Introduction
Gastric cancer (GC) has the fourth highest incidence
rate of human malignant diseases, and causes the second highest
number of cancer-related mortalities worldwide (1). Approximately one million new cases are
diagnosed each year (2,3), yet early diagnosis of GC in clinical
practice remains a challenge. Although gastrectomy remains the
mainstay treatment for GC, the prognosis for patients at an
advanced stage remains very poor (4,5). With a
more complete understanding of the functions of oncogenesis,
development and prognosis associated with GC, significant
improvements in diagnosis and treatment are possible.
microRNAs (miRNAs) are a category of small,
single-stranded endogenous RNA molecules that function as
post-transcriptional regulators of gene expression, regulating
>30% of the protein-coding genes in the human genome (6–8).
Increasing evidence indicates that miRNAs are involved in important
biological processes associated with apoptosis, proliferation,
differentiation, metastasis, angiogenesis and immune responses, the
disruption of which may be crucial to cancer initiation,
progression and treatment outcomes. miRNA has been demonstrated to
be associated with a number of cancer types, including GC, and
plays an important role in the disease (9–15).
miRNA-425-5p (miR-425-5p), which is mapped to human
chromosome 3, has been confirmed to have an abnormally high
expression in GC (16). In the
present study, the expression of miR-425-5p was analyzed in human
GC cell lines, and in vitro and in vivo experiments
were performed to investigate the associations with local tumor
invasion and distant spreading. The aim of the current study was to
explore the potential mechanism of miR-425-5p in GC.
Materials and methods
Cell lines and reagents
Human gastric adenocarcinoma cell lines, SGC-7901,
MKN-28, MKN-45 and BGC-823, and a human immortal gastric mucosal
epithelial cell line, GES-1, were cultured at the Fourth Affiliated
Hospital of Hebei Medical University (Shijiazhuang, China). A human
gastric adenocarcinoma cell line, AGS, was obtained from Shanghai
Institute of Biochemistry and Cell Biology (Shanghai, China). The
cell lines, with the exception of the AGS cells, were grown in
Dulbecco's modified Eagle's medium (Gibco Life Technologies,
Carlsbad, CA, USA) supplemented with 10% (vol/vol) fetal bovine
serum (FBS), 50 U/ml penicillin and 50 µg/ml streptomycin. The AGS
cells were grown in F-12K medium (Sigma-Aldrich, St. Louis, MO,
USA) supplemented with 10% (vol/vol) FBS, 50 U/ml penicillin and 50
µg/ml streptomycin. The cell lines were incubated at 37°C in a
humidified 5% (vol/vol) CO2 atmosphere. The present
study was approved by the Ethics Committee of the Fourth Affiliated
Hospital of Hebei Medical University.
Analysis of miRNA expression using
TaqMan reverse transcription-quantitative polymerase chain reaction
(RT-qPCR)
Total RNA was extracted from the cells using TRIzol®
reagent (Invitrogen Life Technologies, Carlsbad, CA, USA),
according to the manufacturer's instructions. The expression of
mature miRNA was assayed using a TaqMan miRNA assay (Applied
Biosystems Life Technologies, Foster City, CA, USA) specific for
hsa-miR-425-5p. Briefly, 10 ng total RNA was reverse transcribed to
cDNA with specific stem-loop reverse transcription primers (Applied
Biosystems Life Technologies). RT-qPCR was performed using an
Applied Biosystems 7900 Real-time PCR system and a TaqMan Universal
PCR Master Mix (Applied Biosystems Life Technologies). The
predesigned TaqMan® MicroRNA assays included the primer sets and
TaqMan® probe that were used. The thermal cycling conditions used
for PCR were as follows: 95°C for 10 min, followed by 40 cycles at
95°C for 15 sec and 60°C for 60 sec (17,18).
Small nuclear U6 single nucleotide RNA (Applied Biosystems Life
Technologies) was used as an internal control, and the relative
expression was calculated using the comparative threshold cycle
method.
Transient transfection and RNA
extraction
Oligonucleotides of hsa-miR-425-5p mimics,
miR-425-5p inhibitor and a negative control were purchased from
Ambion Life Technologies (Carlsbad, CA, USA). Transfection of
BGC-823 cells (which were selected due to their moderate expression
level of miR-425-5p and superior growth ability comparing with
other cell lines) with the oligonucleotides was performed using
Lipofectamine® 2000 (Invitrogen Life Technologies) at a final
concentration of 100 nM. Small molecule RNAs (≤200 nucleotides)
were extracted from the cell lines using a mirVanaTM miRNA kit
(Ambion Life Technologies), according to the manufacturer's
instructions.
