Introduction
Diabetic retinopathy (DR) is one of the major
microvascular complications of diabetes mellitus (DM), including
type 1 diabetes (T1D) and type 2 diabetes (T2D). It is one of the
leading causes of blindness in patients with DM worldwide,
particularly in industrialized countries (1,2). In
addition to hypertension and the duration of diabetes, chronic
hyperglycaemia has been considered to be the major risk factor for
the development of microvascular complications in patients with
diabetes (3). Pedigree studies have
indicated the presence of genetic influences in the development of
these complications (4,5). Recently, genome-wide linkage analyses
have begun to identify possible genetic associations with DR. A
number of candidate genes have been investigated, including
vascular endothelial growth factor (6), monocyte chemoattractant protein-1
(7), vitamin D receptor (8), aldehyde dehydrogenease 2 (9) and aldose reductase [ALR; also known as
aldoketo reductase family 1, member B1 (ALDR1)] (10,11).
ALR is the first and rate-limiting enzyme in the
human polyol pathway, which occurs under hyperglycemic conditions
(12). Excessive flux of glucose via
the polyol pathway has been implicated in the pathogenesis of
diabetic microvascular complications (13). The ALR gene, located on chromosome
7q35, extends over ~18 kb and consists of 10 exons that give rise
to a 1,384-nucleotide mRNA sequence, which encodes a 316-amino-acid
protein of 35,858 Da (14). It has
been reported as a candidate locus for diabetic nephropathy
(15,16). Previous candidate-gene studies and
meta-analyses have examined the hypothesis that two common ALR gene
variants, including 106C/T and (CA)n, are associated with the risk
of DR (17–20). (CA)n is a microsatellite variant
located 2.1 kb upstream of the ALR transcription initiation site
that has been intensively studied. It has been demonstrated that
(CA)n may cause increased enzyme levels or activity, thus
contributing to diabetic nephropathy and DR (21–23). The
Z-2 allele of the (CA)n ALR variant was determined to be associated
with DR in Caucasian patients with T1D (24), in Chinese patients with T2D (25) and in Tunisian individuals (26). However, negative results were also
reported in populations from Brazil (27) and Korea (28). For the Z+2 allele, significant
associations were detected in Chinese (29), Tunisian (26) and Australian (24) populations, but not in Korean
(28), Caucasian (30) and Brazilian (27) individuals.
Considering the relatively small sample size in
individual studies and controversial results among others, the
present study aimed to provide a more accurate evaluation of the
association between (CA)n ALR variants and the risk of DR. A
meta-analysis was performed to assess the association between ALR
(CA)n variation (Z, Z-2/Z+2, Z-4/Z+4, Z-6/Z+6) and the
susceptibility to DR.
Materials and methods
Search strategy
The meta-analysis of the present study followed the
Cochrane collaboration definition (31) and PRISMA 2009 guidelines for
meta-analyses and systematic reviews (32). A literature search was performed
using the PubMed, Embase, Chinese National Knowledge Infrastructure
(CNKI) and Cochrane Library databases. Studies investigating
associations between ALR (CA)n variations and the risk for DR were
identified using the following keywords: ‘aldose reductase’, ‘ALR’,
‘ALDR1’, ‘ADR’, ‘AR’, ‘aldoketo reductase family 1’, ‘AKR1B1’,
‘polymorphism’, ‘variant’, ‘diabetic retinopathy’ and ‘DR’. No
language restriction was applied. The search included articles
published until August 10, 2018. Additional eligible studies were
retrieved manually.
Studies were included if the following applied: i) A
case-control design that included diabetes with retinopathy and
diabetes without retinopathy, separately; ii) adequate data to
calculate odds ratios (ORs) and 95% confidence intervals (CIs);
iii) evaluation of the association between ALR (CA)n variation and
DR risk.
The following exclusion criteria were applied:
Studies were excluded if they: i) Did not include the genetic
association of ALR (CA)n variation and risk of DR; ii) were
duplicates of studies, or letters, dissertations, abstracts or
reviews; iii) lacked sufficient data for the determination of
genotype frequencies.
Data extraction and quality
assessment
The following terms were extracted from each
eligible study: First author, year, age, percentage of males,
disease duration, glycated hemoglobin, type of DM (T1D or T2D),
number of patients with diabetes with retinopathy or without
retinopathy. Two authors (YX and YB) independently extracted the
above information. Any disagreement was resolved by discussion. The
quality of each study was assessed by using the Newcastle-Ottawa
Scale (NOS) (33). The NOS uses a
‘star’ rating system to judge the quality. The total score ranged
from 0 (lowest quality) to 8 (highest quality). Studies with a
score of ≥6 were considered as being of high quality.
Statistical analysis
Statistical analyses were performed using STATA 12.0
software (StataCorp) and Revman 5 (Cochrane Collaboration). ORs
with 95% CIs were used to assess the strength of associations
between the allelic model of (CA)n variation and the risk of DR. A
Z-test was used to determine the pooled ORs. The
heterogeneity among studies was evaluated by Cochrane's Q-statistic
and I2 statistics. A random-effects model was used in
case of significant heterogeneity (I2>50%, or
P<0.05). Otherwise, the fixed-effects model was used.
Sensitivity analyses were also performed using the fixed-effects
model or random-effects model. A funnel plot in addition to an
Egger's and Begg's test was used to assess potential publication
bias. P<0.05 was considered to indicate statistically
significant differences.
Results
Characteristics of the published
studies
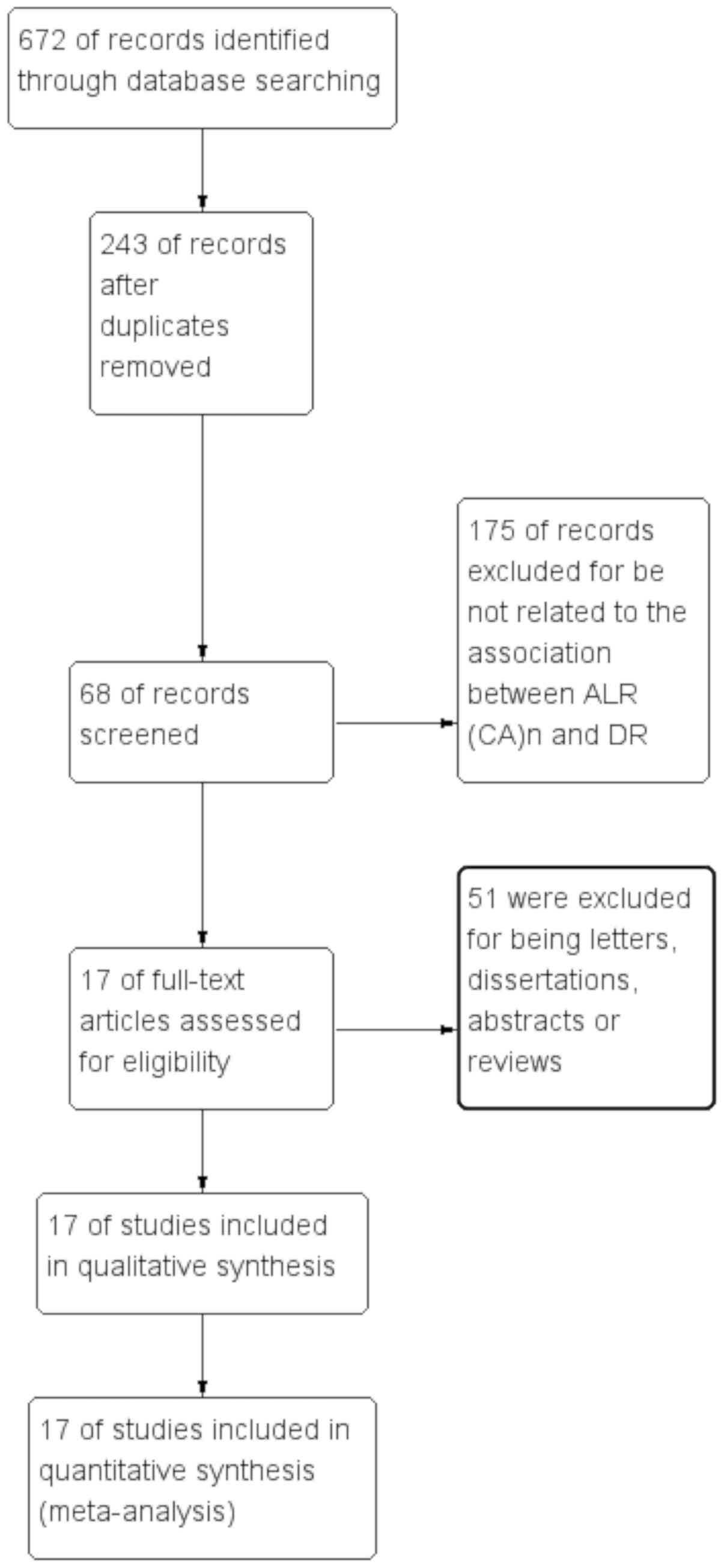
As presented in Fig.
