Introduction
Bone is an organ that has the potential for
persistent regeneration into adult life and is the only tissue that
undergoes constant remodeling throughout life (1). Efficacious bone regeneration could
influence the management of bone and musculoskeletal-associated
disorders (2,3). While most fractures recover
spontaneously, problems in this regenerative process may result in
a variety of diseases.
Mesenchymal stem cells (MSCs) are non-hematopoietic
stem cells, which have the ability to differentiate into tissues of
mesenchymal, as well as non-mesenchymal origin. For example, MSCs
are able to differentiate into osteoblastic, chondrogenic and
adipogenic lineages (4–6). In the last few years, MSCs have
attracted increasing attention for their potential use in
identifying the differentiation signaling pathways, facilitating
tissue engineering and in their roles as gene vectors and
immunomodulators in autoimmune diseases (7–9). In
addition to bone marrow, MSCs have been successfully isolated from
other tissues, such as adipose-derived mesenchymal stem cells
(ADSCs) (10,11). The osteogenesis of ADSCs is a complex
process, which is controlled by a series of endogenous and
environmental factors and signaling pathways (12,13).
However, its effect and the underlying mechanisms involved in the
process of bone regeneration remains unclear.
Although thousands of microRNAs (miRNAs) and mRNAs
have been identified and deposited in several public databases,
such as GENCODE (https://www.gencodegenes.org/), NONCODE (http://www.noncode.org/) and LNCipedia (https://lncipedia.org/), the functional
characterization of these is still in its initial stages. So far,
only a number of miRNAs and mRNAs have been functionally well
explored. It is hypothesized that functionally associated miRNAs
and mRNAs may often be involved in several physiological processes;
however, their involvement in the osteogenesis of ADSCs has not yet
been completely investigated.
Since miRNAs regulate gene expression via
post-transcriptional inhibition or degradation of mRNAs, the
potential interaction between differentially expressed miRNAs and
mRNAs in the ADSC-derived osteoblasts was analyzed. In the current
study, a global network using data from the National Center for
Biotechnology Information Gene Expression Omnibus (NCBI GEO,
http://www.ncbi.nlm.nih.gov/geo/)
identified numerous miRNAs and potential mRNA targets, which were
predicted to participate in the osteogenic differentiation of
ADSCs. Furthermore, dihydropyrimidinase like 3 (DPYSL3), a novel
key regulator of osteogenic differentiation, was identified, which
may present a potential therapeutic target in the management of
bone regeneration-associated diseases.
Materials and methods
Raw data
GEO is a public functional genomics data repository
supporting minimum information about a microarray experiment
(MIAME)-compliant data submissions. Tools are designed to query and
download gene expression profiles for further research. In the
present study, human miRNA expression data based on the
Agilent-031181 Unrestricted_Human_miRNA_V16.0_Microarray was
downloaded from NCBI GEO (GSE72429) and alterations in the miRNA
expression profile during osteogenic differentiation were analyzed
in human ADSCs using a microarray-based approach. In addition,
human mRNA data based on the [HuGene-1_1-st] Affymetrix Human Gene
1.1 ST Array was downloaded from NCBI GEO (GSE37329) with the
purpose of screening for the genes involved in the osteogenic
differentiation of ADSCs.
Screening of differentially expressed
miRNAs (DEMis) and mRNAs (DEMs)
Two sets of microarrays were used to identify DEMis
and DEMs following the osteogenesis of ADSCs. The Benjamini and
Hochberg (using the R package ‘limma’) (14) method was used to adjust the P-value
for the correction of false positive outputs, while logFC was used
to represent the fold change of down- or upregulated genes in ADSCs
prior to and following the induction of osteogenic differentiation.
The DEMis and DEMs were selected when P<0.05 and
|logFC|>1.5.
Prediction of target mRNAs of
DEMis
The target genes of the DEMis from GSE72429 were
predicted using TargetScan (http://www.targetscan.org), an online database for
predicting miRNA targets (15). The
predicted target genes were aligned with the DEMs to obtain an
intersection for further analysis.
Functional and pathway enrichment
analysis
To assess functional enrichment, gene ontology (GO)
Biological Processes term and Kyoto Encyclopedia of Genes and
Genomes (KEGG, http://www.genome.jp/kegg/) pathway analyses of mRNAs
in the miRNA-mRNA network were performed using the Database for
Annotation, Visualization, and Integration Discovery (DAVID,
http://david.ncifcrf.gov/). GO and KEGG
pathway enrichment analysis for DEMis was performed using the
Functional Enrichment analysis tool (FunRich, http://www.funrich.org/), a stand-alone software tool
used primarily for functional enrichment and interaction network
analysis of genes and proteins. These analyses provide a
comprehensive set of functional annotation tools for investigators
to understand the biological meaning behind large lists of genes
(16).
Protein-protein interaction (PPI)
network and module selection
The STRING database (http://string-db.org/) was used for the analysis of
PPIs, including direct and indirect associations (17), and Cytoscape (http://www.cytoscape.org/) was used for the visual
exploration of biomolecule interaction networks (18) The DEMs were mapped in STRING to
evaluate the PPI information and visualized using Cytoscape.
Cell culture, induction of
differentiation and transfection
Human ADSCs were purchased from ScienCell Research
Laboratories, Inc. (San Diego, CA, USA) and routinely maintained in
Dulbecco's modified Eagle's medium with high glucose (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (GE Healthcare Life Sciences, Logan, UT, USA) at 37°C
in a humidified atmosphere of 5% CO2. The culture medium
was refreshed every other day. For differentiation, the cells were
cultured in osteogenic medium consisting of standard growth medium
supplemented with 10 mM dexamethasone, 0.2 mM L-ascorbic acid, 10
mM β-glycerophosphate and 10 mM 1,25-dihydroxyvitamin D3
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Cells cultured in
a normal medium were used as a control. Cells were then seeded in
6-well plates at a density of 2×105 cells/ml per well
and transfection was subsequently performed using Lipofectamine
3000 (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. After 48 h of transfection, the cells
were collected and used for further experimentation. The short hair
RNA plasmids (pGPU6/sh-GFP, pGPU6/sh-DPYSL3-1 and
pGPU6/sh-DPYSL3-2) and overexpression lentivruses (LV/GFP and
LV/DPYSL) were chemically synthesized by Shanghai GenechemCo.,
Ltd., (Shanghai, China).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Tripure isolation reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was used for the extraction of total RNA from
cell-monolayers, according to the manufacturer's instructions. The
first cDNA strands were synthesized using oligodT primers and
Revoscript™ Reverse Transcription PreMix (Intron Biotechnology,
Inc., Seongnam, Korea). Analysis of the expressed mRNA was
performed using Brilliant II SYBR® Green QPCR Master Mix
(Agilent Technologies, Inc., Santa Clara, CA, USA) and a Light
Cycler Nano Machine (Roche Applied Science, Rotkreuz, Switzerland).
