Introduction
An estimated 240 million individuals worldwide are
chronically infected with hepatitis B virus (HBV) (1). Large-scale studies have demonstrated
that effective anti-viral therapy may inhibit HBV replication and
hence remission of the liver disease, thereby preventing associated
morbidity and mortality (2). In
China, current treatments for chronic hepatitis B (CHB) include
interferon (IFN)-α and nucleos(t)ide analogs (NAs). Five NA drugs,
namely lamivudine (LAM), adefovir (ADV), telbivudine, entecavir
(ETV) and tenofovir (TDF), have been approved for the treatment of
CHB. A problem with NA therapy is that a long duration of treatment
leads to an increased risk of drug resistance, which subsequently
results in treatment failure. Although LAM and ADV are not
recommended as first-line therapy in CHB, they are administered to
numerous patients with CHB, either as a monotherapy or in
combination, due to reasons including low cost and availability
(3). As a result, drug resistance
remains a great challenge in HBV treatment.
LAM was the first nucleoside analogue approved for
the treatment of CHB. Resistance to LAM has been reported in ~20%
of anti-viral-naive patients after one year and up to 70% of
patients after five years of treatment (4). It has been indicated that LAM
resistance is mostly associated with the rtM204I/V mutation in the
tyrosine-methionine-aspartate-aspartate (YMDD) motif of the C
domain of the polymerase gene. Several types of rescue therapy have
been applied in patients with LAM-resistant CHB, yielding
virological and biochemical improvements (5). A number of studies have reported good
efficacy of LAM/ADV combination as a rescue therapy against LAM
resistance (6,7). However, ADV resistance was encountered
in ~18% of the patients who had presented with resistance to LAM
after 48 weeks of ADV administration (8). ADV-resistance has been associated with
the rtN236T mutation in the D domain and/or the rtA181V/T mutation
in the B domain. However, the underlying mechanisms of the
development of ADV resistance due to LAM/ADV combination therapy in
patients with LAM resistance have remained elusive.
Although HBV is a DNA virus, it uses pre-genomic RNA
as a replicative intermediate, similar to RNA viruses. The lack of
proof-reading capacity of the HBV reverse transcriptase (RT) and a
high replication rate result in a significantly increased frequency
of viral genomic mutations (9).
Consequently, the virus circulates as quasispecies (QS), a spectrum
of genetically distinct but highly similar viral variants, and
their evolution and distribution require to be monitored to allow
for proper chronic infection management (10). QS may contribute to viral
pathogenesis, and may have an influence on anti-viral treatment and
an involvement in drug resistance, as has been reported previously
for RNA viruses, including hepatitis C virus (HCV) and human
immunodeficiency virus (HIV) (11,12).
Several studies have indicated that the HBV QS complexity may be
associated with the virological response to NAs (13–16).
However, the association between HBV QS and NA treatment is
currently not well-defined, particularly in case of LAM/ADV
combination therapy for LAM resistance.
Due to the overlap between the open reading frames
of the RT and the HBV surface antigen (HBsAg), mutations in the RT
domain frequently affect the amino acid sequence of the HBsAg,
leading to immune evasion (17).
Chen et al (18) reported
changes in nucleotide or amino acid sequences in the HBsAg region
in LAM and/or ADV resistance in LAM/ADV-treated patients. Dynamic
changes in QS within the S region may also have important roles.
However, the impact of QS of the S region on LAM/ADV combination
therapy in LAM-resistant patients has largely remained to be
elucidated. The present study aimed to investigate the evolution of
HBV QS within the RT and S regions during six months of LAM/ADV
combination anti-viral therapy in patients with YMDD mutation and
LAM resistance, and to explore its effects on drug resistance and
anti-viral efficacy.
Materials and methods
Study design
A total of 16 patients with CHB who received LAM/ADV
combination as rescue therapy due to LAM resistance were enrolled
at the First Affiliated Hospital of Anhui Medical University
(Hefei, China) between July 2010 and August 2014. All patients were
initially administered oral doses of LAM at 100 mg per day (Glaxo
Smith Kline Pharmaceuticals, Co., Ltd., Suzhou, China) and LAM plus
ADV at 10 mg per day (Glaxo Smith Kline Co., Ltd.), after the
detection of LAM resistance-associated HBV variants. All patients
received combination therapy for at least six months.
Patients who met the following inclusion criteria
were enrolled: Age, 18–60 years; presence of HBsAg at least six
months prior to LAM treatment; elevated HBV DNA ≥105
copies/ml and serum alanine aminotransferase (ALT) levels higher
than twice the upper limit of normal prior to LAM treatment; no
history of receiving NAs or IFN treatment at any time prior to LAM
treatment; and YMDD mutation confirmed by direct sequencing. The
exclusion criteria were as follows: Co-infection with HIV, HCV or
hepatitis D virus; liver diseases due to other causes (e.g.,
autoimmune liver disease, alcoholic hepatitis or drug-induced
hepatitis); and poor compliance during treatment.
Anti-viral efficacy was evaluated at six months from
the start of LAM/ADV combination treatment. Patients who had
undetectable HBV DNA levels (HBV DNA <1,000 copies/ml) at six
months were defined as responders (RS), while patients who did not
meet this criterion were considered non-RS (NRS).
At baseline and at six months, serum samples were
obtained from all 16 patients and stored at −80°C. Liver
biochemistry, HBV serology and HBV DNA levels were determined at
the First Affiliated Hospital of Anhui Medical University (Hefei,
China), as described previously (19).
Molecular cloning and sequencing
HBV DNA was extracted from serum samples (100 µl)
using the QIAamp blood mini kit (cat. no. 40725; Qiagen GmbH,
Hilden, Germany) following the manufacturer's instructions and
eluted in 30 µl distilled water. The nucleotide sequences of the
HBV RT region were determined using a nested polymerase chain
reaction (PCR) and direct sequencing. DNA was amplified using 5
U/µl Taq DNA polymerase (Takara Biotechnology Co., Ltd., Dalian,
China), 5 µl HBV DNA template, 1 µl each primer and 10 mM each dNTP
in a final volume of 30 µl. A PCR reactor (Biometra TRIO 48;
Biometra biomedizinische Analytik GmbH, Jena, Germany) was used.
Primer sequences for the first round of PCR were forward,
5′-CCTGCTGGTGGCTCCAGTTCMG-3′ (nt 58–79) and reverse,
5′-AGGAGTTCCGCAGTATGGATCGGCAG-3′ (nt 1,286–1,260). The first round
of PCR analysis was performed under the following conditions:
initial denaturation at 95°C for 7 min, 35 cycles of denaturation
at 95°C for 30 sec, annealing at 57°C for 30 sec and extension at
72°C for 90 sec, followed by a final elongation step (7 min at
72°C). The primers used for the second round of PCR were forward,
5′-CCATATCGTCAATCTTMTCGA-3′ (nt 113–134) and reverse,
5′-CCAGTGGGGGTTGCGTCAGCAA-3′ (nt 1,208–1,186). The parameters for
the second round were the same as the first. If the HBV DNA levels
were <10,000 copies/ml, nested PCR was used to amplify the HBV
RT region; if the HBV levels were >10,000 copies/ml, however,
only the second round of PCR was performed. PCR products were
resolved in 2% agarose gels by electrophoresis with ethidium
bromide staining. A fragment of ~1,100 bp containing the entire RT
region was amplified. PCR products were purified from the gels by
using the Agarose Gel DNA Purification kit (Takara Bio Inc.,
Dalian, China) according to the manufacturer's instructions.
Purified PCR products were cloned into the pMD18-T
vector (Takara Bio Inc.,) following the manufacturer's protocols.
Positive clones were sequenced with two primers specific for the
vector, RV-M and M13-47, on an ABI sequencing system (Applied
Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). For
each sample, ~25 clones were sequenced.
