Introduction
Atherosclerotic cardiovascular disease and its
clinical complications, such as myocardial infarction and ischemic
stroke, are the world's first and third causes of death,
respectively, causing 247.9 deaths/100,000 persons in 2013,
representing 84.5% of cardiovascular deaths and 28.2% of all-cause
mortality (1). Atherosclerosis can
lead to lipid accumulation, extracellular matrix protein
deposition, and calcification in the intima and media of the
arteries causing arterial stiffness and reducing arterial
elasticity (2). Oxidized low-density
lipoprotein (ox-LDL) is able to induce apoptosis of endothelial
cells (ECs) and is considered as the major risk factors for
atherosclerosis ((3). In response to
oxidized low-density lipoproteins (oxLDL), endothelial cells
express a range of chemokines and adhesion molecules that
contribute to leukocyte recruitment, adherence, and migration into
the subendothelium (4), the first
stage in the development of atherosclerosis (5). Recently, increasing evidence has
indicated that microRNAs (miRNAs) serve an important role in
atherosclerosis development and progress (6).
miRNAs, widely found in plants and animals,
including humans, are a group of non-coding, single-stranded RNA
molecules that participate in sequence-specific
post-transcriptional regulation of gene expression (7–9). miRNAs
have emerged as crucial players in many biological processes, and
changes in their expression or function are associated with
numerous human diseases (10). It
has been reported that miRNAs regulate several cellular and
molecular biological processes related to the development of
atherosclerosis ranging from interacting with risk factors to
initiating the development, promoting the progression and causing
the rupture of atherosclerotic plaques (11). miRNA (miR)-144-5p has been studied in
several diseases, including cancer (12–14),
chronic periodontitis (15) and
depressive disorders (16). However,
the specific function and mechanism of action of miR-144-5p in
atherosclerosis remain unclear.
SMAD proteins are intracellular mediators of the
transforming growth factor-β family. Smad1/5/8 play an important
role in angiogenesis. Phosphorylation and activation of the
transcription factors SMAD1/5/8 results in the promotion of
angiogenesis. The SMAD1/5/8 signaling pathway, which inhibits
extracellular matrix deposition, promotes endothelial cell
proliferation and migration (17). A
previous study has reported that the balance of
proliferation/apoptosis of vascular smooth muscle cells in
atherosclerosis is modulated by long non-coding RNA-MEG3 via the
regulation of the miR-26a/Smad1 axis (18). In addition, activation of CD137
signaling promotes angiogenesis in atherosclerosis by modulating
the endothelial Smad1/5-nuclear factor of activated T cells pathway
(19). In a previous study using
bioinformatics, it was predicted that SMAD1 was a direct target
gene of miR-144-5p (20).
The present study aimed to investigate the role of
miR-144-5p in atherosclerosis and to further explore the molecular
mechanism of action of miR-144-5p. It was hypothesized that
miR-144-5p alleviates ox-LDL-induced HUVEC proliferation, invasion
and migration by targeting SMAD1, and further implicates the
potential therapeutic targets to reverse atherosclerosis.
Materials and methods
Cell culture and cell
transfection
HUVECs were acquired from the Shanghai Institute of
Life Sciences, Chinese Academy of Sciences. Culture media for
HUVECs contained routine medium 199 (Gibco; Thermo Fisher
Scientific, Inc.) with 10% FBS (unless otherwise stated) (Gibco;
Thermo Fisher Scientific, Inc.), 1% penicillin/streptomycin (Gibco;
Thermo Fisher Scientific, Inc.), 1% endothelial cell growth
supplement (Sigma-Aldrich; Merck KGaA) and 10 ng/ml epidermal
growth factor in a 5% CO2 humidified atmosphere at 37°C.
To mimic atheroprone conditions, HUVECs were exposed to
proatherogenic oxLDL (25 µg/ml; Biomedical Technologies S.L.).
HUVECs (5.0×104 cells/well) were
transfected with 100 nM miR-144-5p mimic or 100 nM mimic control
for 48 h at 37°C using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. After 48 h of transfection, the transfection
efficiency was detected using reverse transcriptase quantitative
PCR (RT-qPCR).
MTT assay
Cell proliferation was detected by MTT assay. HUVECs
(2.0×103 cells/well) were plated into 96-well plates and
cultured at 37°C for 12, 24 or 48 h, then 20 µl MTT (5 mg/ml;
Sigma-Aldrich, Merck KGaA) was added to medium. Following 4-h
incubation, 150 µl DMSO was used to dissolve the formazan crystals.
The absorbance was measured at a wavelength of 490 nm using a
microplate reader.
Dual-luciferase reporter assay
TargetScan (v7.2) bioinformatics software
(http://www.targetscan.org/vert_72/)
was used to predict target genes of miR-144-5p. The 3′-untranslated
region (UTR) of SMAD1 was cloned into the luciferase reporter
vector psiCHECK-2 (Promega Corporation) according to the
manufacturer's instruction. Briefly, 500 ng of each reporter
construct [wild-type (WT) or mutant 3′-UTR of SMAD1 or the
psiCHECK-2 vector] and miR-144-5p mimic or mimic control were
co-transfected into pre-confluent (60–70%) HUVECs plated in 24-well
plate using Lipofectamine 2000 for 48 h at 37°C. Then relative
luciferase activity was detected with a microplate reader
(Molecular Devices, LLC). Renilla luciferase was used for
normalization. Each sample was repeated three times.
