1. Introduction
Gastric cancer (GC), despite declining to the fifth
most commonly diagnosed malignancy, remains the third most common
cause of cancer deaths worldwide and thus a significant global
cancer burden (1). Patients
typically present with disease that appears at an advanced stage
accompanied by extensive invasion and lymph node involvement.
Limited therapeutic methods, the development of drug resistance,
local recurrence and distant metastasis are all challenging the
prognosis of GC patients.
miRNAs
In recent years, we have seen significant progress
in understanding that miRNAs (miRs) act as major intrinsic factors
of gene expression. They have extended our knowledge of how
morphogenesis and differentiation are regulated in cellular
processes. miRs are single-stranded, 19–22 nucleotide long
molecules, and are evolutionarily conserved across species
(2,3). To date, as many as 2588 miRs encoded
by the human genome have been confirmed (4,5),
including the miR-1, miR-206 and miR-133 family, which is observed
playing a crucial role in myogenesis. miRs can be transcribed by
ribonuclease II (RNase II) or ribonuclease III (RNase III) into
primary miRs (pri-miRs), several kilobases in length, which are
cleaved by the RNase III enzyme Drosha into shorter hairpin
structures called precursor-miRs (pre-miRs) that are 70–100 bases
long (6). Pre-miRs have a short
stem and a two-nucleotide 3′ overhang which is recognised by the
nuclear transport receptor exportin 5 (EXP5), and exported from the
nucleus to the cytoplasm by Ran-GTP- and EXP5-dependent mechanisms
(7,8). In the cytoplasm, another RNase III
enzyme, Dicer, further processes the pre-miR into a miR-miR* duplex
approximately 22 bases long (9).
The double-stranded RNA duplex is then loaded into the Argonaute
(AGO) protein and further processed, generally, causing the miR* to
be expelled, which results in a functional RNA-induced silencing
complex (RISC), which can base pair to a target mRNA, mostly
through seed-matched sites located in 3′ untranslated regions
(3′-UTR) of the mRNA, and induce its degradation or silencing
(10–13). miRs have numerous high and low
affinity targets, averaging 300 conserved targets per miRs family
(11). Although the effects of an
individual miR on a specific mRNA target may be relatively modest,
the combined effects of a miR on multiple targets functioning
within a common pathway can be synergistic, and make fine-scale
adjustments on cell proliferation, motility, apoptosis and
cell-fate decisions (14–17).
2. miRs in GC
miRs exhibit varied expression patterns, from
uniform during development to relatively stage-specific and/or
tissue specific (2,3), which suggests miRs profiling could
distinguish cancer from normal tissues or between different cancers
(18–21). miRs are now increasingly being
examined by miRs microarray and bioinformatics to analyze their
correlation with progression and prognosis of GC.
High-throughput studies on miRs in GC
(Table I)
In 160-paired samples of non-tumour mucosa and
cancer, 22 microRNAs were upregulated and 13 were downregulated in
GC (22). Liu et al screened
miR expression in serum samples pooled from 20 patients and 20
controls by Solexa sequencing, and found 19 serum miRs were
markedly upregulated in the GC patients compared to the controls.
The qRT-PCR analysis further identified a profile of five serum
miRs as a biomarker for GC detection (23). Of note, Zhou et al using
qRT-PCR based Exiqon panel analysed a total of 33 miRs abnormally
expressed in GC patient serum, and identified a new five-miR
signature in the peripheral plasma which was supposed to serve as a
non-invasive biomarker in detection of GC (24). In the study of Lo et al, the
signature of aberrant miRs expression was established by screening
and analysis using bioinformatics in Taiwanese patients (25). Kim et al further identified
upregulated miRs associated with chemosensitivity (26). Using both frozen tissue samples and
fresh blood samples, Yan et al confirmed 7 upregulated and 5
down-regulated miRs that could be used to discriminate between GC
with and without recurrence (27).
Yu et al analyzed the global miR expression
profiles of 9 gastric cancer cell lines and 6 normal gastric mucosa
lines using miR microarrays. Seventeen miRs were upregulated in
gastric cancer cell lines and 146 miRs were downregulated compared
to normal gastric mucosa. To validate the micro-array findings,
qPCR was performed on 9 gastric cancer cell lines, 6 normal gastric
mucosa, and a further 40 gastric cancer tissues and matched
adjacent non-cancerous tissues for 41 candidate miRs. The
expression levels of the 41 miRNAs using qPCR showed a great deal
of variation when compared to microarray data, indicating the high
heterogeneity of cancer tissues relative to cancer cell lines
(28). Chen et al
demonstrated that, apart from the commonly altered miRs, GC also
has a specific miR expression pattern different from oesophageal
adenocarcinoma (20). Among these
studies, miR-1, miR-133 and miR-206 are consistently deregulated
(18–20,22–27,29,30)
and associate with histology pattern (22), tumour stage (23), chemosensitivity (26) and progression (23). In the molecular classification of GC
set by The Cancer Genome Atlas (TCGA) datasets, miRs from 295 GC
tissues and 33 adjacent non-malignant samples were sequenced, and
according to the heatmap analysis, the miR-1, -133 and -206 family
was among the most commonly and significantly downregulated miRs
(31). In the remainder of this
review, we will discuss the functional role of the miR-1, -133 and
-206 family with particular emphasis on GC. We will also discuss
the expression and regulation of the miR-1, -133 and -206 family in
GC, and their potential roles in clinical application.
miR-1, -133 and -206 family
The miR-1, -133 and -206 family was initially
discovered through their role in the direction of the development
of mammalian skeletal and cardiac muscles. Recent studies on cancer
illustrate their deregulation in cancer development, in which
typically they function as tumour suppressors, with some evidence
of their oncogenic role in different cell environments (35,36).