1, 672 articles met the inclusion criteria and the full
articles were retrieved from the PubMed, Embase, CNKI and Cochrane
Library databases after the initial search. A total of 243
publications were excluded due to being duplicates. After screening
the titles and abstracts, 175 publications were excluded for not
documenting the association between ALR (CA)n and DR. In addition,
51 studies were excluded for being letters, dissertations,
abstracts or reviews. Finally, 17 articles were included in the
present meta-analysis (24–30,34–43). The
characteristics of all studies included are summarized in Tables I and SI. The NOS scores of all eligible studies
ranged from 6 to 8 stars, indicating high methodological quality of
all of the studies. Among them, 12 studies were performed with
Asian populations (25,26,28,29,34–38,40–42) and
5 studies were performed with Caucasian populations (24,27,30,39,43).
Furthermore, 4 articles included patients with T1D (24,29,39,43) and
11 studies included patients with T2D (25,26,28,30,34,35,37,38,40–42). The
type of DM of the subjects in 2 remaining articles was not
specified (27,36).
 | Table I.Characteristics of eligible studies
included in the meta-analysis. |
Table I.
Characteristics of eligible studies
included in the meta-analysis.
| First author | Year | Nationality | Age, years
(DR+/DR-) | M, % (DR+/DR-) | Duration, years
(DR+/DR-)a | HbA1c, %
(DR+/DR-) | Type of DM | DR+ | DR- | NOS | (Refs.) |
|---|
| Kao | 1999 | Australian |
14.8±14.4/14.3±13.8 | 40.2/51.5 |
7.2±7.3/6.0±6.7 |
8.6±8.6/8.7±8.6 | T1D | 67 | 97 | 8 | (24) |
| Ko | 1995 | Chinese |
33.2±7.3/31.5±8.1 | 54.5/45.5 |
5.0±3.8/15.6±4.5 |
6.8±2.0/6.7±1.7 | T1D | 22 | 22 | 8 | (29) |
| Park | 2002 | Korean |
61.1±8.7/62.4±8.5 | 53.9/34.2 |
15.2±5.0/14.4±4.2 |
9.4±2.3/8.9±2.0 | T2D | 89 | 38 | 8 | (28) |
| Petrovic | 2005 | Caucasian |