Sequence analysis
BioEdit 7.0.9 (20)
was used to assemble sequences. Multiple alignments were performed
on all sequences using Clustal X 2.0 (21). RDP3.0 software was used to detect
potential recombinant sequences (22). QS complexity and diversity were
determined and used to evaluate the dynamics of viral
heterogeneity. QS complexity was measured via the Shannon entropy
(Sn), which refers to the number of variants identified in a single
sample. The Sn was calculated at the nucleotide and amino acid
level using the following formula: Sn=-∑i(pi × lnpi)]/lnN, where pi
is the frequency of each clone in the QS population and N is the
total number of clones (23). Three
parameters were used to assess QS diversity: The mean genetic
distance (d), the number of synonymous substitutions per synonymous
site (dS), and the number of non-synonymous substitutions per
non-synonymous site (dN). d was calculated at the nucleotide and
the amino acid level. d at the nucleotide level was calculated
using Tamura's three-parameter model, whereas that at the amino
acid level was calculated using the Jones-Taylor-Thornton model
with Mega 7.0 software. The dS and dN values were calculated under
the modified Nei-Gojobori model with Jukes-Cantor correction using
Mega 7.0 software (24).
HBV genotyping and phylogenetic
analysis
In the present study, the HBV genotype was assigned
by phylogenetic analysis of the S gene. Sequences were aligned with
the Clustal W program. Phylogenetic trees were constructed by
neighbour-joining analysis with 1,000 replicates of the bootstrap
resampling test using Mega 7.0. Phylogenetic trees were constructed
at baseline and six months of LAM/ADV combination treatment using
all groups of sequences at the nucleotide and the amino acid
level.
Statistical analysis
Continuous variables were expressed as the median
and range. Multiple samples of the measurement data between
different groups and time-points were compared by using the
Kruskal-Wallis test, and the Mann-Whitney U-test was applied as a
post-hoc test when appropriate. Categorical data were examined
using the Chi-square test or Fisher's exact test. A two-tailed
P<0.05 was considered to indicate a statistically significant
difference. Statistical analyses were performed using SPSS 17.0
(SPSS, Inc., Chicago, IL, USA).
Results
Demographic, clinical and laboratory
data
The demographic data and clinical features of the
patients are provided in Table I.
Based on the serum HBV DNA levels, seven patients were identified
to respond to LAM/ADV combination treatment, while nine were NRS
