Introduction
Diabetic nephropathy (DN) is a risk factor for
end-stage renal disease and cardiovascular diseases, which are
associated with high morbidity and mortality (1). Microvascular lesions may lead to
impaired blood flow and contribute to damage to the target organs
(2). Therefore, it is important to
establish a novel therapeutic method for patients with DN and
improve their prognosis.
Podocytes act as the most important barrier to
urinary protein loss through the formation and maintenance of foot
processes and interposed slit-diaphragms (3). Podocytes serve an important role in DN.
A multitude of factors, including oxidative stress and inflammatory
response in diabetes cause abnormalities in podocytes that limit
their ability to self-repair and regenerate (4,5).
Furthermore, diabetes may cause podocyte foot process detachment,
hypertrophy and loss (4,5). Therefore, podocyte injury is widely
considered as a key factor in determining the prognosis of DN
(4,5).
MicroRNAs (miRNAs/miRs) are endogenous small
non-coding RNAs with a length of 19–25 nucleotides, and are
involved in various cellular processes (6–8). They
are reported to function by binding to the 3′-untranslated region
(3′-UTR) of target genes, resulting in the degradation of mRNAs
(9–11). Recent studies have demonstrated that
miRNAs targeting podocyte-associated genes may be potential
candidates for antiapoptotic therapies for DN (12,13). A
previous study demonstrated that miR-25 inhibited high glucose
(HG)-induced apoptosis in renal tubular epithelial cells by
reducing the production of reactive oxygen species, and decreasing
the activity and cleavage of caspase-3 (14). Another study reported that the
inhibition of miR-377 expression ameliorated inflammation and
improved insulin sensitivity (15).
These results suggest that miRNAs serve important roles in
regulating the DN process. Therefore, further investigations
regarding the influence of miRNAs and their targets are a promising
perspective. A previous study has demonstrated that miR-145-5p is
enriched in diabetic patients, and may represent a novel candidate
biomarker (16).
The Notch signaling pathway is associated with
vascular defects, including abnormal structure and leakiness
(17,18). Under normal circumstances, activation
of Notch receptors releases a signal in the form of the Notch
intracellular domain (NICD). When NICD enters the nucleus, its
binding to chorionic somatomammotropin hormone like 1 may trigger
an allosteric alteration and, later, NICD transfers into the
nucleus and activates the gene transcription of hes family bHLH
transcription factor 1 (Hes1) and hes related family bHLH
transcription factor with YRPW motif 1 (Hey1) (19), which are associated with cell
differentiation, proliferation and apoptosis (20,21).
Based on the aforementioned information, the present
study aimed to investigate the role of miR-145-5p and Notch1 in DN.
Furthermore, their interactions and potential mechanism in
HG-induced podocytes was studied. This may provide a theoretical
basis for clinical treatment.
Materials and methods
Podocyte cell culture and treatment
groups
Mouse podocytes (cat. no. M1710) and all cell
culture media, FBS and other supplements were purchased from
ScienCell Research Laboratories, Inc. Mouse podocytes were cultured
according to the manufacturer's protocols as follows: Mouse
podocytes were maintained in RPMI-1640 medium supplemented with 10%
FBS, 100 U/ml penicillin, 100 mg/ml streptomycin and 10 U/ml mouse
recombinant γ-interferon (Sigma-Aldrich; Merck KGaA) with 5%
CO2 at 33°C. Subsequently, cells were cultured in fresh
DMEM-F12 supplemented with 10% FBS, 100 U/ml penicillin, and 100
mg/ml streptomycin at 37°C with 5% CO2 for 10–14 days
(22). Prior to the functional
experiments, expression levels of the podocyte differentiation
marker synaptopodin were detected by immunofluorescence staining
(data not shown). Subsequently, cells were cultured in RPMI-1640
medium supplemented with 10% FBS at 37°C with 5% CO2
containing 25 mM glucose (HG) or 5 mM glucose [normal glucose (NG)]
for 12, 24, 48 and 72 h. The 293T cells (Cell Bank of Type Culture
Collection of Chinese Academy of Sciences) were cultured as
described previously (23).