A 35 cycle-thermal program was conducted, consisting of
denaturation at 95°C for 15 sec, annealing at 90°C for 15 sec and
extension at 72°C for 15 sec. The primer pairs used for PCR were as
follows: DPYSL3, forward, 5′-CCTCGGCATAGATGGAACC-3′, and reverse,
5′-TCTGGGCAGTGCTGAAGGT-3′; runt-related transcription factor 2
(RUNX2), forward, 5′-GCCGGGAATGATGAGAACTA-3′, and reverse,
5′-GGACCGTCCACTGTCACTTT-3′; (Osterix), forward,
5′-GGCGTCCTCCCTGCTTGA-3′, and reverse, 5′-TGCTTTGCCCAGAGTTGTTG-3′;
alkaline phosphatase (ALP), forward, 5′-GACAAGAAGCCCTTCACTGC-3′,
and reverse, 5′-AGACTGCGCCTGGTAGTTGT-3′; GAPDH, forward,
5′-TGAACGGGAAGCTCACTGG-3′, and reverse, 5′-TCCACCACCCTGTTGCTGTA-3′.
The data were analyzed using the 2−ΔΔCq relative
expression method (19). Each
experiment was performed in triplicate.
Western blot analysis
Western blot analysis was performed as described
previously (20). Protein extracts
were prepared from ADSCs cells using RIPA buffer (Invitrogen;
Thermo Fisher Scientific. Inc.) and protein concentration was
determined using the BCA Protein Assay kit (Thermo Fisher
Scientific, Inc.). Samples were combined with 2X SDS loading
buffer, boiled for 10 min and protein (20 µg) was loaded onto a 10%
or 4-20% gradient SDS-PAGE gel. Proteins were transferred to a PVDF
membrane (EMD Millipore, Billerica, MA, USA) over 2 h at 350 mA,
and membranes were incubated in Odyssey Blocking Buffer (LI-COR
Biosciences, Lincoln, NE, USA) for 2 h at room temperature.
Following incubation with primary antibodies overnight at 4°C, the
blots were washed three times in TBS containing 0.1% Tween-20 for
15 min and then incubated with peroxidase- or IRDye-conjugated
secondary antibody (1:5,000; cat. no. 4418; Cell Signaling
Technology, Inc., Danvers, MA, USA) for 1 h in TBS, 0.1% Tween-20
at room temperature. Immunoreactivity was detected using an ECL
reagent with a LI-COR imaging system. Primary antibodies were as
follows: GAPDH (dilution, 1:1,000; cat. no. 8884; Cell Signaling
Technology, Inc.), DPYSL3 (1:100; cat. no. sc-100323; Santa Cruz
Biotechnology, Inc., Dallas, TX USA).
Alizarin red staining (ARS)
ARS was performed at 14 days following osteogenic
induction to detect the osteoblast calcification following the
indicted treatments according to the manufacturer's instructions.
Cells in 24-well plates were washed with PBS, fixed in 95% ethanol
for 10 min at room temperature, washed with distilled water, and
stained at room temperature for 30 min using alizarin red solution
1 g Tris and 0.1 g alizarin red (Bio Basic Inc., Markham, ON,
Canada) in 100 ml ultrapure water]. Following washing with
distilled water twice, the cells were photographed using an optical
microscope.
ALP activity
An ALP assay kit (Beyotime Institute of
Biotechnology, Hangzhou, China) was used to measure the ALP
activity, according to the manufacturer's instructions. Total
protein was extracted using radioimmunoprecipitation assay lysis
buffer (included in the ALP assay kit) and the protein
concentration was determined using a bicinchoninic acid assay. The
cell lysate and substrate (obtained from the ALP assay kit) were
added to a 96-well plate and incubated for 10 min at 37°C.
Following the addition of stop solution, the absorbance was
determined at 405 nm using an ELISA microplate reader.
Statistical analysis
Quantitative data were reported as the mean ±
standard deviation. Statistical analysis was performed using
one-way analysis of variance followed by a Bonferroni test for
multiple groups, or a Student's t-test for the analysis of
differences between two groups (using SPSS 13.0 software; SPSS,
Inc., Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
DEMs and DEMis identification
Alterations in miRNA and mRNA expression profile
during osteogenic differentiation were analyzed in human ADSCs
using a microarray-based approach (GSE72429 and GSE37329). A total
of 16 DEMis were observed to be important in this physiological
process, while 185 DEMs were identified The predicted target genes
were aligned with the DEMs and a number of DEMs were potentially
regulated by >1 DEMis, according to the prediction result. These
key genes are known to be involved in the osteogenic
differentiation of ADSCs (Tables I
and II). Among the DEMis, six
miRNAs (miR-143-3p, miR-135a-5p, miR-31-5p, miR-22-3p, miR-193b-3p
and let-7i-5p) were highly associated with the osteogenesis of
ADSCs.
 | Table I.List of identified DEMis. |
Table I.
List of identified DEMis.
| miRNA | P-value |
Log2FC | Regulation | Number of targets
DEMs |
|---|
| miR-210 | 0.03826 | 5.27376 | Up | 0 |
| miR-143 | 0.00904 | 4.60661 | Up | 29 |
| miR-7 | 0.00275 | 4.16496 | Up | 0 |
| miR-100 | 0.00975 | 3.97587 | Up | 2 |
| let-7i | 0.00945 | 3.86616 | Up | 17 |
| miR-487a | 0.00953 | 3.82785 | Up | 3 |
| miR-502-3p | 0.00977 | 3.66819 | Up | 6 |
| miR-193b | 0.00961 | 3.55430 | Up | 9 |
| miR-22 | 0.00994 | 1.45696 | Up | 17 |
| miR-31 | 0.04184 | 1.03064 | Up | 18 |
| miR-642b | 0.03651 | −1.03150 | Down | 0 |
| miR-1181 | 0.00443 | −1.06189 | Down | 0 |
| miR-1275 | 0.01455 | −1.15712 | Down | 0 |
| miR-762 | 0.00403 | −1.39160 | Down | 0 |
| miR-135a | 0.02155 | −1.87443 | Down | 21 |
| miR-629 | 0.00609 | −5.71026 | Down | 0 |
 | Table II.Expressed mRNAs targeted by
miRNAs. |
Table II.