Wound healing assay
To detect the migratory ability of HUVECs, wound
healing assays were performed. HUVECs (3×105 per well)
were seeded in 6-well plates with human endothelial serum-free
culture medium (cat. no. 11111044; Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 0.5% FBS and cultured until 90% confluent;
the cell monolayer was subsequently scraped with a pipette tip. The
cells were washed three times with PBS to wipe off cell debris and
the cells were cultured in human endothelial serum-free culture
medium without FBS. Images were captured at 0 and 24 h using a
Olympus IX71 Inverted Microscope (Olympus Corporation;
magnification, ×100).
Transwell invasion assay
Transwell invasion assays were performed with using
a Transwell chamber (8 µm pore size; Corning, Inc.). Cells
(2×104 cells/well) were collected and re-suspended in
human endothelial serum-free culture medium (cat. no. 11111044;
Gibco; Thermo Fisher Scientific, Inc.) containing 0.5% FBS and
plated on the top of polycarbonate Transwell filter pre-coated with
Matrigel (BD Biosciences). Complete culture medium (DMEM, Gibco;
Thermo Fisher Scientific, Inc.) containing 10% FBS was added to the
lower chamber. After incubation for 24 h at 37°C, the non-invasive
free cells were carefully removed with cotton swab from the top
chamber. The invaded cells on the lower membranes were fixed with
4% paraformaldehyde for 20 min at room temperature and stained by
0.1% crystal violet for 15 min at room temperature. Cells were
counted in five random fields for each chamber at a magnification
of ×100 under Olympus IX71 Inverted Microscope (Olympus
Corporation) to evaluate invasive capacity.
RT-qPCR
Total RNA was extracted from HUVECs
(5×104 cells/well) using TRIzol® (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. To determine the expression levels of miR-144-5p and
SMAD1 mRNA, cDNA was synthesized using PrimeScript Reverse
Transcriptase Reagent kit (Takara Biotechnology Co., Ltd.)
according to the manufacturer's instruction. qPCR was performed
with the SYBR Premix Ex Taq kit (Takara Biotechnology Co., Ltd.)
according to the manufacturer's protocol. U6 and GAPDH were used as
the endogenous controls. The thermocycling conditions were as
follows: 95°C for 5 min, followed by 38 cycles of denaturation at
95°C for 15 sec and annealing/elongation at 60°C for 30 sec. The
primers used were as follows: U6, forward
5′-GCTTCGGCAGCACATATACTAAAAT-3′, reverse
5′-CGCTTCACGAATTTGCGTGTCAT-3′; GAPDH, forward
5′-CTTTGGTATCGTGGAAGGACTC-3′, reverse 5′-GTAGAGGCAGGGATGATGTTCT-3′;
SMAD1, forward 5′-ACAGTCTGTGAACCATGGATTTGA-3′, reverse
5′-TGAGGTGAACCCATTTGAGTAAGAA-3′. The experiments were repeated in
triplicate, and the 2−ΔΔCq method was used to calculate
the relative gene expression and normalized to the internal
reference gene U6 or GAPDH, respectively (21).
Western blotting
HUVECs (5×104 cells/well) were lysed
using RIPA Lysis Buffer (Gibco; Thermo Fisher Scientific, Inc.)
with protease inhibitor PMSF (Shanghai Biocolor BioScience &
Technology Co., Ltd.). BCA Protein Assay kit (Thermo Fisher
Scientific, Inc.) was used to measure protein concentrations. A
total of 20 µg of protein was subjected to 10% SDS-PAGE
electrophoresis and transferred to PVDF membranes. The membranes
were blocked with 5% Difco™ skim milk (cat. no. 232100, BD
Biosciences) at room temperature for 1 h. Subsequently, the
membranes were probed with primary antibodies against SMAD1 (cat.
no. 6944; 1:1,000; Cell Signaling Technology, Inc.), phosphorylated
(p)-SMAD1 (cat. no. 5753; 1:1,000; Cell Signaling Technology,
Inc.), SMAD5 (cat. no. 12534; 1:1,000; Cell Signaling Technology,
Inc.), p-SMAD5 (cat. no. ab92698; 1:1,000; Abcam), SMAD8 (cat. no.
sc-293413; 1:1,000; Santa Cruz Biotechnology, Inc.), p-SMAD8 (cat.
no. sc-12353, 1:1,000; Santa Cruz Biotechnology, Inc.), and GAPDH
(cat. no. 5174; 1:1,000; Cell Signaling Technology, Inc.), at 4°C
overnight. The next day, the membrane was washed three times with
PBS-Tween-20 (0.05%) buffer and then incubated with horse radish
peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody
(cat. no. 7074; 1:1,000; Cell Signaling Technology, Inc.) or mouse
IgGκ light chain binding protein-HRP (cat. no. sc-516102; 1:1,000;
Santa Cruz Biotechnology, Inc.) for 1 h at room temperature. The
residue antibody solution was completely washed off with PBST, and
protein bands were visualized using RapidStep™ ECL Reagent (cat.
no. 345818; Merck KGaA). GAPDH served as loading control for
normalization. Blot bands were semi-quantitively analyzed using
ImageJ version 4.0 (National Institutes of Health).
