Introduction
Hepatocellular carcinoma (HCC) is the third leading
cause of cancer mortality worldwide. This malignancy occurs more
often in men than in women, with the highest incidence rates
reported in East Asia (1). Surgery
remains the most effective treatment for HCC. However, only 20% of
HCC patients are suitable for surgery, and recurrence in resected
patients is as high as 70%. The overall 5-year survival for
resected patients is ≤19% (2).
Therefore, the development of new therapeutics to prevent
recurrence is necessary. To protect against recurrence, tumor
antigen-specific immunotherapy is an attractive strategy (3). T cell-based immunotherapies for HCC
have used a few tumor-associated antigens (TAAs) and their
epitopes, including AFP (4), GPC
(5), NY-ESO-1 (6), SSX-2 (7), MAGE-A (8), and TERT (9). However, clinical data for only
alpha-fetoprotein (AFP) have been reported (10,11). A
prerequisite for the successful development of T cell-based
immunotherapeutic approaches is the identification and
characterization of immune responses to TAAs. Many TAAs have been
identified during the last 2 decades using various techniques.
These include T cell epitope cloning, MHC peptide elution,
differential gene expression analysis, and serologic expression
cloning (SEREX) (12–15).
Among these methods, the serologic analysis of
recombinant cDNA expression libraries (SEREX) has provided a
powerful approach to identify immunogenic tumor antigens. To date,
over 2,500 tumor antigens have been identified from a variety of
tumors using SEREX. These antigens can be classified into several
categories, including mutational antigens (5,10),
differentiation antigens (10,14),
overexpressed antigens (15), and
cancer/testis (CT) antigens (16,17).
Cancer/testis (CT) antigens are immunogenic protein
antigens with expression restricted to the testis and a wide range
of human tumor types that elicit both humoral and cellular immune
responses in cancer patients (17).
They are considered ideal targets for vaccine-based immunotherapy,
and to date, more than 100 CT antigens, including MAGE, NY-ESO-1,
GAGE, BAGE, LAGE, and SSX2, have been identified (18). CT antigens are divided between those
that are encoded on the X chromosome (CT-X antigens) and those that
are not (non-X CT antigens) (19).
Many CT antigens show heterogeneous expression patterns within the
same tumor tissue (20). Additional
CT antigens are needed for the development of polyvalent cancer
vaccines designed to overcome the limited frequency and
heterogeneity of CT antigen expression. CT antigens identified by
SEREX to date include MAGE-A (21),
NY-ESO-1 (22), SSX2 (23), SCP1 (24), NY-SAR-35 (25), SLCO6A1 (26), and BCP-20 (27). In our study, we performed SEREX
analysis to screen a testicular cDNA library with the aim of
isolating CT antigens in the sera of HCC patients. We isolated 2
previously defined CT antigens, AKAP3 and CTp11 (28,29).
Materials and methods
Human tissues, sera, and cell lines
Human tumor tissues and sera were obtained from the
Department of Pathology and the Korean National Biobank, Pusan
National University Hospital after diagnosis and staging. The
tissues were frozen in liquid nitrogen and stored at −80°C until
use. The human hepatoma cell lines SNU-354, SNU-398, SNU-423,
SNU-449, SNU-475, and HepG2; human colon cancer cell lines SNU-C1,
SNU-C2A, SNU-C4, and SNU-C5; and the human ovarian cancer cell
lines SNU-8 and SNU-840 were obtained from the Korean Type Culture
Collection (KTCC) and the American Type Culture Collection (ATCC).
All cell lines were maintained in RPMI-1640 (Gibco-BRL Life
Technologies Inc., Grand Island, NY, USA) medium supplemented with
10% fetal bovine serum, 2 mM L-glutamine, 100 U/ml penicillin, and
100 μg/ml streptomycin.
Total RNA extraction from tissues and
cell lines
Total RNA was isolated from human tissue samples and
human tumor cell lines using the standard TRIzol reagent (Life
Technologies, Gaithersburg, MD, USA) and RNA isolation kit (RNeasy
Maxi kit, Qiagen) following manufacturer’s instructions. Normal
tissue total RNA was purchased from Clontech Laboratories, Inc.
(Palo Alto, CA, USA) and Ambion, Inc. (Austin, TX, USA). Total RNA
from several cancer cell lines used in this experiment was obtained
from the Ludwig Institute for Cancer Research (LICR), New York
Branch at the Memorial Sloan-Kettering Cancer Center.
Preparation of cDNA library and sera
Poly(A)+ RNA from normal testis was purchased from
Clontech Laboratories, Inc. Five micrograms of mRNA was used to
construct a cDNA library in the ZAP Express vector (Stratagene, La
Jolla, CA, USA), following the manufacturer’s instructions. The
library contained ~1,000,000 recombinants, and was used for
immunoscreening without prior amplification. Sera from 3 HCC
patients were pooled and absorbed. The pooled serum was diluted
1:200 (final dilution, 1:600 for each serum) in Tris-buffered
saline (TBS) containing 1% BSA. The patients ages ranged from 74–79
years, all 3 were male, and all patients had stage I/II HCC. In
addition to this pooled serum, all sera from HCC patients and
healthy individuals in this study were diluted 1:200. To remove
serum antibodies that react with Escherichia
coli/bacteriophage-related antigens, sera were absorbed against
E. coli/bacteriophage lysates as described by Lee et
al (25).