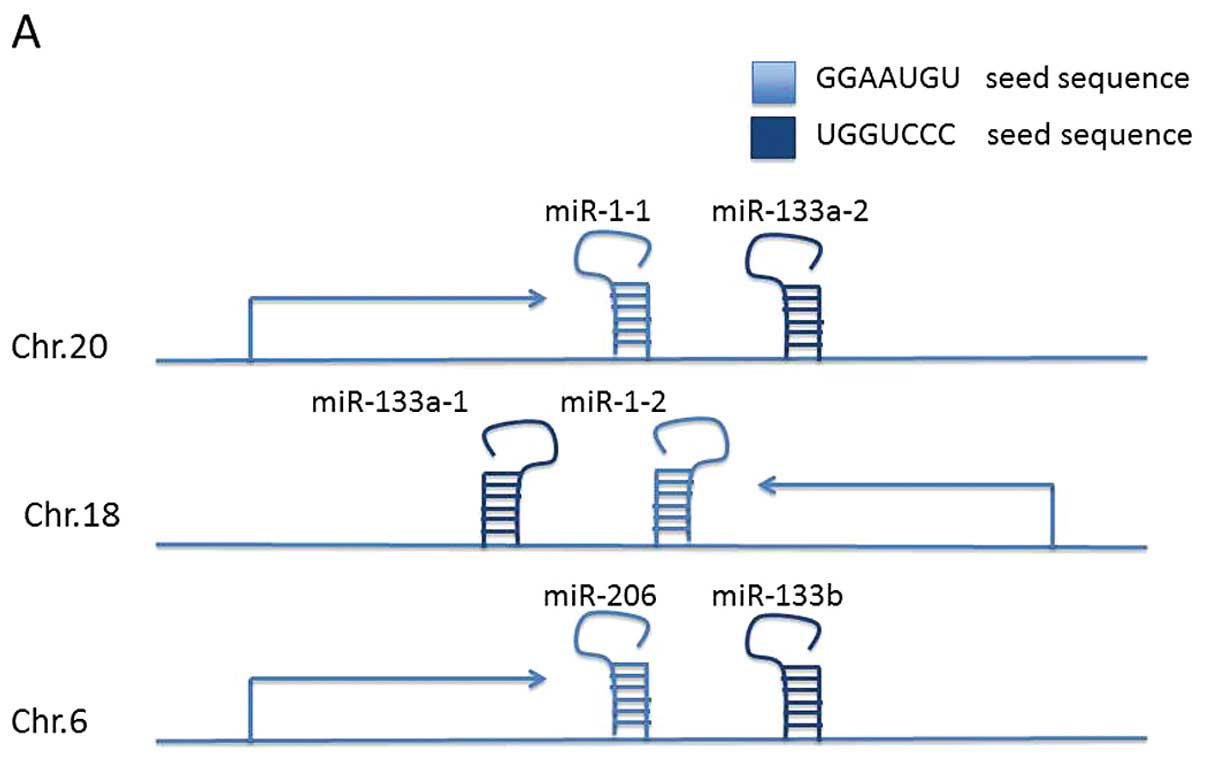
The miR-1, -133 and -206 family is located at three
different loci at chromosomes 20q13.33 (miR-1-1/miR-133a-2),
18q11.2 (miR-1-2/miR-133a-1) and 6p12.2 (miR-206/miR-133b)
(Fig. 1A) (36). The two mature miR-1 isomers have
identical sequences, as do the two miR-133a isomers. The mature
miR-133 isomers (A and B) are also highly similar, differing only
at the 3′-terminal base, with miR-133a-1/2 terminating G-3′ and
miR-133b with A-3′, respectively (37). The miR-206 sequence differs from the
miR-1 sequence by four nucleotides. Due to their close locations at
distinct loci, miR-1/133a, miR-206/133b are constituted as
clustered miRNAs. Since the seed sequence is the determining base
sequence that decides which mRNA is targeted and degraded. miR-1-1,
miR-1-2, miR-206 (miR-1/206) and miR-133a-1, miR-133a-2 and
miR-133b (miR-133a/133b) are functionally classified into two
groups based on their seed sequence (Fig. 1B).
miR-1, -133 and -206 expression profiles
in GC
Changes in the expression of miR-1, -133 and -206
have been documented in various types of cancer, including cancer
of the lung (39–41), breast (42,43),
prostate (44–46), colon (47,48),
oesophagus (49,50) and hepatocarcinoma (51).
miR-1
Several studies have shown that miR-1 was
significantly downregulated in GC cells and tissue samples when
compared to the adjacent non-tumour tissues (20,26,52,53).
Through microarray, Kim et al examined miR-1 expression in
biopsy samples collected prior to chemotherapy, from 90 gastric
cancer patients who were treated with cisplatin/fluorouracil (CF)
and from 34 healthy volunteers, was downregulated in GC by more
than four-fold, the result was also confirmed by qPCR. Further
analysis of miRNA predictors for response to CF therapy showed that
miR-1 was one of 37 miRNAs unique to the chemosensitivity signature
(26). Chen et al applied 2
independent microarray platforms in 3 advanced gastric
adenocarcinomas (stages III and IV) and 3 non-tumour histologically
normal tissue samples, and also found miR-1 was among the most
significantly down-regulated miRs (20). Analyzing data from the TCGA
database, miR-1 was widely suppressed across EBV, Genome Stable
(GS) and Chromosome Instability (CIN) three different subtypes of
GC (EBV subtype typically with PIK3CA mutation, GS subtype
characterized by remarkable mutations of RHOA and CDH1, and CIN
subgroup featured with obvious TP53 mutation and RTK-RAS mutation).
Tsai et al demonstrated downregulation of miR-1 directly
modulated endothelin-1 (END1) expression in GC, and promoted cell
growth, metastasis, angiogenesis, and ultimately suppressed
apoptosis. Transfecting GC cell lines with miR-1 plasmids reduced
cell proliferation and clonogenic survival of GC cells (52). Han et al using immunoblotting
confirmed MET was a direct target for miR-1 in GC and transfection
of miR-1 mimics exhibited the same negative regulation on cell
proliferation and motility as silencing MET expression (53). Taken together, miR-1 was
downregulated in GC, and overexpressed miR-1 in GC cells can
inhibit cell growth, clonality and migration ability. However, its
correlation with clinicopathological characteristics is not yet
explained, and the molecular mechanism remains to be
elucidated.
miR-206
Unlike miR-1, few microarray studies showed
alteration in miR-206 expression in GC patients. Upregulation of
miR-206 differed from miR-1 and tended to serve as a
chemoresistance indicator (26).
However, when detected alone, repressed miR-206 expression in GC
tissues and GC cell lines are consistent in several studies
(54–58). Lin et al compared miR-206
expression in primary GC tissues with those in normal adjacent
mucosa from 30 patients, miR-206 expression was found to be
significantly decreased in 30 of the GC samples (54). Furthermore, Yang et al
validated this observation in a larger population which included 98
paired samples, and clinicopathological analysis revealed that
tumours with low miR-206 expression were more prone to having lymph
node metastasis (P=0.01), presence of venous invasion (P=0.008),
and hematogenous recurrence (P=0.01), and tended to occur in a
worse stage (P=0.03) than the tumours with a high miR-206
expression. GC patients with low miR-206 expression also had
shorter overall survival than those with a high miR-206 expression
(P=0.02). Multivariate analysis showed that miR-206 expression was
an independent prognostic factor for patients with GC as well,
which strongly suggested that the downregulation of miR-206 was
significantly linked with tumour progression and its potential role
served as a prognostic marker in gastric cancer (55). Correlation between downregu-lation
of miR-206 with lymph node metastasis, local invasion, and advanced
TNM staging was also found by Ren et al via qPCR detection.