Flow cytometry apoptosis assay
The Annexin V-FITC Apoptosis Detection kit I (BD
Biosciences) was used to detect cell apoptosis, according to the
manufacturer's protocol. A total of 100,000 suspended cells were
washed twice with PBS, collected, centrifuged with 1,000 × g for 5
min at 20°C, and re-suspended in 100 µl of FITC-binding buffer.
Subsequently, ~5 µl ready-to-use Annexin V-FITC (BD Biosciences)
and 5 µl propidium iodide (PI) were added. Cells were incubated for
30 min at room temperature in the dark. Annexin V-FITC and PI
fluorescence were assessed by BD FACSCalibur flow cytometer (BD
Biosciences). The percentages of cells in early apoptosis (Annexin
V+/PI−) and late apoptosis (Annexin
V+/PI+) were calculated.
Statistical analysis
All data are presented as the mean ± SD from three
independent experiments in triplicate. Student's t-test or one-way
ANOVA followed by Tukey's post hoc test was used for biostatistical
analysis, using SPSS 19.0 (IBM Corp.). P<0.05 was considered to
indicate a statistically significant difference.
Results
Effects of miR-144-5p on HUVEC
proliferation and apoptosis
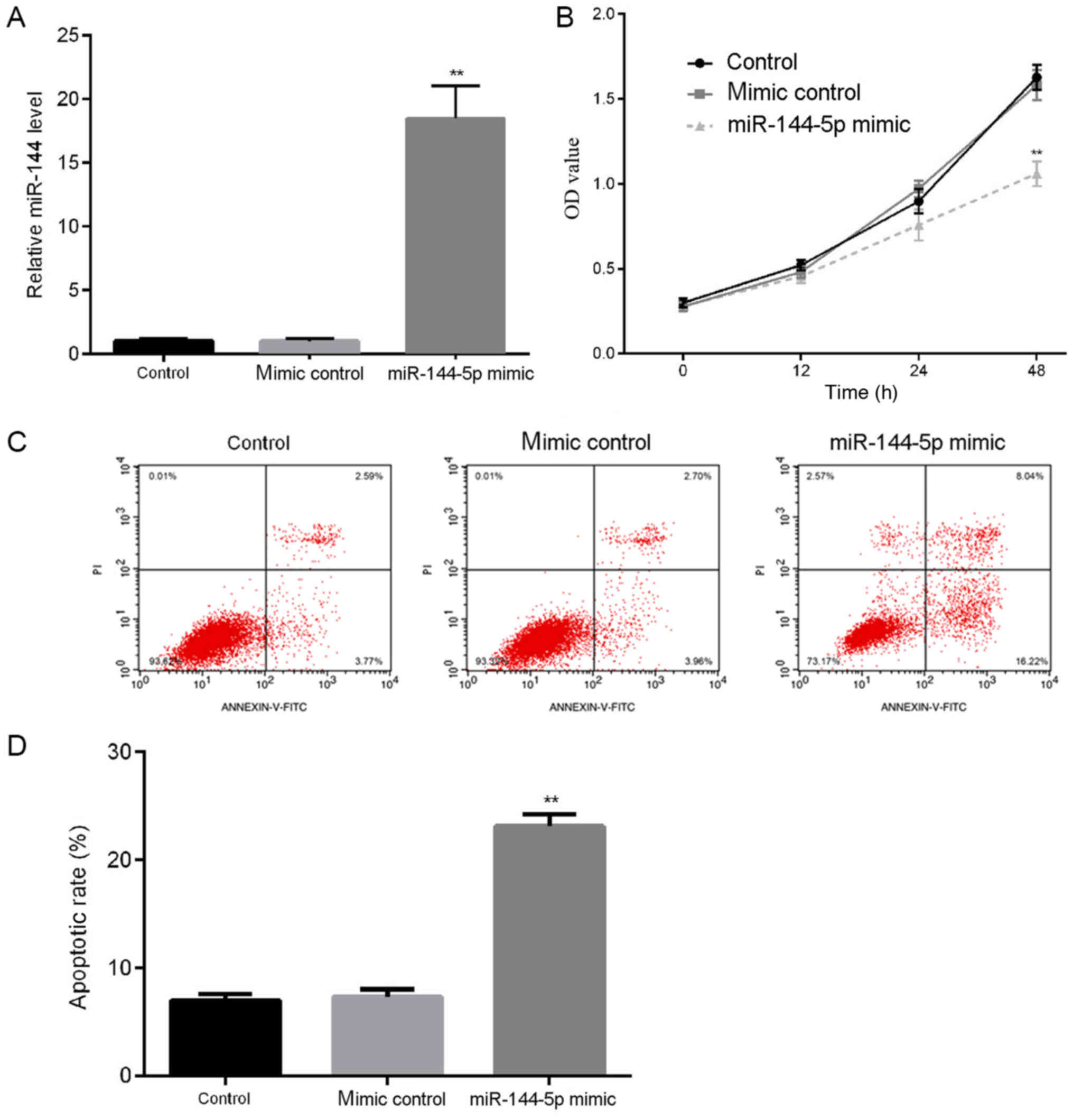
Apoptosis of endothelial cells serves an important
role in the occurrence and development of atherosclerosis (22). To study the role of miR-144-5p in
atherosclerosis, firstly the effects of miR-144-5p on HUVEC
proliferation and apoptosis were explored. HUVECs were transfected
with mimic control or miR-144-5p mimic for 48 h. RT-qPCR was used
to detect the transfection efficiency, and the results demonstrated
that compared with the control group, transfection with miR-144-5p
mimic significantly increased the level of miR-144-5p in HUVECs
(Fig. 1A). Results from the MTT
assay demonstrated that miR-144-5p mimic transfection reduced cell
proliferation of HUVECs (Fig. 1B).