and two experienced a virological breakthrough. Sex, age, HBeAg
status, genotype, drug resistance under LAM treatment, YMDD
pattern, ALT levels and HBV DNA levels in RS and NRS were
comparable at baseline (P>0.05). No significant difference in
ALT levels was identified in either group at months three and six,
but HBV DNA levels in RS were lower than those in NRS (median,
<3.00 vs. 4.20 lg copies/ml, P=0.018; median, <3.00 vs. 4.23
lg copies/ml, P=0.001, respectively) at months three and six
(Table I). In the NRS group, 8
patients switched to ETV plus ADV, and only one patient continued
to use LAM/ADV. At the 9-month follow-up, one patient decided to
discontinue the treatment. At the 12-month follow-up, eight
patients exhibited a virological response. Therefore, no further
research was performed on these patients.
 | Table I.Clinical characteristics of the RS
and NRS groups. |
Table I.
Clinical characteristics of the RS
and NRS groups.
| Characteristic | RS (n=7) | NRS (n=9) |
χ2/Z-value | P-value |
|---|
| Sex (M/F) | 7/0 | 8/1 | 1.202a | 0.273 |
| Age (years) | 35 (27–48) | 33 (25–48) | −0.742b | 0.458 |
| Drug resistance to
LAM (months) | 18 (12–84) | 12 (7–36) | −1.456b | 0.145 |
| YMDD mutation
pattern (YVDD/YIDD) | 3/4 | 4/5 | 0.004a | 0.949 |
| HBeAg status
(+/−) | 4/3 | 8/1 | 2.155a | 0.142 |
| HBV genotype
(B/C) | 3/4 | 4/5 | 0.004a | 0.949 |
| ALT (U/l) |
|
|
|
|
| Baseline | 148 (61–1439) | 96 (22–856) | −1.429b | 0.153 |
| Month 3 | 30 (21–120) | 45 (29–88) | −0.477b | 0.633 |
| Month 6 | 32 (12–86) | 44 (19–141) | −0.370b | 0.711 |
| AST (U/l) |
|
|
|
|
| Baseline | 70 (40–755) | 57 (20–399) | −1.538b | 0.124 |
| Month 3 | 30 (21–56) | 31 (22–52) | −0.477b | 0.633 |
| Month 6 | 26 (21–61) | 29 (16–53) | −0.106b | 0.916 |
| HBV DNA (lg
copies/ml) |
|
|
|
|
| Baseline | 5.85
(3.69–6.60) | 6.85
(4.47–8.61) | −1.217b | 0.223 |
| Month 3 | <3.00
(<3.00–4.52) | 4.20
(3.00–6.79) | −2.376b | 0.018 |
| Month 6 | All
<3.00 | 4.23
(3.06–8.12) | −3.481b | <0.001 |
Viral QS in the RT region and S
region
A total of 608 sequences (range, 21–26 per serum
sample) were generated and analyzed. The characteristics of QS were
analyzed for the RT and S gene region in each patient.
In the RT region, no significant difference in the
QS composition was identified between RS and NRS at baseline, at
either the nucleotide or the amino acid level. Compared with the QS
composition prior to treatment, no change was observed at six
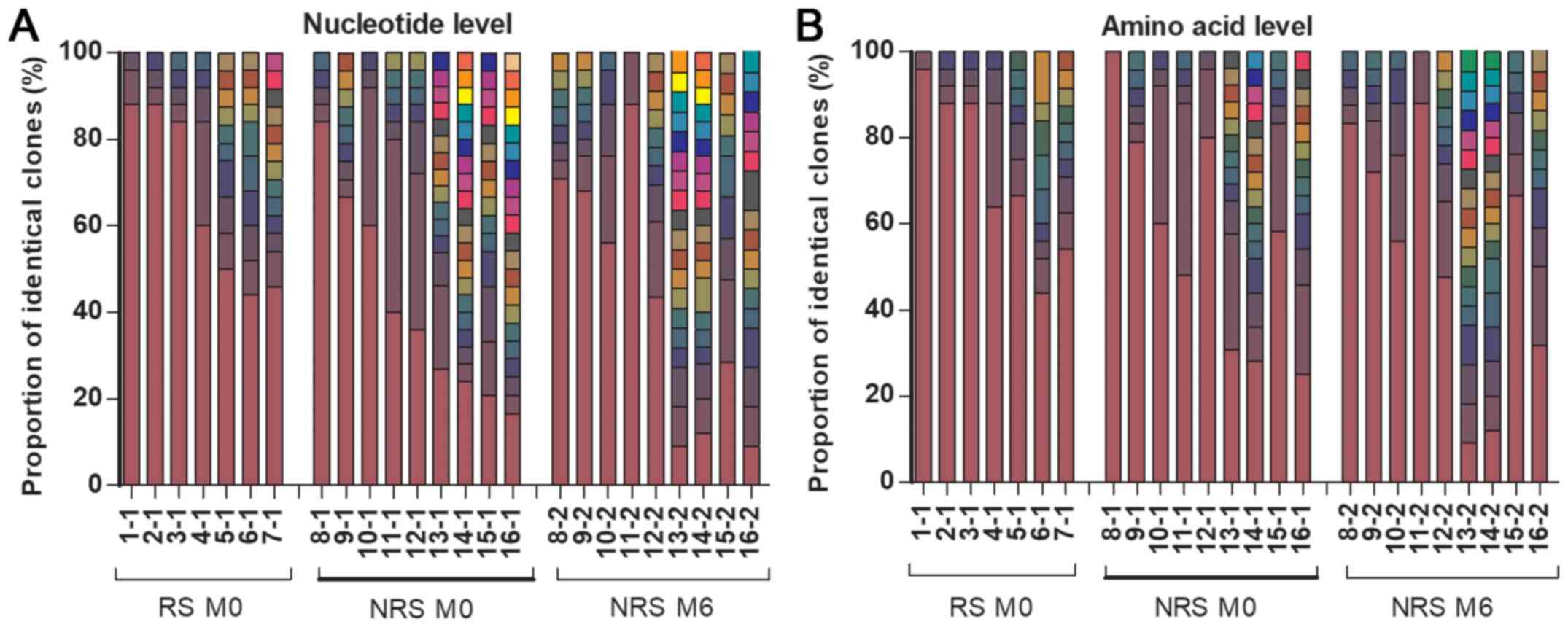
months, not even in the NRS (P>0.05; Fig. 1). In the RT region, the number of QS