Flow cytometry assay
At 48 h post-transfection with miR-425-5p mimics and
the negative control, BGC-823 cells were collected by
trypsinization and washed with phosphate-buffered saline (PBS). For
cell-cycle analysis, the cells were fixed with 70% ethanol and
stored overnight at 4°C. On the following day, the fixed cells were
washed with PBS, treated with 50 µg/ml RNase A (Sangon Biotech Co.,
Ltd., Shanghai, China) and stained with 50 µg/ml propidium iodide
for 30 min in the darkness. The stained cells were analyzed using
flow cytometry (FACS Calibur™; Becton Dickinson, Franklin Lakes,
NJ, USA).
Cell proliferation assay
At 24 h post-transfection with miR-425-5p mimics,
miR-425-5p inhibitor and the negative control, BGC-823 cells
(3×103 cells/well) were seeded into 96-well plates and
incubated at 37°C for 24 h. Cell proliferation was assessed by a
water-soluble tetrazolium salt (WST) assay using Cell Counting
Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto,
Japan). After incubation for 24 h, 20 µl CCK-8 reagent were added
to each well and incubated at 37°C for 2 h. Spectrometric
absorbance at 450 nm was measured using a multilabel counter
microplate reader (Safire; Tecan Austria GmbH, Grödig, Austria).
Experiments were performed in triplicate.
Soft agar colony formation assay
At 24 h post-transfection, BGC-823 cells were
resuspended in 0.5% soft agar in RPMI 1640 medium (Invitrogen Life
Technologies, Gaithersburg, MD, USA) containing 10% FBS, and
layered onto 0.6% solidified agar in RPMI 1640 medium containing
10% FBS in six-well plates (1×103 cells/well). The
plates were incubated for three weeks. Following staining with 0.2%
p-iodonitrotetrazolium violet (Sangon Biotech Co., Ltd.), the
colonies containing ≥50 cells were counted.
Haptotactic migration and Matrigel
chemoinvasion assays
A Transwell insert (24-well; pore size, 8 µm;