Immunoscreening of cDNA library
Immunoscreening of the cDNA library was performed as
previously described (15,22). Briefly, E. coli XL1 blue MRF
cells were transfected with the recombinant phages, plated at a
density of approximately 5,000 pfu/150-mm plate (NZCYM-IPTG agar),
incubated for 8 h at 37°C, and then transferred to nitrocellulose
filters (PROTRAN BA 85, 0.45 μm; Schleicher & Schuell, Keene,
NH, USA). Then the filters were incubated with a 1:200 dilution of
patient sera, which had been preabsorbed to E. coli-phage
lysate. The serum-reactive clones were detected with an
AP-conjugated secondary antibody and visualized by incubation with
5-bromo-4-chloro-3-indolyl-phosphate/nitroblue tetrazolium
(BCIP/NBT). After screening, the isolated positive clones were
removed from the plate and preserved in suspension medium (SM)
buffer with 25 μl of chloroform. Positive phages were mixed with a
helper phage to co-infect XL-1 Blue MRF, and they were rescued into
pBluescript phagemid forms by in vivo excision. The excised
phagemids were transformed into the host bacteria (XLOLR) to
multiply for plasmid extraction and stock creation. The size of the
inserted cDNA was determined primarily by double restriction enzyme
digestion with EcoRI and XhoI. The cDNA was
commercially sequenced (Macrogen, Korea).
RT-PCR
The cDNA used as templates for RT-PCR reactions was
prepared with 1 μg of total RNA using the Superscript first strand
synthesis kit (Invitrogen Life Technologies, Carlsbad, CA, USA).
The PCR primers specific for AKAP3 and CTp11 were
CTAACTTCGGCCTTCCCAGA (forward)/AGTGG GGTTGCCGATTACAG (reverse) and
GGCGGGGTGAA GAGGAGCGT (forward)/ACACAGCCATCAGCTTCTC AAACTT
(reverse), respectively. The cDNA templates were normalized based
on the amplification of GAPDH. For PCR, a 20-μl reaction mixture,
including 2 μl of cDNA, 0.2 mM dNTPs, 1.5 mM MgCl2, 0.25
μM gene-specific forward and reverse primers, and 3 U of Taq DNA
polymerase (Solgent, Daejun, Korea), was preheated to 94°C for 5
min, followed by 35 cycles of 94°C for 30 sec, 60°C for 30 sec, and
72°C for 1 min, with a final elongation step of 72°C for 5 min.
Amplified PCR products were analyzed on 1.5% agarose gels stained
with ethidium bromide.
Phage plaque assay
To determine the immunoreactivity of the isolated
clones, a phage plaque assay was performed as described previously
(16). Briefly, phage from a
positive clone were mixed with -ZAP clone without an insert as an
internal negative control at a ratio 1:1, and then transfected into
E. coli. Plaques were blotted onto nitrocellulose membranes
and washed. Next, they were incubated with sera at 4°C for 16 h.
The spots were determined to be positive only if the tested clones
were clearly distinguishable from the negative phage.
Results
Identification of HCC antigens by
SEREX
A testis cDNA expression library of
~2×105 clones was immunoscreened with pooled sera from
HCC patients. Three clones representing 4 genes were isolated, and
the antigens were AKAP3, CTp11, and UBQLN3. These 3 genes are
associated with testis specific expression in the UniGene database.
Comparison of the cDNA sequences encoding the antigens to those
deposited in cancer immunome database showed that AKAP3 had been
previously identified as NY-TLU-37 in lung cancer patients
(26), while CTp11 and UBQLN3 had
not been previously reported (Table
I).
 | Table IHCC antigens by SEREX. |
Table I
HCC antigens by SEREX.