and dominant strains exhibited no significant difference between RS
or NRS at baseline and NRS at six months, at either the nucleotide
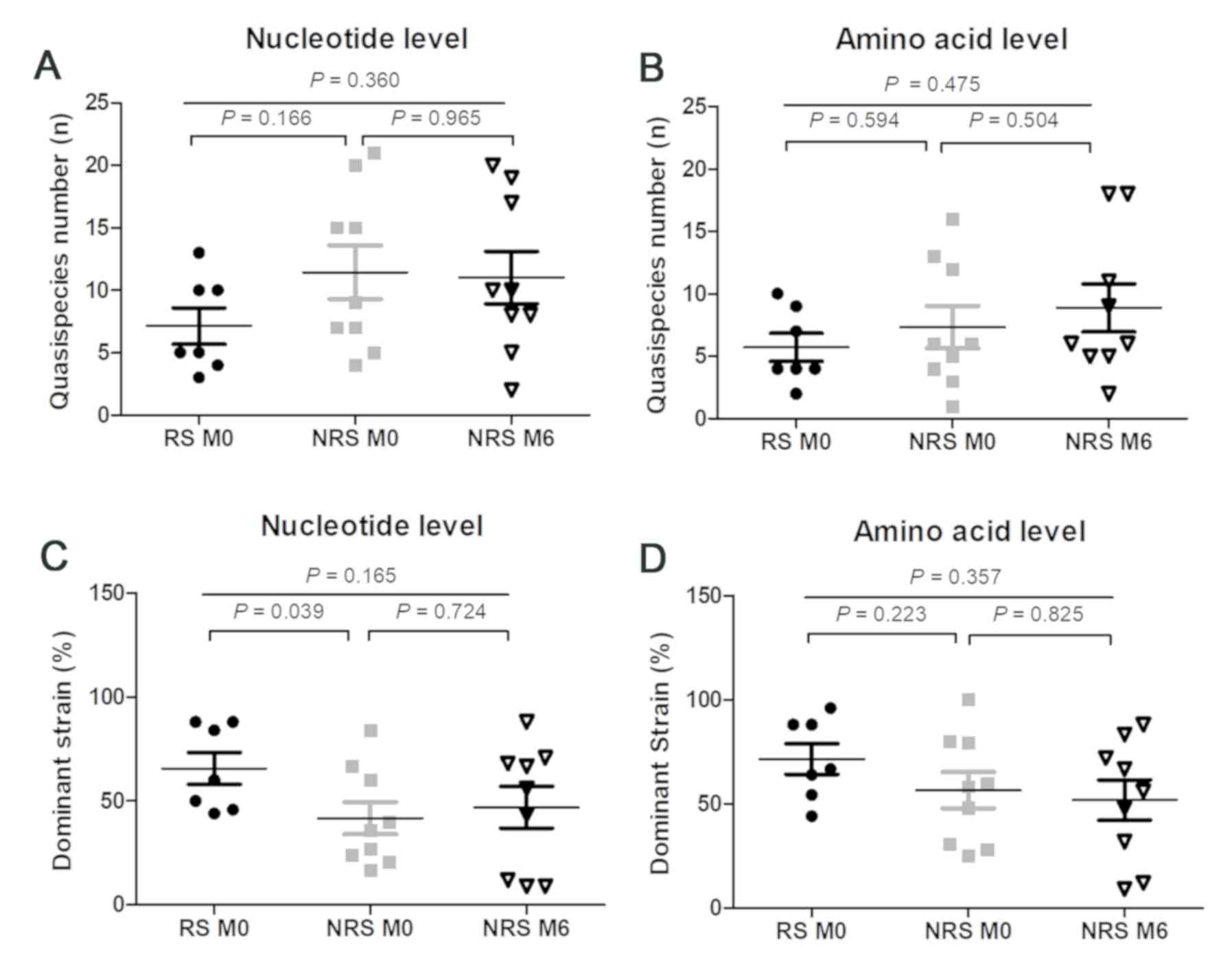
or the amino acid level (all P>0.05; Fig. 2). The number of QS in RS was also
similar to that in NRS at the baseline, at the nucleotide and the
amino acid level (P=0.166 and 0.594, respectively; Fig. 2A and B). The frequency of the
dominant strain in RS was significantly higher than that in NRS at
the nucleotide level (P=0.039), but indistinguishable at the amino
acid level (P=0.223; Fig. 2C and D).
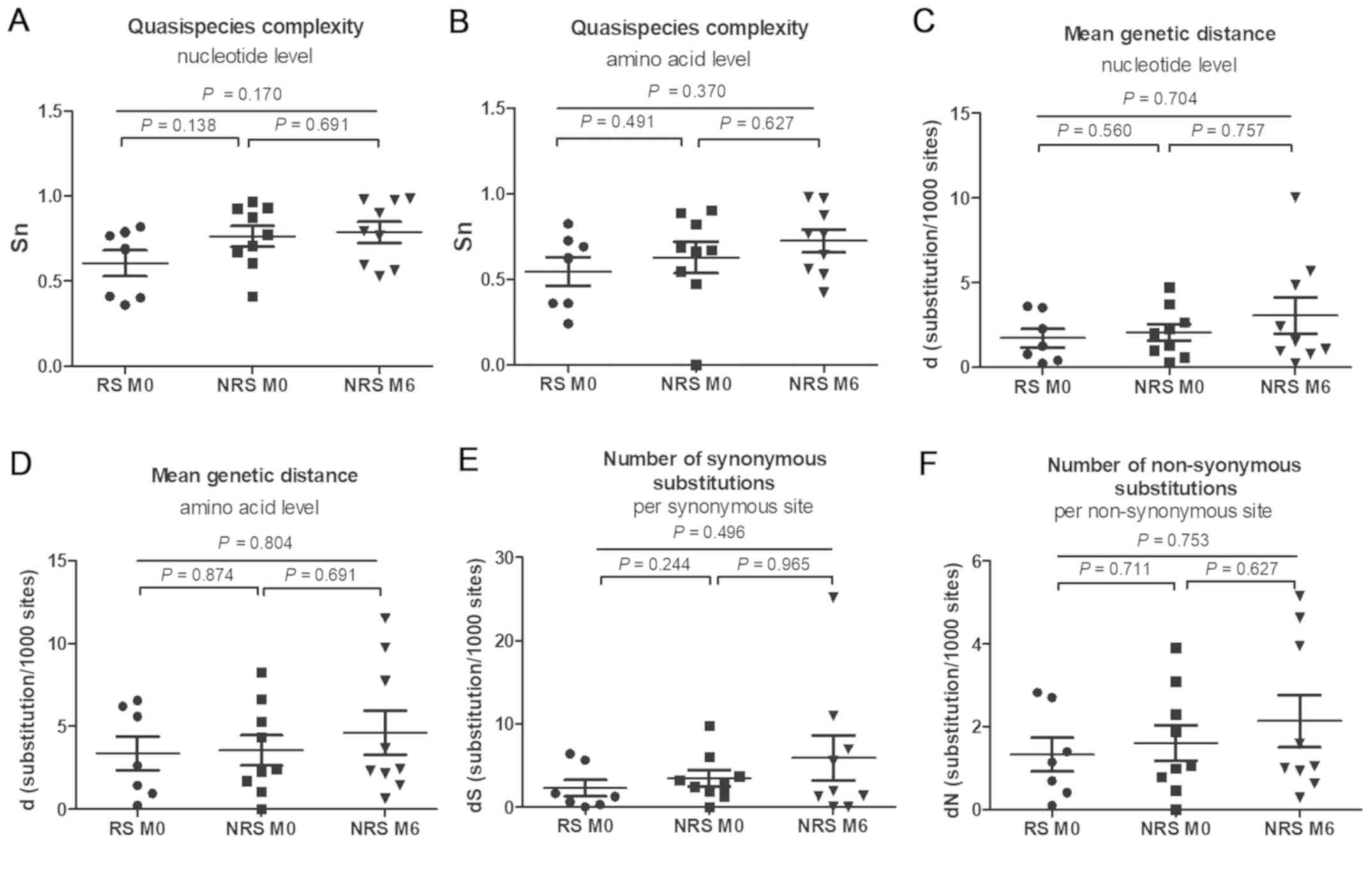
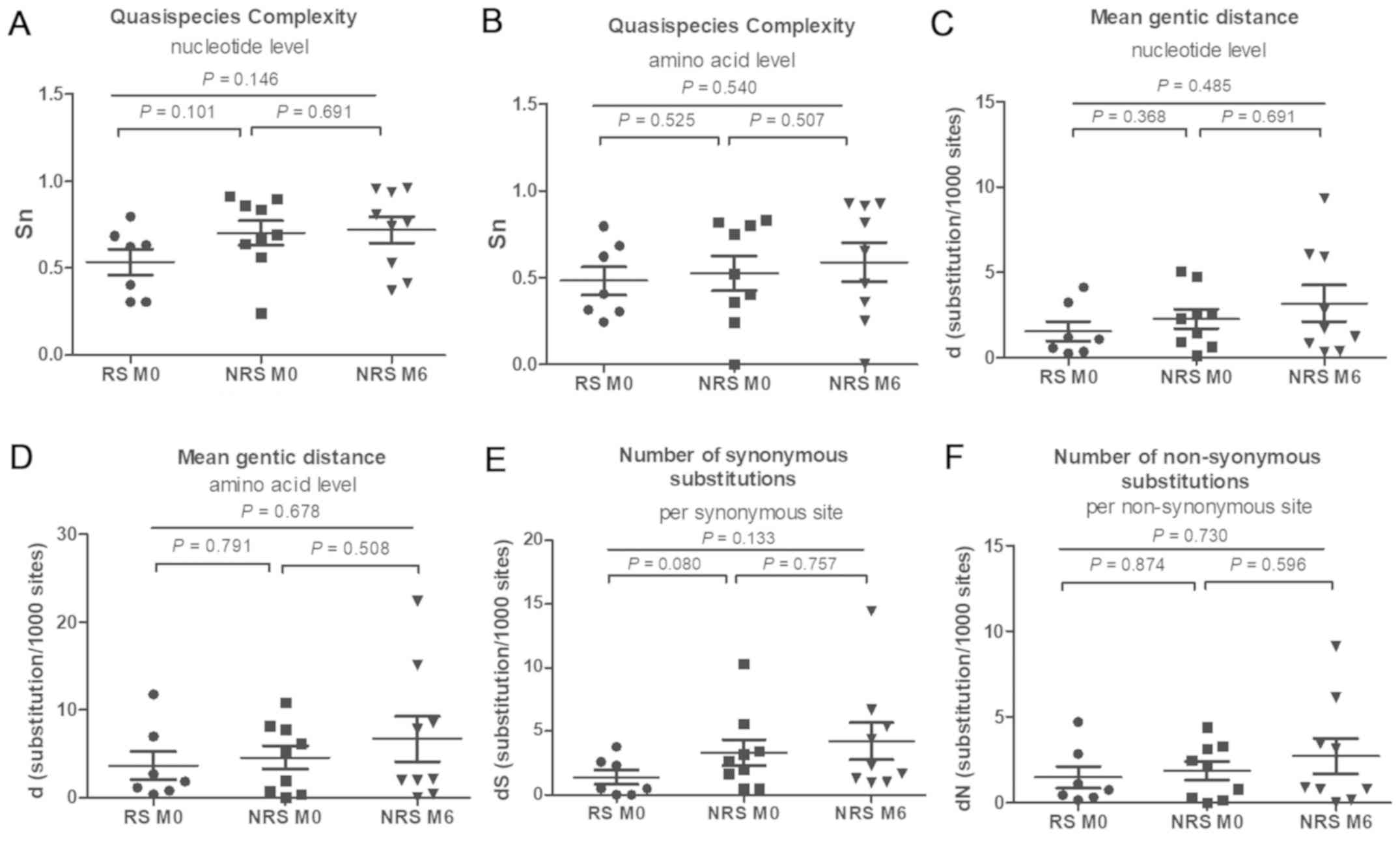
QS complexity and diversity was not significantly different between
RS and NRS at baseline and NRS at six months, at the nucleotide and
the amino acid level in the RT region (all P>0.05; Fig. 3). QS complexity at baseline exhibited
no significant difference between RS and NRS at the nucleotide and
the amino acid level (P=0.138 and 0.491, respectively; Fig. 3A and B). In the NRS group, the QS
complexity at the baseline [0.7734 (0.4110–0.9680) and 0.6699
(0.0000–0.9045) at the nucleotide and amino acid level,
respectively] was not significantly different from that at six
months [0.7961 (0.5294–0.9856) and 0.7654 (0.4235–0.9822) at the
nucleotide and amino acid level, respectively; P=0.691 and 0.627,
respectively]. The QS diversity was not significantly different
between the two groups at baseline (P>0.05; Fig. 3C-F). No significant difference in d,
dS and dN between the baseline and month six was identified in the
NRS group (P>0.05).
Similar results to those for the RT region were
obtained for the S region. The QS composition was similar to that
of the RT region (data not shown). The number of QS and dominant
strains exhibited no significant difference between RS and NRS at
the baseline and NRS at six months, at either the nucleotide or
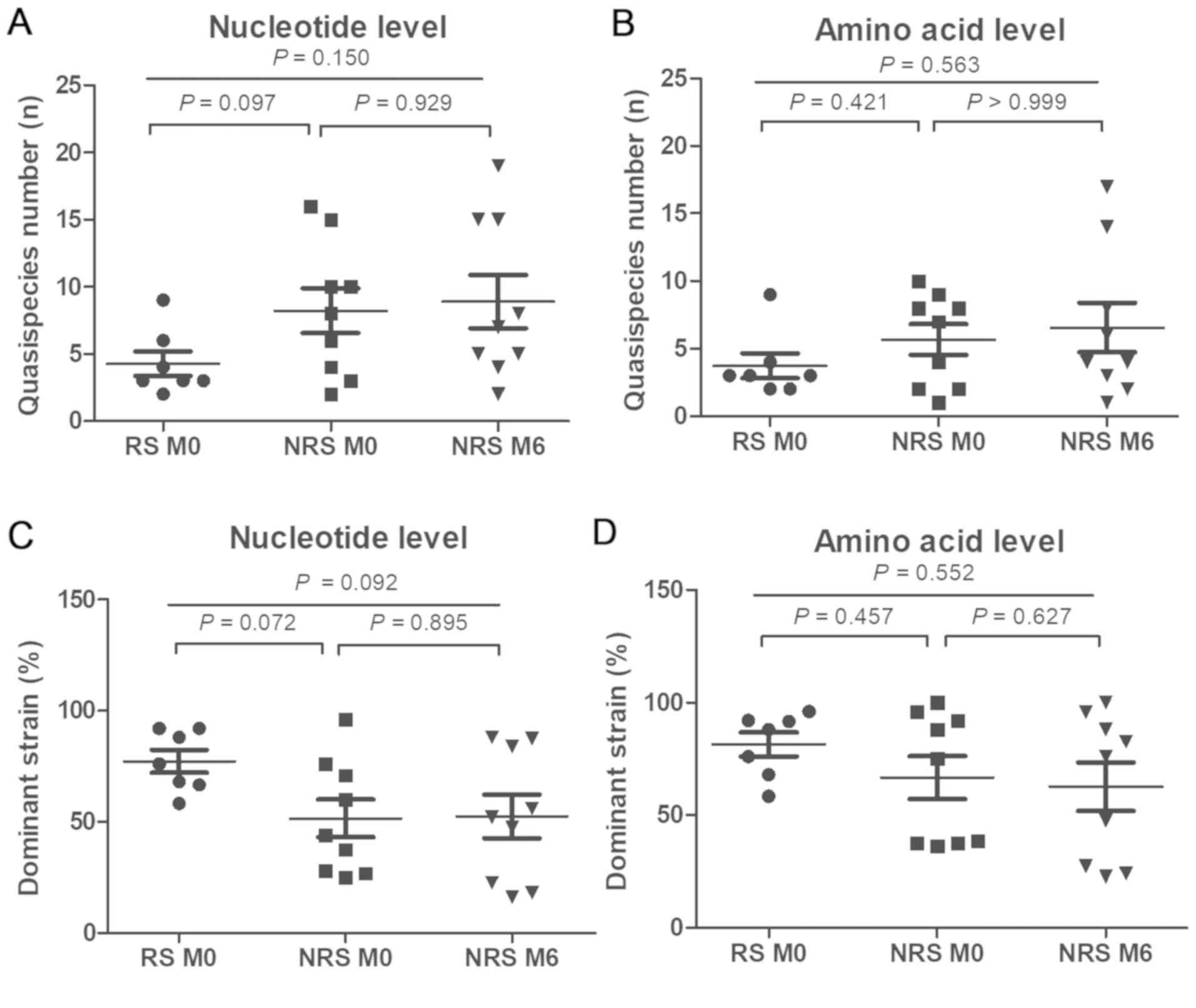
amino acid level in the S region (all P>0.05; Fig. 4). The number of QS and dominant
strains in RS was not significantly different from that in NRS, at
the nucleotide and the amino acid level (P>0.05; Fig. 4). Furthermore, the number of QS and
dominant strains for NRS at the nucleotide and the amino acid level
were similar between the baseline and at six months (P>0.05;
Fig. 4). QS complexity and diversity
in the S region were not significantly different between RS and NRS
at the baseline and NRS at six months, at the nucleotide and the
amino acid level (all P>0.05; Fig.