In addition, to determine whether miR-144-5p regulated the
apoptosis of HUVECs, flow cytometric analyses were performed to
detect cell apoptosis. The results indicated that miR-144-5p mimic
transfection significantly increased the apoptotic rates of HUVECs
compare with the control (Fig. 1C and
D).
Effects of miR-144-5p on HUVEC
migration and invasion
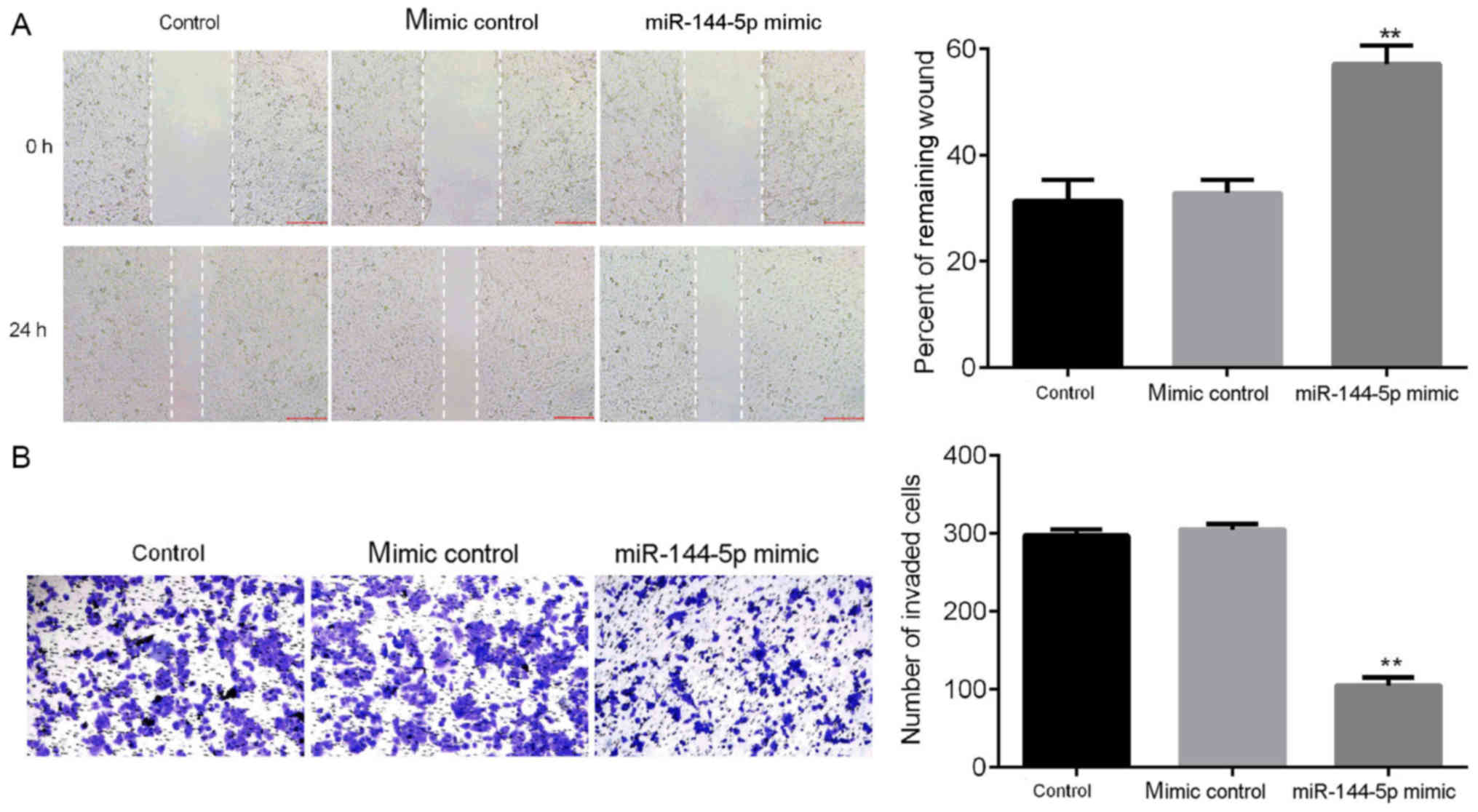
To further examine the effects of miR-144-5p on the
biological function of HUVECs, wound healing and Matrigel assays
were performed (Fig. 2). The wound
healing assay results demonstrated that miR-144-5p mimic suppressed
cell migration compared with control groups (Fig. 2A). Matrigel assay results indicated
that miR-144-5p mimic significantly inhibited the invasive ability
of HUVECs (Fig. 2B). Taken together,
these results showed that miR-144-5p inhibited the invasive and
migratory potential of HUVECs.