Corning, Inc., Corning, NY, USA) was used to determine the effects
of miR-425-5p on GC cell migration and invasion in vitro.
Briefly, transfected cells were starved overnight in serum-free
medium. A total of 3×104 cells in serum-free medium were
added to the upper chamber. The lower chamber was filled with 10%
FBS, which functioned as the chemoattractant. The reaction system
was incubated for 48 h for the migration assay and 72 h for the
invasion assay. For the invasion assay, the inserts had been
previously coated with extracellular matrix gel (BD Biosciences,
Bedford, MA, USA). On completion of the experiments, cells on the
upper surface of the membrane were removed and cells on the lower
surface were fixed and stained with 0.1% crystal violet (Sangon
Biotech Co., Ltd.). Five visual fields were randomly selected from
each insert and counted under a BX51 Olympus light microscope
(Olympus Corporation, Tokyo, Japan).
Metastasis assay in vivo
Female BALB/c mice weighing 15–16 g (Shanghai
Laboratory Animal Center of China, Shanghai, China) were randomly
divided into three groups. BGC-823 cells (1×106
cells/mouse) were stably transfected with miR-425-5p inhibitor,
wild type or the negative control and were injected into
4–5-week-old female BALB/C nude mice through the tail vein. The
mice were housed for six weeks following the injection and were
then euthanized by cervical dislocation. The lungs were dissected
and examined histologically. Briefly, formalin-fixed,
paraffin-embedded tissues were stained with hematoxylin and eosin.
Following staining, the tissues were observed under a light
microscope and the number of metastases was counted.
Statistical analysis
Results are expressed as the mean ± standard
deviation from three separate experiments. The statistical
significance of the data was analyzed using the Student's t-test