5). No significant difference was observed in QS complexity and
diversity between RS and NRS at baseline (P>0.05; Fig. 5). In the NRS group, the QS complexity
and diversity did not significantly differ between baseline and six
months (P>0.05).
Mutations within the HBV RT and S
region
Mutated residues and mutation frequencies at
different locations of HBV RT in clones of the RS and NRS group are
summarized in Table II. Sequence
analysis revealed that almost all clones had rtM204V/I resistance
mutations prior to LAM/ADV combination rescue treatment, and the
frequency of this mutation was not significantly different between
the RS and NRS [167/173 (96.5%) vs. 221/223 (99.1%), P=0.069].
However, in NRS, the frequency of this mutation at baseline was
higher than that at six months of combination treatment [221/223
(99.1%) vs. 190/212 (89.6%), P<0.001].
 | Table II.Mutation residues and frequencies at
positions within the RT region of hepatitis B virus for common
antiviral drug-associated resistance variants. |
Table II.
Mutation residues and frequencies at
positions within the RT region of hepatitis B virus for common
antiviral drug-associated resistance variants.
|
| Frequency of
mutations by number of clones (%)a |
| Frequency of
mutations by number of samples (%)b |
|
|---|
|
|
|
|
|
|
|---|
| RT region
mutations | Baseline | Month 6 | P-value | Baseline | Month 6 | P-value |
|---|
|
Lamivudine-associated resistance |
|
|
|
|
|
|
| L80I/V |
|
| 0.957c | 28.6 |
| 0.276c |
| RS | 28.9 |
|
|
|
|
|
|
NRS | 29.1 | 11.3 |
<0.001d | 55.6 | 33.3 | 0.341d |
| L82M/F |
|
| 0.129 |
|
| 0.112 |
| RS | 0 |
|
| 0 |
|
|
|
NRS | 0.9 | 0 | 0.102 | 22.2 | 0 | 0.082 |
| L91I |
|
| <0.001 |
|
| 0.055 |
| RS | 26.6 |
|
| 28.6 |
|
|
|
NRS | 0 | 0.9 | 0.089 | 0 | 11.1 | 0.229 |
| T128A |
|
| <0.001 |
|
| 0.375 |
| RS | 28.3 |
|
| 28.6 |
|
|
|
NRS | 11.2 | 11.8 | 0.849 | 11.1 | 11.1 | 1.000 |
| N139D |
|
| 0.189 |
|
| 0.187 |
| RS | 0.6 |
|
| 14.3 |
|
|
|
NRS | 0 | 0 | 1.000 | 0 | 0 | 1.000 |
| R153Q |
|
| 0.129 |
|
| 0.273 |
| RS | 0 |
|
| 0 |
|
|
|
NRS | 0.9 | 0 | 0.102 | 11.1 | 0 | 0.229 |
| L180M |
|
| 0.038 |
|
| 0.377 |
| RS | 39.3 |
|
| 57.1 |
|
|
|
NRS | 49.8 | 57.1 | 0.127 | 77.8 | 77.8 | 1.000 |
| M204V |
|
| 0.453 |
|
| 0.614 |
| RS | 39.3 |
|
| 42.9 |
|
|
|
NRS | 43.0 | 45.3 | 0.639 | 55.6 | 55.6 | 1.000 |
| M204I |
|
| 0.816 |
|
| 0.949 |
| RS | 57.2 |
|
| 57.1 |
|
|
|
NRS | 56.1 | 44.3 | 0.015 | 55.6 | 55.6 | 1.000 |
| M204I/V |
|
| 0.069 |
|
| 1.000 |
| RS | 96.5 |
|
| 100 |
|
|
|
NRS | 99.1 | 89.6 | <0.001 | 100 | 100.0 | 1.000 |
| L229V/W/M/F |
|
| <0.001 |
|
| 0.614 |
| RS | 36.4 |
|
| 57.1 |
|
|
|
NRS | 19.7 | 17.0 | 0.459 | 44.4 | 44.4 | 1.000 |
| S256G/C |
|
| 0.944 |
|
| 0.849 |
| RS | 11.0 |
|
| 14.3 |
|
|
|
NRS | 10.8 | 10.8 | 0.977 | 11.1 | 11.1 | 1.000 |
| Adefovir-associated
resistance |
|
|
|
|
|
|
| V84M |
|
| <0.001 |
|
| 0.187 |
| RS | 14.5 |
|
| 14.3 |
|
|
|
NRS | 0.0 | 8.5 | <0.001 | 0 | 11.1 | 0.229 |
| N118H/D/T |
|
| 0.411 |
|
| 0.684 |
| RS | 14.5 |
|
| 14.3 |
|
|
|
NRS | 11.7 | 11.3 | 0.912 | 22.2 | 11.1 | 0.524 |
| A181T |
|
| 0.068 |
|
| 0.187 |
| RS | 1.2 |
|
| 14.3 |
|
|
|
NRS | 0 | 0 | 1.000 | 0 | 0 | 1.000 |
| S213T |
|
| 0.004 |
|
| 0.273 |
| RS | 0.0 |
|
| 0 |
|
|
|
NRS | 3.1 | 1.4 | 0.223 | 11.1 | 11.1 | 1.000 |
| I233L |
|
| 0.283 |
|
| 0.273 |
| RS | 0.0 |
|
| 0 |
|
|
|
NRS | 0.4 | 0 | 0.247 | 11.1 | 0 | 0.229 |
| N236T |
|
| 1.000 |
|
| 1.000 |
| RS | 0 |
|
| 0 |
|
|
|
NRS | 0 | 0 | 1.000 | 0 | 0 | 1.000 |
| P237T |
|
| 1.000 |
|
| 1.000 |
| RS | 0.0 |
|
| 0 |
|
|
|
NRS | 0.0 | 0.5 | 0.230 | 0 | 11.1 | 0.229 |
|
Lamivudine+adefovir-associated
resistance |
|
|
|
|
|
|
| rtV191I |
|
| <0.001 |
|
| 0.375 |
| RS | 15.0 |
|
| 28.6 |
|
|
|
NRS | 0.9 | 0.5 | 0.588 | 11.1 | 11.1 | 1.000 |
| V207I/M |
|
| 0.004 |
|
| 0.273 |
| RS | 0 |
|
| 0 |
|
|
|
NRS | 3.1 | 2.8 | 0.850 | 11.1 | 22.2 | 0.524 |
| Q215P/S/H |
|
| <0.001 |
|
| 0.187 |
| RS | 14.5 |
|
| 14.3 |
|
|
|
NRS | 0 | 0.0 | 1.000 | 0 | 0 | 1.000 |
|
Entecavir-associated resistance |
|
|
|
|
|
|
| rtI169T |
|
| 0.129 |
|
| 0.273 |
| RS | 0 |
|
| 0 |
|
|
|
NRS | 0.9 | 0.9 | 0.959 | 11.1 | 11.1 | 1.000 |
| T184L/S/A |
|
| <0.001 |
|
| 0.112 |
| RS | 0 |
|
| 0 |
|
|
|
NRS | 11.7 | 15.6 | 0.234 | 22.2 | 22.2 | 1.000 |
| S202G |
|
| <0.001 |
|
| 0.273 |
| RS | 0 |
|
| 0 |
|
|
|
NRS | 9.4 | 7.5 | 0.485 | 11.1 | 11.1 | 1.000 |
| Other possible
associated resistance |
|
|
|
|
|
|
| M271L |
|
| 0.129 |
|
| 0.273 |
| RS | 0 |
|
| 0 |
|
|
|
NRS | 0.9 | 0 | 0.102 | 11.1 | 0 | 0.229 |
| W284e |
|
| 0.100 |
|
| 0.187 |
| RS | 2.3 |
|
| 14.3 |
|
|
|
NRS | 0 | 0 | 1.000 | 0 | 0 | 1.000 |
| N337H/T |
|
| <0.001 |
|
| 0.684 |
| RS | 14.5 |
|
| 14.3 |
|
|
|
NRS | 0.9 | 1.4 | 0.611 | 22.2 | 22.2 | 1.000 |
Prior to combination treatment, the rtA181V mutation
was identified in one patient in the RS group (two in 24 clones),
and no ADV resistance mutation (rtN236T) was detected. Common and
frequent mutations identified in the cohort of the present study
included rtL80V/I, rtV84M, rtL91I, rtN118H/D/T, rtT128A, rtL180M,
rtT184L/S/A, rtV191I, rtS202G, rtV207I/M, rtS213T, rtQ215P/S/H,
rtL229V/M/W/F, rtS256G/C, and rtN337H/T, as presented in Table II. In addition, a stop codon
mutation at site 284 was identified in one patient in the RS group
(4 in 25 clones).