SMAD1 is the direct target gene of
miR-144-5p
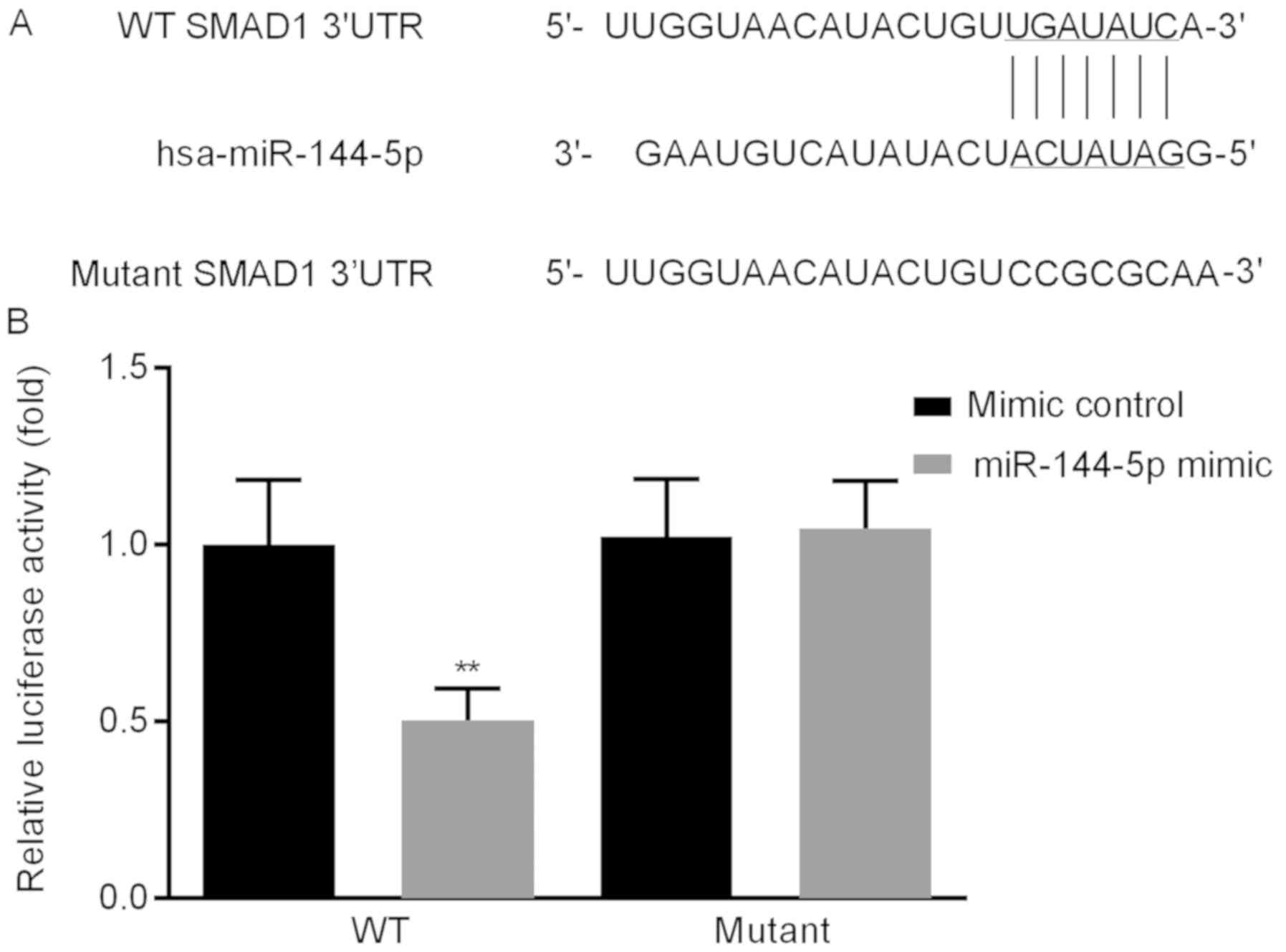
To investigate the underlying mechanism of
miR-144-5p on HUVECs, a putative biological target gene of
miR-144-5p was identified and verified; the online bioinformatics
tool TargetScan was used to predict target genes. The results
identified a miR-144-5p target site in the SMAD1 3′UTR (Fig. 3A). Dual-luciferase reporter assay
results revealed that miR-144-5p mimic reduced the luciferase
activity of the WT SMAD1 3′ UTR, but miR-144-5p mimic did not
inhibit that of the reporter fused to the mutant SMAD1 3′ UTR
(Fig. 3B). Taken together, these
results demonstrated that SMAD1 was a direct target gene of
miR-144-5p.