Expressed mRNAs targeted by
miRNAs.
| Gene | Gene title | P-value |
Log2FC | Targeted by |
|---|
| PPARGC1A | PPARG coactivator 1
alpha | 0.0036 | 2.35 | let-7i-5p,
miR-193b-3p, miR-31-5p, miR-22-3p, miR-487a-3p |
| DPYSL3 | Dihydropyrimidinase
like 3 | 0.0173 | −2.07 | miR-143-3p,
let-7i-5p, miR-22-3p, miR-31-5p, |
| ADAMTS6 | ADAM
metallopeptidase with thrombospondin type 1 motif 6 | 0.0015 | −1.72 | miR-143-3p,
let-7i-5p, miR-135a-5p, miR-22-3p |
| CNR1 | Cannabinoid
receptor 1 | 0.0067 | 1.69 | miR-143-3p,
miR-487a-3p, let-7i-5p |
| EDN1 | Endothelin 1 | 0.0002 | 2.40 | miR-143-3p,
let-7i-5p miR-135a-5p |
| FBN2 | Fibrillin 2 | 0.0028 | −1.68 | miR-143-3p,
miR-22-3p miR-502-3p |
| LGR4 | Leucine rich repeat
containing G protein-coupled receptor 4 | 0.0030 | −2.62 | let-7i-5p,
miR-193b-3p miR-487a-3p |
| NTNG1 | Netrin G1 | 0.0128 | −1.56 | miR-22-3p,
miR-502-3p miR-135a-5p |
| MMP16 | Matrix
metallopeptidase 16 | 0.0225 | −2.90 | miR-135a-5p,
miR-31-5p miR-193b-3p |
miRNA-mRNA and PPI networks
To speculate on the function of mRNAs targeted by
miRNAs, a network among miRNAs and mRNAs was constructed and
visualized. As shown in Fig. 1A, the
miRNA-mRNA network consisted of 72 mRNA and nine miRNA nodes.
Bioinformatics analysis highlighted six miRNAs among the network
(miR-143-3p, miR-135a-5p, miR-31-5p, miR-22-3p, miR-193b-3p and
let-7i-5p; Fig. 1B) that exhibited
>15 predicted DEM targets. The PPI network of DEMs was
illustrated in order to select significant genes (Fig. 1C).
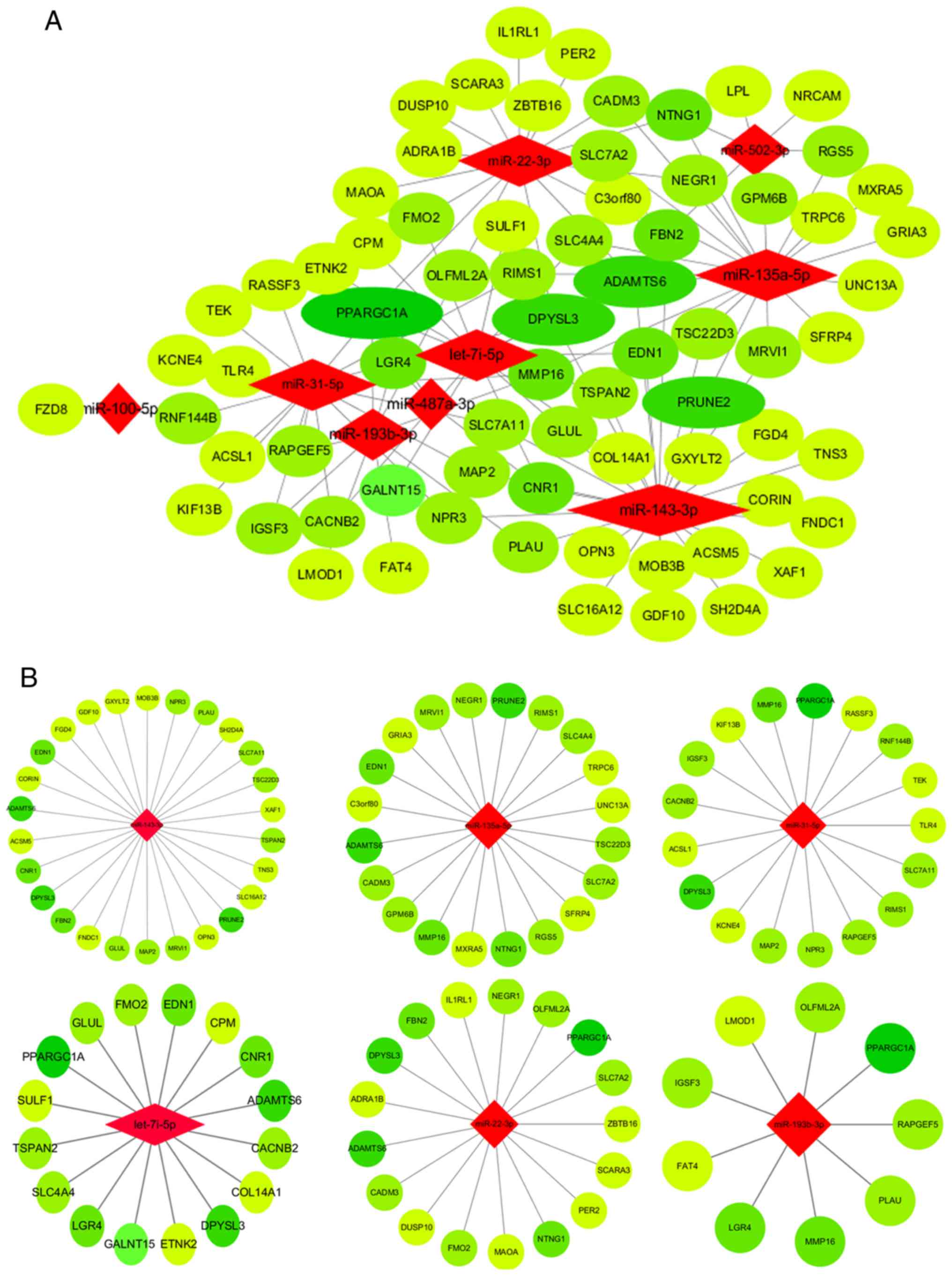 | Figure 1.miRNA-mRNA network, PPI network and
hub genes. (A) Core miRNA-mRNA network. Red diamonds represent
miRNAs. Light green circles represent upregulated and dark green
circles represent downregulated genes; the size of the circles
reflects the fold change in mRNA expression following osteoblastic
induction; the larger the size, the greater the fold change. (B)
Six miRNAs (miR-143-3p, miR-135a-5p, miR-31-5p, let-7i-5p,
miR-22-3p and miR-193b-3) and their potential target DEMs. Red
diamonds represent miRNAs. Light green circles represent
upregulated and dark green circles represent downregulated genes.