In the present study, the most prevalent mutations
were those in the S region (sP120Q, sQ129R, sT131I, sM133I, sG145R,
sY161F/H, sI195M/T, sW196S/L, sW199C/L and sM213I/T; Table III). Furthermore, three out of five
stop codons in the S region (sS34*, sW74*, sW172*, sW182*, sW196*)
were identified in the RT region; the stop codons have been
associated with NAs resistance, also affected the HBsAg reading
frame (rtA181T/sW172*, rtV191I/sW182* and rtM204I/sW196*).
 | Table III.Mutation residue frequencies at
different positions of the HBV S region. |
Table III.
Mutation residue frequencies at
different positions of the HBV S region.
|
|
| Frequency of clones
mutations (%)a |
| Frequency of
individuals mutations (%)b |
|
|---|
|
|
|
|
|
|
|
|---|
| S region
mutations | Response | Baseline | Month 6 | P-value | Baseline | Month 6 | P-value |
|---|
| Stop codon
mutation |
|
|
|
|
|
|
|
|
S34*e | RS | 0 |
| 0.283c | 0 |
| 0.273c |
|
| NRS | 0.4 | 0 | 0.247d | 11.1 | 0 | 0.229d |
|
W74*e | RS | 0 |
| 0.283c | 0 |
| 0.273c |
|
| NRS | 0.4 | 0 | 0.247d | 11.1 | 0 | 0.229d |
|
W172*e | RS | 1.2 |
| 0.068c | 14.3 |
| 0.187c |
|
| NRS | 0 | 0 | 1.000d | 0 | 0 | 1.000d |
|
W182*e | RS | 15.0 |
|
<0.001c | 14.3 |
| 0.375c |
|
| NRS | 0.9 | 0.5 | 0.588d | 11.1 | 11.1 | 1.000d |
|
W196*e | RS | 14.5 |
|
<0.001c | 14.3 |
| 0.849c |
|
| NRS | 0.4 | 5.7 | 0.001d | 11.1 | 11.1 | 1.000d |
| Other
mutations |
|
|
|
|
|
|
|
|
P120Q | RS | 0 |
| 1.000c | 0 |
| 1.000c |
|
| NRS | 0 | 0.5 | 0.230d | 0 | 11.1 | 0.229d |
|
Q129R | RS | 11.0 |
|
<0.001c | 14.3 |
| 0.187c |
|
| NRS | 0 | 0 | 1.000d | 0 | 0 | 1.000d |
|
T131I | RS | 9.2 |
|
<0.001c | 14.3 |
| 0.187c |
|
| NRS | 0 | 0 | 1.000d | 0 | 0 | 1.000d |
|
M133I | RS | 0 |
| 1.000c | 0 |
| 1.000c |
|
| NRS | 0 | 0.5 | 0.230d | 0 | 11.1 | 0.229d |
|
G145R | RS | 0 |
| 0.129c | 0 |
| 0.273c |
|
| NRS | 0.9 | 0 | 0.102d | 11.1 | 0 | 0.229d |
|
Y161F/H | RS | 0 |
|
<0.001c | 0 |
| 0.112c |
|
| NRS | 12.6 | 10.4 | 0.476d | 22.2 | 11.1 | 0.524d |
|
I195M/T | RS | 53.8 |
| 0.034c | 57.1 |
| 0.949c |
|
| NRS | 43.0 | 45.3 | 0.639d | 55.6 | 55.6 | 1.000d |
|
W196S/L | RS | 42.8 |
| 0.030c | 57.1 |
| 0.949c |
|
| NRS | 56.1 | 38.7 | 0.002d | 55.6 | 55.6 | 1.000d |
|
W199C/L | RS | 0.6 |
| 0.053c | 14.3 |
| 0.849c |
|
| NRS | 3.1 | 1.4 | 0.223d | 11.1 | 11.1 | 1.000d |
|
M213I/T | RS | 18.5 |
|
<0.001c | 28.6 |
| 0.513c |
|
| NRS | 43.5 | 37.3 | 0.185d | 44.4 | 44.4 | 1.000d |
Phylogenetic analysis
A phylogenetic tree was constructed with the maximum
likelihood algorithm by aligning the HBV RT and S regions. To keep
the phylogenetic tree concise, identical sequences were discarded
and all QS from a patient were retained. As expected, the sequences
from individual patients clustered together. While it was not
possible to accurately calculate the mean branch lengths for the RS
and NRS, they appeared similar (data not shown).
Discussion
Despite the availability of first-line NA therapy
with a high resistance gene barrier, LAM has been widely used in
China for CHB treatment (25). As a
result, a large number of LAM-resistant patients have emerged, and
NA resistance remains a serious problem in China (25). As a rescue treatment for
LAM-resistant patients, in addition to TDF, LAM/ADV combination
treatment is provided to a large number of patients due to economic
reasons and availability (26).
In vivo and in vitro studies have suggested that ADV
is effective in patients with LAM resistance arising from YMDD
mutation (5–7). The present study indicated that LAM/ADV
combination treatment suppresses viral replication in certain
LAM-resistant CHB patients with YMDD mutation within six months.
However, 9 patients had a poor response at six months and 2 even
experienced a virological breakthrough. The reason for the poor
response to LAM/ADV combination treatment in cases with LAM
resistance had remained elusive and required to be investigated.
Earlier studies reported that HBV DNA levels are correlated with
anti-viral efficacy. According to the Roadmap Concept (report of an
international workshop), HBV DNA levels at week 24 are an important
indicator for anti-viral efficacy and drug management (27). In the present study, patients were
divided into two groups based on their virological response to
combined ADV treatment at six months. No significant differences in
age, sex, drug resistance to LAM, YMDD pattern, HBV genotype, ALT
levels and HBeAg status were identified between these groups. HBV
DNA levels at month three were revealed to be better at predicting
the response to treatment for combination treatment than that at
month six. In NRS, HBV DNA decreased significantly in the first
three months and then plateaued. It remains elusive whether viral
QS are associated with the efficacy of combination therapy. Since
the RT region of HBV polymerase is the target of NAs and the S
region overlaps with the RT region, the dynamic changes of HBV QS
within the RT and S regions were investigated during LAM/ADV
combination treatment to study their effects on anti-viral efficacy
and drug resistance.