65.6±9.7/71.3±7.0 | 49/49.4 |
18.7±9.1/16.8±6.8 |
8.7±1.8/8.4±1.7 | T2D | 124 | 81 | 8 | (30) |
| Richeti | 2007 | Brazilian | NA | NA | NA | NA | NA | 62 | 66 | 6 | (27) |
| Zghal-Mokni | 2005 | Tunisian | NA | NA | NA | NA | T2D | 47 | 28 | 6 | (26) |
| Li | 2001 | Chinese |
56.5±7.2/55.4±9.7 | NA |
12.5±10.3/12.5±11.4 | NA | T2D | 105 | 40 | 6 | (34) |
| Zou | 2000 | Chinese |
45.88±10.15/48.06±7.99 | 46/53 |
2.01±2.02/15.32±4.81 |
9.46±2.37/9.19±2.48 | T2D | 100 | 132 | 8 | (37) |
| Chen | 2013 | Chinese |
58.25±8.40/58.66±8.06 | NA |
16.13±4.23/16.29±3.09 |
8.63±2.4/8.62±1.95 | NA | 164 | 158 | 7 | (36) |
| Zhang | 2008 | Chinese |
59.6±11.3/58.7±13.1 | 58.9/54.2 |
3.4±1.3/14.4±3.1 |
8.00±1.41/7.65±2.04 | T2D | 39 | 35 | 8 | (35) |
| Liu | 1999 | Chinese |
60.7±12.2/57.8±12.5 | 46.1/59.7 |
8.9±7.8/3.9±4.9 | NA | T2D | 78 | 139 | 7 | (25) |
| Wang | 2003 | Chinese |
62.0±9.6/53.4±13.0 | 41.7/41.5 |
9.4±6.9/4.4±4.6 |
8.8±2.2/7.7±1.9 | T2D | 187 | 551 | 8 | (38) |
|
Kumaramanickavel | 2003 | Indian | NA | 79/63 | 16/19.6 | NA | T2D | 105 | 109 | 7 | (42) |
| Isermann | 2000 | German | NA | NA | NA | NA | T1D | 170 | 84 | 5 | (43) |
| Demaine | 2000 | British | NA | 48.4/37.1 |
24.2±7.4/33.4±10.2 | NA | T1D | 159 | 70 | 7 | (39) |
| Ikegishi | 1999 | Japanese |
47.8±11.9/49.3±11.5 | 62.9/52.9 |
10.3±5.1/15.8±5.4 |
7.9±1.1/7.6±1.7 | T2D | 27 | 34 | 8 | (41) |
| Ichikawa | 1999 | Japanese | 54.4±15.7 | NA | 14.4±7.4/NA | 9.3±2.9/NA | T2D | 30 | 57 | 7 | (40) |
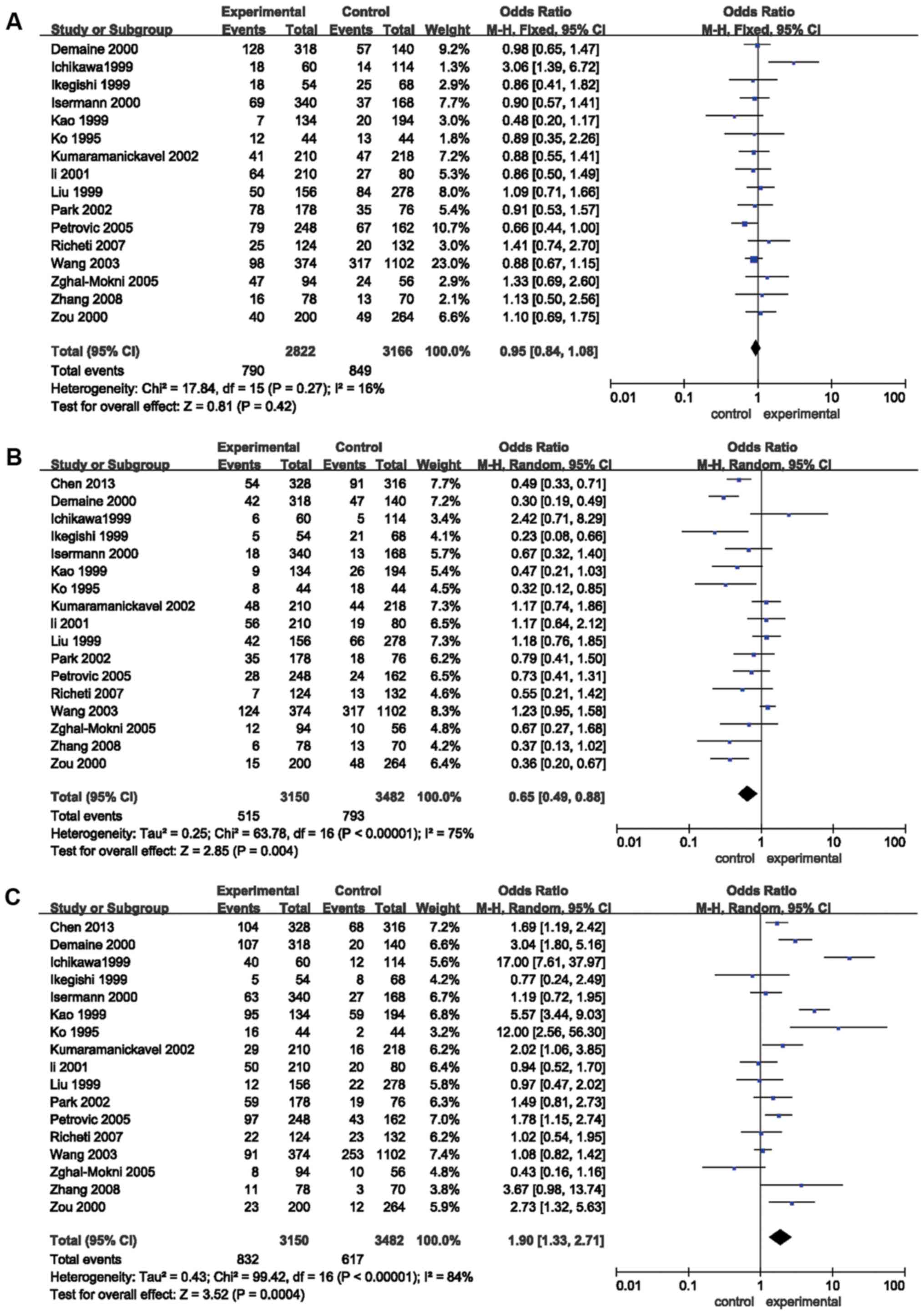
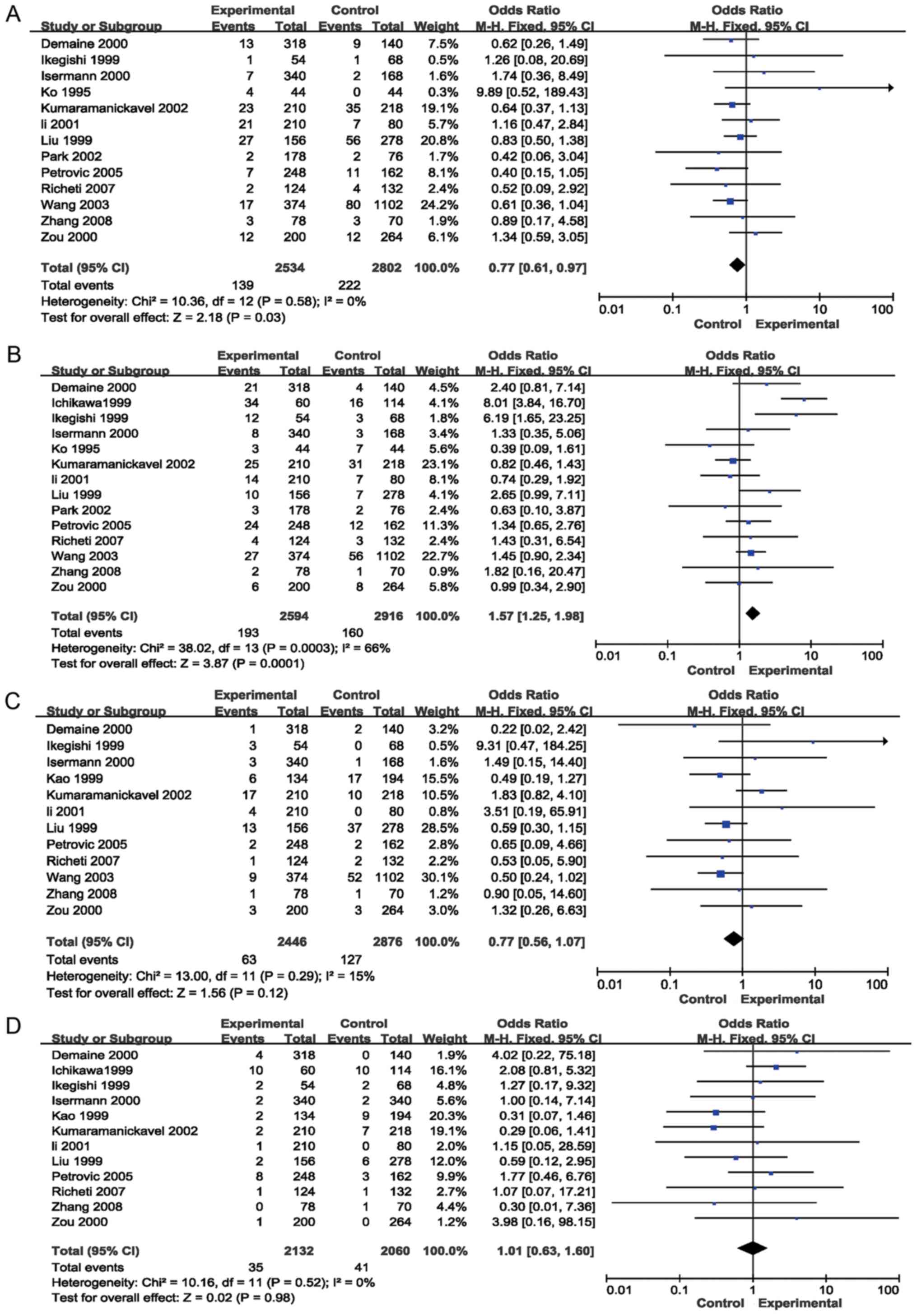
Results of the meta-analysis
The results of the meta-analysis preformed in the
present study are provided in Table
II and Figs. 2 and 3. The pooled risk estimates indicated that
the Z+2 allele was associated with a decreased risk of DR (OR=0.65,
95% CI: 0.49–0.88, P=0.004; Fig.