(C) PPI network of DEMs. Red circles represent upregulated and
green circles represent downregulated genes, the lighter the color,
the greater the fold change. miRNA, microRNA; PPI, protein-protein
interaction; DEMs, differentially expressed mRNAs. |
Functional prediction of miRNAs based
on the miRNA-mRNA network
To further understand the function and mechanism of
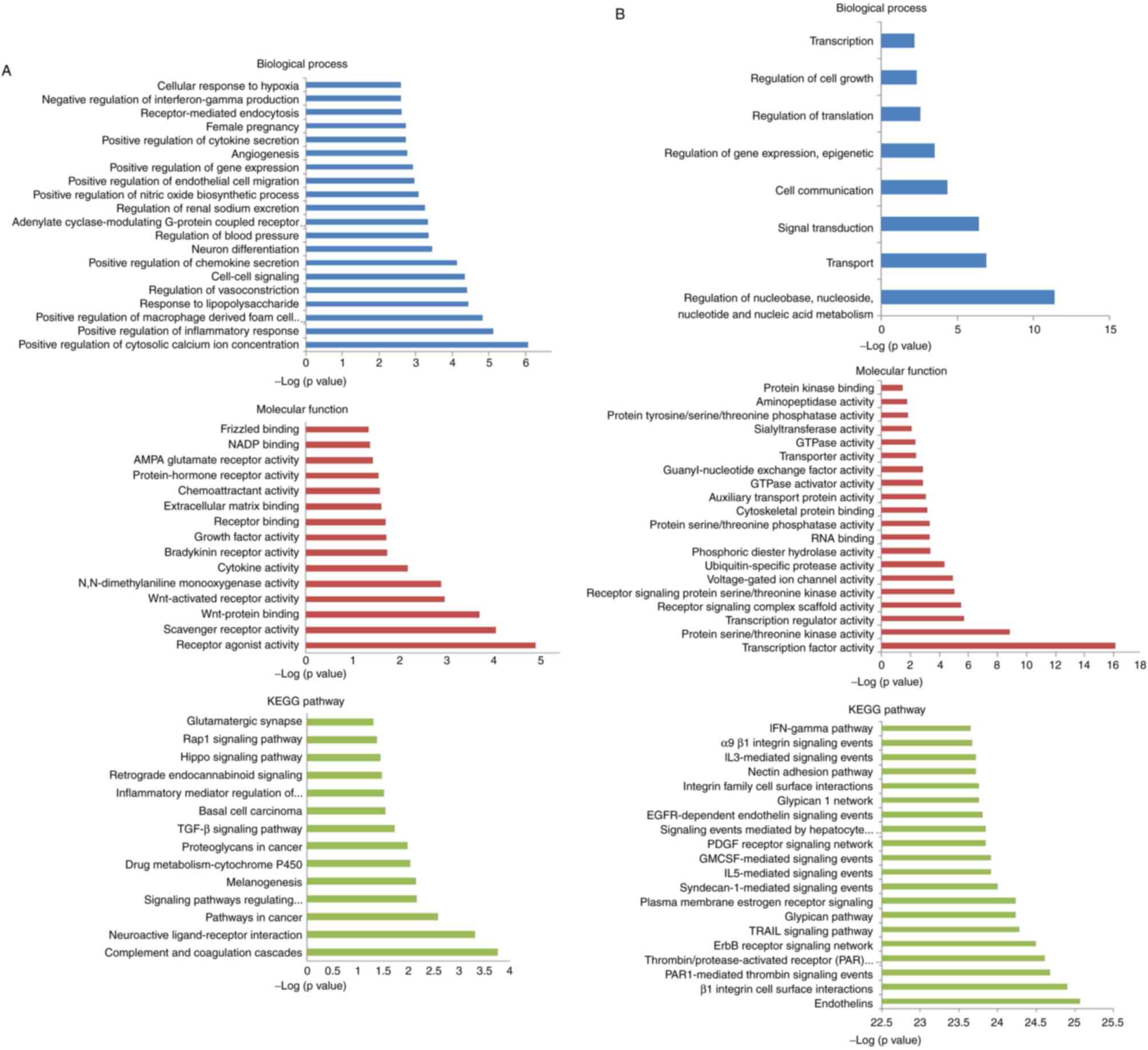
identified DEMs and DEMis, functional and pathway enrichment
analyses, including GO and KEGG, were performed using DAVID. For
DEMs analysis, the results of GO analysis revealed 105 enriched GO
terms in the ‘Biological Process’ category, particularly in the
regulation of cytosolic calcium ion concentration, 16 enriched
terms in the ‘Molecular Function’ category and 14 enriched terms in
the in the ‘KEGG pathway’ category, according to P<0.05 and
Benjamini corrected P<0.05 (Fig.
2A). The top significant terms in the Biological Processes,
Molecular Function and KEGG pathway categories of DEMis are shown
in Fig. 2B.
DPYSL3 downregulation in the process
of the osteogenesis of ADSCs
In order to explore the possible target genes
involved in the osteogenesis of ADSCs from the aforementioned
miRNA-mRNA network, the mechanisms of miRNA mediation of downstream
target genes were analyzed. It was identified that
dihydropyrimidinase like 3 (DPYSL3) may be modulated by four
differential DEMis, and was in the top 10 downregulated genes.