| Gene name | Unigene cluster | Chromosome
location | Number of
redundancies | Previously identified
by SEREX |
|---|
| AKAP3 | Hs.98397 | 12p13.3 | 2 | Y |
| SPANXA2 | Hs.711784 | Xq27.1 | 1 | N |
| UBQLN3 | Hs.189184 | 11p15 | 1 | N |
mRNA expression in normal tissues,
tumors, and cancer cell lines
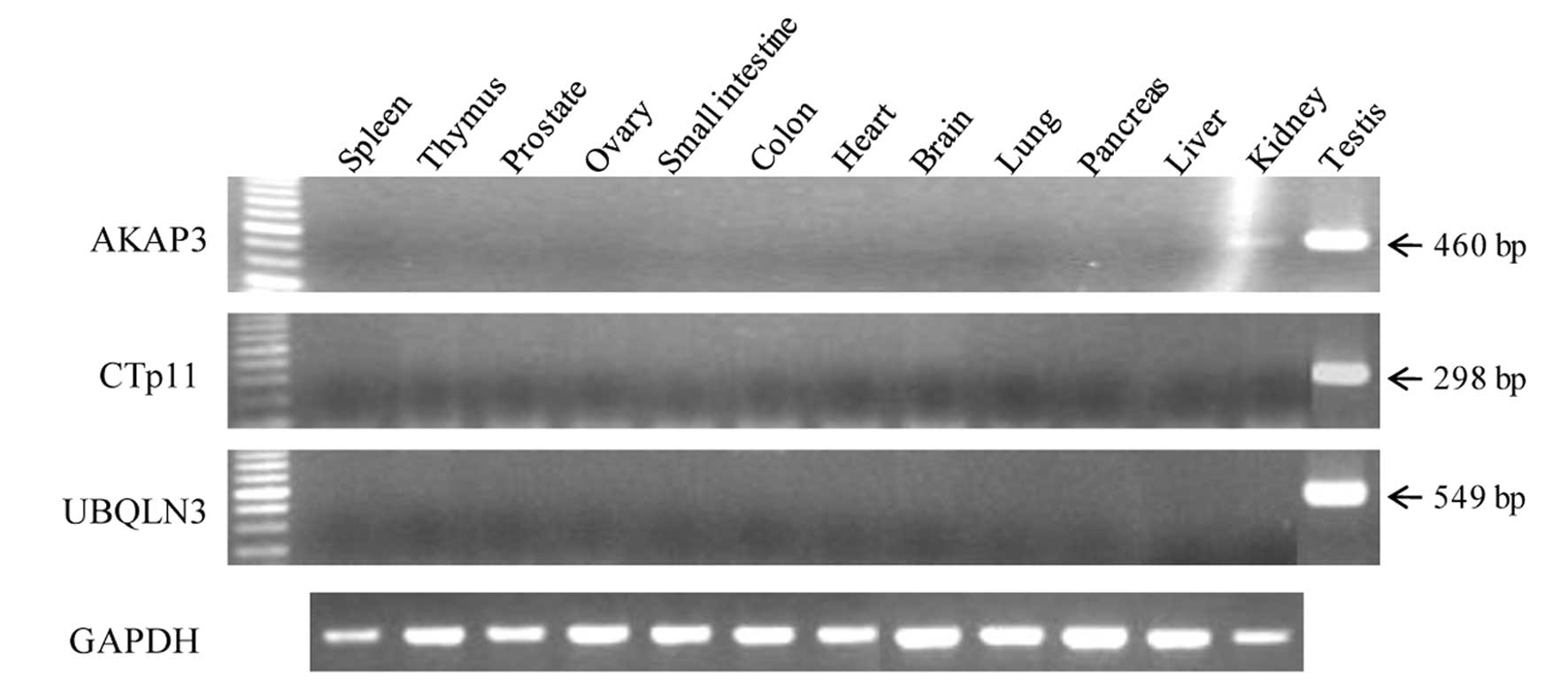
To investigate the restricted expression of the
mRNAs of the 3 genes in normal adult tissues, we performed
conventional RT-PCR. As shown in Fig.
1, AKAP3 was strongly expressed in testis, but was weakly
expressed in brain and kidney. In addition, expression of CTp11 and
UBQLN3 mRNAs was restricted to the testis. The AKAP3 gene was
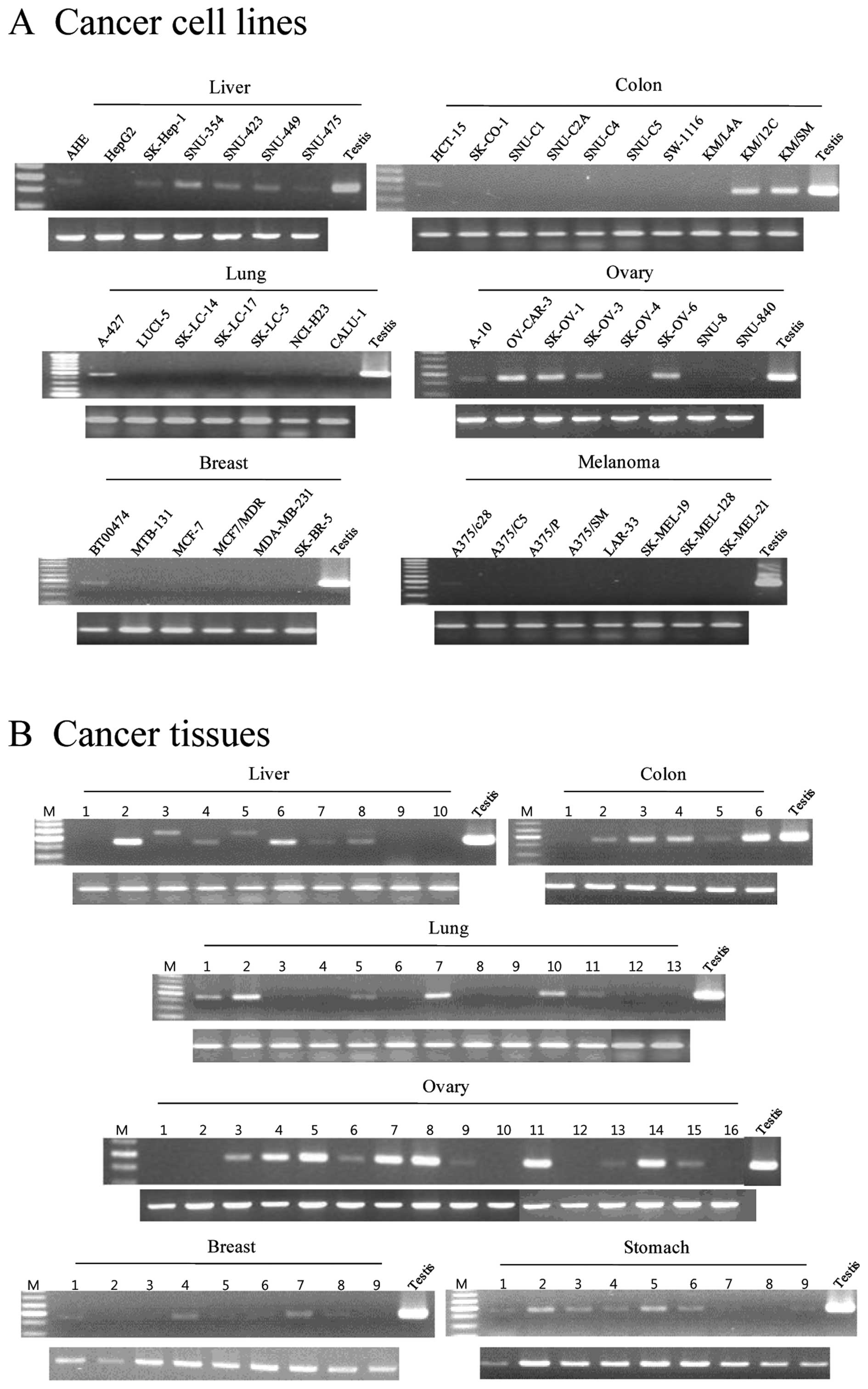
