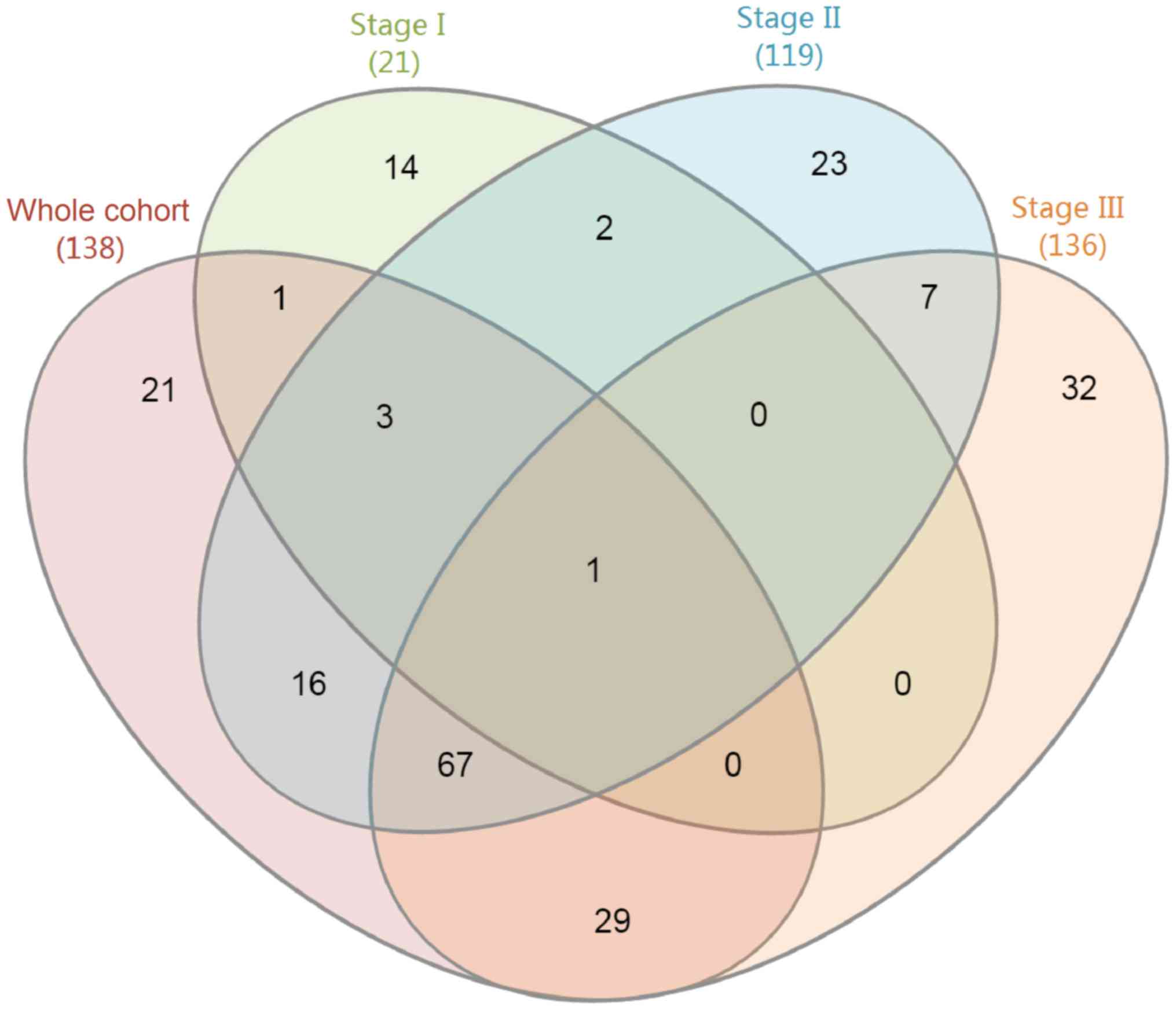
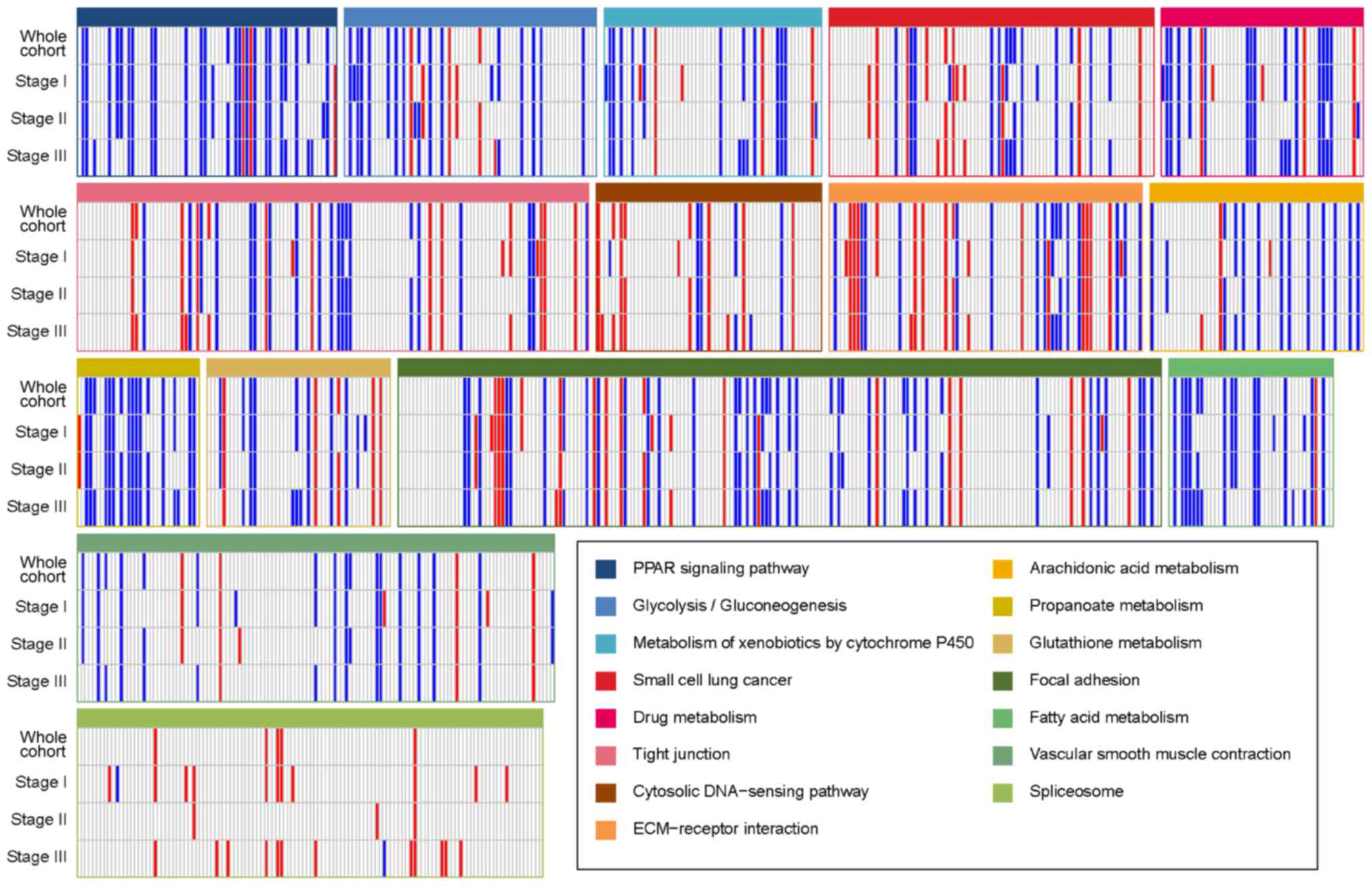
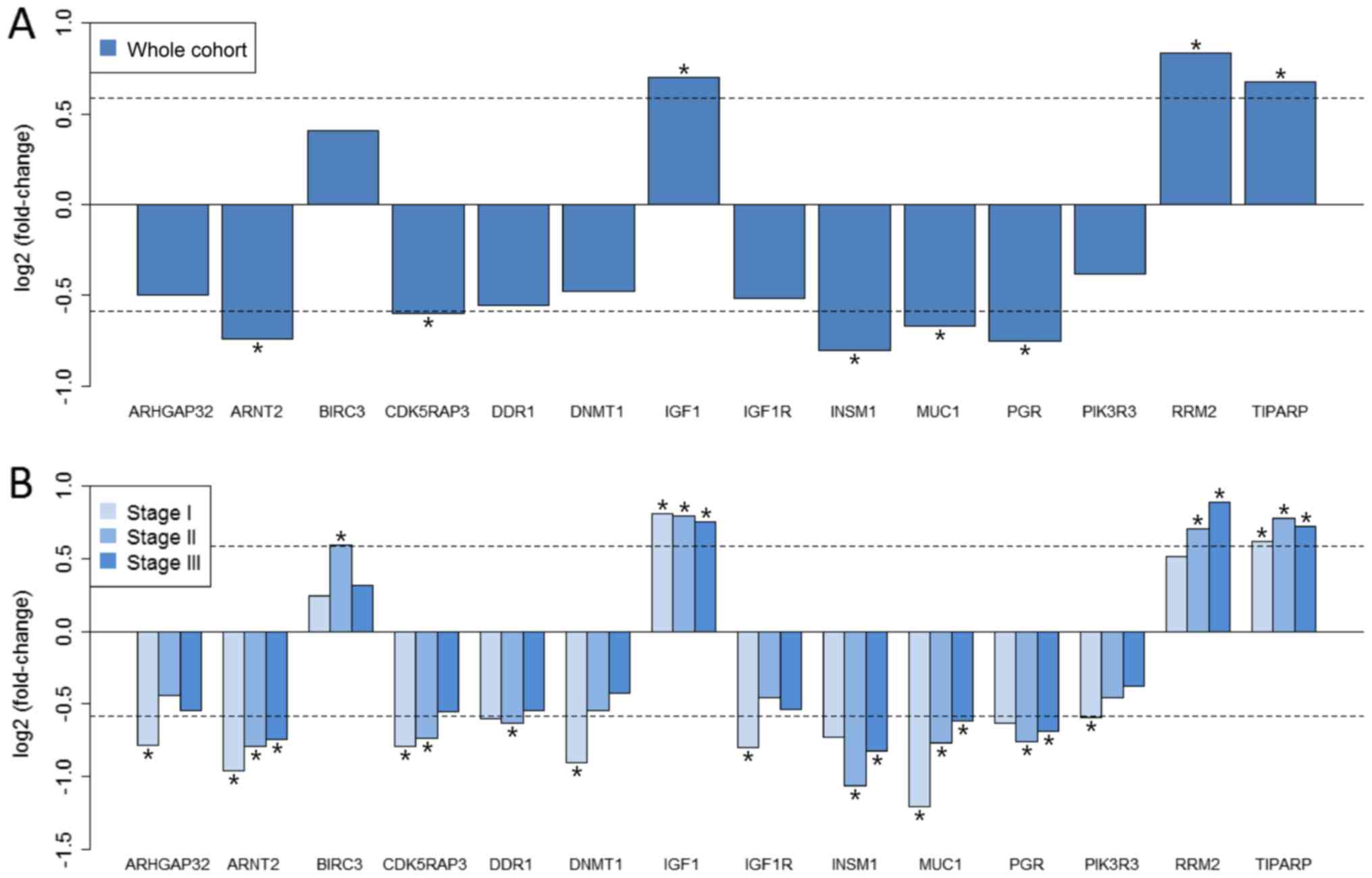
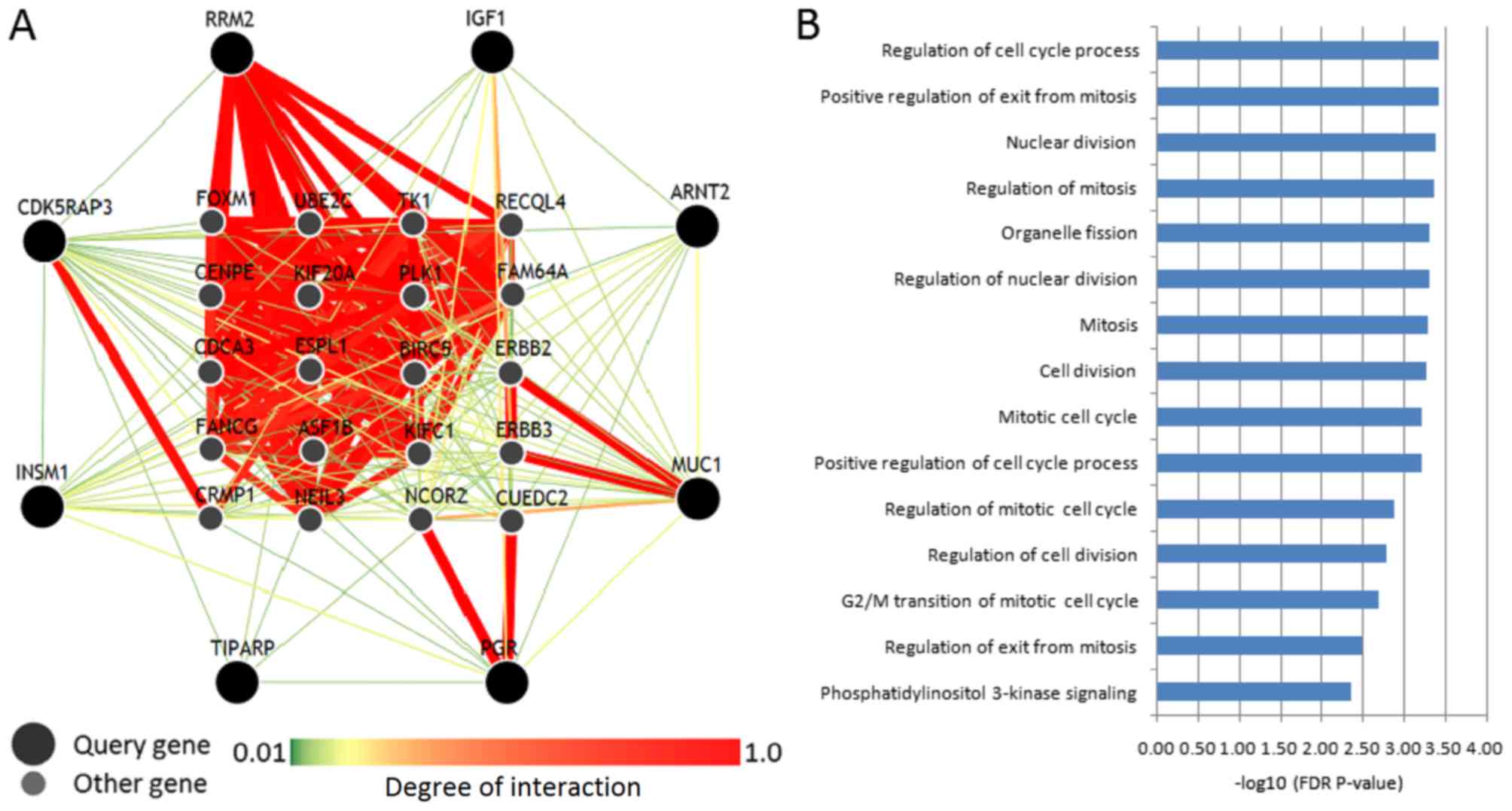
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Global Burden of Disease Cancer
Collaboration, . Fitzmaurice C, Allen C, Barber RM, Barregard L,
Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, et al:
Global, regional, and national cancer incidence, mortality, years
of life lost, years lived with disability, and disability-adjusted
life-years for 32 cancer groups, 1990 to 2015: A systematic
analysis for the global burden of disease study. JAMA Oncol.