Lipofectamine transfection
miR-145-5p mimics, miR-145-5p inhibitor and
scrambled controls were applied to create miR-145-5p overexpression
and knockdown in podocytes. Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) was used for cell
transfection. Briefly, Lipofectamine® 2000 was diluted
and the podocytes were then added to the diluted solution. The
solution was added into 6-well plates and incubated for 6 h at 37°C
with 5% CO2. Subsequently, 10 nM miRNA duplexes and
miRNA transfection reagent were mixed and added to 100 ml miRNA
transfection medium for 45 min at 20°C. Finally, the medium was
changed to conventional medium and cultured for a further 48 h.
After 48 h of transfection, the total RNA and protein were
extracted from the cells and tested using RT-qPCR and western
blotting, respectively.
TUNEL staining
Cell apoptosis was examined by TUNEL staining using
a DeadEnd™ fluorometric TUNEL system kit (Promega Corporation),
according to the manufacturer's protocol as previously described
(24). Cells (5×104/well)
were first washed using saline. Subsequently, the cells were fixed
with 4% neutral formaldehyde in PBS at room temperature for 15 min.
Then, the slides were washed three times with PBS, and with 3%
H2O2 in methanol for 10 min at room
temperature. After treatment with 0.2% Triton X-100 in PBS for 15
min at room temperature, the cell slides were immersed into 100 ml
equilibration buffer at room temperature (22°C) for 15 min,
followed by incubation in 5% bovine serum albumin (BSA; Thermo
Fisher Scientific, Inc.) for 30 min at room temperature. After a
terminal deoxynucleotidyl transferase buffer pre-incubation for 10
min at room temperature, slides were incubated in the TUNEL
reaction mixture for 60 min at 37°C and sealed using fluorescent
mounting medium (Beyotime Institute of Biotechnology). Fluorescence
microscopy was used for detection of the fluorescence of apoptotic
cells and data analysis. The size (mm2) of each area
containing TUNEL-positive cells was measured with a micro ruler at
×200 magnification. After the measurement, the number of apoptotic
cells was counted by two technicians in three high-power fields
(magnification, ×400) under a light microscope.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
To detect and compare gene expression, RT-qPCR was
performed. Total RNA was extracted from podocytes using TRIzol
reagent (Sigma-Aldrich; Merck KGaA). Then, the RNA concentration
was determined by measuring the absorbance (A) at 260 and 280 nm,
and a A260/A280 ratio of 1.8–2.0 indicated acceptable purity.
Briefly, 2 µg total RNA was reverse-transcribed to cDNA using
PrimeScript™ RT Master Mix (Takara Bio, Inc.) using the
temperature protocol of 42°C for 60 min followed by 70°C for 5 min.
RT-qPCR was performed on an ABI 7500 Real-Time PCR system
thermocycler using SYBR® Premix Ex Taq™
(Takara Bio, Inc.). The PCR conditions consisted of an initial
denaturation step of 94°C for 2 min, followed by 30 cycles of 94°C
for 30 sec, 59°C for 30 sec, 72°C for 2 min and a final elongation
step at 72°C for 10 min. The primers used for amplification are
shown in Table I, and were
synthesized by Takara Biotechnology Co., Ltd. The results were
analyzed using the 2−ΔΔCq method (25). GAPDH was used as an internal
control.
 | Table I.Primer sequences for reverse
transcription-quantitative PCR analysis. |
Table I.