Therefore DPYSL3 was selected as the target gene in the
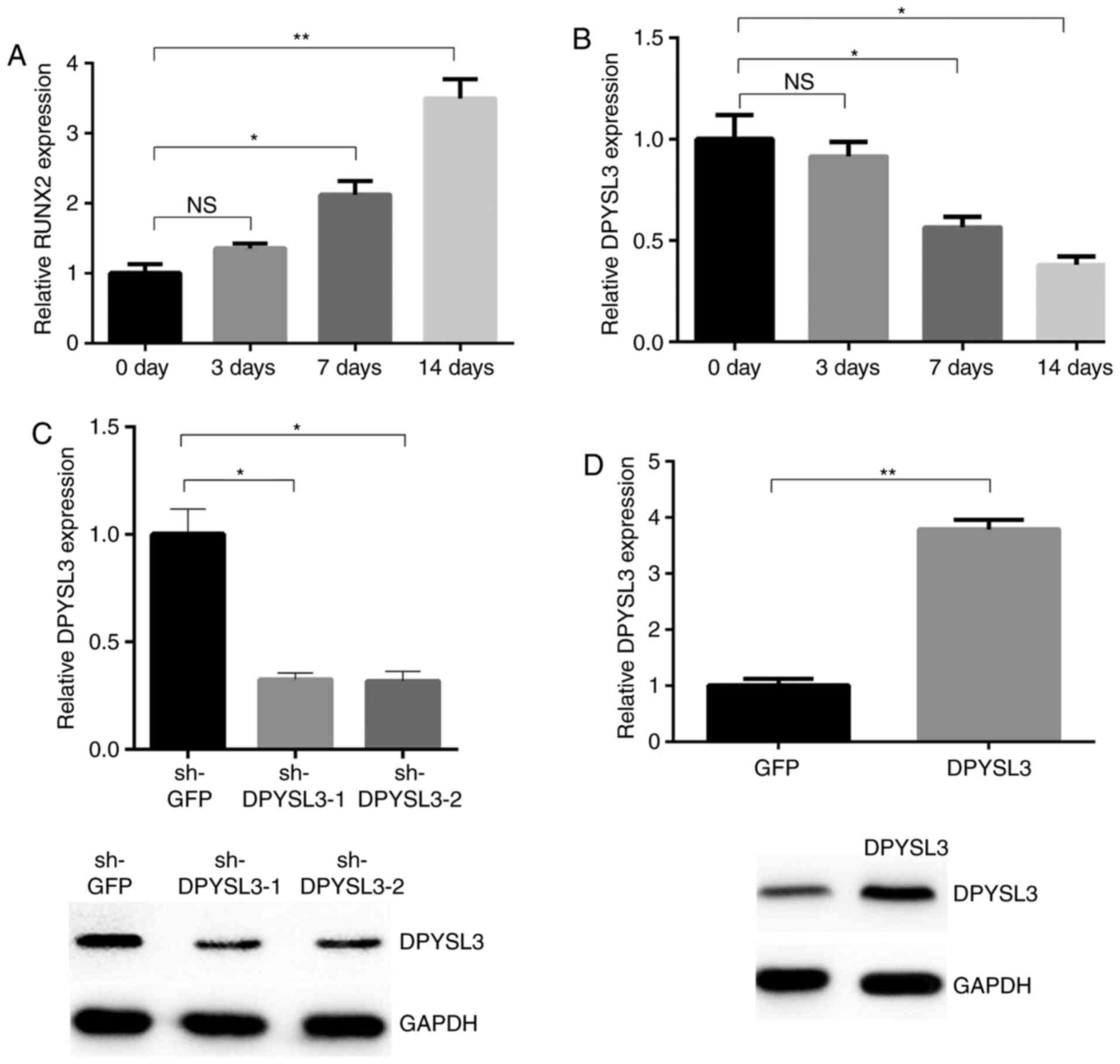
osteogenesis of ADSCs. Firstly, to verify the osteogenesis model of
ADSCs, the expression of RUNX2, a marker of osteogenesis, was
examined in ADSCs. It was observed that the expression of this gene
was increased in the process of osteogenesis of ADSCs (Fig. 3A). RT-qPCR results indicated that the
expression of DPYSL3 was decreased in the process of osteogenesis
of ADSCs (Fig. 3B). Significant
differences were observed following 7 and 14 days but not following
3 days of osteogenesis, when compared with the control group.
Identified effects for silencing or
overexpression of DPYSL3
To determine the functional role of DPYSL3 in the
ADSC osteogenesis process, knockdown of DPYSL3 expression in CRC
cells was achieved using two independent lentiviral-mediated short
hairpin RNAs (shRNAs). Compared with the sh-GFP control, DPYSL3
levels in the sh-DPYSL3-1 and sh-DPYSL3-2 groups were significantly
reduced (Fig. 3C). Furthermore,
ectopic expression of DPYSL3 could increase DPYSL3 expression
level. ADSCs were infected with lentiviral vectors containing
DPYSL3. Overexpression of DPYSL3 was associated with increased
DPYSL3 expression, both at the mRNA and protein levels (Fig. 3D). These results indicate that the
silencing or overexpression of DPYSL3 may decrease or increase the
DPYSL3 levels, respectively.
DPYSL3 is the target gene in the
process of osteogenesis of ADSCs
Further investigation into the functional role of
DPYSL3 in the process of osteogenesis of ADSCs was performed.
Following treatment with osteogenic medium for 14 days, it was
observed that the silencing of DPYSL3 expression significantly
promoted the increased expression of osteoblast
differentiation-associated genes, RUNX2, OSX and ALP (Fig. 4A). By contrast, overexpression of
DPYSL3 significantly reduced the expression levels of these genes
(Fig. 4B). Western blot analysis
demonstrated that overexpression or silencing of DPYSL3 could
decrease or increase the protein levels of RUNX2 following
induction of differentiation using osteogenic medium for 14 days,
respectively (Fig. 4C). Furthermore,
ALP activity demonstrated a significant increase or decrease
following silencing or overexpression of the DPYSL3 gene,
respectively (Fig. 4D). These
findings were supported by the ARS results at 14 days indicating
the same tendency (Fig. 4E).