In vitro and in vivo studies also demonstrated
miR-206 may mediate the anti-metastatic effect by targeting
metastasis regulatory genes STC2, HDAC4, KLF4, IGF1R, FRS2, SFRP1,
BCL2, BDNF, and K-ras, which were drastically down-regulated by
stable expression of exogenous miR-206 in GC cell lines (56). Zhang et al showed miR-206
expression in metastatic lesions was more decreased than those in
the corresponding primary tumour samples (57). Presented analyses proved that
miR-206 acted as a tumour suppressor in GC, and considering its
role in predicting prognosis of GC patients, strategies regarding
the regulation of miR-206 in GC treatment might be useful.
miR-133
Data mining in miRNomes across TCGA datasets showed
miR-133a and miR-133b as well as miR-1 were consistently
downregulated in GC (31). This
result was confirmed in GC cell lines and primary GC tissues by
both microarray and qPCR methods. The level of miR-133 in GC and
corresponding non-tumour tissues was detected in three different
groups of GC patients independently (59–61).
Results further revealed that miR-133 reduction is more likely to
perform worse in tumour size, invasion depth and peripheral organ
metastasis. Dysfunction of miR-133 was an independent prognosis
factor for overall survival (60,61).
Moreover, by luciferase assay, miR-133 was proved to target 3′UTR
of EGFR and HER-2, which may cut cell growth signals off, and
inhibits cell growth ultimately promoting apoptosis (59). These related research results of
miR-133 in GC as a whole have elucidated a novel mechanism for
oncogene inhibition by miRNA-mediated pathways and offer new
avenues for GC treatment.
Circulating miRs in GC patients
Although data has helped to form a better
understanding of the alteration of microRNAs in GC samples as well
as in patient serum, the consistency between tissues and blood
samples, the source of generation and storage of circulating miRs,
and their roles in the mechanism of GC still need further
confirmation.
Circulating microRNAs were previously reported to be
stored in exosomes and released from normal tissues or tissues
affected by diseases (62–64). Containing miRs, exosomes were
secreted into circulation and transferred to target cells in either
normal or pathologic conditions (65). Therefore, the finding of
tumour-derived miRs in plasma or serum could support the
application of circulating miRs in disease early-detection since
circulating miRs are protected from degradation by ribonucleases in
blood and thereby can be stably detected (64), and could serve as low-invasive
useful biomarkers for various cancers (66–68),
including GC (69,70). However, these researchers showed
inconsistent results on the use of miRs for GC detection (71).
Liu et al (23) identified a profile of five serum
microRNAs (miR-1, -20a, -27a, -34, and -423-5p) as biomarkers for
GC detection by Solexa sequencing. Huang et al collected 82
blood samples from patients who were diagnosed with metastatic or
recurrent GC before first-line chemotherapy. After performing
qRT-PCR assay, they demonstrated that patients with higher serum
miR-1 expression levels tended to have a higher rate of liver
metastasis. Patients with higher serum miR-1 expression levels also
showed a high potential of chemoresistance, with the partial
response rates of 11.1%, whilst those in the patients with low
miR-1 expression was 23.1% (P=0.048) (72). However, Cai et al analyzed 90
plasma samples from GC patients randomly divided into training and
testing groups, the results did not show any aberrant conditions
for miR-1 (73). Furthermore, in
the study of Liu et al (60), 305 cases of diagnosed gastric
adenocarcinoma from TCGA data set were enrolled to detect miR
biomarkers for GC diagnostic and prognostic purposes. The results
showed miR-133b, miR-133a-2, and miR-1-2 levels were significantly
negative related with race, tumour pathology, and tumour stage
(P<0.05). Therefore, the feasibility of using circulating
microRNAs for the early detection or for predicting chemotherapy
effect and prognosis of GC remains to be established.
3. miR-1, -133 and -206 are regulated by
both transcription and by epigenetic regulation
Studies have identified a number of transcription
factors that positively or negatively regulate miR-1, -133 and -206
expressions. Independent upstream enhancers have been identified
for each pair of genes, and these independent enhancers allow the
different isomer genes to be independently expressed under cell
specific regulation. MEF2 and EVI1 were reported as enhancers to
regulate mir-1-2/133-1-a expression (74,75).
Either KLF4 or AGO2 may serve as an enhancer acting on the promoter
zone of the miR-206 gene separately (76,77).
The myogenic transcription factors myogenin and myogenic
differentiation 1 (MyoD) (78,79),
as well as Carm1/Prmt4 (80), bind
to regions upstream of the miR-1 and miR-133 stem loop, thereby
providing a molecular explanation for the observed induction during
myogenesis. However, activation of HMOX1 (81) or NRF2 (82) signalling attenuates miR-1 and
miR-206 expression, promoting cellular proliferation and
tumorigenesis. Although the mTOR (83) and the ERK1/2 (84) signalling pathways negatively
regulate expression of miR-1 and miR-133 indirectly in myogenesis,
their functions in cancer need further investigation.
In recent years, epigenetic factors such as DNA
methylation and histone modifications have increasingly been linked
to regulation of the miR-1, -133 and -206 family. Datta et
al showed in 2008 that epigenetic drugs 5-azacytidine (DNA
hypomethylating agent) and/or trichostatin A (histone deacetylase
inhibitor) differentially regulated expression of a few miRs,
particularly miR-1-1, in hepatocellular carcinoma (HCC) cells. The
CGI spanning exon 1 and intron 1 of miR-1-1 was methylated in HCC
cell lines and in primary human HCCs but not in matching liver
tissues. The miR-1-1 gene was hypomethylated and activated in
DNMT1−/− HCT 116 cells but not in DNMT3B null cells, indicating a
key role for DNMT1 in its methylation (85). DNA methylation at the miR-1-1/133a-2
promoter correlated highly with invasive capacity of colorectal
carcinoma (CRC) cell lines and played a critical role in colorectal
cancer metastasis by silencing TAGLN2 (86). Tsai et al demonstrated that
DNA hypermethylation contributed to the silenced miR-1 expression
in GC cells (52).