2B). However, the Z-2 allele was significantly associated with
an increased risk of DR (OR=1.90, 95% CI: 1.33–2.71, P=0.0004;
Table II, Fig. 2C). Significant associations were also
detected between the Z+4 (OR=0.77; 95% CI, 0.61–0.97, P=0.03;
Fig. 3A) and Z-4 (OR=1.57; 95% CI,
1.25–1.98; P=0.0001; Fig. 3B)
alleles and the risk for DR. However, no association was detected
between the Z, Z+6 or Z-6 alleles and the risk of DR (P>0.05;
Table II; Figs. 2A, 3C and
D, respectively). Stratification analyses based on ethnicity
were then performed. The results indicated that the Z-2 allele was
significantly associated with DR in Asian (OR=1.82, 95% CI:
1.16–2.86, P=0.009) and Caucasian (OR=2.08, 95% CI: 1.14–3.79,
P=0.02) populations. Z+2 was significantly associated with DR in
Caucasian individuals (OR=0.50, 95% CI: 0.34–0.73, P=0.0004), but
not in Asian patients (P=0.07). Furthermore, Z-4 was significantly
associated with DR in Asian populations (OR=1.57, 95% CI:
1.22–2.03, P=0.0005), but not in Caucasian individuals (P=0.09). In
addition, the Z+4 allele was not significantly associated with DR
in Asians or Caucasians (P>0.05). Stratification analyses based
on the type of DM (T1D and T2D) were also performed. The results
revealed that the Z-2 allele was associated with the risk of DR in
patients with T1D (OR=3.42, 95% CI: 1.46–8.04, P=0.005) and T2D
(OR=1.66, 95% CI: 1.05–2.63, P=0.03). The Z+2 allele was associated
with the risk of DR in patients with T1D (OR=0.39, 95% CI:
0.27–0.57, P<0.00001), but not in patients with T2D (P>0.05).
Furthermore, the Z+4 (OR=0.74; 95% CI, 0.57–0.96; P=0.02) and Z-4
(OR=1.62; 95% CI, 1.27–2.08; P=0.0001) alleles were associated with
the risk of DR in patients with T1D, but not in patients with T2D
(P>0.05; Table II).
 | Table II.Pooled ORs and 95% CIs of the
association between (CA)n of aldose reductase gene and diabetic
retinopathy. |
Table II.
Pooled ORs and 95% CIs of the
association between (CA)n of aldose reductase gene and diabetic
retinopathy.
|
|
| Numbers | Test of
association |
| Test of
heterogeneity |
|---|
|
|
|
|
|
|
|
|---|
|
Allele/subgroup | Number of
studies | Cases | Controls | OR (95% CI) | P-value | Model | P-value | I2
(%) |
|---|
| Z |
|
|
|
|
|
|
|
|
|
Total | 16 | 2,822 | 3,166 | 0.95 (0.84,
1.08) | 0.42 | F | 0.27 | 16 |
|
Asian | 11 | 1,658 | 2,370 | 1.00 (0.86,
1.16) | 0.98 | F | 0.38 | 6 |
|
Caucasian | 5 | 1,164 | 796 | 0.86 (0.69,
1.06) | 0.16 | F | 0.21 | 32 |
|
T1D | 4 | 836 | 546 | 0.88 (0.67,
1.15) | 0.34 | F | 0.56 | 0 |
|
T2D | 11 | 1,862 | 2,488 | 0.95 (0.83,
1.10) | 0.51 | F | 0.71 | 29 |
| Z+2 |
|
|
|
|
|
|
|
|
|
Total | 17 | 3,150 | 3,482 | 0.65 (0.49,
0.88) | 0.004 | R | <0.00001 | 75 |
|
Asian | 12 | 1,986 | 2,686 | 0.73 (0.52,
1.02) | 0.07 | R | <0.00001 | 75 |
|
Caucasian | 5 | 1,164 | 796 | 0.50 (0.34,
0.73) | 0.0004 | F | 0.17 | 38 |
|
T1D | 4 | 836 | 546 | 0.39 (0.27,
0.57) | <0.00001 | F | 0.32 | 14 |
|
T2D | 11 | 1,862 | 2,488 | 0.82 (0.60,
1.12) | 0.22 | R | 0.0009 | 66 |
| Z-2 |
|
|
|
|
|
|
|
|
|
Total | 17 | 3,150 | 3,482 | 1.90 (1.33,
2.71) | 0.0004 | R | <0.00001 | 84 |
|
Asian | 12 | 1,986 | 2,686 | 1.82 (1.16,
2.86) | 0.009 | R | <0.00001 | 83 |
|
Caucasian | 5 | 1,164 | 796 | 2.08 (1.14,
3.79) | 0.02 | R | <0.0001 | 86 |
|
T1D | 4 | 836 | 546 | 3.42 (1.46,
8.04) | 0.005 | R | <0.0001 | 87 |
|
T2D | 11 | 1,862 | 2,488 | 1.66 (1.05,
2.63) | 0.03 | R | <0.00001 | 93 |
| Z+4 |
|
|
|
|
|
|
|
|
|
Total | 13 | 2,534 | 2,802 | 0.77 (0.61,
0.97) | 0.03 | F | 0.58 | 0 |
|
Asian | 9 | 1,504 | 2,200 | 0.81 (0.62,
1.05) | 0.11 | F | 0.53 | 0 |
|
Caucasian | 4 | 1,030 | 602 | 0.61 (0.35,
1.06) | 0.08 | F | 48 | 0 |
|
T1D | 3 | 702 | 352 | 1.09 (0.54,
2.21) | 0.80 | R | 13 | 51 |
|
T2D | 9 | 1,708 | 2,318 | 0.74 (0.57,
0.96) | 0.02 | F | 0.65 | 0 |
| Z-4 |
|
|
|
|
|
|
|
|
|
Total | 14 | 2,594 | 2,916 | 1.57 (1.25,
1.98) | 0.0001 | R | 0.0003 | 66 |
|
Asian | 10 | 1,564 | 2,314 | 1.57 (1.22,
2.03) | 0.0005 | R | <0.0001 | 76 |
|
Caucasian | 4 | 1,030 | 602 | 1.57 (0.94,
2.62) | 0.09 | F | 0.84 | 0 |
|
T1D | 3 | 702 | 352 | 1.29 (0.66,
2.53) | 0.45 | R | 0.13 | 50 |
|
T2D | 11 | 1,768 | 2,432 | 1.62 (1.27,
2.08) | 0.0001 | R | 0.0001 | 73 |
| Z+6 |
|
|
|
|
|
|
|
|
|
Total | 12 | 2,446 | 2,876 | 0.77 (0.56,
1.07) | 0.12 | F | 0.29 | 15 |
|
Asian | 7 | 1,282 | 2,080 | 0.85 (0.59,
1.23) | 0.39 | F | 0.11 | 42 |
|
Caucasian | 5 | 1,164 | 796 | 0.54 (0.26,
1.09) | 0.08 | F | 0.85 | 0 |
|
T1D | 3 | 792 | 502 | 0.52 (0.23,
1.16) | 0.11 | F | 0.51 | 0 |
|
T2D | 8 | 1,530 | 2,242 | 0.85 (0.59,
1.21) | 0.36 | F | 0.16 | 33 |
| Z-6 |
|
|
|
|
|
|
|
|
|
Total | 12 | 2,132 | 2,060 | 1.01 (0.63,
1.60) | 0.98 | F | 0.52 | 0 |
|
Asian | 7 | 968 | 1,092 | 1.02 (0.56,
1.85) | 0.96 | F | 0.38 | 6 |
|
Caucasian | 5 | 1,164 | 968 | 0.99 (0.48,
2.06) | 0.98 | F | 0.44 | 0 |
|
T1D | 3 | 792 | 674 | 0.71 (0.27,
1.85) | 0.48 | F | 0.28 | 22 |
|
T2D | 8 | 1,216 | 1,254 | 1.12 (0.65,
1.93) | 0.67 | F | 0.45 | 0 |
Heterogeneity
Statistically significant heterogeneity across
studies was observed when analyzing the Z+2 (I2=75%;
P<0.00001), Z-2 (I2=84%; P<0.00001) and Z-4
(I2=66%; P=0.0003) alleles (Table II). Significant heterogeneity
regarding Z+2 was primarily identified in the studies by Zou et
al (37), Wang et al
(38), Liu et al (25), Demaine et al (39), Li et al (34) and Ichikawa et al (40). Removal of these studies from the
meta-analysis resulted in 40% (P=0.08) heterogeneity. Significant
heterogeneity regarding Z-2 was primarily present in the studies by
Kao et al (24), Ko et
al (29), Demaine et al
(39), Zghal-Mokni et al
(26), and Ichikawa et al
(40). Removal of these studies from
the meta-analysis resulted in 36% (P=0.10) heterogeneity.