Most of the previous studies investigating this
matter suggested that HBV QS heterogeneity in the RT and/or S
regions may be a predictor of the anti-viral efficacy of NAs
(13–15,28–30). In
the present study, however, the complexity and diversity of HBV QS,
whether in the RT or S region, were identified to be comparable
between RS and NRS at baseline, suggesting that the QS
characteristics of patients with CHB at baseline were not
associated with the virological response in this cohort of
patients. This result echoed those from previous studies on
patients with CHB receiving NAs therapy. Chen et al
(13) and Liu et al (14) investigated the HBV QS heterogeneity
at baseline and identified no association with the response to NA
treatment. Other studies obtained opposite results. For instance, a
study by Han et al (29)
using the massively parallel ultra-deep DNA pyrosequencing method
suggested that the HBV QS heterogeneity at baseline is a predictor
for the efficacy of LAM. In the present study, no statistically
significant difference in QS heterogeneity, in either RT or the S
region, was identified between the baseline and six months of
treatment in NRS. This was also in line with the baseline results
of a study by Lavocat et al (31), which obtained no significant
difference at the baseline in QS complexity or diversity between
treatment groups of patients with incomplete ADV response receiving
TDF monotherapy or emtricitabine in combination. The QS complexity
increased at week 12, and the complexity and diversity decreased at
week 48 in the two treatment groups (31). Whether HBV QS heterogeneity in the
present cohort had a trend similar to that reported by Lavocat
et al (31), which exhibited
an early rise followed by a decline, has not been explored in the
present study. The present study also indicated that the QS
composition of RS was not significantly different from that of the
NRS at baseline. Although the number of RT region dominant strains
at the nucleotide level in the RS group was relatively higher than
that in the NRS at baseline, the number of QS in RS was not
significantly higher than that in NRS. Furthermore, analysis of the
phylogenetic tree indicated that, regardless of the presence of LAM
resistance, at six months after combination treatment with ADV, QS
components and complexity were not significantly changed.
Therefore, the baseline QS had no significant predictive value
regarding the efficacy of LAM/ADV combination therapy regarding
YMDD mutant-associated LAM resistance. In the present study, a YMDD
mutation was detected in all patients, and the Y-valine (V)DD and
Y-isoleucine (I)DD mutation rates were also similar. During LAM/ADV
combination rescue therapy, although the frequency of YMDD mutation
declined, it did not disappear in NRS after six months of
treatment. In a previous study, Ijaz et al (32) indicated that no reversion to
wild-type HBV occurred in patients with LAM-resistant HBV receiving
LAM/ADV combination therapy. In addition to the emergence of the
YMDD mutation, other LAM-associated mutations were also detected in
the clonal analysis, e.g. rtA181T was detected in one patient in
the RS group. The rtA181T mutation has been suggested as an
atypical mutation associated with LAM and ADV resistance (33). This patient developed a virological
breakthrough during the follow-up (data not shown). The ADV
resistance- associated rtN236T mutation was not detected at all in
the present study. Other mutations, including rtV84M, rtN118H/D/T,
rtI169T, rtT184A/L/S, rtV191I, rtV207I/M, rtS213T, rtQ215P/S/H,
rtI233L, rtP237H/T and rtS202G, which have been previously reported
to be associated with ADV or ETV resistance (34,35),
were detected at baseline or after six months of LAM/ADV treatment.
In addition, the substitution mutation rtA194T, which has been
associated with TDF resistance (36), was not detected. Hence, these results
suggest that TDF, but not ADV or ETV, should be considered for the
treatment of patients with LAM resistance. In addition to these
well-documented major and compensatory mutations, 2 more mutations
(rtS256G/C and rtM337H/T) were detected at a high rate.
Furthermore, rtW284* was detected in one RS patient (4 of 25
clones) at baseline. The significance of these mutations requires
to be investigated in follow-up studies. However, in the present
study, no definite association between certain mutations and a
patient's response to combination therapy was established.
Since the polymerase gene overlaps with the S gene,
a mutation in the nucleoside may result in amino acid mutations in
the RT and S genes. Similar to previous studies, the present study
suggested that truncation mutations (rtA181T/sW172*, rtM204I/sW196*
and rtV191I/sW182*) occurred at the highest frequency (37,38).
Other studies have indicated that these three mutants may hamper
virion secretion, leading to HBsAg protein retention and decreased
serum HBV DNA levels. In the present study, most S region mutations
were located in the ‘a’ determinant and were detected in NRS rather
than in RS (17). In contrast to
previous studies, a number of S gene mutations (e.g. sP120T and
sG145R) that may be associated with LAM resistance (39), were not detected in the present
study. However, the significance of these mutations has remained to
be assessed in detail.
Of note, the present study had certain limitations.
First, the sample size was relatively small. Furthermore, the
present study only cloned HBV DNA samples obtained at baseline and
six months after LAM/ADV rescue treatment. HBV DNA from other
follow-up time-points were not cloned due to lack of available
serum samples. In addition, the cloning strategy applied in the
present study was expensive and labor-intensive, and ultra-deep
pyrosequencing may be a good alternative, but cloning/sequencing is
considered as the gold standard technique for assessment of viral
QS (14). However, the major aim of
the present study was to assess the QS of patients with
drug-resistance, which is different from that of other studies on
treatment-naïve patients.
In conclusion, the present study indicated that in
CHB patients with LAM resistance, the baseline HBV QS within the RT
and S regions was not significantly different between RS and NRS
during rescue treatment with LAM/ADV. The baseline viral QS of HBV
in patients with YMDD mutations may not predict the efficacy of
combination therapy with LAM/ADV in LAM-resistant patients. In
addition, no specific HBV mutant was selected during rescue
treatment with LAM/ADV for LAM resistance. Finally, LAM resistance
may occur in HBV variants with cross-resistance to other NAs,
including Telbivudine Entecavir and ADV (when rt180 mutation
occurs), so rescue treatment with TDF rather than ADV should be
recommended. Future large-scale studies using samples collected
prior to and after therapy with NAs are required to clarify the
resistance-associated dynamics of viral QS.
Acknowledgements
Not applicable.
Funding
The current study was supported by the National
Science Foundation of China (grant no. 81273142) and the Provincial
Natural Science Foundation (grant no. 1608085MH162).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZZ conceived the study, performed the literature
search and edited the manuscript. CW, SY, YZ, MZ, LL and CH
performed the experiments. CW, CH and ZZ analyzed the data and
wrote the manuscript. XL, CH and JL reviewed the manuscript. XL was
responsible for the acquisition of funding, collection of data and
general supervision of the research group. XL, JL and ZZ provided
reagents, materials and analysis tools. CW, JL and ZZ interpreted
the data.
Ethics approval and patient consent
The study was approved by the Ethics Committee of
Anhui Medical University (Hefei, China) and written informed
consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ott JJ, Stevens GA, Groeger J and Wiersma
ST: Global epidemiology hepatitis B virus infection: New estimates
of age-specific HBsAg seroprevalence and endemicity. Vaccine.
30:2212–2219. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lavanchy D: Hepatitis B virus
epidemiology, disease burden, treatment, and current and emerging
prevention and control measures. J Viral Hepat. 11:97–107. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Geretti AM, Patel M, Sarfo FS, Chadwick D,
Verheyen J, Fraune M, Garcia A and Phillips RO: Detection of highly
prevalent hepatitis B virus coinfection among HIV-seropositive
persons in Ghana. J Clin Microbiol. 48:3223–3230. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lok AS, Lai CL, Leung N, Yao GB, Cui ZY,
Schiff ER, Dienstag JL, Heathcote EJ, Little NR, Griffiths DA, et
al: Long-term safety of lamivudine treatment in patients with
chronic hepatitis B. Gastroenterology. 125:1714–1722. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim HJ, Park JH, Park DI, Cho YK, Sohn CI,
Jeon WK and Kim BI: Rescue therapy for lamivudine-resistant chronic
hepatitis B: Comparison between entecavir 1.0 mg monotherapy,
adefovir monotherapy and adefovir add-on lamivudine combination
therapy. J Gastroenterol Hepatol. 25:1374–1380. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Perrillo R, Hann HW, Mutimer D, Willems B,
Leung N, Lee WM, Moorat A, Gardner S, Woessner M, Bourne E, et al:
Adefovir dipivoxil added to ongoing lamivudine in chronic hepatitis
B with YMDD mutant hepatitis B virus. Gastroenterology. 126:81–90.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vassiliadis TG, Giouleme O, Koumerkeridis
G, Koumaras H, Tziomalos K, Patsiaoura K, Grammatikos N,
Mpoumponaris A, Gkisakis D, Theodoropoulos K, et al: Adefovir plus
lamivudine are more effective than adefovir alone in
lamivudine-resistant HBeAg-chronic hepatitis B patients: A 4-year
study. J Gastroenterol Hepatol. 25:54–60. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee YS, Suh DJ, Lim YS, Jung SW, Kim KM,
Lee HC, Chung YH, Lee YS, Yoo W and Kim SO: Increased risk of
adefovir resistance in patients with lamivudine-resistant chronic
hepatitis B after 48 weeks of adefovir dipivoxil monotherapy.