Recently, adding 5-Azacytidine, histone methylation
inhibitor DZNep or histone deacetylation (HDAC) inhibitor SAHA in
GC cell culture respectively, Liu et al found that added
DZNep and SAHA treatment consistently increased the expression of
miR-133b/a-3p in GC cell lines. A ChIP assay further quantified the
histone epigenetic modification levels in genomic regulatory
regions of miR-133b and miR-133a-1. GC cell lines demonstrated
reduced levels of H3K4me3 and H3 acetylation in miR-133a-1 promoter
region, both linked to transcriptional activation; whereas levels
of H3K27me3, linked to transcriptional repression, in miR-133a-1
promoter region, were significantly upregulated in GC cell lines
(60).
In physiological muscle differentiation, miR-1, -133
and -206 influence a plethora of cellular cues, leading to cell
cycle arrest and terminal differentiation by silencing the
expression of Pax7, the early activator of myogenic commitment, or
by affecting histone deacetylase-4 (HDAC4) and DNA polymerase-α
(87). It is widely accepted that
inappropriate deacetylation by the HDAC family, including HDAC4, is
a mechanism leading to increase in growth rate and cellular
proliferation (88). However, HDAC4
playing out its DNA binding function is Mef2c- and Mef2d-dependent.
Notch3 is paradoxically upregulated during the early stages of
differentiation by an enhancer that requires both MyoD and
activated Notch1, while Notch3 itself strongly inhibits the
myogenic transcription factor Mef2c, which induces microRNAs miR-1
and miR-206 (89). Noteworthy,
Notch3 and HDAC4 have been confirmed as direct targets of miR-1 and
miR-206 (90–92). Singh et al found that loss of
NRF2 decreased the expression of the HDAC4, forced overexpression
of HDAC could repress expression of miR-1 and miR-206, which
function as a regulatory feedback loop that repressed HDAC4
expression (82). Moreover, Nasser
et al found that repressed miR-1 was also activated in lung
cancer cells upon treatment with a histone deacetylase inhibitor
(91). Although only a few studies
have shown the regulation of the miR-1, -133 and -206 family in
cancer, they do suggest these factors are executing a vital role in
maintaining levels of miR-1, -133 and -206 in cancer cells. In
summary, epigenetic control of miR expression, including the miR-1,
-133 and -206, is an important tool for the cells to acquire
correct fate decision. Transcription factors together with
epigenetic modulation demonstrate the complex network of factors
controlling the miR-1, -133 and -206 family. In addition, the
interactions of miR-1, -133 and -206 with their targets show large
amount of complexity of the reciprocal communication between the
miR-1, -133 and -206 family and their regulators and targets.
4. Target genes and pathways involved in the
miR-1, -133 and -206 family regulation in GC
Online databases, such as TargetScan, and miRWalk,
provide plausible targets of the miR-1, -133 and -206 family and
their involvement in variable effects in signal pathways. But these
computational target genes are known to show false-negative and
false-positive results when compared to those results from
laboratory techniques (93).
miRTarBase (94), a new database,
collected miRNA-target interactions that are validated
experimentally by reporter assay, western blot, microarray and
next-generation sequencing experiments. According to its
collections, the miR-1, -133 and -206 family may regulate several
pathways in cancer, such as DNA replication, cell cycle, cell
junction, p53 and VEGF signalling pathways. In GC, MET, which is a
major participant in regulation of cell growth, migration and
clonogenic survival, was twice confirmed to have a negative
correlation with both miR-1 (53)
and miR-206 (95), their direct
interactions elucidated by reporter assay were reported in lung
cancer (91) and rhabdomyosarcoma
(96). Zhang et al explained
downregulated miR-206 enhanced MET expression via upregulation PAX3
(57), and miR-206-PAX3-MET
signalling is critical to GC metastasis (57).
Another study proved miR-206 targeted cyclinD2
(CCND2) directly, downregulated miR-206 lead to increased CCND2
level, thus promoting cell growth and colony forming ability in GC
cells with a G0/G1 cell cycle arrest abolished. Gain of function
studies revealed that miR-206 reduced GC cell proliferation at
least partially through targeting CCND2 (97). Shi et al (58) detected miR-206 and CCND2 mRNA
expression levels by qRT-PCR in 220 match-pairs of GC and adjacent
non-cancerous tissues, and showed that the expression levels of
miR-206 and CCND2 mRNA were, respectively, reduced and markedly
elevated in GC tissues, when compared with the adjacent
non-cancerous tissues (both P<0.001). Notably, the expression
levels of miR-206 in GC tissues were negatively correlated with
those of CCND2 mRNA, significantly (r= −0.463, P<0.001). Further
analyses displayed that low miR-206 expression and high CCND2
expression, alone or in combination, were all significantly
associated with great depth of invasion, positive lymph node and
distant metastases, and advanced TNM stage of GC (all P<0.05).
The researchers also found that the overall survivals of the
patients with low miR-206 expression and high CCND2 expression
were, respectively, shorter than those with high miR-206 expression
and low CCND2 expression. In addition, miR-206-low/CCND2-high
expression was associated with a significantly worst overall
survival of all miR-206/CCND2 groups (P<0.001). Furthermore,
multivariate analysis identified miR-206 and/or CCND2 expression as
independent prognostic factors for overall survival in patients
with gastric cancer (58).
Cell division cycle 42 (CDC42), an important member
of the Ras homolog (Rho) family, also known as PAK activating
factor, is considered to be involved in regulating cell cycle
progression, migration, cell cytoskeleton organisation, and cell
differentiation. A study demonstrated that CDC42 was a direct and
functional target gene of miR-133. Downregulation of miR-133
contributed to elevated expression of CDC42, since a body of
evidence indicates that CDC42/PAK pathway plays an important role
in tumour growth, invasion and metastasis, as a target of miR-133,
overexpression of miR-133 downregulated CDC42 expression and PAK
activation, and inhibited cancer cell proliferation and migration
(61).
Restoration of miR-133b/a-3p expression suppressed
cell proliferation and promoted cell apoptosis as observed in the
study of Liu et al (60), to
explain this phenomenon, after exploring the putative target of
miR-133 using TargetScan database, Mcl-1 and Bcl-xL 3′-UTR were
assembled into the XbaI site of the pGL3-promoter construct,
luciferase assay confirmed that miR-133 could directly bind to
3′-UTR of Mcl-1 and Bcl-xL, and the knock-down of Mcl-1 and Bcl-xL
markedly suppresses tumour growth (60).