expressed broadly and frequently in various cancer cell lines and
cancer tissues, including HCC (6/7 and 5/10), colon cancer (3/10
and 5/6), lung cancer (4/7 and 6/13), ovarian cancer (6/8 and
11/16), and breast cancer (3/6 and 6/9), respectively (Fig. 2). In addition, AKAP3 mRNA was
detected in melanoma cancer cell lines (1/8), renal cancer cell
lines (2/2), leukemia cancer cell lines (2/3), sarcoma cell lines
(2/3), and a glioma cell line (1/2), but it was not detected in
prostate cancer cell lines and small cell lung cancer cell lines
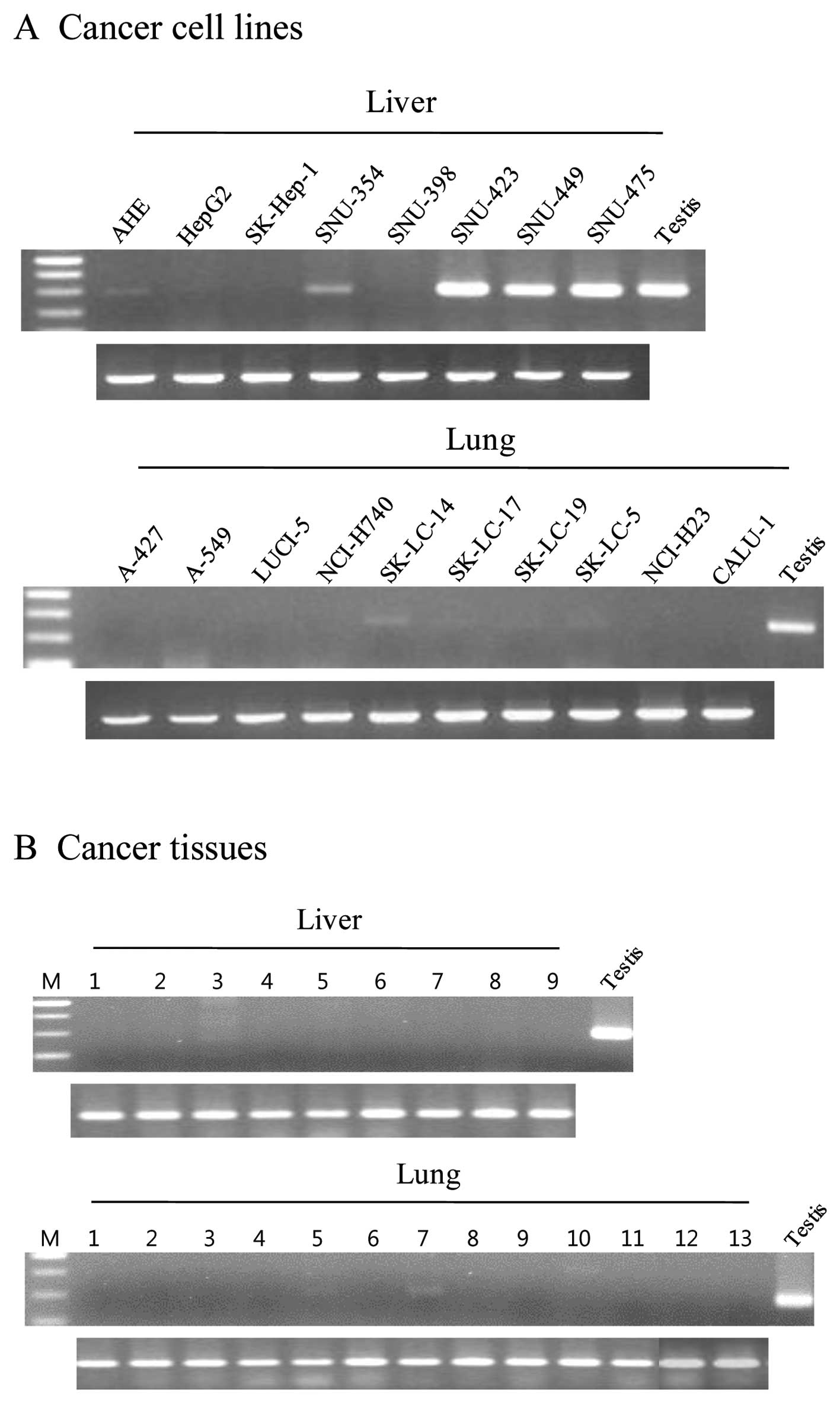
(Table II). In addition, CTp11 was
expressed in HCC cell lines and cancer tissues (5/8 and 1/9,
respectively) and lung cancer cell lines and cancer tissues (4/10
and 2/13, respectively) (Fig. 3).