Furthermore, the significant heterogeneity in Z-4 was mainly due to
the study by Ichikawa et al (40); upon removal of this study from the
meta-analysis, heterogeneity was reduced to 27% (P=0.17) (Table SII). In the subgroup analysis,
significant heterogeneity across studies was identified regarding
the Z+2, Z-2 and Z-4 alleles of Asian patients with T2D (Table II).
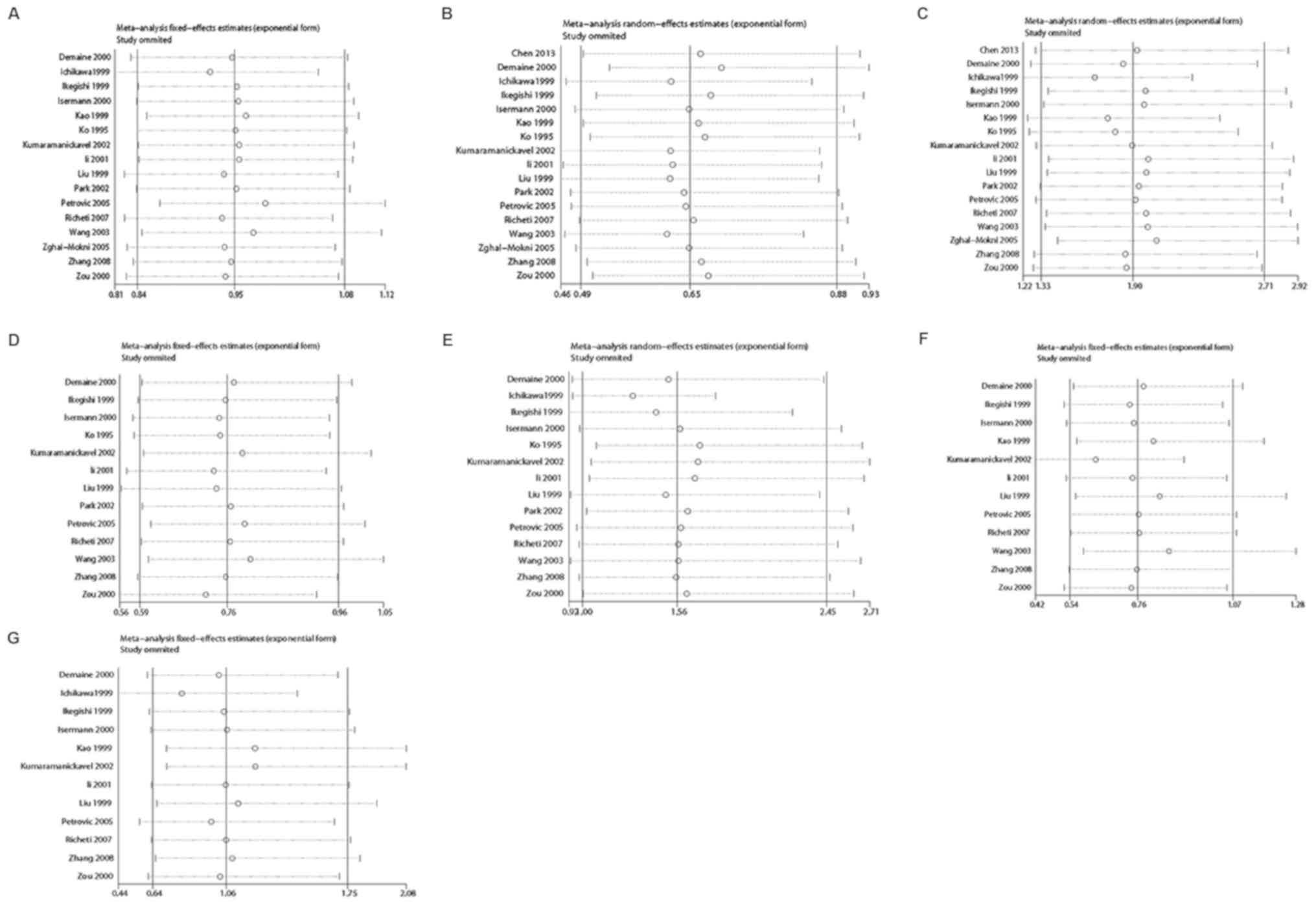
Sensitivity analysis and publication
bias
In the sensitivity analysis, the influence of a
single study on the overall risk estimate was determined by
removing one article at a time. The results revealed that the ORs
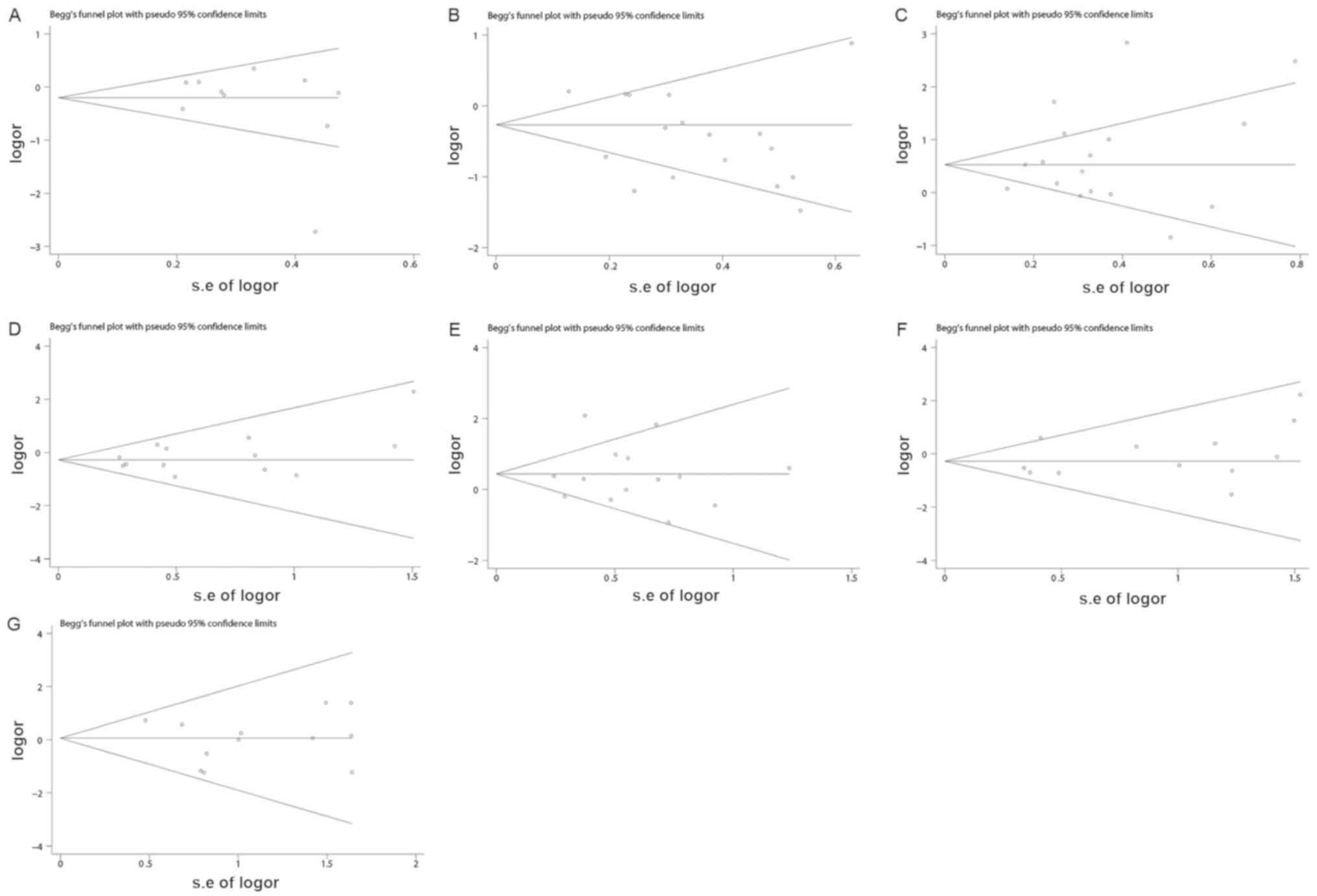
were not significantly altered in each genotype (Fig. 4). Begg's funnel plots and Egger's
test were performed to assess publication bias. Some outliers were
observed on the Begg's funnels plots, which may be due to the
missed researches including articles with negative results and the
small sample sizes of the studies. However, the results of Egger's
test demonstrated that P>0.05, which indicated the missed
researches and the small sample sizes studies had no effects on the
results. Therefore, the results indicated that there was no
publication bias for the comparison of Z, Z+2, Z-2, Z+4, Z-4, Z+6
and Z-6 (Table III and Fig. 5).