3:524–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
O'Shaughnessy J: Extending survival with
chemotherapy in metastatic breast cancer. Oncologist. 10 Suppl
3:S20–S29. 2005. View Article : Google Scholar
|
|
5
|
Huang CS, Lin CH, Lu YS and Shen CY:
Unique features of breast cancer in Asian women-breast cancer in
Taiwan as an example. J Steroid Biochem Mol Biol. 118:300–303.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Reddy SM, Barcenas CH, Sinha AK, Hsu L,
Moulder SL, Tripathy D, Hortobagyi GN and Valero V: Long-term
survival outcomes of triple-receptor negative breast cancer
survivors who are disease free at 5 years and relationship with low
hormone receptor positivity. Br J Cancer. 118:17–23. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Giuliano AE, Connolly JL, Edge SB,
Mittendorf EA, Rugo HS, Solin LJ, Weaver DL, Winchester DJ and
Hortobagyi GN: Breast Cancer-Major changes in the American Joint
Committee on Cancer eighth edition cancer staging manual. CA Cancer
J Clin. 67:290–303. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Locke WJ and Clark SJ: Epigenome
remodelling in breast cancer: Insights from an early in vitro model
of carcinogenesis. Breast Cancer Res. 14:2152012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Harvell DM, Kim J, O'Brien J, Tan AC,
Borges VF, Schedin P, Jacobsen BM and Horwitz KB: Genomic
signatures of pregnancy-associated breast cancer epithelia and
stroma and their regulation by estrogens and progesterone. Horm
Cancer. 4:140–153. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mobasheri MB, Shirkoohi R, Zendehdel K,
Jahanzad I, Talebi S, Afsharpad M and Modarressi MH: Transcriptome
analysis of the cancer/testis genes, DAZ1, AURKC, and TEX101, in
breast tumors and six breast cancer cell lines. Tumour Biol.
36:8201–8206. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nogales-Cadenas R, Cai Y, Lin JR, Zhang Q,
Zhang W, Montagna C and Zhang ZD: MicroRNA expression and gene
regulation drive breast cancer progression and metastasis in PyMT
mice. Breast Cancer Res. 18:752016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lang JE, Scott JH, Wolf DM, Novak P, Punj
V, Magbanua MJ, Zhu W, Mineyev N, Haqq CM, Crothers JR, et al:
Expression profiling of circulating tumor cells in metastatic
breast cancer. Breast Cancer Res Treat. 149:121–131. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li WX, He K, Tang L, Dai SX, Li GH, Lv WW,
Guo YC, An SQ, Wu GY, Liu D, et al: Comprehensive tissue-specific
gene set enrichment analysis and transcription factor analysis of
breast cancer by integrating 14 gene expression datasets.
Oncotarget. 8:6775–6786. 2016.
|
|
14
|
Miyoshi K and Hennighausen L:
Beta-catenin: A transforming actor on many stages. Breast Cancer
Res. 5:63–68. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bartkova J, Horejsí Z, Koed K, Krämer A,
Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et
al: DNA damage response as a candidate anti-cancer barrier in early
human tumorigenesis. Nature. 434:864–870. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu H, Chen Y, Liang J, Shi B, Wu G, Zhang
Y, Wang D, Li R, Yi X, Zhang H, et al: Hypomethylation-linked
activation of PAX2 mediates tamoxifen-stimulated endometrial
carcinogenesis. Nature. 438:981–987. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:(Database Issue).
D991–D995. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pedraza V, Gomez-Capilla JA, Escaramis G,
Gomez C, Torné P, Rivera JM, Gil A, Araque P, Olea N, Estivill X
and Fárez-Vidal ME: Gene expression signatures in breast cancer
distinguish phenotype characteristics, histologic subtypes, and
tumor invasiveness. Cancer. 116:486–496. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Desmedt C, Giobbie-Hurder A, Neven P,
Paridaens R, Christiaens MR, Smeets A, Lallemand F, Haibe-Kains B,
Viale G, Gelber RD, et al: The Gene expression Grade Index: A
potential predictor of relapse for endocrine-treated breast cancer
patients in the BIG 1–98 trial. BMC Med Genomics. 2:402009.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lopez FJ, Cuadros M, Cano C, Concha A and
Blanco A: Biomedical application of fuzzy association rules for
identifying breast cancer biomarkers. Med Biol Eng Comput.