Taken together, these data imply that the miR-1,
-133 and -206 family functions as pleiotropic modulators of cell
proliferation, invasion and cell cycle arrest by targeting various
genes in GC. Based on computational analysis of the selective
maintenance or avoidance of miRNA complementary sites during
evolution and experimental identification of messages destabilised
or those preferentially associated with argonaute proteins in the
presence of a miRNA, large-scale approaches for studying the
regulatory effects of miRs have revealed important insights into
target recognition and function (98). However, only a few of these gene
expression changes have been explained by predicted direct binding
of the miR-1, -133 and -206 family to corresponding mRNAs,
suggesting that the majority of these proteomic effects may result
indirectly. Thereby, with the aid of 'in silico' data,
further studies of the miR-1, -133 and -206 family in GC are
warranted due to their potential ability in suppressing and
inhibiting cancer cells.
5. Conclusions
The miR-1, -133 and -206 family has been found to be
repressed by hypermethylation of promoter or negatively regulated
transcription factors in GC. Their deregulation results in general
change in gene expression buried in diverse pathways directly or
indirectly, and affect cell growth, migration, cell cycle arrest
and apoptosis to a variable extent. The miR-1, -133 and -206 family
may serve as biomarker for GC diagnosis, progression, prognosis and
potential therapeutic targets. By increasing our understanding of
the functional role of miR-1, -133 and -206, their roles in
suppressing tumorigenesis and metastasis have been evidenced,
however, more efforts are required to illuminate the mechanism
behind these effects before early detection, or therapy for
knocking-in or -out these miRs, are performed.
Acknowledgments
We thank Cancer Research Wales for supporting our
work. This project was also funded by Natural Science Foundation of
China (81374016) and Beijing Municipal Science and Technology
Commission (D141100000414002).
References
|
1
|
International Agency for Research on
Cancer: GLOBOCAN 2012: Estimated Cancer Incidence, Mortality and
Prevalence Worldwide in 2012. http://globocan.iarc.fr/Pages/fact_sheets_cancer.asp.
|
|
2
|
Ambros V: microRNAs: Tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar
|
|
3
|
Meltzer PS: Cancer genomics: Small RNAs
with big impacts. Nature. 435:745–746. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kozomara A and Griffiths-Jones S: miRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res. 42:D68–D73. 2014. View Article : Google Scholar :
|
|
5
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:6972015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S, et al: The nuclear RNase III
Drosha initiates microRNA processing. Nature. 425:415–419. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yi R, Qin Y, Macara IG and Cullen BR:
Exportin-5 mediates the nuclear export of pre-microRNAs and short
hairpin RNAs. Genes Dev. 17:3011–3016. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lund E, Güttinger S, Calado A, Dahlberg JE
and Kutay U: Nuclear export of microRNA precursors. Science.
303:95–98. 2004. View Article : Google Scholar
|
|
9
|
Zhang H, Kolb FA, Brondani V, Billy E and
Filipowicz W: Human Dicer preferentially cleaves dsRNAs at their
termini without a requirement for ATP. EMBO J. 21:5875–5885. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ameres SL and Zamore PD: Diversifying
microRNA sequence and function. Nat Rev Mol Cell Biol. 14:475–488.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ørom UA, Nielsen FC and Lund AH:
MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and
enhances their translation. Mol Cell. 30:460–471. 2008. View Article : Google Scholar
|
|
13
|
Tay Y, Zhang J, Thomson AM, Lim B and
Rigoutsos I: MicroRNAs to Nanog, Oct4 and Sox2 coding regions
modulate embryonic stem cell differentiation. Nature.
455:1124–1128. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim VN: MicroRNA biogenesis: Coordinated
cropping and dicing. Nat Rev Mol Cell Biol. 6:376–385. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chivukula RR and Mendell JT: Circular
reasoning: microRNAs and cell-cycle control. Trends Biochem Sci.
33:474–481. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lynam-Lennon N, Maher SG and Reynolds JV:
The roles of microRNA in cancer and apoptosis. Biol Rev Camb Philos
Soc. 84:55–71. 2009. View Article : Google Scholar
|
|
18
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li X, Zhang Y, Zhang Y, Ding J, Wu K and
Fan D: Survival prediction of gastric cancer by a seven-microRNA
signature. Gut. 59:579–585. 2010. View Article : Google Scholar
|
|
20
|
Chen Z, Saad R, Jia P, Peng D, Zhu S,
Washington MK, Zhao Z, Xu Z and El-Rifai W: Gastric adenocarcinoma
has a unique microRNA signature not present in esophageal
adenocarcinoma. Cancer. 119:1985–1993. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Moreira FC, Assumpção M, Hamoy IG, Darnet
S, Burbano R, Khayat A, Gonçalves AN, Alencar DO, Cruz A, Magalhães
L, et al: MiRNA expression profile for the human gastric antrum
region using ultra-deep sequencing. PLoS One. 9:e923002014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ueda T, Volinia S, Okumura H, Shimizu M,
Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al:
Relation between microRNA expression and progression and prognosis
of gastric cancer: A microRNA expression analysis. Lancet Oncol.
11:136–146. 2010. View Article : Google Scholar
|
|
23
|
Liu R, Zhang C, Hu Z, Li G, Wang C, Yang
C, Huang D, Chen X, Zhang H, Zhuang R, et al: A five-microRNA
signature identified from genome-wide serum microRNA expression
profiling serves as a fingerprint for gastric cancer diagnosis. Eur
J Cancer. 47:784–791. 2011. View Article : Google Scholar
|
|
24
|
Zhou X, Zhu W, Li H, Wen W, Cheng W, Wang
F, Wu Y, Qi L, Fan Y, Chen Y, et al: Diagnostic value of a plasma
microRNA signature in gastric cancer: A microRNA expression
analysis. Sci Rep. 5:112512015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lo SS, Hung PS, Chen JH, Tu HF, Fang WL,
Chen CY, Chen WT, Gong NR and Wu CW: Overexpression of miR-370 and
downregulation of its novel target TGFβ-RII contribute to the
progression of gastric carcinoma. Oncogene. 31:226–237. 2012.