However, CTp11 was either not expressed or infrequently expressed
in various tumors and cancer cell lines, including breast cancer,
colon cancer, and ovary cancer (Table
II). These results indicate that AKAP3 is a CT antigen that is
frequently expressed in a variety of cancers, including HCC.
 | Table IISummary of expression of AKAP3 and
CTp11 mRNA. |
Table II
Summary of expression of AKAP3 and
CTp11 mRNA.
| AKAP3 | CTp11 |
|---|
|
|
|
|---|
| Cell lines | Tissues | Cell lines | Tissues |
|---|
| Tumor type | (Positive/total) | (Positive/total) |
|---|
| HCC | 6/7 | 5/10 | 5/8 | 1/9 |
| Colon cancer | 3/10 | 5/6 | 1/10 | 0/6 |
| Lung cancer | 4/7 | 6/13 | 4/10 | 2/13 |
| Ovary cancer | 6/8 | 11/16 | 1/8 | 0/15 |
| Breast cancer | 3/6 | 6/9 | 0/6 | 0/9 |
| Melanoma | 1/8 | ND | 0/9 | ND |
| Stomach cancer | ND | 8/9 | ND | 0/9 |
| Small cell lung
cancer | 0/5 | ND | 0/5 | ND |
| Renal cell
cancer | 2/2 | ND | 0/2 | ND |
| Prostate
cancer | 0/3 | ND | 0/3 | ND |
| Leukemia | 2/3 | ND | 0/3 | ND |
| Sarcoma | 2/3 | ND | 2/3 | ND |
| Glioma | 1/2 | ND | 0/2 | ND |
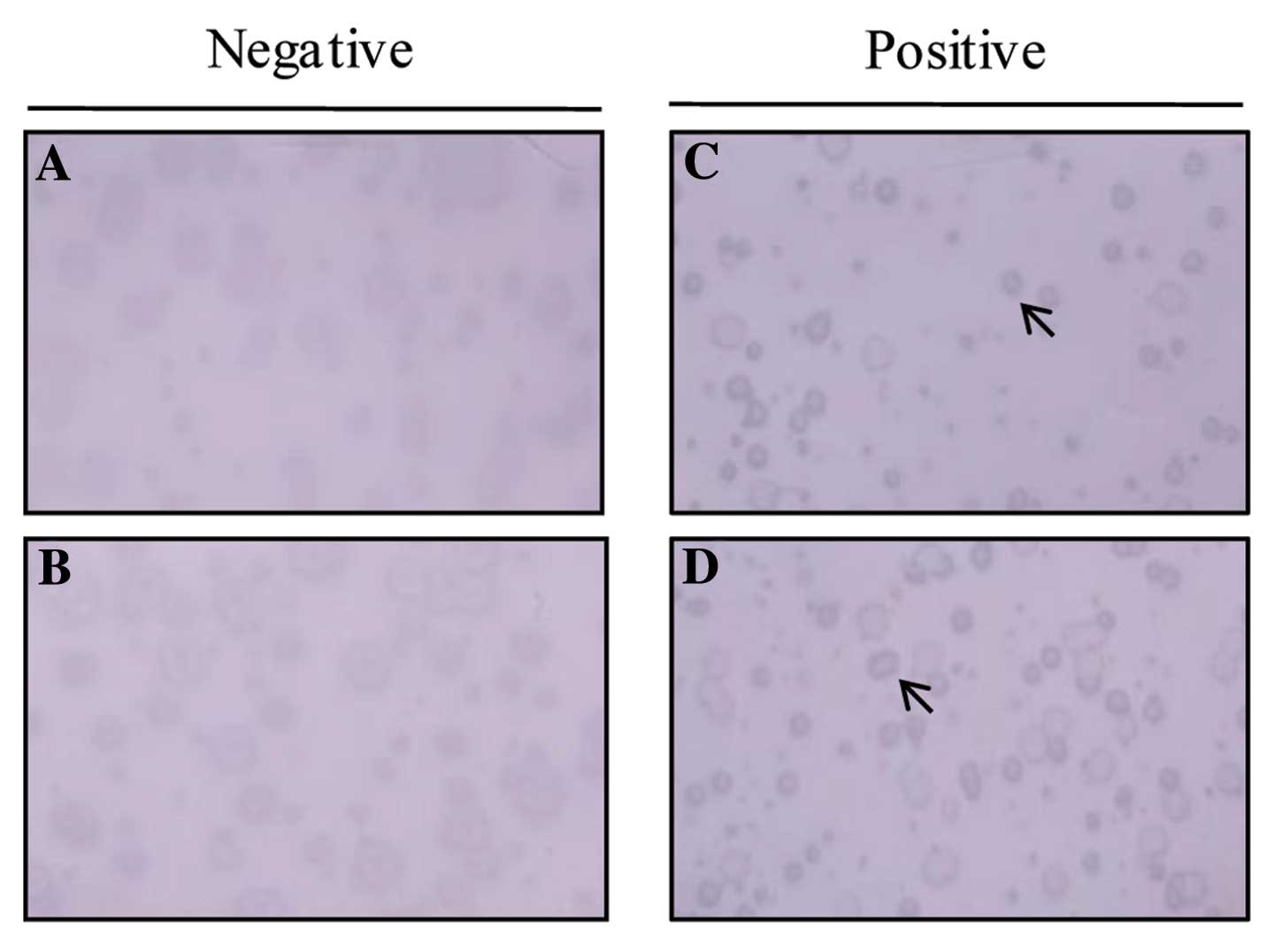
Seroreactivity of the isolated
antigens
To determine whether immune recognition of AKAP3,
CTp11, and UBQLN3 proteins was cancer-related, allogeneic sera
samples obtained from 27 normal blood donors and 27 patients with
HCC were tested for reactivity by phage plaque assay. A
representative phage plaque assay from the sera of HCC patients is
shown in Fig. 4. Fifteen of 27
(63%) sera samples from HCC patients and 8/27 (30%) samples from
normal patients were reactive against AKAP3. In contrast, 1 of 27
HCC cancer sera samples reacted with CTp11 and UBQLN3, while none
with sera samples from normal individuals reacted with CTp11 and
UBQLN3. Although 8/27 heath donors were positive for AKAP3
reactivity, 63% recognition by sera from HCC patients indicates
that it may be an immunogenic tumor antigen.
Discussion
To identify additional immunoreactive antigens in
HCC, a SEREX analysis was performed using a testis cDNA library and
sera from HCC patients. Three distinct antigens were isolated,
AKAP3, CTp11, and UBQLN3. UBQLN3 encodes an ubiquitin-like protein
specifically expressed in the testis that has been proposed to
regulate cell cycle progression during spermatogenesis (30). Conventional RT-PCR demonstrated
strong UBQLN3 mRNA expression in testis; however, transcripts
encoding UBQLN3 were not detected in cancer cell lines and tumor
tissues (data not shown).