50:981–990. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Clarke C, Madden SF, Doolan P, Aherne ST,
Joyce H, O'Driscoll L, Gallagher WM, Hennessy BT, Moriarty M, Crown
J, et al: Correlating transcriptional networks to breast cancer
survival: A large-scale coexpression analysis. Carcinogenesis.
34:2300–2308. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Aswad L, Yenamandra SP, Ow GS, Grinchuk O,
Ivshina AV and Kuznetsov VA: Genome and transcriptome delineation
of two major oncogenic pathways governing invasive ductal breast
cancer development. Oncotarget. 6:36652–36674. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Carvalho BS and Irizarry RA: A framework
for oligonucleotide microarray preprocessing. Bioinformatics.
26:2363–2367. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li WX, Dai SX, Wang Q, Guo YC, Hong Y,
Zheng JJ, Liu JQ, Liu D, Li GH and Huang JF: Integrated analysis of
ischemic stroke datasets revealed sex and age difference in
anti-stroke targets. PeerJ. 4:e24702016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li WX, Dai SX, Liu JQ, Wang Q, Li GH and
Huang JF: Integrated analysis of Alzheimer's disease and
schizophrenia dataset revealed different expression pattern in
learning and memory. J Alzheimers Dis. 51:417–425. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Greene CS, Krishnan A, Wong AK, Ricciotti
E, Zelaya RA, Himmelstein DS, Zhang R, Hartmann BM, Zaslavsky E,
Sealfon SC, et al: Understanding multicellular function and disease
with human tissue-specific networks. Nat Genet. 47:569–576. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu Z, Zhan Y, Tu Y, Chen K, Liu Z and Wu
C: PDZ and LIM domain protein 1(PDLIM1)/CLP36 promotes breast
cancer cell migration, invasion and metastasis through interaction
with a-actinin. Oncogene. 34:1300–1311. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dong H, Claffey KP, Brocke S and Epstein
PM: Inhibition of breast cancer cell migration by activation of
cAMP signaling. Breast Cancer Res Treat. 152:17–28. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cacace A, Sboarina M, Vazeille T and
Sonveaux P: Glutamine activates STAT3 to control cancer cell
proliferation independently of glutamine metabolism. Oncogene.
36:2074–2084. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Apostoli AJ, Roche JM, Schneider MM,
SenGupta SK, Di Lena MA, Rubino RE, Peterson NT and Nicol CJ:
Opposing roles for mammary epithelial-specific PPARg signaling and
activation during breast tumour progression. Mol Cancer. 14:852015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
El-Rayes BF, Ali S, Heilbrun LK, Lababidi
S, Bouwman D, Visscher D and Philip PA: Cytochrome p450 and
glutathione transferase expression in human breast cancer. Clin
Cancer Res. 9:1705–1709. 2003.PubMed/NCBI
|
|
35
|
Leung T, Rajendran R, Singh S, Garva R,
Krstic-Demonacos M and Demonacos C: Cytochrome P450 2E1 (CYP2E1)
regulates the response to oxidative stress and migration of breast
cancer cells. Breast Cancer Res. 15:R1072013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Davydov DR, Davydova NY, Rodgers JT,
Rushmore TH and Jones JP: Toward a systems approach to the human
cytochrome P450 ensemble: Interactions between CYP2D6 and CYP2E1
and their functional consequences. Biochem J. 474:3523–3542. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li Y, Steppi A, Zhou Y, Mao F, Miller PC,
He MM, Zhao T, Sun Q and Zhang J: Tumoral expression of drug and
xenobiotic metabolizing enzymes in breast cancer patients of
different ethnicities with implications to personalized medicine.