View Article : Google Scholar
|
|
26
|
Kim CH, Kim HK, Rettig RL, Kim J, Lee ET,
Aprelikova O, Choi IJ, Munroe DJ and Green JE: miRNA signature
associated with outcome of gastric cancer patients following
chemotherapy. BMC Med Genomics. 4:792011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yan Z, Xiong Y, Xu W, Gao J, Cheng Y, Wang
Z, Chen F and Zheng G: Identification of hsa-miR-335 as a
prognostic signature in gastric cancer. PLoS One. 7:e400372012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yu BQ, Su LP, Li JF, Cai Q, Yan M, Chen
XH, Yu YY, Gu QL, Zhu ZG and Liu BY: microrna expression signature
of gastric cancer cells relative to normal gastric mucosa. Mol Med
Rep. 6:821–826. 2012.PubMed/NCBI
|
|
29
|
Li X, Zhang Y, Zhang H, Liu X, Gong T, Li
M, Sun L, Ji G, Shi Y, Han Z, et al: miRNA-223 promotes gastric
cancer invasion and metastasis by targeting tumor suppressor
EPB41L3. Mol Cancer Res. 9:824–833. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Darnet S, Moreira FC, Hamoy IG, Burbano R,
Khayat A, Cruz A, Magalhães L, Silva A, Santos S, Demachki S, et
al: High-throughput sequencing of miRNAs reveals a tissue signature
in gastric cancer and suggests novel potential biomarkers.
Bioinform Biol Insights. 9(Suppl 1): 1–8. 2015.PubMed/NCBI
|
|
31
|
Cancer Genome Atlas Research Network:
Comprehensive molecular characterization of gastric adenocarcinoma.
Nature. 513:202–209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu G, Qin XQ, Guo JJ, Li TY and Chen JH:
AKT/ERK activation is associated with gastric cancer cell
resistance to paclitaxel. Int J Clin Exp Pathol. 7:1449–1458.
2014.PubMed/NCBI
|
|
33
|
Pennarun B, Meijer A, de Vries EG,
Kleibeuker JH, Kruyt F and de Jong S: Playing the DISC: Turning on
TRAIL death receptor-mediated apoptosis in cancer. Biochim Biophys
Acta. 1805:123–140. 2010.
|
|
34
|
Tchernitsa O, Kasajima A, Schäfer R, Kuban
RJ, Ungethüm U, Györffy B, Neumann U, Simon E, Weichert W, Ebert
MP, et al: Systematic evaluation of the miRNA-ome and its
downstream effects on mRNA expression identifies gastric cancer
progression. J Pathol. 222:310–319. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sempere LF, Freemantle S, Pitha-Rowe I,
Moss E, Dmitrovsky E and Ambros V: Expression profiling of
mammalian microRNAs uncovers a subset of brain-expressed microRNAs
with possible roles in murine and human neuronal differentiation.
Genome Biol. 5:R132004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nohata N, Hanazawa T, Enokida H and Seki
N: microRNA-1/133a and microRNA-206/133b clusters: Dysregulation
and functional roles in human cancers. Oncotarget. 3:9–21.
2012.PubMed/NCBI
|
|
37
|
Mitchelson KR and Qin WY: Roles of the
canonical myomiRs miR-1, -133 and -206 in cell development and
disease. World J Biol Chem. 6:162–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hilmarsdottir B, Briem E, Bergthorsson JT,
Magnusson MK and Gudjonsson T: Functional Role of the microRNA-200
Family in Breast Morphogenesis and Neoplasia. Genes (Basel).
5:804–820. 2014.
|
|
39
|
Mataki H, Enokida H, Chiyomaru T, Mizuno
K, Matsushita R, Goto Y, Nishikawa R, Higashimoto I, Samukawa T,
Nakagawa M, et al: Downregulation of the microRNA-1/133a cluster
enhances cancer cell migration and invasion in lung-squamous cell
carcinoma via regulation of Coronin1C. J Hum Genet. 60:53–61. 2015.
View Article : Google Scholar
|
|
40
|
Liu L, Shao X, Gao W, Zhang Z, Liu P, Wang
R, Huang P, Yin Y and Shu Y: MicroRNA-133b inhibits the growth of
non-small-cell lung cancer by targeting the epidermal growth factor
receptor. FEBS J. 279:3800–3812. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sun C, Liu Z, Li S, Yang C, Xue R, Xi Y,
Wang L, Wang S, He Q, Huang J, et al: Down-regulation of c-Met and
Bcl2 by microRNA-206, activates apoptosis, and inhibits tumor cell
proliferation, migration and colony formation. Oncotarget.
6:25533–25574. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Beltran AS, Russo A, Lara H, Fan C,
Lizardi PM and Blancafort P: Suppression of breast tumor growth and
metastasis by an engineered transcription factor. PLoS One.
6:e245952011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ge X, Lyu P, Cao Z, Li J, Guo G, Xia W and
Gu Y: Overexpression of miR-206 suppresses glycolysis,
proliferation and migration in breast cancer cells via PFKFB3
targeting. Biochem Biophys Res Commun. 463:1115–1121. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chang YS, Chen WY, Yin JJ,
Sheppard-Tillman H, Huang J and Liu YN: EGF receptor promotes
prostate cancer bone metastasis by downregulating miR-1 and
activating TWIST1. Cancer Res. 75:3077–3086. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kojima S, Chiyomaru T, Kawakami K, Yoshino
H, Enokida H, Nohata N, Fuse M, Ichikawa T, Naya Y, Nakagawa M, et
al: Tumour suppressors miR-1 and miR-133a target the oncogenic
function of purine nucleoside phosphorylase (PNP) in prostate
cancer. Br J Cancer. 106:405–413. 2012. View Article : Google Scholar :
|
|
46
|
Tao J, Wu D, Xu B, Qian W, Li P, Lu Q, Yin
C and Zhang W: microRNA-133 inhibits cell proliferation, migration
and invasion in prostate cancer cells by targeting the epidermal
growth factor receptor. Oncol Rep. 27:1967–1975. 2012.PubMed/NCBI
|
|
47
|
Xu L, Zhang Y, Wang H, Zhang G, Ding Y and
Zhao L: Tumor suppressor miR-1 restrains epithelial-mesenchymal
transition and metastasis of colorectal carcinoma via the MAPK and
PI3K/AKT pathway. J Transl Med. 12:2442014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Oberg AL, French AJ, Sarver AL,
Subramanian S, Morlan BW, Riska SM, Borralho PM, Cunningham JM,
Boardman LA, Wang L, et al: miRNA expression in colon polyps
provides evidence for a multihit model of colon cancer. PLoS One.