Primer sequences for reverse
transcription-quantitative PCR analysis.
| Complementary
DNA | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| miR-145-5p |
GTCCAGTTTTCCCAGGAATCC |
TCGCTTCGGCAGCACATAT |
| NICD |
GTGGATGACCTAGGCAAGTCG |
GTCTCCTCCTTGTTGTTCTGC |
| Bcl-2 |
CGGAGGCTGGGATGCCTTTG |
TTTGGGGCAGGCATGTTGAC |
| Bax |
GCCCTTTTGCTTCAGGGTTT |
TCCAATGTCCAGCCTTTG |
| Caspase-3 |
TACAGGAACAGACCATAATACC |
AGACCAGTGCTCACAAGGAAC |
| Hes1 |
CACGACACCGGACAAACCA |
GCCGGGAGCTATCTTTCTTAAGTG |
| Hey1 |
AAGACGGAGAGGCATCATCGAG |
CAGATCCCTGCTTCTCAAAGGCAC |
| U6 |
CTCGCTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT |
| GAPDH |
ACAACTTTGGTATCGTGGAAGG |
GCCATCACGCCACAGTTTC |
For measurement of miR-145-5p expression, the
Taqman™ MicroRNA Reverse Transcription kit (Thermo
Fisher Scientific, Inc.) and Taqman® Universal Master
Mix II (Thermo Fisher Scientific, Inc.) were used for reverse
transcription and RT-qPCR, respectively. The PCR conditions
consisted of an initial denaturation step of 94°C for 2 min,
followed by 30 cycles of 94°C for 30 sec, 59°C for 30 sec, 72°C for
2 min and a final elongation step at 72°C for 10 min. U6 was used
as an internal control.
Western blot analysis
Western blot analysis was conducted as previously
described (26). Briefly, podocytes
were washed with saline and harvested by scraping the culture
dishes followed by RIPA buffer (Thermo Fisher Scientific, Inc.).
The buffer was cooled for 40 min in an ice bath and centrifuged for
20 min at 1,000 × g at room temperature. After the supernatant was
discarded, the protein concentration was assessed using a
Bicinchoninic Acid protein assay. Then, 30 µg/lane proteins were
transferred to 10% PVDF membranes, and the membranes were blocked
by treatment with 5% non-fat milk for 2 h at 37°C. After rinsing,
the membranes were incubated at 4°C overnight with primary
antibodies. The primary polyclonal antibodies were as follows: NICD
(1:200 dilution; cat. no. ab8925; Abcam); Hes1 (1:500 dilution;
cat. no. ab71559; Abcam), Hey1 (1:500 dilution; cat. no. ab22614;
Abcam); cleaved caspase-3 (1:3,000 dilution; cat. no. ab49822;
Abcam), Bcl-2 (1:3,000 dilution; cat. no. ab692; Abcam) and Bax
(1:3,000 dilution; cat. no. ab77566; Abcam) and GAPDH (loading
control; 1:3,000 dilution; cat. no ab9485; Abcam). After washing
with Tris buffer solution, secondary rabbit anti-primary IgG
conjugated with horseradish peroxidase (1:1,500; cat. no. ab6721
Abcam). was added to the solutions and incubated for 1 h at room
temperature. Binding was detected using Western
Lightning™ Plus ECL reagent (Thermo Fisher Scientific,
Inc). Densitometric analysis was performed for semi-quantification
of the blots using ImageJ software (version 1.8; National
Institutes of Health).
Flow cytometry
Flow cytometry was conducted as previously described
(27). Briefly, podocytes
(5×104/well) were incubated with fully-supplemented
RPMI-1640 for 24 h following transfection with miR-145-5p mimics,
inhibitor or scrambled control. Subsequently, for apoptosis
measurements, podocytes were collected, washed with PBS,
resuspended in 100 µl 1X binding buffer and stained with 5 µl
Annexin V and 5 µl propidium iodide (PI; Becton, Dickinson and
Company) at room temperature for 15 min in the dark. Fixed cells
(1×104/well) were rehydrated in PBS for 10 min and
subjected to PI/RNase (10 µg/ml) staining at 4°C for 20 min A flow
cytometer was utilized to evaluate the apoptotic levels and cell
cycle distribution in each sample according to the manufacturer's
protocol. The results were analyzed using a BD FACScan™
flow cytometer and BD FACSDiva™ software (version 1.2; BD
Biosciences).