miR-144-5p inhibited SMAD1/5/8 pathway
in HUVECs
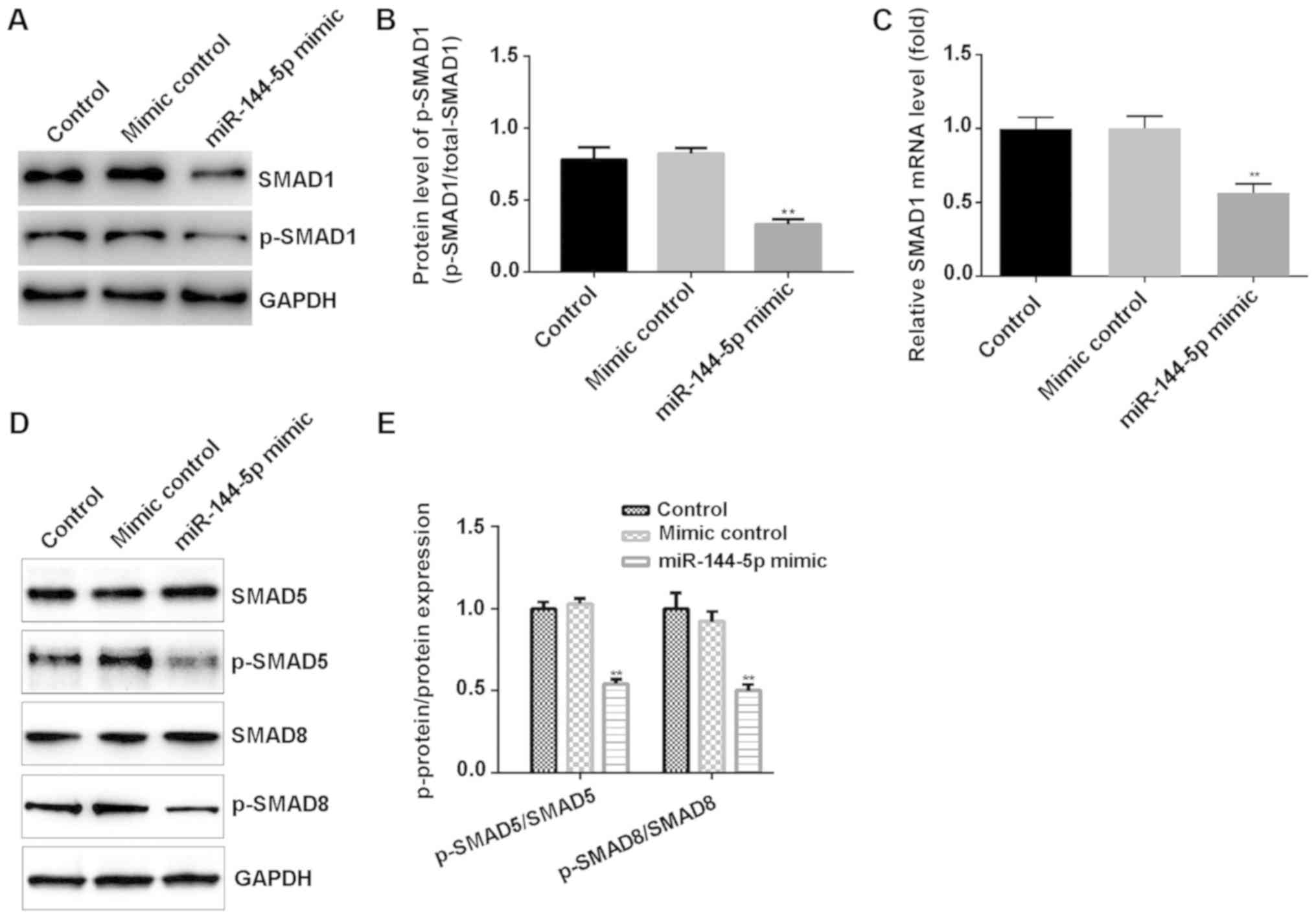
To investigate the specific mechanism of action of
miR-144-5p on HUVECs, the expression levels of proteins within the
SMAD1/5/8 signaling pathway were examined. Western blot assay
suggested that miR-144-5p mimic markedly decreased the protein
level of SMAD1 and p-SMAD1 (Fig.
4A), and significantly reduced the ratio of p-SMAD1/SMAD1
(Fig. 4B). RT-qPCR assay results
showed that miR-144-5p mimic decreased the relative mRNA expression
of SMAD1 in HUVECs (Fig. 4C).
Moreover, western blotting indicated that miR-144-5p mimic
significantly decreased p-SMAD5/8 protein expression in HUVECs
(Fig. 4D and E).
Discussion
In this present study, the primary aim was to
explore the effect of miR-144-5p on HUVECs in vitro, to
determine the role of miR-144-5p in atherosclerosis. It was
demonstrated that miR-144-5p mimic transfection could suppress
proliferation and induce apoptosis in HUVECs. It was also found
that miR-144-5p mimic suppressed HUVEC migration and invasion.
Furthermore, it was found that SMAD1 was a direct target of
miR-144-5p, and miR-144-5p mimic suppressed the SMAD1/5/8 pathway
in HUVECs.
miRNAs are a group of small, non-coding RNAs
(9). Recent reports have identified
multiple miRNAs that are involved in the pathogenesis of
atherosclerosis (23–29). Vascular endothelial cells are crucial
barriers in the vascular lumen (30). They maintain the stability of
hemodynamics and material exchange, secrete inflammatory cytokines
and regulate blood pressure through vasodilation and contraction
factors (31). Previous studies have
demonstrated that various miRNAs have different roles in the injury
of vascular endothelial cells in both normal and tumor tissues. For
example, previous research indicated that miRNA-129-1 and miRNA-133
promote vascular endothelial cell proliferation (32). However, the specific function and
mechanism of action of miR-144-5p on HUVECs remain unclear.