Hepatology. 43:1385–1391. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou B, Dong H, He Y, Sun J, Jin W, Xie Q,
Fan R, Wang M, Li R, Chen Y, et al: Composition and interactions of
hepatitis B virus quasispecies defined the virological response
during telbivudine therapy. Sci Rep. 5:171232015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Domingo E and Gomez J: Quasispecies and
its impact on viral hepatitis. Virus Res. 127:131–150. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fan X, Mao Q, Zhou D, Lu Y, Xing J, Xu Y,
Ray SC and Di Bisceglie AM: High diversity of hepatitis C viral
quasispecies is associated with early virological response in
patients undergoing antiviral therapy. Hepatology. 50:1765–1772.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sherman KE, Rouster SD, Stanford S,
Blackard JT, Shire N, Koziel M, Peters M and Chung RT;: AIDS
Clinical Trials Group 5071 Study Team: Hepatitis C virus (HCV)
quasispecies complexity and selection in HCV/HIV-coinfected
subjects treated with interferon-based regimens. J Infect Dis.
201:712–719. 2010.PubMed/NCBI
|
|
13
|
Chen L, Zhang Q, Yu DM, Wan MB and Zhang
XX: Early changes of hepatitis B virus quasispecies during
lamivudine treatment and the correlation with antiviral efficacy. J
Hepatol. 50:895–905. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu F, Chen L, Yu DM, Deng L, Chen R,
Jiang Y, Chen L, Huang SY, Yu JL, Gong QM and Zhang XX:
Evolutionary patterns of hepatitis B virus quasispecies under
different selective pressures: Correlation with antiviral efficacy.
Gut. 60:1269–1277. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Peveling-Oberhag J, Herrmann E,
Kronenberger B, Farnik H, Susser S, Sarrazin C, Zeuzem S and
Hofmann WP: Dynamics of hepatitis B virus quasispecies
heterogeneity and virologic response in patients receiving
low-to-moderate genetic barrier nucleoside analogs. J Viral Hepat.
20:234–239. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yin F, Wu Z, Fang W, Wu C, Rayner S, Han
M, Deng F, Du R, Liu J, Wang M, et al: Resistant mutations and
quasispecies complexity of hepatitis B virus during telbivudine
treatment. J Gen Virol. 96:3302–3312. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sheldon J and Soriano V: Hepatitis B virus
escape mutants induced by antiviral therapy. J Antimicrob
Chemother. 61:766–768. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen CH, Lee CM, Tung WC, Wang JH, Hung
CH, Hu TH, Wang JC, Lu SN and Changchien CS: Evolution of
full-length HBV sequences in chronic hepatitis B patients with
sequential lamivudine and adefovir dipivoxil resistance. J Hepatol.
52:478–485. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu H, Wang C, Zhang Y, Wei S, Li X and
Zhang Z: Prediction model for sustained hepatitis B e antigen
seroconversion to peginterferon alfa-2a in chronic hepatitis B. J
Gastroenterol Hepatol. 31:1963–1970. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hall TA: BioEdit: A user-friendly
biological sequence alignment editor and analysis program for
Windows 95/98/NT. Nucl Acids Symp Ser. 41:95–98. 1999.
|
|
21
|
Thompson JD, Gibson TJ, Plewniak F,
Jeanmougin F and Higgins DG: The CLUSTAL_X windows interface:
Flexible strategies for multiple sequence alignment aided by
quality analysis tools. Nucleic Acids Res. 25:4876–4882. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Martin D and Rybicki E: RDP: Detection of
recombination amongst aligned sequences. Bioinformatics.
16:562–563. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Domingo E, Martin V, Perales C,
Grande-Pérez A, Garcia-Arriaza J and Arias A: Viruses as
quasispecies: Biological implications. Curr Top Microbiol Immunol.
299:51–82. 2006.PubMed/NCBI
|
|
24
|
Kumar S, Stecher G and Tamura K: MEGA7:
Molecular evolutionary genetics analysis version 7.0 for bigger
datasets. Mol Biol Evol. 33:1870–1874. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng X, Wang J and Yang D: Antiviral
therapy for chronic hepatitis B in China. Med Microbiol Immunol.
204:115–120. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee HJ, Kim SJ, Kweon YO, Park SY, Heo J,
Woo HY, Hwang JS, Chung WJ, Lee CH, Kim BS, et al: Evaluating the
efficacy of switching from lamivudine plus adefovir to tenofovir
disoproxil fumarate monotherapy in lamivudine-resistant stable
hepatitis B patients. PLoS One. 13:e01905812018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Keeffe EB, Zeuzem S, Koff RS, Dieterich
DT, Esteban-Mur R, Gane EJ, Jacobson IM, Lim SG, Naoumov N,
Marcellin P, et al: Report of an international workshop: Roadmap
for management of patients receiving oral therapy for chronic
hepatitis B. Clin Gastroenterol Hepatol. 5:890–897. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cheng Y, Guindon S, Rodrigo A and Lim SG:
Increased viral quasispecies evolution in HBeAg seroconverter
patients treated with oral nucleoside therapy. J Hepatol.
58:217–224. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Han Y, Gong L, Sheng J, Liu F, Li XH, Chen
L, Yu DM, Gong QM, Hao P and Zhang XX: Prediction of virological
response by pretreatment hepatitis B virus reverse transcriptase
quasispecies heterogeneity: The advantage of using next-generation
sequencing. Clin Microbiol Infect. 21:797.e1–e8. 2015. View Article : Google Scholar
|
|
30
|
Chen L, Gan QR, Zhang DQ, Yao LF, Lin RS,
Li Q, Lin MH, Yu DM, Zhang XX and Pan C: Increased intrahepatic
quasispecies heterogeneity correlates with off-treatment sustained
response to nucleos(t)ide analogues in e antigen-positive chronic
hepatitis B patients. Clin Microbiol Infect. 22:201–207. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lavocat F, Dény P, Pichoud C, Al Hawajri
N, Kitrinos K, Borroto-Esoda K and Zoulim F: Similar evolution of
hepatitis B virus quasispecies in patients with incomplete adefovir
response receiving tenofovir/emtricitabine combination or tenofovir
monotherapy. J Hepatol. 59:684–695. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ijaz S, Arnold C, Dervisevic S, Mechurova
J, Tatman N, Tedder RS and Naoumov NV: Dynamics of
lamivudine-resistant hepatitis B virus during adefovir monotherapy
versus lamivudine plus adefovir combination therapy. J Med Virol.
80:1160–1170. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Villet S, Pichoud C, Billioud G, Barraud
L, Durantel S, Trépo C and Zoulim F: Impact of hepatitis B virus
rtA181V/T mutants on hepatitis B treatment failure. J Hepatol.
48:747–755. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lapiński TW, Pogorzelska J and Flisiak R:
HBV mutations and their clinical significance. Adv Med Sci.
57:18–22. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang ZH, Wu CC, Chen XW, Li X, Li J and
Lu MJ: Genetic variation of hepatitis B virus and its significance
for pathogenesis. World J Gastroenterol. 22:126–144. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sheldon J, Camino N, Rodés B,
Bartholomeusz A, Kuiper M, Tacke F, Núñez M, Mauss S, Lutz T,
Klausen G, et al: Selection of hepatitis B virus polymerase
mutations in HIV-coinfected patients treated with tenofovir.
Antivir Ther. 10:727–734. 2005.PubMed/NCBI
|
|
37
|
Coppola N, Onorato L, Minichini C, Di
Caprio G, Starace M, Sagnelli C and Sagnelli E: Clinical
significance of hepatitis B surface antigen mutants. World J
Hepatol. 7:2729–2739. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang ML and Tang H: Nucleos(t)ide
analogues causes HBV S gene mutations and carcinogenesis.
Hepatobiliary Pancreat Dis Int. 15:579–586. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Amini-Bavil-Olyaee S, Vucur M, Luedde T,
Trautwein C and Tacke F: Differential impact of immune escape
mutations G145R and P120T on the replication of
lamivudine-resistant hepatitis B virus e antigen-positive and
-negative strains. J Virol. 84:1026–1033. 2010. View Article : Google Scholar : PubMed/NCBI
|