Sci Rep. 7:47472017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Braak Ter B, Siezen CL, Lee JS, Rao P,
Voorhoeve C, Ruppin E, van der Laan JW and van de Water B:
Insulin-like growth factor 1 receptor activation promotes mammary
gland tumor development by increasing glycolysis and promoting
biomass production. Breast Cancer Res. 19:142017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shi J, Aronson KJ, Grundy A, Kobayashi LC,
Burstyn I, Schuetz JM, Lohrisch CA, SenGupta SK, Lai AS,
Brooks-Wilson A, et al: Polymorphisms of insulin-like growth factor
1 pathway genes and breast cancer risk. Front Oncol. 6:1362016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
MacPherson L, Tamblyn L, Rajendra S,
Bralha F, McPherson JP and Matthews J:
2,3,7,8-Tetrachlorodibenzo-p-dioxin poly(ADP-ribose) polymerase
(TiPARP, ARTD14) is a mono-ADP-ribosyltransferase and repressor of
aryl hydrocarbon receptor transactivation. Nucleic Acids Res.
41:1604–1621. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
McKay J, Tenet V, Franceschi S, Chabrier
A, Gheit T, Gaborieau V, Chopin S, Avogbe PH, Tommasino M, Ainouze
M, et al: Immuno-related polymorphisms and cervical cancer risk:
The IARC multicentric case-control study. PLoS One.
12:e01777752017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Maltepe E, Keith B, Arsham AM, Brorson JR
and Simon MC: The role of ARNT2 in tumor angiogenesis and the
neural response to hypoxia. Biochem Biophys Res Commun.
273:231–238. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Qin XY, Wei F, Yoshinaga J, Yonemoto J,
Tanokura M and Sone H: siRNA-mediated knockdown of aryl hydrocarbon
receptor nuclear translocator 2 affects hypoxia-inducible factor-1
regulatory signaling and metabolism in human breast cancer cells.
FEBS Lett. 585:3310–3315. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang E, Hu XF and Xing PX: Advances of
MUC1 as a target for breast cancer immunotherapy. Histol
Histopathol. 22:905–922. 2007.PubMed/NCBI
|
|
45
|
Varga Z, Lebeau A, Bu H, Hartmann A,
Penault-Llorca F, Guerini-Rocco E, Schraml P, Symmans F, Stoehr R,
Teng X, et al: An international reproducibility study validating
quantitative determination of ERBB2, ESR1, PGR, and MKI67 mRNA in
breast cancer using MammaTyper®. Breast Cancer Res.
19:552017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Putluri N, Maity S, Kommagani R, Creighton
CJ, Putluri V, Chen F, Nanda S, Bhowmik SK, Terunuma A, Dorsey T,
et al: Pathway-centric integrative analysis identifies RRM2 as a
prognostic marker in breast cancer associated with poor survival
and tamoxifen resistance. Neoplasia. 16:390–402. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shah KN, Wilson EA, Malla R, Elford HL and
Faridi JS: targeting ribonucleotide reductase M2 and NF-κB
activation with didox to circumvent tamoxifen resistance in breast
cancer. Mol Cancer Ther. 14:2411–2421. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hiraki M, Maeda T, Bouillez A, Alam M,
Tagde A, Hinohara K, Suzuki Y, Markert T, Miyo M, Komura K, et al:
MUC1-C activates BMI1 in human cancer cells. Oncogene.
36:2791–2801. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Raina D, Uchida Y, Kharbanda A, Rajabi H,
Panchamoorthy G, Jin C, Kharbanda S, Scaltriti M, Baselga J and
Kufe D: Targeting the MUC1-C oncoprotein downregulates HER2
activation and abrogates trastuzumab resistance in breast cancer
cells. Oncogene. 33:3422–3431. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ishizawar RC, Miyake T and Parsons SJ:
c-Src modulates ErbB2 and ErbB3 heterocomplex formation and
function. Oncogene. 26:3503–3510. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Dominguez-Brauer C, Thu KL, Mason JM,
Blaser H, Bray MR and Mak TW: Targeting mitosis in cancer: Emerging
strategies. Mol Cell. 60:524–536. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen L, Jenjaroenpun P, Pillai AM, Ivshina
AV, Ow GS, Efthimios M, Zhiqun T, Tan TZ, Lee SC, Rogers K, et al:
Transposon insertional mutagenesis in mice identifies human breast
cancer susceptibility genes and signatures for stratification. Proc
Natl Acad Sci USA. 114:E2215–E2224. 2017. View Article : Google Scholar : PubMed/NCBI
|