for independent samples, and statistical analyses were performed
using SPSS software (version 15.0; SPSS, Inc., Chicago, IL, USA). A
two-tailed value of P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of miR-425-5p is
upregulated in GC cell lines
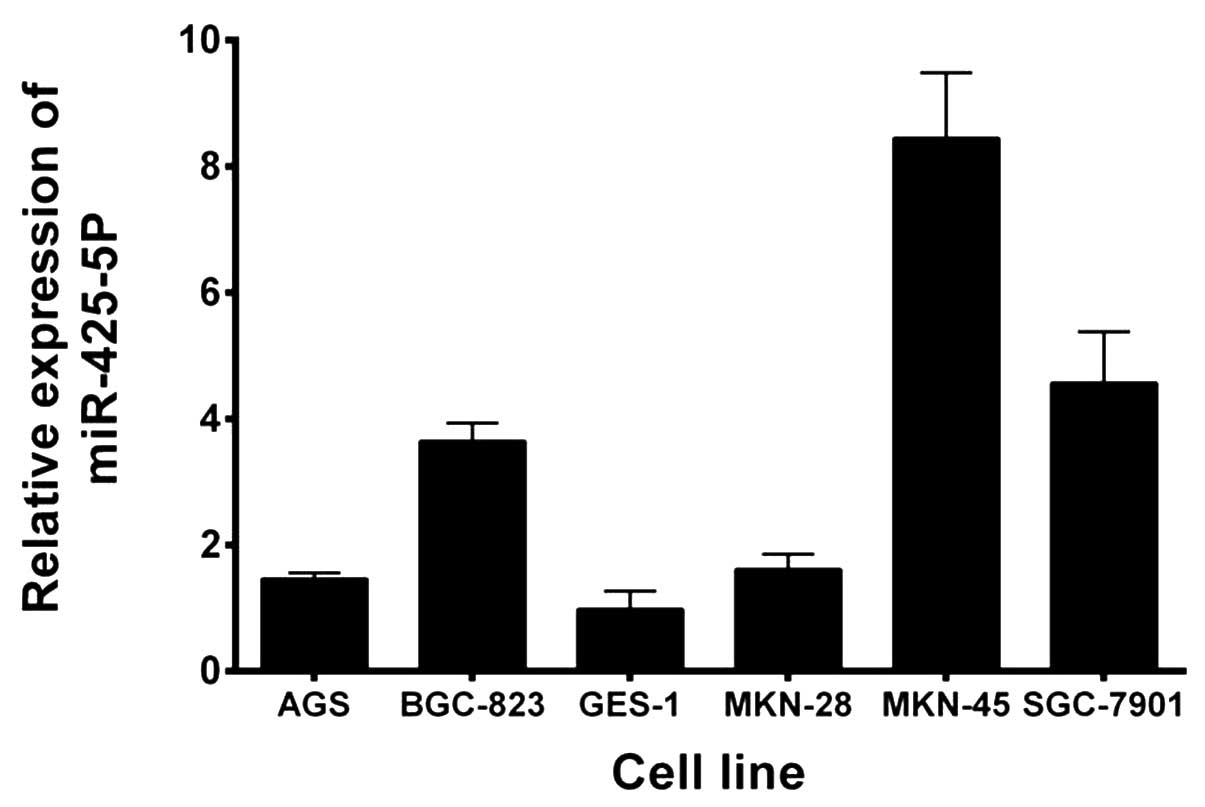
Expression levels of miR-425-5p were evaluated using
RT-qPCR in five GC cell lines and a GES-1 cell line. As shown in
Fig. 1, miR-425-5p expression was
significantly upregulated in all the GC cell lines when compared
with the GES-1 cell line. Based on these results and those of a
previous study (16), BGC-823 cells
were selected for further study.
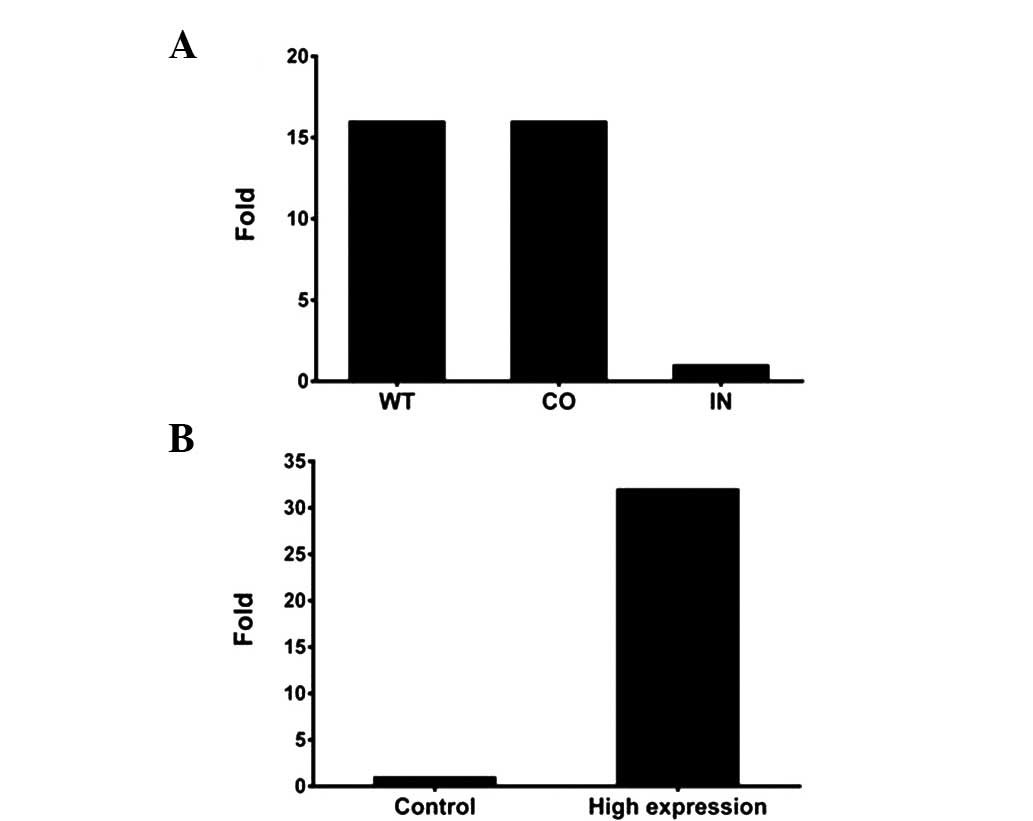
Assessment of the transient
transfection efficiency for miR-425-5p mimics and inhibitor
Transfection efficiency was monitored using RT-qPCR
analysis. As shown in Fig. 2,
BGC-823 cells transfected with the miR-425-5p inhibitor exhibited
significantly lower miR-425-5p expression levels when compared with
the wild-type (WT) cells or those transfected with a negative
control, while cells transfected with miR-425-5p mimics exhibited
significantly higher miRNA expression levels when compared with the
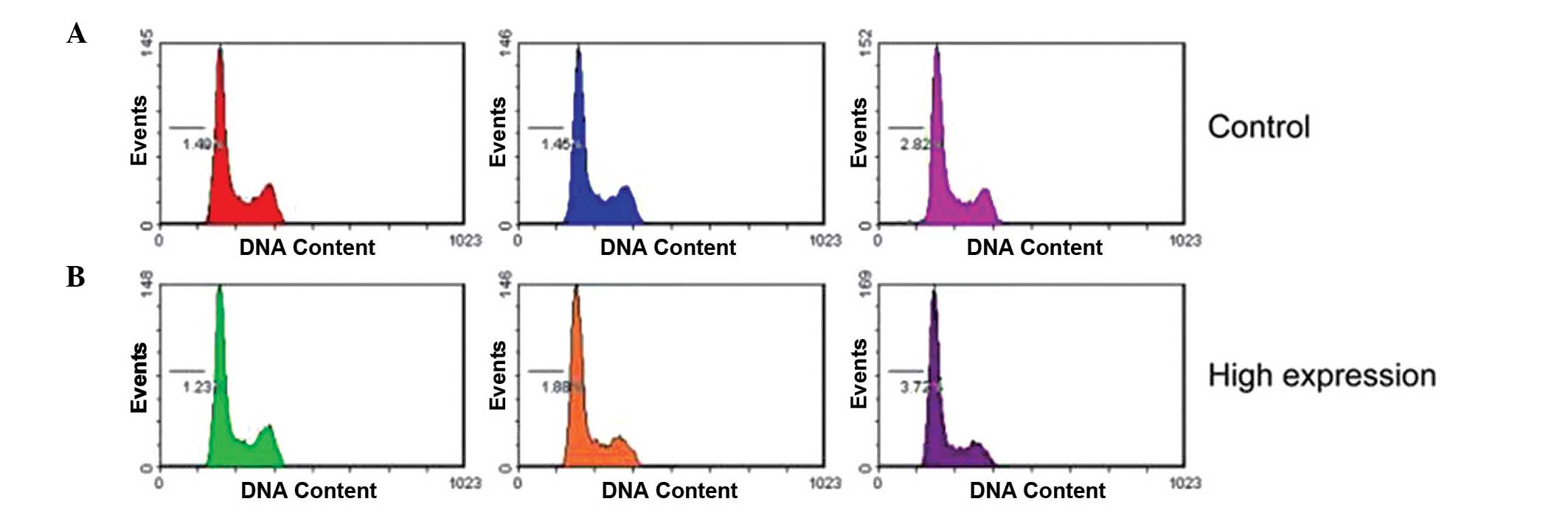
negative control cells. In addition, transfection with miR-425-5p
mimics did not affect the cell-cycle profile, as demonstrated by
flow cytometry (Fig. 3).
Downregulation of miR-425-5p
expression inhibits GC cell proliferation
Since miR-425-5p expression was shown to be
significantly upregulated in GC tissues and cell lines, the miRNA
may function as a tumor promoter. Thus, the effect that low
expression levels of miR-425-5p has on cell growth was examined in
BGC-823 cells. Synthetic miR-425-5p mimics, miR-425-5p inhibitor
and a negative control were transfected into BGC-823 cells, and
miR-425-5p expression was confirmed using RT-qPCR analysis.