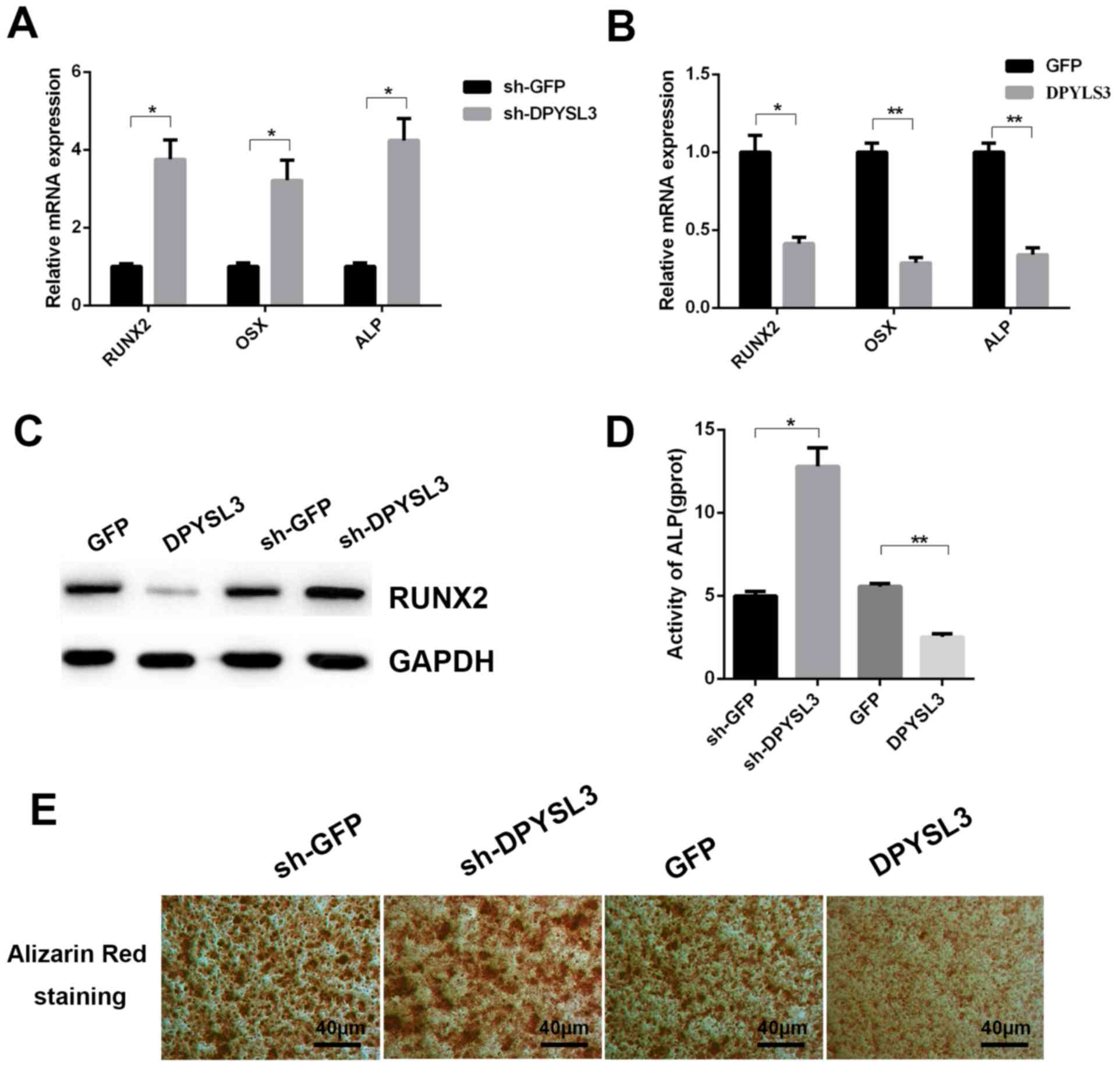 | Figure 4.DPYSL3 inhibits osteoblastic
differentiation. Human ADSCs were transfected with (A) DPYSL3 shRNA
or (B) LV-DPYSL3, respectively for 48 h, and the mRNA expression of
osteoblastic differentiation-associated genes, RUNX2, OSX and ALP
was determined by RT-qPCR. (C) Western blot analysis detecting
RUNX2 expression in DPYSL3 silence or overexpression in ADSCs
cells. (D) Alkaline phosphatase activity in the differentiated
ADSCs. (E) Alizarin red staining of differentiated ADSCs.
*P<0.05 and **P<0.01 vs. the control group. DPYSL3,
dihydropyrimidinase like 3; ADSCs, LV-DPYSL3, overexpressed
dihydropyrimidinase like 3; adipose-derived stem cells; shRNA,
short hairpin RNA; RUNX2, runt-related transcription factor 2; OSX,
Osterix; RT-qPCR, reverse transcription-quantitative polymerase
chain reaction; ALP, alkaline phosphatase; sh-, small hairpin. |
Discussion
MSCs are bone marrow stromal cells that are capable
of differentiating into osteogenic, chondrogenic, adipogenic or
myogenic cell lineages (21–23). Osteoblastic differentiation is a
complex, closely regulated process that is critical for proper bone
formation and is influenced by a variety of endogenous and
environmental factors, including bone morphogenetic proteins,
peroxisome proliferator-activated receptor γ, RUNX2 and
Wnt/β-Catenin (24,25). However, its effect and the underlying
mechanisms involved in the progression of bone regeneration remain
unclear.
Developments in high-throughput technology have
provided a large amount of osteogenesis profiles, which provide
information that enables the investigation of osteoblastic
differentiation. Bioinformatics analysis of the data has indicated
that specific genes with aberrant expression may be important for
osteoblastic differentiation or function as potential biomarkers
for osteogenesis diagnosis and prognosis (26–29). In
particular, miRNAs have been reported to be involved in the
regulation of cellular behavior, including differentiation and
development, metabolism, proliferation, apoptosis, viral infection
and tumorigenesis (30–32). Dysregulation of miRNA expression may
result in various pathological states (33). Recently, certain miRNAs were
identified as regulators of post-transcriptional gene expression
and were therefore considered important for osteogenesis (34,35).
In previous studies, GO and pathway analyses were
useful tools for analyzing biological functions, which were
enriched among differentially expressed coding-genes (36,37). GO
analysis is used to probe the roles of differentially expressed
genes, and annotate described genes and gene products distributed
among all organisms. KEGG is a database resource that integrates
genomic, chemical and systemic functional information; KEGG pathway
analysis can provide promising and more biologically meaningful
results, including information regarding molecular interactions and
cellular processes (38). Gene
catalogs from fully sequenced genomes are linked to higher-level
systemic functions of the cell, the organism and the ecosystem
(39,40). In the GO analysis, the term of
regulation of cytosolic calcium ion concentration ranks greatly
statistics significance, proving the reliability of this study. In
addition, the molecular function analysis of the Wnt-activated
receptor and the Wnt-protein binding indicated significant
involvement in the osteoblastic differentiation (P<0.05,
Benjamini corrected P<0.05). Pathway analysis further
demonstrated that 14 pathways were enriched. The transforming
growth factor beta and the ras-proximate-1 signaling pathway, as
well as the signaling pathways regulating pluripotency of stem
cells, have been observed to serve important roles in the
osteogenesis of ADSCs (41,42).
Emerging evidence has demonstrated that miRNAs have
displayed superior potential as diagnostic and prognostic
biomarkers due to their close association between mRNA expression
and function (43,44). Recently, previous studies have
identified several miRNA-focused signatures that may advance the
diagnoses of diseases, including ankylosing spondylitis, oral
squamous cell carcinoma and multiple myeloma (45–47).