CTp11 was previously reported as a CT antigen
isolated from a melanoma cell line by differential display
(29). The gene encodes an 11-kDa
protein and is located on chromosome Xq26.3-Xq27.1. CTp11 mRNA was
expressed in 25–30% of the melanoma and bladder carcinoma cell
lines tested, while only 2 of the 29 other tumor cell lines were
positive. Recent studies reported a high frequency of CTp11
expression in 56.2% of HCC tissues and 31.1% of PBMC from HCC
patients (31). In our study, CTp11
was expressed in 62% of HCC cell lines and 11% of HCC tissues.
Although the frequency of CTp11 expression in our study differed
from the previous results, we suggest the possibility of CTp11
antigen as a new potential candidate for HCC immunotherapy that
will need to be further analyzed.
In addition, AKAP3 was previously reported as a CT
antigen (28). A previous study
showed that AKAP3 was a sperm protein, and that the mRNA was only
expressed in the testis (32). This
gene encodes a member of the A-kinase anchor proteins (AKAPs), and
is only expressed in testis. The protein is localized to the ribs
of the fibrous sheath in the principal piece of the sperm tail. It
may function as a regulator of both motility- and head-associated
functions, such as capacitation and the acrosome reaction (33). Two groups previously showed high
AKAP3 mRNA expression in ovarian cancer, and expression was
correlated to the histological grade and clinical stage of the
tumors (28,34). These previous reports showed AKAP3
mRNA expression in 58 and 28% of ovarian cancer specimens.
We demonstrated similar findings; high AKAP3 mRNA
expression was observed in ovarian tumors (69%) and ovarian cancer
cell lines (75%), suggesting that AKAP3 is a new independent
prognostic factor for ovarian cancer patients. AKAP3 mRNA was
broadly and frequently expressed in various tumors and cancer cell
lines, including HCC (Fig. 2,
Table II). In our study, AKAP3 was
isolated for the first time from HCC patients by SEREX, was highly
expressed in HCC tumors (5/10) and HCC cell lines (6/7), and
anti-AKAP3 antibody was detected in 15 of 27 HCC sera samples by
phage plaque analysis. These findings suggested that AKAP3 is a
favorable molecular marker for prognosis and diagnosis in HCC
patients. To address this possibility, we are examining AKAP3 mRNA
expression in additional sera samples from HCC patients and
investigating whether expression is correlated with the
histological grade and clinical stage of tumors.