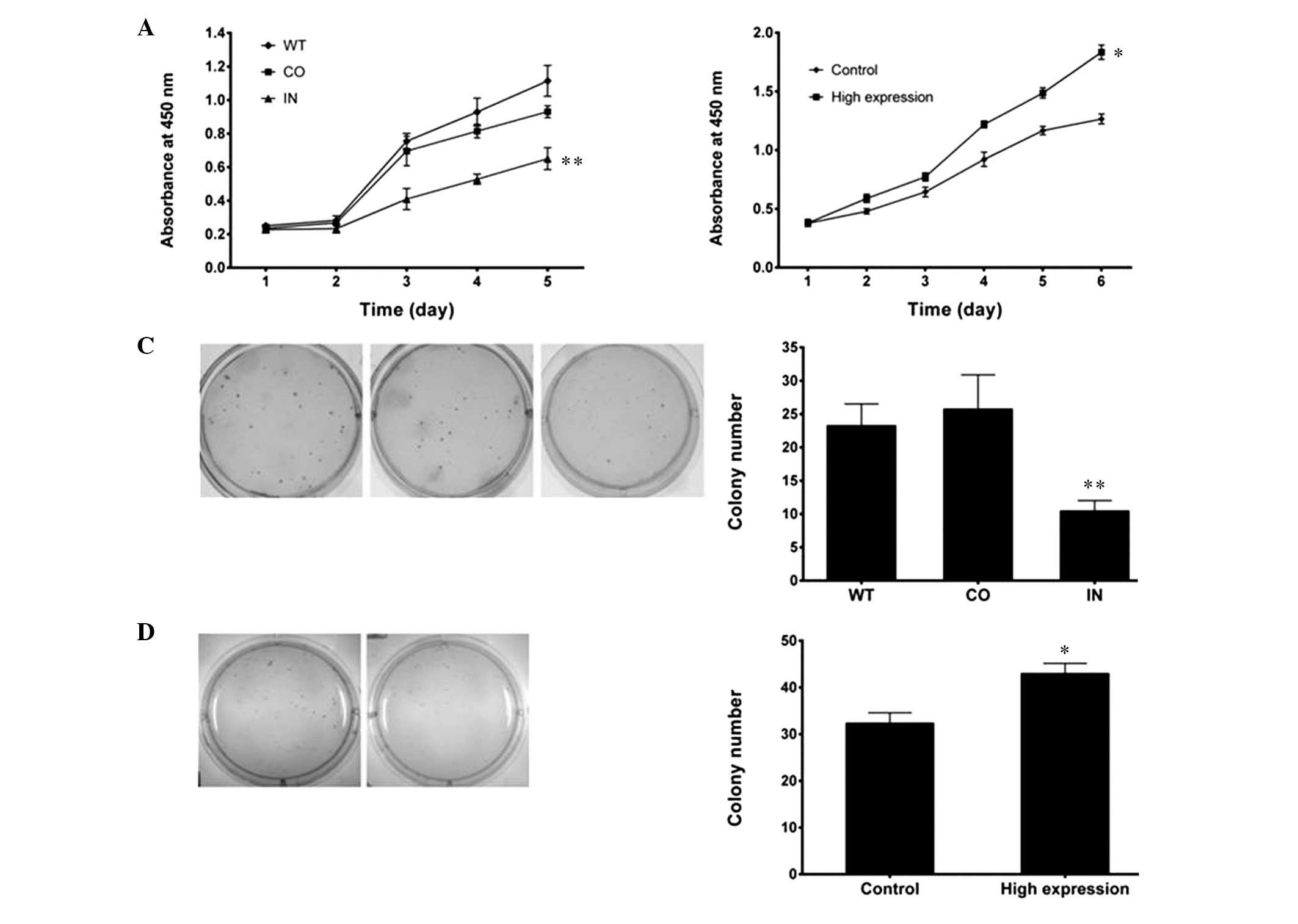
A WST cell-growth assay revealed significant
inhibition of cell growth in the miR-425-5p inhibitor
transfectants, as compared with the cells transfected with the
negative control and the WT cells (P<0.01; Fig. 4A). Significant promotion of cell
growth in the miR-425-5p mimic transfectants was shown, as compared
with the control (P<0.05; Fig.
4B). In order to further characterize the effects of miR-425-5p
on cell growth, a colony formation assay was performed in the
transfected cells. The number of colonies formed from BGC-823 cells
transfected with the miR-425-5p inhibitor was less than half of
that formed from the negative control and WT cells (P<0.01;
Fig. 4C). BGC-823 cells transfected
with the miR-425-5p mimics (high expression) displayed more
colonies than the negative control (P<0.05; Fig. 4D). These results demonstrated that
miR-425-5p promoted tumorigenicity in GC cells in vitro.