 | Table III.Egger's linear regression test for
funnel plot asymmetries of (AC)n in the aldose reductase gene. |
Table III.
Egger's linear regression test for
funnel plot asymmetries of (AC)n in the aldose reductase gene.
| Allele | P-value | 95% CI |
|---|
| Z | 0.281 |
−0.8225458–2.62835 |
| Z+2 | 0.084 |
−4.256574–0.3006727 |
| Z-2 | 0.319 |
−1.63998–4.707237 |
| Z+4 | 0.267 |
−0.5488476–1.791998 |
| Z-4 | 0.967 |
−2.481549–2.578416 |
| Z+6 | 0.340 |
−0.7528741–1.981472 |
| Z-6 | 0.658 |
−2.021688–1.334716 |
Discussion
ALR is an NADPH-dependent enzyme that exhibits
glucose-reducing activity (44).
Elevated glucose levels may enhance ALR activity in the case of a
hyperglycaemic event, which then increases the flux of glucose
through the polyol pathway (45).
This pathway has been revealed to serve an important role in
diabetic complications, including nephropathy and retinopathy.
Furthermore, significantly elevated ALR mRNA and protein levels
have been detected in the peripheral blood monocytes of patients
with DR (46). Therefore, inhibition
of ALR expression may offer a therapeutic strategy for patients
suffering from DR.
Recently, two common variants in the ALR gene,
including a (CA)n dinucleotide repeat microsatellite and a 106C/T
polymorphism, have been examined for their genetic association with
DR in several populations (36,38). The
(CA)n variant, located 2.1 kb upstream of the transcription
initiation site of ALR, may alter the secondary structure of DNA
(47) or RNA (48), consequently affecting splicing and
the generation of non-coding RNA. Multiple alleles have been
identified for the (CA)n variation, including Z, Z+2, Z-2, Z+4,
Z-4, Z+6, Z-6, Z+8, Z-8, Z+10 and Z-10. Z, Z+2 and Z-2 were the
most common alleles identified as (CA)n variations. Furthermore,
the Z-2 allele has been reported to be linked to increased
expression of the ALR gene in the peripheral mononuclear cells of
patients with diabetes or hyperglycaemia (49). In vitro gene reporter assays
have revealed that the susceptibility polymorphism Z-2 increases
transcriptional activity (49). In
addition, the Z-2 allele has been associated with the
susceptibility to DR (24). Other
studies have also indicated that the Z+2 allele serves a protective
role against DR (24,26,29).
However, the results indicating the association between the Z-2 or
Z+2 and DR were inconsistent and inconclusive. Among the 17 studies
included in the present analysis, only 5 (29,36,37,39,41)
reported significant associations between Z+2 and DR, and the 12
remaining studies indicated negative results. In addition,
regarding Z-2, 7 studies reported a significant association with DR
(24,29,30,36,37,39,40,42).
However, the 10 remaining studies indicated no association between
Z-2 and DR. The meta-analysis of the present study demonstrated
that the Z-2 allele is likely to be a genetic risk factor
(OR=1.90), while the Z+2 may exert a protective effect to reduce
the susceptibility to DR (OR=0.65). However, a certain
heterogeneity regarding Z+2 and Z-2 alleles was identified among
the results of individual studies.
To determine the role of the genetic background and
types of DM in DR, a subgroup analysis based on ethnicity and type
of DM was performed in the present study. Subgroup analysis with
stratification by ethnicity revealed that Z+2 exerted protective
effects for Caucasian patients with DR only. Furthermore, Z-2 was
demonstrated to be a risk factor for DR in Asian as well as
Caucasian populations. These inconsistent results in different
populations may indicate the important role of the genetic
background in the pathogenesis of DR (50–53). In
addition, subgroup analysis with stratification by type of DM
revealed that Z-2 was associated with DR in T1D and T2D.
Furthermore, Z+2 was associated with DR in T1D, but not in T2D. The
prevalence of DR was higher in patients with T1D, with
sight-threatening retinopathy reported to be up to 2.5× more common
than in those with T2D (54).
However, despite including a larger number of patients with T2D,
the meta-analysis of the present study only demonstrated a slight
association between Z-2 and DR in T2D. This may indicate the
importance of a larger sample size to provide a more accurate
evaluation of the association between gene polymorphisms and
disease susceptibility.
A significant association between Z+4 or Z-4 and DR
was determined in the present meta-analysis. Based on a larger
number of patients, the Z+4 allele was determined to be a
protective factor for DR, whereas the Z-4 allele was determined to
be a risk factor for DR. However, none of the studies included
detected any significant associations between Z+4 and DR.
Furthermore, only one study reported a significant association
between Z-4 and DR (41). These
results may indicate the importance of larger sample sizes to
provide a more accurate evaluation of the genetic association and
increased calculation power in the combined analysis. Subgroup
analysis based on ethnicity and type of DM revealed that Z-4 may be
a risk factor for DR in Asian populations, but not in Caucasians.
In addition, Z+4 and Z-4 were only determined to be associated with
DR in patients with T2D. To the best of our knowledge, the present
study is the first to identify a significant association between
Z+4 or Z-4 and the susceptibility for DR by performing a
meta-analysis. However, to fully confirm these genetic
associations, larger numbers of case-control studies with more
patients are necessary.
The present meta-analysis had various limitations.
First, the sample size in the present meta-analysis was relatively
small. Although 17 studies with 1,575 cases and 1,741 controls were
included, the sample size in the subgroup analysis, particularly in
the Caucasian and T1D subgroups, was insufficient. Furthermore,
only the genetic association between alleles of the (CA)n variation
and DR were assessed. No data on the genotype and haplotype were
provided. In addition, multiple factors, including genetic and
environmental factors, or the interaction between the two factors
and other unknown risk factors, were not considered in the
pathogenesis of DR. Furthermore, the studies included in the
present meta-analysis had been performed in either Asian or
Caucasian populations, but not in any subjects of other
ethnicities. To more precisely assess the association between (CA)n
polymorphisms and DR, analysis of multiple populations is
necessary. Finally, the information on the cases was relatively
limited, and the present analysis did not assess the approach (or
medication) to control glucose due to lack of data in the
individual studies.