The correlation between AKAP3 mRNA expression and
anti AKAP3 IgG was not evaluated due to a lack of paired samples;
therefore, this should be investigated in a future study.
Nonetheless, AKAP3 recognition by sera from HCC patients and
healthy individuals indicates that AKAP3 is an immunogenic tumor
antigen in HCC patients.
In conclusion, we reported for the first time the
isolation of AKAP3 and CTp11 CT antigens from HCC patient sera by
SEREX. We demonstrated high AKAP3 mRNA expression and high
seroreactivity, which may have a favorable impact on diagnosis and
immunotherapy of HCC patients.
Acknowledgements
The study was supported by a grant from Pusan
National University (2011–2012).
References
|
1
|
Schutte K, Bornschein J and Malfertheiner
P: Hepatocellular carcinoma - epidemiological trends and risk
factors. Dig Dis. 27:80–92. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Butterfield LH: Recent advances in
immunotherapy for hepatocellular cancer. Swiss Med Wkly. 137:83–90.
2007.PubMed/NCBI
|
|
3
|
Breous E and Thimme R: Potential of
immunotherapy for hepatocellular carcinoma. J Hepatol. 54:830–834.
2011. View Article : Google Scholar
|
|
4
|
Liu Y, Daley S, Evdokimova VN, Zdobinski
DD, Potter DM and Butterfield LH: Hierarchy of alpha fetoprotein
(AFP)-specific T cell responses in subjects with AFP-positive
hepatocellular cancer. J Immunol. 177:712–721. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Komori H, Nakatsura T, Senju S, Yoshitake
Y, Motomura Y, Ikuta Y, Fukuma D, Yokomine K, Harao M, Beppu T, et
al: Identification of HLA-A2- or HLA-A24-restricted CTL epitopes
possibly useful for glypican-3-specific immunotherapy of
hepatocellular carcinoma. Clin Cancer Res. 12:2689–2697. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gehring AJ, Ho ZZ, Tan AT, Aung MO, Lee
KH, Tan KC, Lim SG and Bertoletti A: Profile of tumor
antigen-specific CD8 T cells in patients with hepatitis B
virus-related hepatocellular carcinoma. Gastroenterology.
137:682–690. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bricard G, Bouzourene H, Martinet O,
Rimoldi D, Halkic N, Gillet M, Chaubert P, Macdonald HR, Romero P,
Cerottini JC and Speiser DE: Naturally acquired MAGE-A10- and
SSX-2-specific CD8+ T cell responses in patients with
hepatocellular carcinoma. J Immunol. 174:1709–1716. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zerbini A, Pilli M, Soliani P, Ziegler S,
Pelosi G, Orlandini A, Cavallo C, Uggeri J, Scandroglio R, Crafa P,
et al: Ex vivo characterization of tumor-derived melanoma antigen
encoding gene-specific CD8+ cells in patients with
hepatocellular carcinoma. J Hepatol. 40:102–109. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mizukoshi E, Nakamoto Y, Marukawa Y, Arai
K, Yamashita T, Tsuji H, Kuzushima K, Takiguchi M and Kaneko S:
Cytotoxic T cell responses to human telomerase reverse
transcriptase in patients with hepatocellular carcinoma.
Hepatology. 43:1284–1294. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Butterfield LH, Ribas A, Dissette VB, Lee
Y, Yang JQ, De la Rocha P, Duran SD, Hernandez J, Seja E, Potter
DM, et al: A phase I/II trial testing immunization of
hepatocellular carcinoma patients with dendritic cells pulsed with
four alpha-fetoprotein peptides. Clin Cancer Res. 12:2817–2825.
2006. View Article : Google Scholar
|
|
11
|
Thimme R, Neagu M, Boettler T,
Neumann-Haefelin C, Kersting N, Geissler M, Makowiec F, Obermaier
R, Hopt UT, Blum HE and Spangenberg HC: Comprehensive analysis of
the alpha-fetoprotein-specific CD8+ T cell responses in
patients with hepatocellular carcinoma. Hepatology. 48:1821–1833.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gaugler B, Van den Eynde B, van der
Bruggen P, Romero P, Gaforio JJ, De Plaen E, Lethe B, Brasseur F
and Boon T: Human gene MAGE-3 codes for an antigen recognized on a
melanoma by autologous cytolytic T lymphocytes. J Exp Med.
179:921–930. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
van der Bruggen P, Traversari C, Chomez P,
Lurquin C, De Plaen E, Van den Eynde B, Knuth A and Boon T: A gene
encoding an antigen recognized by cytolytic T lymphocytes on a
human melanoma. Science. 254:1643–1647. 1991.
|
|
14
|
Pascolo S, Schirle M, Guckel B, Dumrese T,
Stumm S, Kayser S, Moris A, Wallwiener D, Rammensee HG and
Stevanovic S: A MAGE-A1 HLA-A A*0201 epitope identified by mass
spectrometry. Cancer Res. 61:4072–4077. 2001.
|
|
15
|
Sahin U, Tureci O, Schmitt H, Cochlovius
B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I and
Pfreundschuh M: Human neoplasms elicit multiple specific immune
responses in the autologous host. Proc Natl Acad Sci USA.