6:e204652011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Du YY, Zhao LM, Chen L, Sang MX, Li J, Ma
M and Liu JF: The tumor-suppressive function of miR-1 by targeting
LASP1 and TAGLN2 in esophageal squamous cell carcinoma. J
Gastroenterol Hepatol. 31:384–393. 2016. View Article : Google Scholar
|
|
50
|
Fu HL, Wu P, Wang XF, Wang JG, Jiao F,
Song LL, Xie H, Wen XY, Shan HS, Du YX, et al: Altered miRNA
expression is associated with differentiation, invasion, and
metastasis of esophageal squamous cell carcinoma (ESCC) in patients
from Huaian, China. Cell Biochem Biophys. 67:657–668. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wei W, Hu Z, Fu H, Tie Y, Zhang H, Wu Y
and Zheng X: MicroRNA-1 and microRNA-499 downregulate the
expression of the ets1 proto-oncogene in HepG2 cells. Oncol Rep.
28:701–706. 2012.PubMed/NCBI
|
|
52
|
Tsai KW, Hu LY, Chen TW, Li SC, Ho MR, Yu
SY, Tu YT, Chen WS and Lam HC: Emerging role of microRNAs in
modulating endothelin-1 expression in gastric cancer. Oncol Rep.
33:485–493. 2015.
|
|
53
|
Han C, Zhou Y, An Q, Li F, Li D, Zhang X,
Yu Z, Zheng L, Duan Z and Kan Q: MicroRNA-1 (miR-1) inhibits
gastric cancer cell proliferation and migration by targeting MET.
Tumour Biol. 36:6715–6723. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lin YH, Park ZY, Lin D, Brahmbhatt AA, Rio
MC, Yates JR III and Klemke RL: Regulation of cell migration and
survival by focal adhesion targeting of Lasp-1. J Cell Biol.
165:421–432. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yang Q, Zhang C, Huang B, Li H, Zhang R,
Huang Y and Wang J: Downregulation of microRNA-206 is a potent
prognostic marker for patients with gastric cancer. Eur J
Gastroenterol Hepatol. 25:953–957. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ren J, Huang HJ, Gong Y, Yue S, Tang LM
and Cheng SY: MicroRNA-206 suppresses gastric cancer cell growth
and metastasis. Cell Biosci. 4:262014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang L, Xia L, Zhao L, Chen Z, Shang X,
Xin J, Liu M, Guo X, Wu K, Pan Y, et al: Activation of PAX3-MET
pathways due to miR-206 loss promotes gastric cancer metastasis.
Carcinogenesis. 36:390–399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Shi H, Han J, Yue S, Zhang T, Zhu W and
Zhang D: Prognostic significance of combined microRNA-206 and
CyclinD2 in gastric cancer patients after curative surgery: A
retrospective cohort study. Biomed Pharmacother. 71:210–215. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhang XT, Zhang Z, Xin YN, Ma XZ and Xuan
SY: Impairment of growth of gastric carcinoma by miR-133-mediated
Her-2 inhibition. Tumour Biol. 36:8925–8930. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Liu Y, Zhang X, Zhang Y, Hu Z, Yang D,
Wang C, Guo M and Cai Q: Identification of miRNomes in human
stomach and gastric carcinoma reveals miR-133b/a-3p as therapeutic
target for gastric cancer. Cancer Lett. 369:58–66. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Cheng Z, Liu F, Wang G, Li Y, Zhang H and
Li F: miR-133 is a key negative regulator of CDC42-PAK pathway in
gastric cancer. Cell Signal. 26:2667–2673. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Rechavi O, Erlich Y, Amram H, Flomenblit
L, Karginov FV, Goldstein I, Hannon GJ and Kloog Y: Cell
contact-dependent acquisition of cellular and viral nonautonomously
encoded small RNAs. Genes Dev. 23:1971–1979. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chitwood DH and Timmermans MC: Small RNAs
are on the move. Nature. 467:415–419. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zen K and Zhang CY: Circulating microRNAs:
A novel class of biomarkers to diagnose and monitor human cancers.
Med Res Rev. 32:326–348. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z,
Wang JF, Zhang Z, Lu S, Huang X, et al: Plasma microRNA panel to
diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin
Oncol. 29:4781–4788. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Schultz NA, Dehlendorff C, Jensen BV,
Bjerregaard JK, Nielsen KR, Bojesen SE, Calatayud D, Nielsen SE,
Yilmaz M, Holländer NH, et al: MicroRNA biomarkers in whole blood
for detection of pancreatic cancer. JAMA. 311:392–404. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Ng EK, Chong WW, Jin H, Lam EK, Shin VY,
Yu J, Poon TC, Ng SS and Sung JJ: Differential expression of
microRNAs in plasma of patients with colorectal cancer: A potential
marker for colorectal cancer screening. Gut. 58:1375–1381. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Tsujiura M, Ichikawa D, Komatsu S,
Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi
K, Fujiwara H, et al: Circulating microRNAs in plasma of patients
with gastric cancers. Br J Cancer. 102:1174–1179. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhu C, Ren C, Han J, Ding Y, Du J, Dai N,
Dai J, Ma H, Hu Z, Shen H, et al: A five-microRNA panel in plasma
was identified as potential biomarker for early detection of
gastric cancer. Br J Cancer. 110:2291–2299. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhu X, Lv M, Wang H and Guan W:
Identification of circulating microRNAs as novel potential
biomarkers for gastric cancer detection: A systematic review and
meta-analysis. Dig Dis Sci. 59:911–919. 2014. View Article : Google Scholar
|
|
72
|
Huang D, Wang H, Liu R, Li H, Ge S, Bai M,
Deng T, Yao G and Ba Y: miRNA27a is a biomarker for predicting
chemosensitivity and prognosis in metastatic or recurrent gastric
cancer. J Cell Biochem. 115:549–556. 2014. View Article : Google Scholar
|
|
73
|
Cai H, Yuan Y, Hao YF, Guo TK, Wei X and
Zhang YM: Plasma microRNAs serve as novel potential biomarkers for
early detection of gastric cancer. Med Oncol. 30:4522013.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Liu N, Williams AH, Kim Y, McAnally J,
Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R and
Olson EN: An intragenic MEF2-dependent enhancer directs
muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad
Sci USA. 104:20844–20849. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Gómez-Benito M, Conchillo A, García MA,
Vázquez I, Maicas M, Vicente C, Cristobal I, Marcotegui N,
García-Ortí L, Bandrés E, et al: EVI1 controls proliferation in
acute myeloid leukaemia through modulation of miR-1–2. Br J Cancer.