Consistent with these previous results, the present study provided
evidence supporting the important role of miR-144-5p in HUVECs; the
data demonstrated that miR-144-5p mimic suppressed the
proliferation, migration and invasion of HUVECs and induced cell
apoptosis.
The pathological role of miR-144-5p was further
explored by identifying its direct target gene. Through
bioinformatics, it was predicted that SMAD1 was a direct target of
miR-144-5p. SMAD1 is a member of the SMAD family, and the family
members are signal transducers and transcriptional modulators that
mediate multiple signaling pathways (20). SMAD1 mediates bone morphogenetic
proteins (BMP) signaling, and BMP stimulation increases SMAD1
phosphorylation, allowing it to form complexes with GAL4 to act as
a functional transcription regulator (33). The BMP-SMAD1/5/8 signaling pathway
participates in multiple biological processes including cell
growth, apoptosis, morphogenesis, differentiation and immune
modulation. A previous study also reported that SMAD1 is a target
gene of miR-144 (20). Ren et
al reported that miR-144 upregulation could decrease the
activity of mTOR signaling pathways and suppress cell proliferation
in osteosarcoma cells (34). In the
present study, it was found that miR-144-5p was associated with the
SMAD signaling pathway. miR-144-5p mimic inhibited the expression
of p-SMAD1/5/8. Consequently, these results further confirm that
miR-144-5p serves a crucial role in HUVECs.
In conclusion, miR-144-5p may modulate HUVECs
proliferation, apoptosis, invasion and migration through affecting
the SMAD signaling pathway by altering the expression of SMAD1, and
thus may participate in the onset and development of
atherosclerosis. Therefore, the data from this present study may
provide a new theoretical basis and strategy for the diagnosis and
treatment of atherosclerosis. However, this study is only a
preliminary investigation in to the role of miR-144-5p in
atherosclerosis. Further studies are needed to better understand
the role of miR-144-5p in atherosclerosis. For example, it would be
interesting to investigate whether SMAD1 upregulation could reverse
the effect of miR-144-5p on HUVECs. How the SMAD1/5/8 pathway is
involved in the effect of miR-144-5p in HUVECs should also be
further explored. Furthermore, the effect of downregulating
miR-144-5p in HUVECs should investigated. Finally, the relationship
between the expression of miR-144-5p and SMAD1, in the context of
the clinical features of atherosclerosis needs to be explored.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZL designed the study and revised the manuscript. WF
and JZ wrote the manuscript and collected the data. YS and RZ
searched the literature and interpreted the data. HZ collected the
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ference BA, Ginsberg HN, Graham I, Ray KK,
Packard CJ, Bruckert E, Hegele RA, Krauss RM, Raal FJ, Schunkert H,
et al: Low-density lipoproteins cause atherosclerotic
cardiovascular disease. 1. Evidence from genetic, epidemiologic,
and clinical studies. A consensus statement from the European
Atherosclerosis Society Consensus Panel. Eur Heart J. 38:2459–2472.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen LY, Leening MJ, Norby FL, Roetker NS,
Hofman A, Franco OH, Pan W, Polak JF, Witteman JC, Kronmal RA, et
al: Carotid intima-media thickness and arterial stiffness and the
risk of atrial fibrillation: The Atherosclerosis Risk in
Communities (ARIC) Study, Multi-Ethnic Study of Atherosclerosis
(MESA), and the Rotterdam Study. J Am Heart Assoc. 5:e0029072016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Trpkovic A, Resanovic I, Stanimirovic J,
Radak D, Mousa SA, Cenic-Milosevic D, Jevremovic D and Isenovic ER:
Oxidized low-density lipoprotein as a biomarker of cardiovascular
diseases. Crit Rev Clin Lab Sci. 52:70–85. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weber C and Noels H: Atherosclerosis:
Current pathogenesis and therapeutic options. Nat Med.
17:1410–1422. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mestas J and Ley K: Monocyte-endothelial
cell interactions in the development of atherosclerosis. Trends
Cardiovasc Med. 18:228–232. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Koroleva IA, Nazarenko MS and Kucher AN:
Role of microRNA in development of instability of atherosclerotic
plaques. Biochemistry (Mosc). 82:1380–1390. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Laffont B and Rayner KJ: MicroRNAs in the
pathobiology and therapy of atherosclerosis. Can J Cardiol.
33:313–324. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Papageorgiou N, Tslamandris S, Giolis A
and Tousoulis D: MicroRNAs in cardiovascular disease: Perspectives
and reality. Cardiol Rev. 24:110–118. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Giral H, Kratzer A and Landmesser U:
MicroRNAs in lipid metabolism and atherosclerosis. Best Pract Res
Clin Endocrinol Metab. 30:665–676. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Matsushita R, Seki N, Chiyomaru T,
Inoguchi S, Ishihara T, Goto Y, Nishikawa R, Mataki H, Tatarano S,
Itesako T, et al: Tumour-suppressive microRNA-144-5p directly
targets CCNE1/2 as potential prognostic markers in bladder cancer.