92:11810–11813. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gure AO, Stockert E, Scanlan MJ, Keresztes
RS, Jager D, Altorki NK, Old LJ and Chen YT: Serological
identification of embryonic neural proteins as highly immunogenic
tumor antigens in small cell lung cancer. Proc Natl Acad Sci USA.
97:4198–4203. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Scanlan MJ, Gure AO, Jungbluth AA, Old LJ
and Chen YT: Cancer/testis antigens: an expanding family of targets
for cancer immunotherapy. Immunol Rev. 188:22–32. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Caballero OL and Chen YT: Cancer/testis
(CT) antigens: potential targets for immunotherapy. Cancer Sci.
100:2014–2021. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Simpson AJ, Caballero OL, Jungbluth A,
Chen YT and Old LJ: Cancer/testis antigens, gametogenesis and
cancer. Nat Rev Cancer. 5:615–625. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim YD, Park HR, Song MH, Shin DH, Lee CH,
Lee MK and Lee SY: Pattern of cancer/testis antigen expression in
lung cancer patients. Int J Mol Med. 29:656–662. 2012.PubMed/NCBI
|
|
21
|
Chen YT, Gure AO, Tsang S, Stockert E,
Jager E, Knuth A and Old LJ: Identification of multiple
cancer/testis antigens by allogeneic antibody screening of a
melanoma cell line library. Proc Natl Acad Sci USA. 95:6919–6923.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen YT, Scanlan MJ, Sahin U, Tureci O,
Gure AO, Tsang S, Williamson B, Stockert E, Pfreundschuh M and Old
LJ: A testicular antigen aberrantly expressed in human cancers
detected by autologous antibody screening. Proc Natl Acad Sci USA.
94:1914–1918. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tureci O, Sahin U, Schobert I, Koslowski
M, Scmitt H, Schild HJ, Stenner F, Seitz G, Rammensee HG and
Pfreundschuh M: The SSX-2 gene, which is involved in the t(X;18)
translocation of synovial sarcomas, codes for the human tumor
antigen HOM-MEL-40. Cancer Res. 56:4766–4772. 1996.PubMed/NCBI
|
|
24
|
Tureci O, Sahin U, Zwick C, Koslowski M,
Seitz G and Pfreundschuh M: Identification of a meiosis-specific
protein as a member of the class of cancer/testis antigens. Proc
Natl Acad Sci USA. 95:5211–5216. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee SY, Obata Y, Yoshida M, Stockert E,
Williamson B, Jungbluth AA, Chen YT, Old LJ and Scanlan MJ:
Immunomic analysis of human sarcoma. Proc Natl Acad Sci USA.
100:2651–2656. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee SY, Williamson B, Caballero OL, Chen
YT, Scanlan MJ, Ritter G, Jongeneel CV, Simpson AJ and Old LJ:
Identification of the gonad-specific anion transporter SLCO6A1 as a
cancer/testis (CT) antigen expressed in human lung cancer. Cancer
Immun. 4:132004.PubMed/NCBI
|
|
27
|
Song MH, Ha JC, Lee SM, Park YM and Lee
SY: Identification of BCP-20 (FBXO39) as a cancer/testis antigen
from colon cancer patients by SEREX. Biochem Biophys Res Commun.
408:195–201. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hasegawa K, Ono T, Matsushita H, Shimono
M, Noguchi Y, Mizutani Y, Kodama J, Kudo T and Nakayama E: A-kinase
anchoring protein 3 messenger RNA expression in ovarian cancer and
its implication on prognosis. Int J Cancer. 108:86–90. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zendman AJ, Cornelissen IM, Weidle UH,
Ruiter DJ and van Muijen GN: CTp11, a novel member of the family of
human cancer/testis antigens. Cancer Res. 59:6223–6229.
1999.PubMed/NCBI
|
|
30
|
Conklin D, Holderman S, Whitmore TE,
Maurer M and Feldhaus AL: Molecular cloning, chromosome mapping and
characterization of UBQLN3 a testis-specific gene that contains a
ubiquitin-like domain. Gene. 249:91–98. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhao L, Mou DC, Peng JR, Huang L, Wu ZA
and Leng XS: Diagnostic value of cancer-testis antigen mRNA in
peripheral blood from hepatocellular carcinoma patients. World J
Gastroenterol. 16:4072–4078. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vijayaraghavan S, Liberty GA, Mohan J,
Winfrey VP, Olson GE and Carr DW: Isolation and molecular
characterization of AKAP110, a novel, sperm-specific protein kinase
A-anchoring protein. Mol Endocrinol. 13:705–717. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Carnegie GK, Means CK and Scott JD:
A-kinase anchoring proteins: from protein complexes to physiology
and disease. IUBMB Life. 61:394–406. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sharma S, Qian F, Keitz B, Driscoll D,
Scanlan MJ, Skipper J, Rodabaugh K, Lele S, Old LJ and Odunsi K:
A-kinase anchoring protein 3 messenger RNA expression correlates
with poor prognosis in epithelial ovarian cancer. Gynecol Oncol.
99:183–188. 2005. View Article : Google Scholar : PubMed/NCBI
|