Luciferase reporter assay
Potential target mRNAs of miR-145-5p were searched
for using TargetScan (version 7.2; http://www.targetscan.org/) (28), miRanda (version 1.0; http://www.microrna.org) and miRDB (version 1.0;
http://mirdb.org/) (29). For the luciferase reporter assay, a
fragment from the 3′-UTR of Notch1, which was predicted to contain
a binding sequence for miR-145-5p, was amplified using the pGL3
luciferase promoter vector (Promega Corporation) Then,
1×105 293 cells were cultured and transfected with the
wild-type or mutated 3′-UTR fragments and miR-145-5p inhibitor or
mimic using Lipofectamine® 2000. After culturing for 48
h, the cells were harvested and lysed. Luciferase activity was
measured using a Dual Luciferase Reporter Assay system (Promega
Corporation). Relative luciferase activity was normalized to that
of Renilla luciferase activity.
Statistical analysis
All experiments were repeated three times and
analyzed using GraphPad Prism 5.1 software (GraphPad Software,
Inc.). Data are expressed as the means ± SD. Statistical analyses
were carried out using one-way ANOVA followed by Tukey's multiple
comparison post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression levels of miR-145-5p are
decreased in HG-induced podocytes
First, in order to investigate the role of
miR-145-5p in HG-treated podocytes, the expression levels of
miR-145-5p were detected using RT-qPCR following incubation with 25
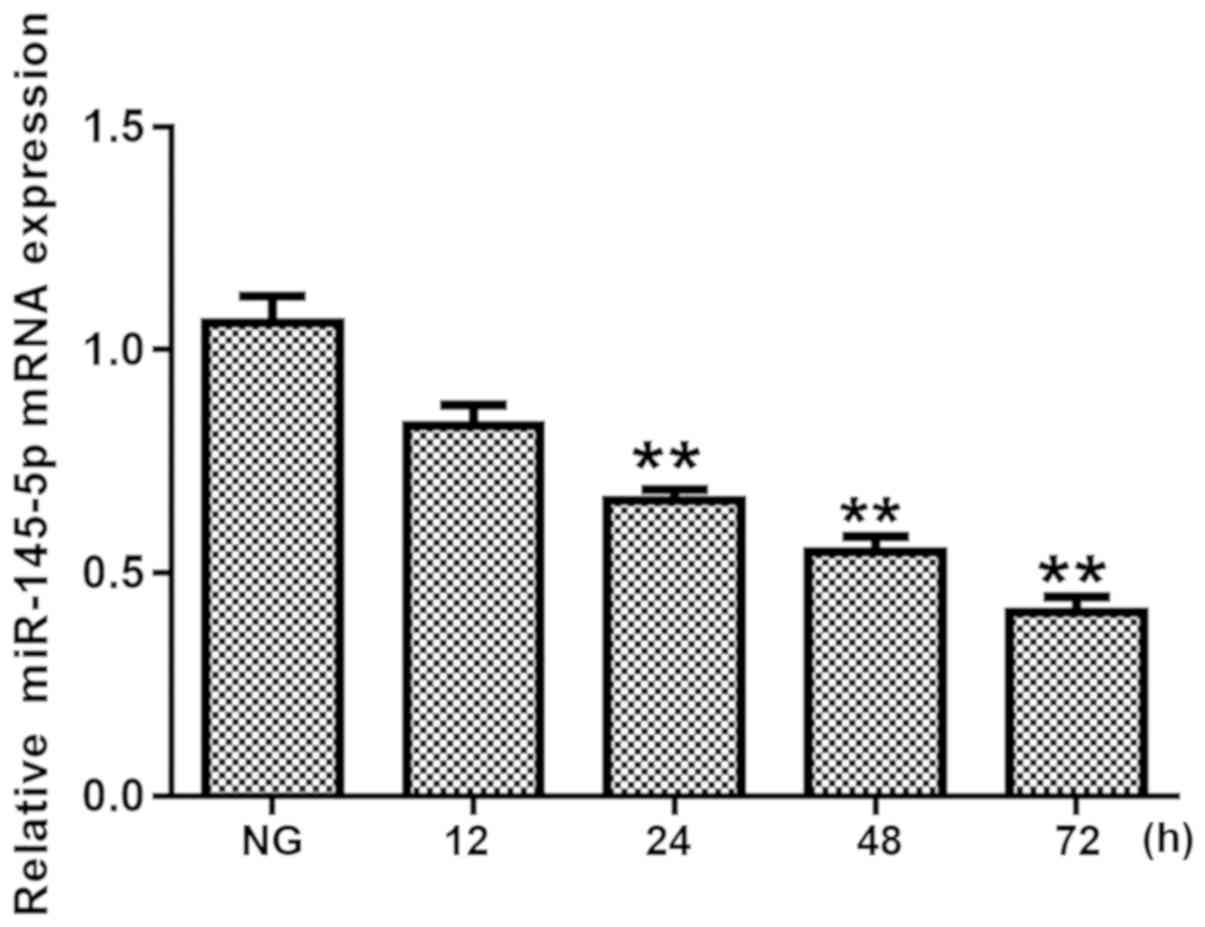
mM glucose for 12, 24, 48 and 72 h. As shown in Fig. 1, the expression levels of miR-145-5p
were significantly decreased in HG-treated podocytes from 24 h
onwards. These data indicate that miR-145-5p serves a critical role
in HG-stimulated podocytes.
miR-145-5p expression is successfully
changed following transfection with miR-145-5p mimic or
inhibitor
miR-145-5p expression was detected in different