103:1292–1296. 2010. View Article : Google Scholar
|
|
76
|
Sharma SB, Lin CC, Farrugia MK, McLaughlin
SL, Ellis EJ, Brundage KM, Salkeni MA and Ruppert JM: MicroRNAs 206
and 21 cooperate to promote RAS-extracellular signal-regulated
kinase signaling by suppressing the translation of RASA1 and
SPRED1. Mol Cell Biol. 34:4143–4164. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Adams BD, Claffey KP and White BA:
Argonaute-2 expression is regulated by epidermal growth factor
receptor and mitogen-activated protein kinase signaling and
correlates with a transformed phenotype in breast cancer cells.
Endocrinology. 150:14–23. 2009. View Article : Google Scholar :
|
|
78
|
Rao PK, Kumar RM, Farkhondeh M,
Baskerville S and Lodish HF: Myogenic factors that regulate
expression of muscle-specific microRNAs. Proc Natl Acad Sci USA.
103:8721–8726. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Tan SB, Li J, Chen X, Zhang W, Zhang D,
Zhang C, Li D and Zhang Y: Small molecule inhibitor of myogenic
microRNAs leads to a discovery of miR-221/222-myoD-myomiRs
regulatory pathway. Chem Biol. 21:1265–1270. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Mallappa C, Hu YJ, Shamulailatpam P, Tae
S, Sif S and Imbalzano AN: The expression of myogenic microRNAs
indirectly requires protein arginine methyltransferase (Prmt)5 but
directly requires Prmt4. Nucleic Acids Res. 39:1243–1255. 2011.
View Article : Google Scholar :
|
|
81
|
Kozakowska M, Ciesla M, Stefanska A,
Skrzypek K, Was H, Jazwa A, Grochot-Przeczek A, Kotlinowski J,
Szymula A, Bartelik A, et al: Heme oxygenase-1 inhibits myoblast
differentiation by targeting myomirs. Antioxid Redox Signal.
16:113–127. 2012. View Article : Google Scholar :
|
|
82
|
Singh A, Happel C, Manna SK,
Acquaah-Mensah G, Carrerero J, Kumar S, Nasipuri P, Krausz KW,
Wakabayashi N, Dewi R, et al: Transcription factor NRF2 regulates
miR-1 and miR-206 to drive tumorigenesis. J Clin Invest.
123:2921–2934. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Sun Y, Ge Y, Drnevich J, Zhao Y, Band M
and Chen J: Mammalian target of rapamycin regulates miRNA-1 and
follistatin in skeletal myogenesis. J Cell Biol. 189:1157–1169.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Feng Y, Niu LL, Wei W, Zhang WY, Li XY,
Cao JH and Zhao SH: A feedback circuit between miR-133 and the
ERK1/2 pathway involving an exquisite mechanism for regulating
myoblast proliferation and differentiation. Cell Death Dis.
4:e9342013. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Datta J, Kutay H, Nasser MW, Nuovo GJ,
Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD, et
al: Methylation mediated silencing of MicroRNA-1 gene and its role
in hepato-cellular carcinogenesis. Cancer Res. 68:5049–5058. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Chen WS, Leung CM, Pan HW, Hu LY, Li SC,
Ho MR and Tsai KW: Silencing of miR-1-1 and miR-133a-2 cluster
expression by DNA hypermethylation in colorectal cancer. Oncol Rep.
28:1069–1076. 2012.PubMed/NCBI
|
|
87
|
Winbanks CE, Beyer C, Hagg A, Qian H,
Sepulveda PV and Gregorevic P: miR-206 represses hypertrophy of
myogenic cells but not muscle fibers via inhibition of HDAC4. PLoS
One. 8:e735892013. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wade PA: Transcriptional control at
regulatory checkpoints by histone deacetylases: Molecular
connections between cancer and chromatin. Hum Mol Genet.
10:693–698. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Gagan J, Dey BK, Layer R, Yan Z and Dutta
A: Notch3 and Mef2c proteins are mutually antagonistic via Mkp1
protein and miR-1/206 microRNAs in differentiating myoblasts. J
Biol Chem. 287:40360–40370. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Furukawa S, Kawasaki Y, Miyamoto M,
Hiyoshi M, Kitayama J and Akiyama T: The miR-1-NOTCH3-Asef pathway
is important for colorectal tumor cell migration. PLoS One.
8:e806092013. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Nasser MW, Datta J, Nuovo G, Kutay H,
Motiwala T, Majumder S, Wang B, Suster S, Jacob ST and Ghoshal K:
Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression
of tumorigenic property of lung cancer cells and their
sensitization to doxorubicin-induced apoptosis by miR-1. J Biol
Chem. 283:33394–33405. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Hudson RS, Yi M, Esposito D, Watkins SK,
Hurwitz AA, Yfantis HG, Lee DH, Borin JF, Naslund MJ, Alexander RB,
et al: MicroRNA-1 is a candidate tumor suppressor and prognostic
marker in human prostate cancer. Nucleic Acids Res. 40:3689–3703.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Alexiou P, Maragkakis M, Papadopoulos GL,
Reczko M and Hatzigeorgiou AG: Lost in translation: An assessment
and perspective for computational microRNA target identification.
Bioinformatics. 25:3049–3055. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Hsu SD, Tseng YT, Shrestha S, Lin YL,
Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al:
miRTarBase update 2014: An information resource for experimentally
validated miRNA-target interactions. Nucleic Acids Res. 42:D78–D85.
2014. View Article : Google Scholar :
|
|
95
|
Zheng Z, Yan D, Chen X, Huang H, Chen K,
Li G, Zhou L, Zheng D, Tu L and Dong XD: MicroRNA-206: Effective
Inhibition of Gastric Cancer Progression through the c-Met Pathway.
PLoS One. 10:e01287512015. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Yan D, Dong XE, Chen X, Wang L, Lu C, Wang
J, Qu J and Tu L: MicroRNA-1/206 targets c-Met and inhibits
rhabdo-myosarcoma development. J Biol Chem. 284:29596–29604. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zhang L, Liu X, Jin H, Guo X, Xia L, Chen
Z, Bai M, Liu J, Shang X, Wu K, et al: miR-206 inhibits gastric
cancer proliferation in part by repressing cyclinD2. Cancer Lett.
332:94–101. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Baek D, Villén J, Shin C, Camargo FD, Gygi
SP and Bartel DP: The impact of microRNAs on protein output.
Nature. 455:64–71. 2008. View Article : Google Scholar : PubMed/NCBI
|