Br J Cancer. 113:282–289. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yamada Y, Arai T, Kojima S, Sugawara S,
Kato M, Okato A, Yamazaki K, Naya Y, Ichikawa T and Seki N:
Regulation of antitumor miR-144-5p targets oncogenes: Direct
regulation of syndecan-3 and its clinical significance. Cancer Sci.
109:2919–2936. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Song L, Peng L, Hua S, Li X, Ma L, Jie J,
Chen D, Wang Y and Li D: miR-144-5p Enhances the radiosensitivity
of non-small-cell lung cancer cells via targeting ATF2. BioMed Res
Int. 2018:51094972018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li J, Wang R, Ge Y, Chen D, Wu B and Fang
F: Assessment of microRNA-144-5p and its putative targets in
inflamed gingiva from chronic periodontitis patients. J Periodontal
Res. 54:266–277. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang X, Sundquist K, Hedelius A, Palmér K,
Memon AA and Sundquist J: Circulating microRNA-144-5p is associated
with depressive disorders. Clin Epigenetics. 7:692015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Goumans MJ, Liu Z and ten Dijke P:
TGF-beta signaling in vascular biology and dysfunction. Cell Res.
19:116–127. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bai Y, Zhang Q, Su Y, Pu Z and Li K:
Modulation of the proliferation/apoptosis balance of cascular
smooth muscle cells in atherosclerosis by lncRNA-MEG3 via
regulation of miR-26a/smad1 axis. Int Heart J. 60:444–450. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Weng J, Wang C, Zhong W, Li B, Wang Z,
Shao C, Chen Y and Yan J: Activation of CD137 signaling promotes
angiogenesis in atherosclerosis via modulating endothelial
Smad1/5-NFATc1 pathway. J Am Heart Assoc. 6:62017. View Article : Google Scholar
|
|
20
|
Peng YG and Zhang L: Baohuoside-I
suppresses cell proliferation and migration by up-regulating
miR-144 in melanoma. Pharm Biol. 56:43–50. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Δ Δ C(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mudau M, Genis A, Lochner A and Strijdom
H: Endothelial dysfunction: The early predictor of atherosclerosis.
Cardiovasc J Afr. 23:222–231. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li L, Li Y and Tang C: The role of
microRNAs in the involvement of vascular smooth muscle cells in the
development of atherosclerosis. Cell Biol Int. 43:1102–1112. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Su Y, Yuan J, Zhang F, Lei Q, Zhang T, Li
K, Guo J, Hong Y, Bu G, Lv X, et al: MicroRNA-181a-5p and
microRNA-181a-3p cooperatively restrict vascular inflammation and
atherosclerosis. Cell Death Dis. 10:3652019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li K, Chen ZT and Qin YW: Expression
profiles of microRNA related to atherosclerosis in patients with
OSA. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 33:304–309.
2019.(In Chinese). PubMed/NCBI
|
|
26
|
Wang Z, Zhang J, Zhang S, Yan S, Wang Z,
Wang C and Zhang X: miR-30e and miR-92a are related to
atherosclerosis by targeting ABCA1. Mol Med Rep. 19:3298–3304.
2019.PubMed/NCBI
|
|
27
|
Cheng Y, Zhou M and Zhou W: MicroRNA-30e
regulates TGF-β-mediated NADPH oxidase 4-dependent oxidative stress
by Snai1 in atherosclerosis. Int J Mol Med. 43:1806–1816.
2019.PubMed/NCBI
|
|
28
|
Yang L and Gao C: miR-590 inhibits
endothelial cell apoptosis by inactivating the TLR4/NF-κB pathway
in atherosclerosis. Yonsei Med J. 60:298–307. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fang M, Li Y, Wu Y, Ning Z, Wang X and Li
X: miR-185 silencing promotes the progression of atherosclerosis
via targeting stromal interaction molecule 1. Cell Cycle.
18:682–695. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Biedermann BC: Vascular endothelial cells:
An interesting immunological barrier. Schweiz Med Wochenschr.
129:1712–1716. 1999.(In German). PubMed/NCBI
|
|
31
|
Tian X, Yu C, Shi L, Li D, Chen X, Xia D,
Zhou J, Xu W, Ma C, Gu L, et al: MicroRNA-199a-5p aggravates
primary hypertension by damaging vascular endothelial cells through
inhibition of autophagy and promotion of apoptosis. Exp Ther Med.
16:595–602. 2018.PubMed/NCBI
|
|
32
|
Soufi-Zomorrod M, Hajifathali A, Kouhkan
F, Mehdizadeh M, Rad SM and Soleimani M: MicroRNAs modulating
angiogenesis: miR-129-1 and miR-133 act as angio-miR in HUVECs.
Tumour Biol. 37:9527–9534. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu F, Hata A, Baker JC, Doody J, Cárcamo
J, Harland RM and Massagué J: A human Mad protein acting as a
BMP-regulated transcriptional activator. Nature. 381:620–623. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ren YF, Zhang TH, Zhong S, Zhao YT and Lv
YN: miR-144 suppresses proliferation and induces apoptosis of
osteosarcoma cells via direct regulation of mTOR expression. Oncol
Lett. 15:1163–1169. 2018.PubMed/NCBI
|