transfection groups. Podocytes were transfected with scrambled
control, miR-145-5p mimic or miR-145-5p inhibitor for 48 h.
Following transfection, the expression levels of miR-145-5p were
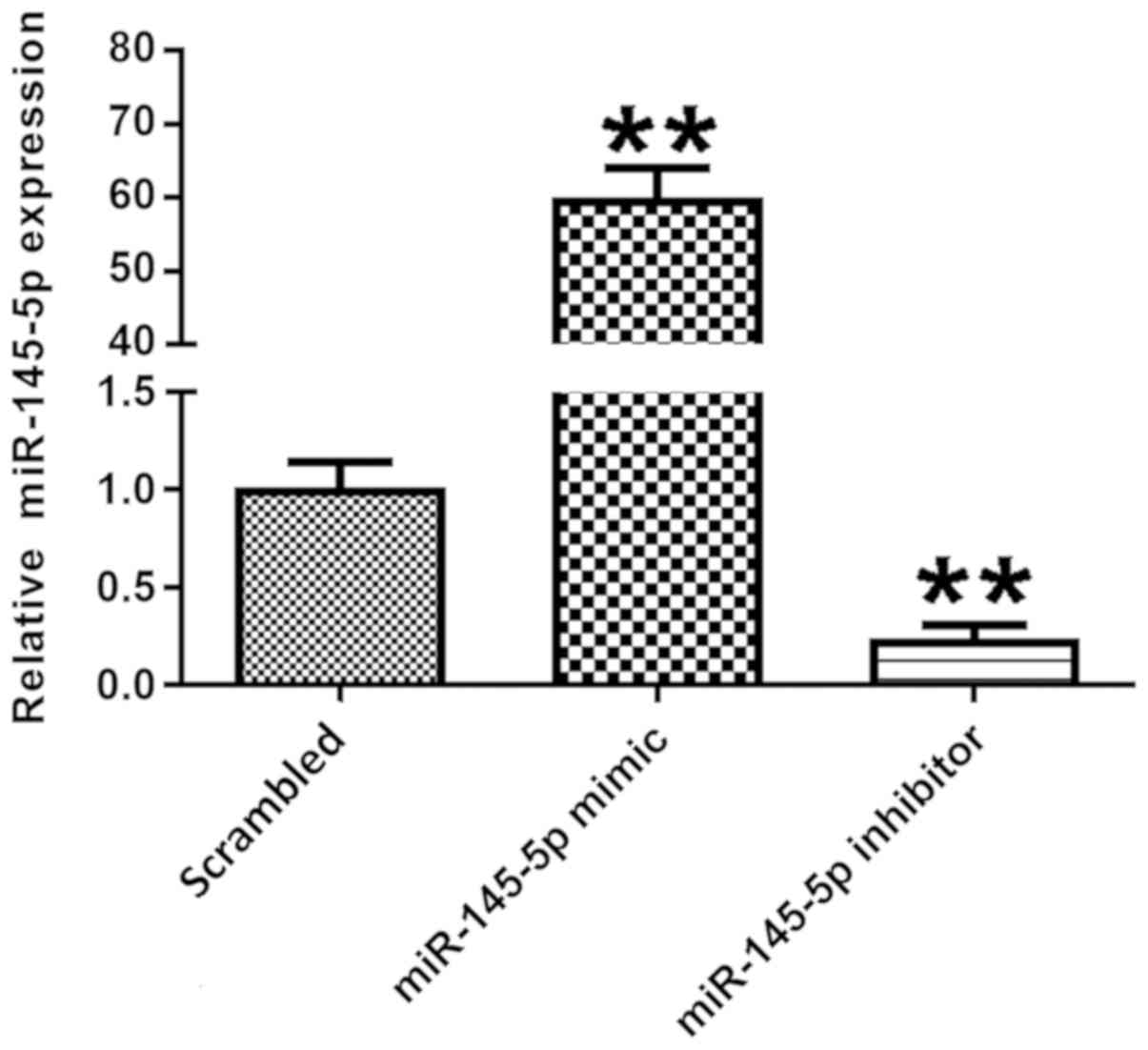
detected via RT-qPCR. Fig. 2 shows
that in the miR-145-5p overexpression group, transfected with
miR-145-5p mimic, the expression levels of miR-145-5p were
significantly higher compared with those in the scrambled control
group. Additionally, in the miR-145-5p knockdown group, transfected
with miR-145-5p inhibitor, miR-145-5p expression was significantly
decreased in the podocytes compared with control. These data
indicate that the transfection was successful.
Overexpression of miR-145 inhibits
HG-induced apoptosis in podocytes
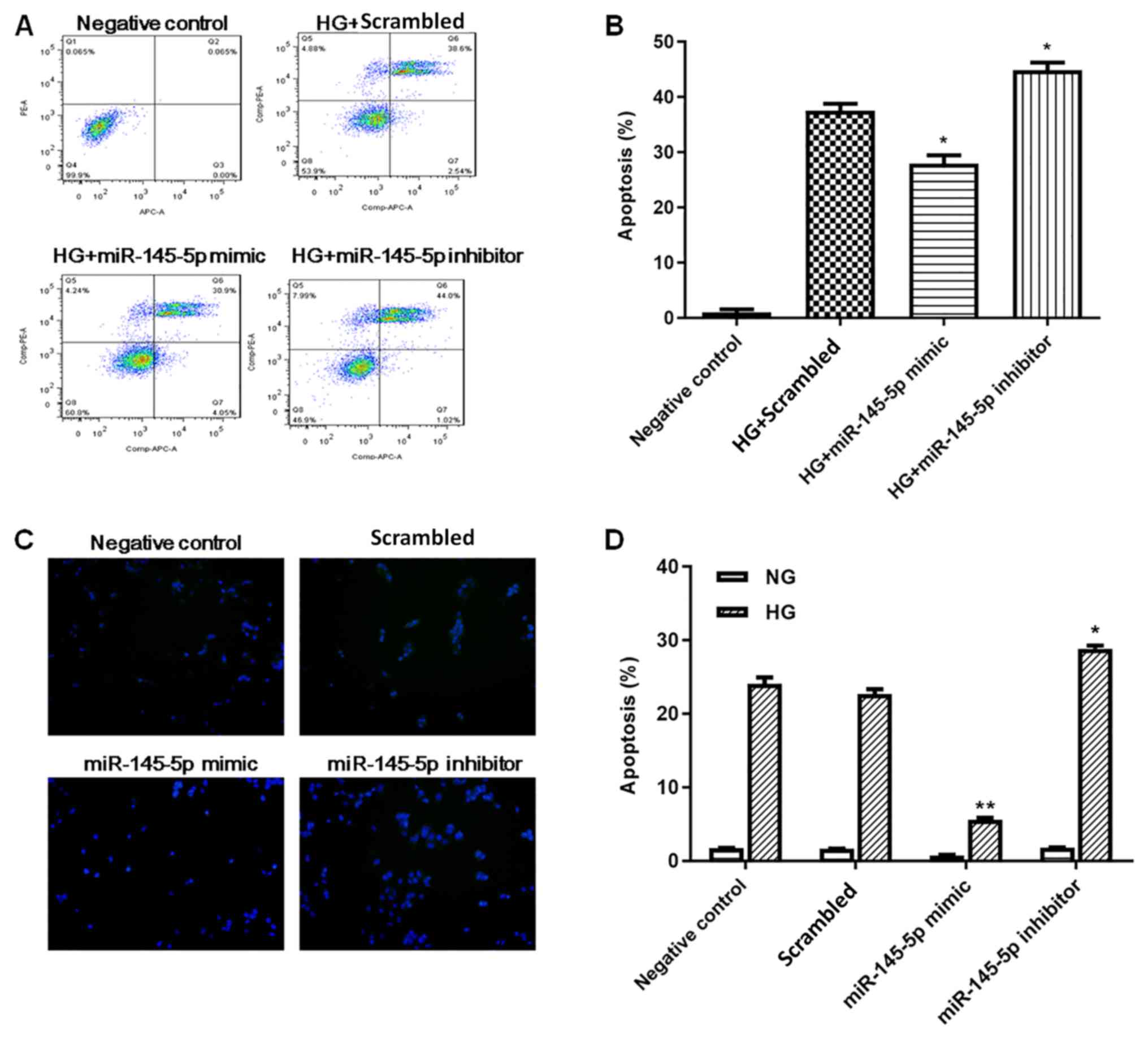
Podocyte apoptosis was measured by flow cytometry
and TUNEL staining. In flow cytometry assays (Fig. 3A and B), HG-treated podocytes
exhibited a significant increase in apoptotic cells compared with
the negative control group. Under HG conditions, the percentage of
apoptotic cells was significantly decreased in the miR-145-5p
overexpression group compared with the scrambled control group,
whereas the percentage of apoptotic cells in the miR-145-5p
inhibitor group was significantly increased. In the TUNEL staining
assay (Fig. 3C and D), the numbers
of apoptotic cells were markedly increased in the HG-induced
podocytes compared with NG podocytes. Under HG conditions, the
number of apoptotic cells was significantly lower in the miR-145-5p
mimic group compared with that in the scrambled control group;
however, in the miR-145-5p inhibitor group, the number of apoptotic
cells was significantly higher than that in the scrambled control
group. Overall, these functional experiments demonstrated that
miR-145-5p attenuated HG-induced cell apoptosis in podocytes.