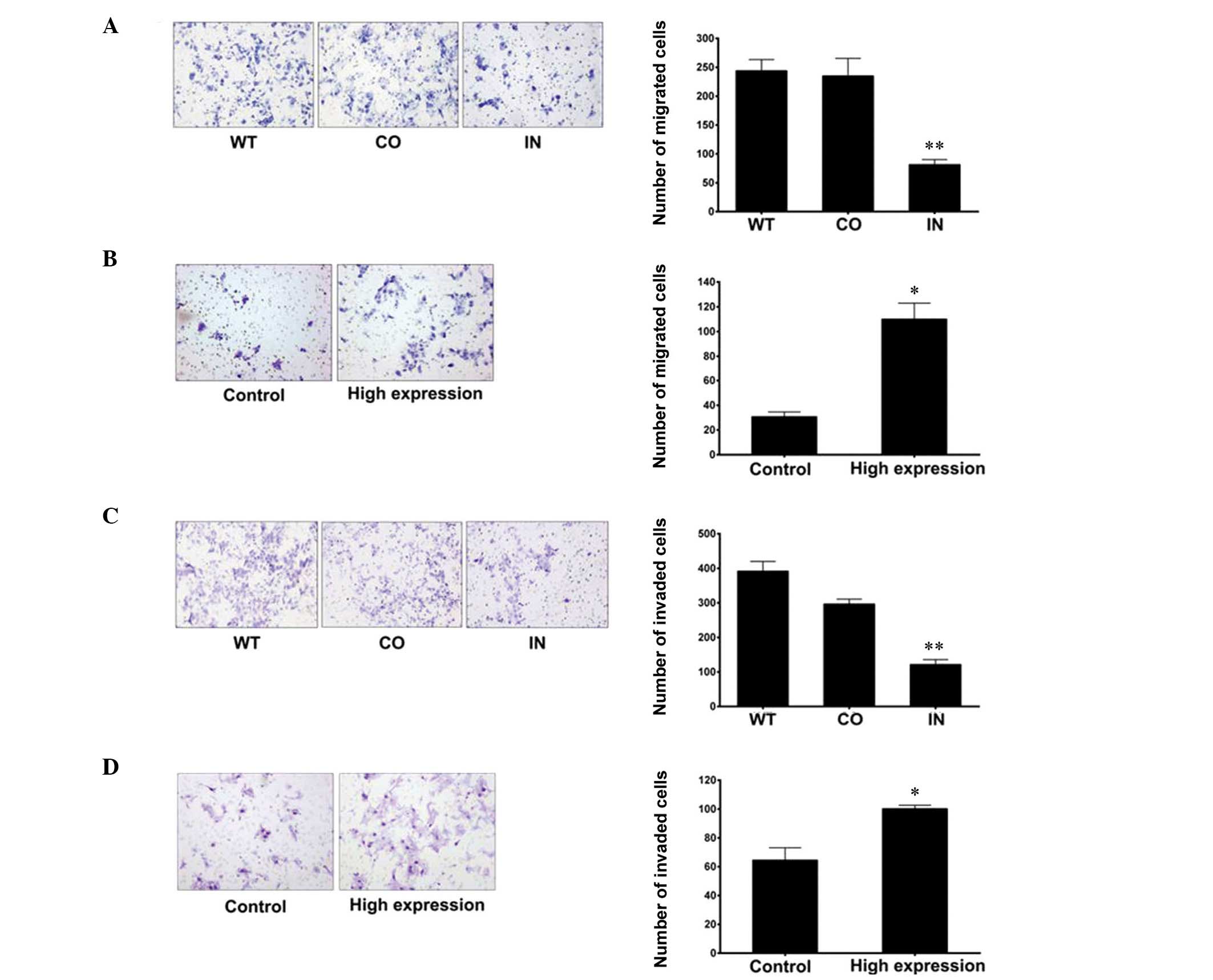
Downregulation of miR-425-5p
expression inhibits GC cell invasion and migration
Migration and invasion assays were performed to
investigate the mechanisms underlying the effects of miR-425-5p on
GC cell migration and invasion. When compared with the control
groups, the results demonstrated that downregulation of miR-425-5p
expression significantly inhibited the migration and invasion of
BGC-823 cells (Fig. 5).
