, Winer E, Harris L and Tuck D: GPHMM: An
integrated hidden Markov model for identification of copy number
alteration and loss of heterozygosity in complex tumor samples
using whole genome SNP arrays. Nucleic Acids Res. 39:4928–4941.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li H, Handsaker B, Wysoker A, Fennell T,
Ruan J, Homer N, Marth G, Abecasis G and Durbin R; 1000 Genome
Project Data Processing Subgroup, : The sequence alignment/map
format and SAMtools. Bioinformatics. 25:2078–2079. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yoon S, Xuan Z, Makarov V, Ye K and Sebat
J: Sensitive and accurate detection of copy number variants using
read depth of coverage. Genome Res. 19:1586–1592. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mortazavi A, Williams BA, McCue K,
Schaeffer L and Wold B: Mapping and quantifying mammalian
transcriptomes by RNA-Seq. Nat Methods. 5:621–628. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bilmes JA: A gentle tutorial of the EM
algorithm and its-application to parameter estimation for Gaussian
mixture and hidden Markov models. Int Comput Sci Inst.
4:1261998.
|
|
26
|
Olshen AB, Venkatraman ES, Lucito R and
Wigler M: Circular binary segmentation for the analysis of
array-based DNA copy number data. Biostatistics. 5:557–572. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Møller MF: A scaled conjugate gradient
algorithm for fast supervised learning. Neural Net. 6:525–533.
1993. View Article : Google Scholar
|
|
28
|
Yu Z, Liu Y, Shen Y, Wang M and Li A:
CLImAT: Accurate detection of copy number alteration and loss of
heterozygosity in impure and aneuploid tumor samples using
whole-genome sequencing data. Bioinformatics. 30:2576–2583. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shah SP, Roth A, Goya R, Oloumi A, Ha G,
Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al: The clonal
and mutational evolution spectrum of primary triple-negative breast
cancers. Nature. 486:395–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Van Loo P, Nordgard SH, Lingjærde OC,
Russnes HG, Rye IH, Sun W, Weigman VJ, Marynen P, Zetterberg A,
Naume B, et al: Allele-specific copy number analysis of tumors.
Proc Natl Acad Sci USA. 107:16910–16915. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Oesper L, Mahmoody A and Raphael BJ:
THetA: Inferring intra-tumor heterogeneity from high-throughput DNA
sequencing data. Genome Biol. 14:R802013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tan R, Wang Y, Kleinstein SE, Liu Y, Zhu
X, Guo H, Jiang Q, Allen AS and Zhu M: An evaluation of copy number
variation detection tools from whole-exome sequencing data. Hum
Mutat. 35:899–907. 2014. View Article : Google Scholar : PubMed/NCBI
|