However, the diagnostic role of miRNAs in the osteogenesis of ADSCs
has not been fully investigated. In order to identify the key
miRNAs involved, which may be used as potential novel biomarkers
for the clinical diagnosis and treatment of the osteogenesis of
ADSCs, the hub nodes and the number of relationship pairs were used
in the present study. Previous studies have demonstrated that hub
nodes, which are characterized by their high degree of connectivity
to other nodes, may be important genes that could prove valuable to
future study (48,49). Generally, miRNAs with multiple
relationship pairs take part in more mRNA interactions, thus
identifying the miRNA as a hub. Hence, miRNAs are important in
network organization (50). In the
current study, six miRNAs (miR-143-3p, miR-135a-5p, miR-31-5p,
miR-22-3p, miR-193b-3p and let-7i-5p) were identified as
topological key nodes, where the node degrees and the number of
miRNA-mRNA pairs were significantly higher compared with other
miRNAs. This indicates that these miRNAs have profound implications
for the osteogenesis of ADSCs and may be considered as key
miRNAs.
Further analyses indicated that one of these
identified genes, DPYSL3, was predicted to be modulated by the
majority of miRNAs and exhibited a greater fold change. These
findings indicate that DPYSL3 may be a critical gene involved in
osteogenesis. However, the contribution of DPYSL3 to osteogenesis
of ADSCs was not evaluated in the present study. By analyzing its
expression in the process of osteogenesis of ADSCs, it was
demonstrated that DPYSL3 expression was gradually decreased, which
indicates that this gene might be involved in the osteogenesis of
ADSCs. The selective knockdown or overexpression of this gene
demonstrated that silencing of DPYSL3 promoted the expression of
osteoblast differentiation-associated genes, while overexpression
of DPYSL3 inhibited the expression of these genes following the
induction by osteogenic medium for 14 days. Simultaneously, ALP
activity exhibited a significant increase or decrease following
silencing or overexpression of DPYSL3, respectively. The results
were confirmed by ARS at 14 days, which demonstrated the same
trend. These results indicated that DPYSL3 may be a target gene
involved in the process of osteogenesis of ADSCs.
In conclusion, the miRNA-mRNA network constructed in
the present study may enable the analysis of miRNA-mRNA-mediated
genes in the development of osteogenesis of ADSCs at a system-wide
level. These results identified DPYSL3 as a novel regulator in
osteogenesis of ADSCs, and may provide novel insight into the
potential of DPYSL3 as a therapeutic target in the management of
bone regeneration-associated diseases.
Acknowledgements
The authors are grateful to Professor Jianjiang Zhao
(Laboratory for Oral Diseases, Department of Oral and Maxillofacial
Surgery, Stomatological Hospital, Southern Medical University,
China) and Professor Qing Li (Guangzhou School of Clinical
Medicine, Southern Medical University, Guangzhou General Hospital
of Guangzhou Military Region, China) for providing help with the
data analysis.
Funding
This study was financially supported by National
Natural Science Foundation of China (81670950) and the Science and
Technology Project of Guangdong Province (201802020018).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BJ, JZ and QL designed the experiments, BJ, ZZ and
XQ carried out the experiments, BJ and XS analyzed the experimental
results. BJ, HC and XZ analyzed the sequencing data and developed
the analytical tools, BJ and XQ wrote the manuscript, and JZ and QL
contributed to finalizing the manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing
interests.
References
|
1
|
Pensak MJ and Lieberman JR: Gene therapy
for bone regeneration. Curr Pharm Des. 19:3466–3473. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Thor A, Palmquist A, Hirsch JM, Rännar LE,
Dérand P and Omar O: Clinical, morphological, and molecular
evaluations of bone regeneration with an additive manufactured
osteosynthesis plate. J Craniofac Surg. 27:1899–1904. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
D'Mello S, Atluri K, Geary SM, Hong L,
Elangovan S and Salem AK: Bone regeneration using Gene-Activated
matrices. AAPS J. 19:43–53. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kristjánsson B and Honsawek S: Mesenchymal
stem cells for cartilage regeneration in osteoarthritis. World J
Orthop. 8:674–680. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Roura S, Gálvez-Montón C, Mirabel C, Vives
J and Bayes-Genis A: Mesenchymal stem cells for cardiac repair: Are
the actors ready for the clinical scenario? Stem Cell Res Ther.
8:2382017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Manatsathit W, Samant H and
Nakayuenyongsuk W: Mesenchymal stem cells for hepatitis B patients
with acute on chronic liver failure-are we there? Hepatology.
66:1705–1706. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Moreira A, Kahlenberg S and Hornsby P:
Therapeutic potential of mesenchymal stem cells for diabetes. J Mol
Endocrinol. 59:R109–R120. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ding SLS, Kumar S and Mok PL: Cellular
reparative mechanisms of mesenchymal stem cells for retinal
diseases. Int J Mol Sci. 18:pii: E1406. 2017. View Article : Google Scholar
|
|
9
|
Roskies MG, Fang D, Abdallah MN,
Charbonneau AM, Cohen N, Jordan JO, Hier MP, Mlynarek A, Tamimi F
and Tran SD: Three-dimensionally printed polyetherketoneketone
scaffolds with mesenchymal stem cells for the reconstruction of
critical-sized mandibular defects. Laryngoscope. 127:E392–E398.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Razmkhah M, Mansourabadi Z, Mohtasebi MA,
Talei AR and Ghaderi A: Cancer and normal adipose-derived
mesenchymal stem cells (ASCs): Do they have differential effects on
tumor and immune cells? Cell Biol Int. 42:334–343. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Carstens MH, Mendieta M, Pérez C,
Villareal E and Garcia R: Assisted salvage of ischemic
fasciocutaneous flap using Adipose-Derived mesenchymal stem cells:
In-Situ revascularization. Aesthet Surg J. 37 Suppl 3:S38–S45.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gao W, Zhang L, Zhang Y, Sun C, Chen X and
Wang Y: Adipose-derived mesenchymal stem cells promote liver
regeneration and suppress rejection in small-for-size liver
allograft. Transpl Immunol. 45:1–7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Calabrese G, Giuffrida R, Forte S, Fabbi
C, Figallo E, Salvatorelli L, Memeo L, Parenti R, Gulisano M and
Gulino R: Human adipose-derived mesenchymal stem cells seeded into
a collagen-hydroxyapatite scaffold promote bone augmentation after
implantation in the mouse. Sci Rep. 7:71102017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dennis GJ Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sun C, Yuan Q, Wu D, Meng X and Wang B:
Identification of core genes and outcome in gastric cancer using
bioinformatics analysis. Oncotarget. 8:70271–70280. 2017.PubMed/NCBI
|
|
18
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chu H, Jia B, Qiu X, Pan J, Sun X, Wang Z
and Zhao J: Investigation of proliferation and migration of tongue
squamous cell carcinoma promoted by three chemokines, MIP-3α,
MIP-1β, and IP-10. Oncotargets Ther. 10:4193–4203. 2017. View Article : Google Scholar
|
|
21
|
He BC, Chen L, Zuo GW, Zhang W, Bi Y,
Huang J, Wang Y, Jiang W, Luo Q, Shi Q, et al: Synergistic
antitumor effect of the activated PPARgamma and retinoid receptors
on human osteosarcoma. Clin Cancer Res. 16:2235–2245. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Aubin JE: Regulation of osteoblast
formation and function. Rev Endocr Metab Disord. 2:81–94. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Deng ZL, Sharff KA, Tang N, Song WX, Luo
J, Luo X, Chen J, Bennett E, Reid R, Manning D, et al: Regulation
of osteogenic differentiation during skeletal development. Front
Biosci. 13:2001–2021. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wagner ER, He BC, Chen L, Zuo GW, Zhang W,
Shi Q, Luo Q, Luo X, Liu B, Luo J, et al: Therapeutic implications
of PPARgamma in human osteosarcoma. PPAR RES. 2010:9564272010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tang N, Song WX, Luo J, Haydon RC and He
TC: Osteosarcoma development and stem cell differentiation. Clin
Orthop Relat Res. 466:2114–2130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao W, Wang D, Zhao J and Zhao W:
Bioinformatic analysis of retinal gene function and expression in
diabetic rats. Exp Ther Med. 14:2485–2492. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Koehler AV, Korhonen PK, Hall RS, Young
ND, Wang T, Haydon SR and Gasser RB: Use of a
bioinformatic-assisted primer design strategy to establish a new
nested PCR-based method for Cryptosporidium. Parasit Vectors.