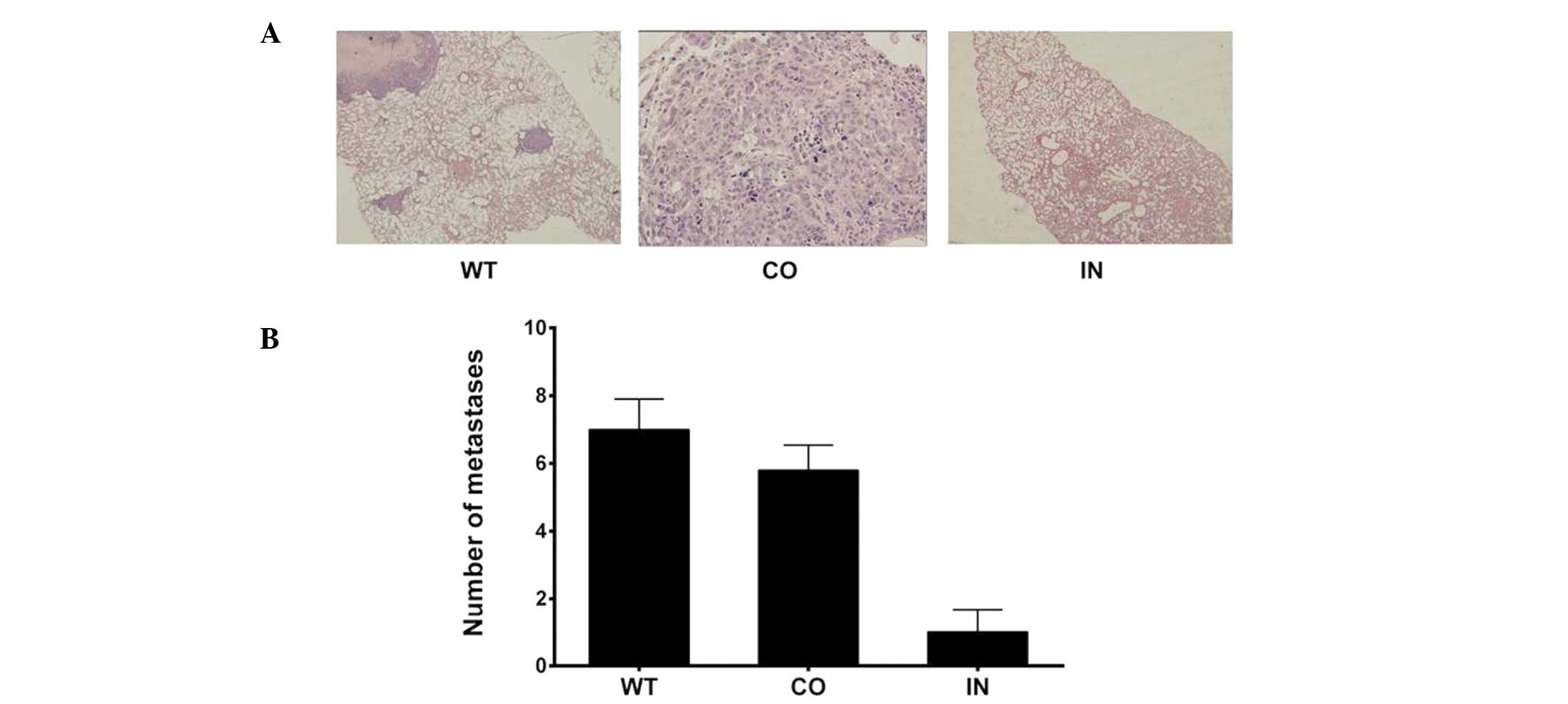
Downregulation of miR-425-5p
expression suppresses pulmonary metastasis in nude mice
Mice were euthanized six weeks after receiving an
injection of transfected cells. The number of lung metastatic nodes
per mouse was markedly lower and the nodes were smaller in the mice
injected with BGC-823 cells that had been transfected with the
miR-425-5p inhibitor, as compared with those in the control and WT
groups (P<0.05; Fig. 6).
Discussion
In recent years, numerous studies have shown that
miRNAs may serve as functional ‘oncogenes’ or ‘tumor suppressor
genes’ by regulating multiple cellular processes associated with
tumorigenesis and cancer progression (19–23).
Although intensive research effort has been conducted in this area,
only a limited number of case studies have identified specific
miRNAs that are involved in human tumor formation (24,25). In
the current study, miR-425-5p expression was shown to be
upregulated in GC cell lines. Furthermore, to the best of our
knowledge, the present study demonstrated for the first time that
miR-425-5p regulates invasion and migration in vitro and
in vivo.
In the present study, the results demonstrated that
miR-425-5p enhanced GC cell proliferation; however, flow cytometric
analysis revealed no effects on the cell cycle and apoptosis. This
is inconsistent with the findings of a recently published study
(26) that outlined the critical
role of nuclear factor-κB-dependent upregulation of miR-425-5p,
which represents a new pathway for the suppression of phosphatase
and tensin activation and the promotion of cell survival upon
interleukin-1β induction. However, different cell lines and
detection methods may cause identical or inconsistent results.
Di Leva et al (17) demonstrated a duality in the
biological role of miR-191/425 clusters in breast cancer. High
levels of estrogen-dependent miR-191/425 were demonstrated to
induce proliferation in estrogen receptor-α-positive cells by
suppressing strong tumor-suppressor genes, such as early growth
response protein 1. However, low levels of miR-191/425 clusters
were shown to be essential for the high expression of important
modulators, such as special AT-rich sequence binding protein 1,
cyclin D2 and fascin actin-bundling protein 1 (FSCN1), which confer
a proliferative advantage to aggressive breast cancer cells.
Upon recourse to bioinformatics technology, the
target gene of miR-425-5p can be detected. To increase the accuracy
of this prediction, genes that were predicted by at least three out
of four databases [DIANA LAB (Athens, Greece), Microcosm Targets
(Hinxton, UK), TargetScan (Whitehead Institute for Biomedical
Research, Cambridge, MA, USA), miRDB (Washington University School
of Medicine, St. Louis, MO, USA)] were selected as putative
targets. Possible target genes, including SSX2IP, MAP3K5, CLCN4 and
FSCN1, and their mechanisms are being further studied by the
authors of the present study. In addition, investigation into the
existence of two-way adjustment mechanisms and therapeutic targets
should be performed in future.