In conclusion, the Z+2 allele may be a protective
factor for DR in Caucasian patients with T1D. Furthermore, Z+4 may
be a protective factor for DR in patients with T2D. However, Z-2
may be a risk factor for DR in all subgroups and Z-4 may be a risk
factor for DR in Asian patients with T2D.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The current study was funded by the Youth Foundation
of the Education Department of Hunan (grant no. 17B035) and the
Foundation of the Education Department of Hunan (grant no.
18C1186).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WSM and YHB participated in data curation,
methodology, software and validation. YX participated in project
administration and the acquisition of funding. WSM wrote the
original manuscript draft. YX was involved with the conception and
design of the current study and revising the manuscript critically
for important intellectual content. YX and YHB reviewed and edited
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ola MS, Nawaz MI, Siddiquei MM, Al-Amro S
and Abu El-Asrar AM: Recent advances in understanding the
biochemical and molecular mechanism of diabetic retinopathy. J
Diabetes Complications. 26:56–64. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chang YC and Wu WC: Dyslipidemia and
diabetic retinopathy. Rev Diabet Stud. 10:121–132. 2012. View Article : Google Scholar
|
|
3
|
Tuuminen R, Haukka J and Loukovaara S:
Poor glycemic control associates with high intravitreal
angiopoietin-2 levels in patients with diabetic retinopathy. Acta
Ophthalmologica. 93:e515–e516. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Grassi MA, Tikhomirov A, Ramalingam S,
Below JE, Cox NJ and Nicolae DL: Genome-wide meta-analysis for
severe diabetic retinopathy. Hum Mol Genet. 20:2472–2481. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Warpeha KM and Chakravarthy U: Molecular
genetics of microvascular disease in diabetic retinopathy. Eye
(Lond). 17:305–311. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gonzalez-Salinas R, Garcia-Gutierrez MC,
Garcia-Aguirre G, Morales-Canton V, Velez-Montoya R,
Soberon-Ventura VR, Gonzalez V, Lechuga R, Garcia-Solis P,
Garcia-Gutierrez DG, et al: Evaluation of VEGF gene polymorphisms
and proliferative diabetic retinopathy in Mexican population. Int J
Ophthalmol. 10:135–139. 2017.PubMed/NCBI
|
|
7
|
Wang W, He M and Huang W: Association of
monocyte chemoattractant protein-1 gene 2518A/G polymorphism with
diabetic retinopathy in type 2 diabetes mellitus: A meta-analysis.
Diabetes Res Clin Pract. 120:40–46. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang Y, Xia W, Lu P and Yuan HJ: The
association between VDR gene polymorphisms and diabetic retinopathy
susceptibility: A systematic review and meta-analysis. Biomed Res
Int. 2016:53052822016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li GY, Li ZB, Li F, Dong LP, Tang L, Xiang
J, Li JM and Bao MH: Meta-analysis on the association of ALDH2
polymorphisms and type 2 diabetic mellitus, diabetic retinopathy.
Int J Environ Res Public Health. 14(pii): E1652017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fujisawa T, Ikegami H, Kawaguchi Y,
Shintani M, Kawabata Y, Ono M, Nishino M, Ogihara T, Okamoto N and
Fukuda M: Genetic susceptibility to diabetic retinopathy: CA repeat
polymorphism of the aldose reductase gene. Folia Jap Ophthalmol
Clin. 52:128–130. 2001.
|
|
11
|
Abhary S, Burdon KP, Laurie KJ, Thorpe S,
Landers J, Goold L, Lake S, Petrovsky N and Craig JE: Aldose
reductase gene polymorphisms and diabetic retinopathy
susceptibility. Diabetes Care. 33:1834–1836. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Crespo I, Giménez-Dejoz J, Porté S,
Cousido-Siah A, Mitschler A, Podjarny A, Pratsinis H, Kletsas D,
Parés X, Ruiz FX, et al: Design, synthesis, structure-activity
relationships and X-ray structural studies of novel
1-oxopyrimido[4,5-c]quinoline-2-acetic acid derivatives as
selective and potent inhibitors of human aldose reductase. Eur J
Med Chem. 152:160–174. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Radha V, Rema M and Mohan V: Genes and
diabetic retinopathy. Indian J Ophthalmol. 50:5–11. 2002.PubMed/NCBI
|
|
14
|
Graham A, Brown L, Hedge PJ, Gammack AJ
and Markham AF: Structure of the human aldose reductase gene. J
Biol Chem. 266:6872–6877. 1991.PubMed/NCBI
|
|
15
|
Li YY, Wang H, Yang XX, Geng HY, Gong G
and Lu XZ: AR C-106T gene polymorphism and diabetic nephropathy in
the Eastern Asians with T2DM: A meta-analysis including 2120
subjects. Diabetes Res Clin Pract. 130:244–251. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gupta B and Singh SK: Association of
aldose reductase gene polymorphism (C-106T) in susceptibility of
diabetic peripheral neuropathy among north Indian population. J
Diabetes Complications. 31:1085–1089. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Abhary S, Hewitt AW, Burdon KP and Craig
JE: A systematic meta-analysis of genetic association studies for
diabetic retinopathy. Diabetes. 58:2137–2147. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Olmos P, Bastías MJ, Vollrath V, Toro L,
Trincado A, Salinas P, Claro JC, López JM, Acosta AM, Miquel JF and
Castro J: C(−106)T polymorphism of the aldose reductase gene and
the progression rate of diabetic retinopathy. Diabetes Res Clin
Pract. 74:175–182. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou M, Zhang P, Xu X and Sun Z: The
relationship between aldose reductase C106T polymorphism and
diabetic retinopathy: An updated meta-analysis. Invest Ophthalmol
Vis Sci. 56:2279–2289. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Song ZD, Tao Y, Han N and Wu YZ:
Association of the aldose reductase-106TT genotype with increased
risk for diabetic retinopathy in the Chinese han population: An
updated meta-analysis. Curr Eye Res. 41:1087–1091. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ng DP, Conn J, Chung SS and Larkins RG:
Aldose reductase (AC)n microsatellite polymorphism and diabetic
microvascular complications in Caucasian Type 1 diabtes mellitus.
Diabetes Res Clin Pract. 52:21–27. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
dos Santos KG, Canani LH, Gross JL,
Tschiedel B, Souto KE and Roisenberg I: The-106CC genotype of the
aldose reductase gene is associated with an increased risk of
proliferative diabetic retinopathy in Caucasian-Brazilians with
type 2 diabetes. Mol Genet Metab. 88:280–284. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Heesom AE, Hibberd ML, Millward A and
Demaine AG: Polymorphism in the 5′-end of the aldose reductase gene
is strongly associated with the development of diabetic nephropathy
in type I diabetes. Diabetes. 46:287–291. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kao YL, Donaghue K, Chan A, Knight J and
Silink M: An aldose reductase intragenic polymorphism associated
with diabetic retinopathy. Diabetes Res Clin Pract. 46:155–160.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu L, Xiang K and Zheng T: A study of
association between polymorphism of aldose reductase gene and
diabetic microangiopathy. Chin J Endocrinol Metab. 15:263–266.