10:5092017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang C, Chen Y, Mao X, Huang Y, Jung SY,
Jain A, Qin J and Wang Y: A bioinformatic algorithm for analyzing
cell signaling using temporal proteomic data. Proteomics.
17:2017.doi: 10.1002/pmic.201600425. View Article : Google Scholar
|
|
29
|
Tsukanov KY, Krasnenko AY, Plakhina DA,
Korostin DO, Churov AV, Druzhilovskaya OS, Rebrikov DV and Ilinsky
VV: A bioinformatic pipeline for NGS data analysis and mutation
calling in human solid tumors. Biomed Khim. 63:413–417. 2017.(In
Russian). View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Irwandi RA and Vacharaksa A: The role of
microRNA in periodontal tissue: A review of the literature. Arch
Oral Biol. 72:66–74. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM
and Zhang GZ: Biological functions of microRNAs: A review. J
Physiol Biochem. 67:129–139. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hamidi-Asl E, Palchetti I, Hasheminejad E
and Mascini M: A review on the electrochemical biosensors for
determination of microRNAs. Talanta. 115:74–83. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jia HY, Chen F, Chen JZ, Wu SS, Wang J,
Cao QY, Chen Z and Zhu HH: MicroRNA expression profiles related to
early stage murine concanavalin A-induced hepatitis. Cell Physiol
Biochem. 33:1933–1944. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Khraiwesh B, Zhu JK and Zhu J: Role of
miRNAs and siRNAs in biotic and abiotic stress responses of plants.
Biochim Biophys Acta. 1819:137–148. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
van Wijnen AJ, van de Peppel J, van
Leeuwen JP, Lian JB, Stein GS, Westendorf JJ, Oursler MJ, Im HJ,
Taipaleenmäki H, Hesse E, et al: MicroRNA functions in osteogenesis
and dysfunctions in osteoporosis. Curr Osteoporos Rep. 11:72–82.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Song C, Zhang J, Liu Y, Pan H, Qi HP, Cao
YG, Zhao JM, Li S, Guo J, Sun HL and Li CQ: Construction and
analysis of cardiac hypertrophy-associated lncRNA-mRNA network
based on competitive endogenous RNA reveal functional lncRNAs in
cardiac hypertrophy. Oncotarget. 7:10827–10840. 2016.PubMed/NCBI
|
|
37
|
Wu Q, Guo L, Jiang F, Li L, Li Z and Chen
F: Analysis of the miRNA-mRNA-lncRNA networks in ER+ and ER-breast
cancer cell lines. J Cell Mol Med. 19:2874–2887. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kanehisa M: Enzyme annotation and
metabolic reconstruction using KEGG. Methods Mol Biol.
1611:135–145. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45(D1): D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wu Y, Zhou J, Li Y, Zhou Y, Cui Y, Yang G
and Hong Y: Rap1A regulates osteoblastic differentiation via the
ERK and p38 mediated signaling. PLoS One. 10:e1437772015.
View Article : Google Scholar
|
|
42
|
Wu M, Chen G and Li YP: TGF-β and BMP
signaling in osteoblast, skeletal development, and bone formation,
homeostasis and disease. Bone Res. 4:160092016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shen X, Zhang Y, Wu X, Guo Y, Shi W, Qi J,
Cong H, Wang X, Wu X and Ju S: Upregulated lncRNA-PCAT1 is closely
related to clinical diagnosis of multiple myeloma as a predictive
biomarker in serum. Cancer Biomark. 18:257–263. 1017. View Article : Google Scholar
|
|
46
|
Li X, Chai W, Zhang G, Ni M, Chen J, Dong
J, Zhou Y, Hao L, Bai Y and Wang Y: Down-Regulation of
lncRNA-AK001085 and its influences on the diagnosis of ankylosing
spondylitis. Med Sci Monit. 23:11–16. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tang H, Wu Z, Zhang J and Su B: Salivary
lncRNA as a potential marker for oral squamous cell carcinoma
diagnosis. Mol Med Rep. 7:761–766. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhou M, Diao Z, Yue X, Chen Y, Zhao H,
Cheng L and Sun J: Construction and analysis of dysregulated
lncRNA-associated ceRNA network identified novel lncRNA biomarkers
for early diagnosis of human pancreatic cancer. Oncotarget.
7:56383–56394. 2016.PubMed/NCBI
|
|
49
|
Zhang Y, Xu Y, Feng L, Li F, Sun Z, Wu T,
Shi X, Li J and Li X: Comprehensive characterization of lncRNA-mRNA
related ceRNA network across 12 major cancers. Oncotarget.
7:64148–64167. 2016.PubMed/NCBI
|
|
50
|
Cao Y, Wang P, Ning S, Xiao W, Xiao B and
Li X: Identification of prognostic biomarkers in glioblastoma using
a long non-coding RNA-mediated, competitive endogenous RNA network.
Oncotarget. 7:41737–41747. 2016. View Article : Google Scholar : PubMed/NCBI
|