In conclusion, the present study demonstrated that
miR-425-5p expression levels are upregulated in GC cell lines. In
addition, miR-425-5p was shown to regulate invasion and migration
in vitro and in vivo. Therefore, the results indicate
that miR-425-5p is an important regulator in the metastasis of GC,
demonstrating the potential application of miR-425-5p in GC
therapy.
Acknowledgements
The authors thank Dr Xiaogang Tan for providing
technical support in the animal experiments.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kamangar F, Dores GM and Anderson WF:
Patterns of cancer incidence, mortality, and prevalence across five
continents: defining priorities to reduce cancer disparities in
different geographic regions of the world. J Clin Oncol.
24:2137–2150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Catalano V, Labianca R, Beretta GD, Gatta
G, de Braud F and Van Cutsem E: Gastric cancer. Crit Rev Oncol
Hematol. 71:127–164. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Parkin DM, Bray F, Ferlay and Pisani P:
Global cancer statistics 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mirnezami AH, Pickard K, Zhang L, Primrose
JN and Packham G: MicroRNAs: key players in carcinogenesis and
novel therapeutic targets. Eur J Surg Oncol. 35:339–347. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ruan K, Fang X and Ouyang G: MicroRNAs:
novel regulators in the hallmarks of human cancer. Cancer Lett.
285:116–126. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim K, Chadalapaka G, Pathi SS, et al:
Induction of the transcriptional repressor ZBTB4 in prostate cancer
cells by drug-induced targeting of microRNA-17-92/106b-25 clusters.
Mol Cancer Ther. 11:1852–1862. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Selcuklu SD, Donoghue MT, Rehmet K, et al:
MicroRNA-9 inhibition of cell proliferation and identification of
novel miR-9 targets by transcriptome profiling in breast cancer
cells. J Biol Chem. 287:29516–29528. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dey N, Das F, Ghosh-Choudhury N, et al:
microRNA-21 governs TORC1 activation in renal cancer cell
proliferation and invasion. PLoS One. 7:e373662012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He HC, Zhu JG, Chen XB, et al:
MicroRNA-23b downregulates peroxiredoxin III in human prostate
cancer. FEBS Lett. 586:2451–2458. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hur K, Toiyama Y, Takahashi M, et al:
MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT)
in human colorectal cancer metastasis. Gut. 62:1315–1326. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ueda T, Volinia S, Okumura H, et al:
Relation between microRNA expression and progression and prognosis
of gastric cancer: a microRNA expression analysis. Lancet Oncol.
11:136–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Di Leva G, Piovan C, Gasparini P, et al:
Estrogen mediated-activation of miR-191/425 cluster modulates
tumorigenicity of breast cancer cells depending on estrogen
receptor status. PLoS Genet. 9:e10033112013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Peng WZ, Ma R, Wang F, Yu J and Liu ZB:
Role of miR-191/425 cluster in tumorigenesis and diagnosis of
gastric cancer. Int J Mol Sci. 15:4031–4048. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sander S, Bullinger L, Klapproth K,
Fiedler K, et al: MYC stimulates EZH2 expression by repression of
its negative regulator miR-26a. Blood. 112:4202–4212. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jazbutyte V and Thum T: MicroRNA-21: from
cancer to cardiovascular disease. Current Drug Targets. 11:926–935.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Uziel T, Karginov FV, Xie S, Parker JS, et
al: The miR-17∼92 cluster collaborates with the Sonic Hedgehog
pathway in medulloblastoma. Proc Natl Acad Sci USA. 106:2812–2817.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bryant RJ, Pawlowski T, Catto JW, Marsden
G, et al: Changes in circulating microRNA levels associated with
prostate cancer. Br J Cancer. 106:768–774. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang M, Gu H, Qian H, Zhu W, et al:
miR-17-5p/20a are important markers for gastric cancer and murine
double minute 2 participates in their functional regulation. Eur J
Cancer. 49:2010–2021. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mavrakis KJ, Wolfe AL, Oricchio E,
Palomero T, et al: Genome-wide RNA-mediated interference screen
identifies miR-19 targets in Notch-induced T-cell acute
lymphoblastic leukaemia. Nat Cell Biol. 12:372–379. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hsu KW, Wang AM, Ping YH, Huang KH, et al:
Downregulation of tumor suppressor MBP-1 by microRNA-363 in gastric
carcinogenesis. Carcinogenesis. 35:208–217. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma J, Liu J, Wang Z, et al:
NF-kappaB-dependent microRNA-425 upregulation promotes gastric
cancer cell growth by targeting PTEN upon IL-1beta induction. Mol
Cancer. 13:402014. View Article : Google Scholar : PubMed/NCBI
|