1999.
|
|
26
|
Zghal-Mokni I, Arfa I, Elloumi-Zghal H,
Abid A, Amrouche-Rached C, Kaabi B, Chakroun S, Blousa-Chabchoub S,
Gaïgi S, Ayed S, et al: Association study between diabetic
retinopathy and aldose reductase gene polymorphism in Tunisians. J
Fr Ophtalmol. 28:386–390. 2005.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Richeti F, Noronha RM, Waetge RT, de
Vasconcellos JP, de Souza OF, Kneipp B, Assis N, Rocha MN, Calliari
LE, Longui CA, et al: Evaluation of AC(n) and C(−106)T
polymorphisms of the aldose reductase gene in Brazilian patients
with DM1 and susceptibility to diabetic retinopathy. Mol Vis.
13:740–745. 2007.PubMed/NCBI
|
|
28
|
Park HK, Ahn CW, Lee GT, Kim SJ, Song YD,
Lim SK, Kim KR, Huh KB and Lee HC: (AC)(n) polymorphism of aldose
reductase gene and diabetic microvascular complications in type 2
diabetes mellitus. Diabetes Res Clin Pract. 55:151–157. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ko BC, Lam KS, Wat NM and Chung SS: An
(A-C)n dinucleotide repeat polymorphic marker at the 5′ end of the
aldose reductase gene is associated with early-onset diabetic
retinopathy in NIDDM patients. Diabetes. 44:727–732. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Petrovic MG, Peterlin B, Hawlina M and
Petrovic D: Aldose reductase (AC)n gene polymorphism and
susceptibility to diabetic retinopathy in type 2 diabetes in
Caucasians. J Diabetes Complications. 19:70–73. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Higgins J and Green S: Cochrane handbook
for systematic reviews of interventions, version 5.1.0 [updated
March 2011]The Cochrane Collaboration. 2011
|
|
32
|
Boccia S: PRISMA: An attempt to improve
standards for reporting systematic review and meta-analysis.
Epidemiol Biostatist Public Health. 6:E3822009.
|
|
33
|
Wells GA, Shea BJ, O'Connell D, Robertson
J, Peterson J, Welch V, Losos M and Tugwell P: The newcastle-ottawa
scale (NOS) for assessing the quality of nonrandomized studies in
meta-analysis. Appl Eng Agric. 18:727–734. 2014.
|
|
34
|
Li QJ, Xie P, Zeng WM and Song HP:
Correlation between (AC)n polymorphism of the aldose reductase gene
and retinopathy in type 2 diabetes mellitus. Chin J Endocrinol
Metab. 17:367–368. 2001.
|
|
35
|
Zhang JH, Wang HY and Li SY: Correlation
between polymorphism of the aldose reductase gene and retinopathy
in type 2 diabetes mellitus. J Shandong Univ. 46:399–406. 2008.
|
|
36
|
Chen S, Liu CS and Wang XJ: The
correlation study between the two gene polymorphism of aldose
reductase gene 5′end and diabetic retinopathy. China Mod Med.
20:13–15. 2013.
|
|
37
|
Zou X and Lu J: Study on the relationship
between the polymorphism of (ac)n in the 5′-end of the ar gene and
the susceptibility of microangopathy in type 2 diabetes mellitus.
Med J Chin Peoples Liberat Army. 25:353–356. 2000.
|
|
38
|
Wang Y, Ng MC, Lee SC, So WY, Tong PC,
Cockram CS, Critchley JA and Chan JC: Phenotypic heterogeneity and
associations of two aldose reductase gene polymorphisms with
nephropathy and retinopathy in type 2 diabetes. Diabetes Care.
26:2410–2415. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Demaine A, Cross D and Millward A:
Polymorphisms of the aldose reductase gene and susceptibility to
retinopathy in type 1 diabetes mellitus. Invest Ophthalmol Vis Sci.
41:4064–4068. 2000.PubMed/NCBI
|
|
40
|
Ichikawa F, Yamada K, Ishiyama-Shigemoto
S, Yuan X and Nonaka K: Association of an (A-C)n dinucleotide
repeat polymorphic marker at the 5′-region of the aldose reductase
gene with retinopathy but not with nephropathy or neuropathy in
Japanese patients with Type 2 diabetes mellitus. Diabet Med.
16:744–748. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ikegishi Y, Tawata M, Aida K and Onaya T:
Z-4 allele upstream of the aldose reductase gene is associated with
proliferative retinopathy in Japanese patients with NIDDM, and
elevated luciferase gene transcription in vitro. Life Sci.
65:2061–2070. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kumaramanickavel G, Sripriya S, Ramprasad
VL, Upadyay NK, Paul PG and Tarun S: Z-aldose reductase allele and
diabetic retinopathy in India. Ophthalmic Genet. 24:41–48. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Isermann B, Susanne Schmidt, Bierhaus A,
Schiekofer S, Borcea V, Ziegler R, Nawroth PP and Ritz E: (CA)(n)
dinucleotide repeat polymorphism at the 5′-end of the aldose
reductase gene is not associated with microangiopathy in
Caucasians, with long-term diabetes mellitus 1. Nephrol Dial
Transplant. 15:918–920. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kaur N and Vanita V: Association of aldose
reductase gene (AKR1B1) polymorphism with diabetic retinopathy.
Diabetes Res Clin Pract. 121:41–48. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Petrash JM, Flath M, Sens D and Bylander
J: Effects of osmotic stress and hyperglycemia on aldose reductase
gene expression in human renal proximal tubule cells. Biochem
Biophys Res Commun. 187:201–208. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Nishimura C, Saito T, Ito T, Omori Y and
Tanimoto T: High levels of erythrocyte aldose reductase and
diabetic retinopathy in NIDDM patients. Diabetologia. 37:328–330.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lu L, Risch E, Deng Q, Biglia N, Picardo
E, Katsaros D and Yu H: An insulin-like growth factor-II intronic
variant affects local DNA conformation and ovarian cancer survival.
Carcinogenesis. 34:2024–2030. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lu L, Katsaros D, Mayne ST, Risch HA,
Benedetto C, Canuto EM and Yu H: Functional study of risk loci of
stem cell-associated gene lin-28B and associations with disease
survival outcomes in epithelial ovarian cance. Carcinogenesis.
33:2119–2125. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Shah VO, Scavini M, Nikolic J, Sun Y, Vai
S, Griffith JK, Dorin RI, Stidley C, Yacoub M, Vander Jagt DL, et
al: Z-2 microsatellite allele is linked to increased expression of
the aldose reductase gene in diabetic nephropathy. J Clin
Endocrinol Metab. 83:2886–2891. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Donaghue KC, Margan SH, Chan AK, Holloway
B, Silink M, Rangel T and Bennetts B: The association of aldose
reductase gene (AKR1B1) polymorphisms with diabetic neuropathy in
adolescents. Diabet Med. 22:1315–1320. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liew G, Klein R and Wong TY: The role of
genetics in susceptibility to diabetic retinopathy. Int Ophthalmol
Clin. 49:35–52. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Cho H and Sobrin L: Genetics of diabetic
retinopathy. Curr Diab Rep. 14:5152014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ju Z, Daichao W, Yan L and Tan SJ:
Association of luteinizing hormone/choriogonadotropin receptor gene
polymorphisms with polycystic ovary syndrome risk: A meta-analysis.
Gynecol Endocrinol. 35:81–85. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Roy MS, Klein R, O'Colmain BJ, Klein BE,
Moss SE and Kempen JH: The prevalence of diabetic retinopathy among
adult type 1 diabetic persons in the United States. Arch
Ophthalmol. 122:546–551. 2004. View Article : Google Scholar : PubMed/NCBI
|