miR-145-5p is a direct target of
Notch1 in podocytes
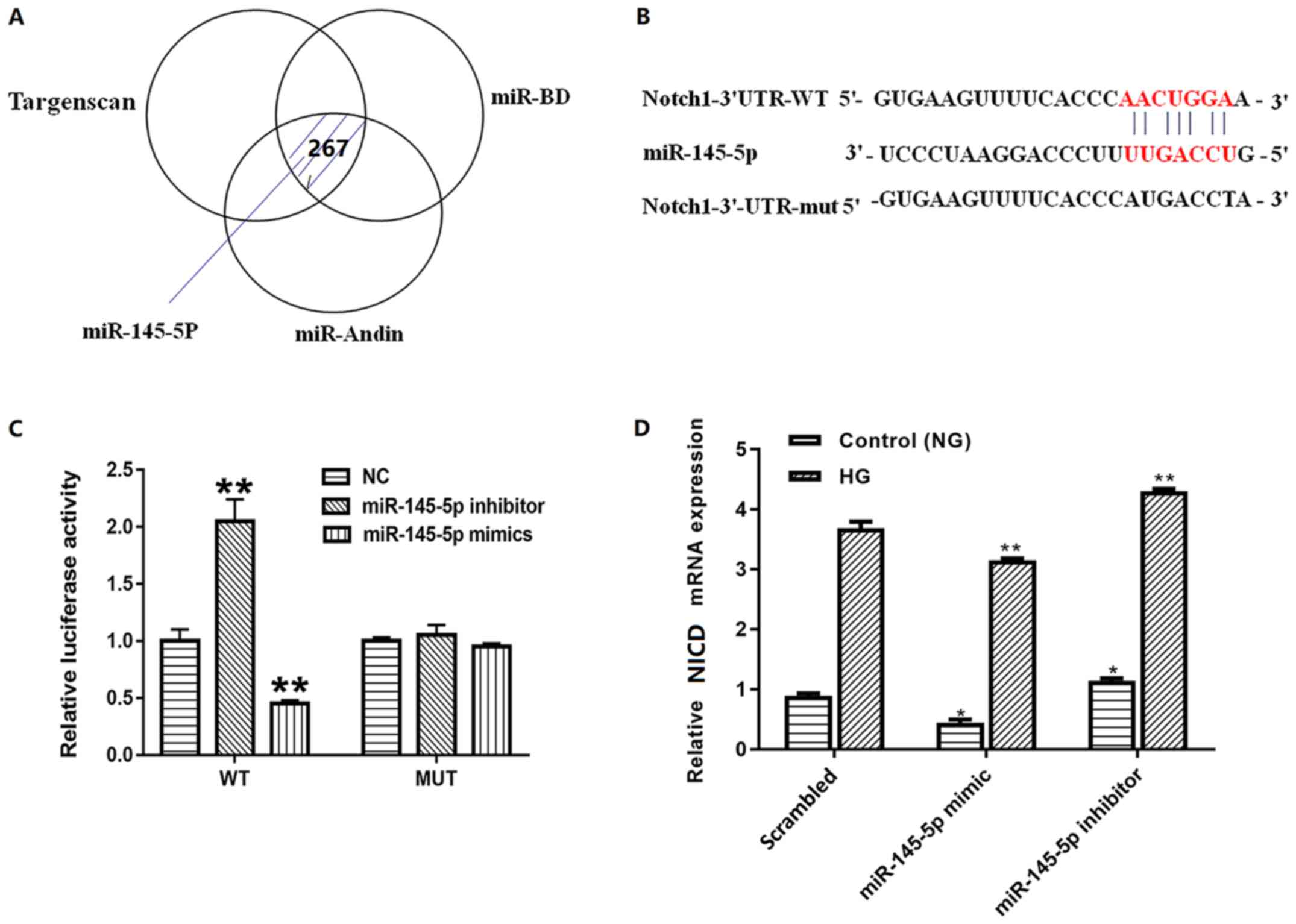
To identify the further mechanisms, the present
study first screened the putative target of miR-145-5p using three
different bioinformatic algorithms (Fig.
4A). Notch1, which is a vital factor in the Notch signaling
pathway and serves an important role in podocyte apoptosis, was
identified as a potential target by all three (Fig. 4B). To confirm the association between
miR-145-5p and Notch1, luciferase reporter constructs containing
wild-type and mutated forms of a putative miR-145-5p binding site
were constructed. Subsequently, wild-type pGL3-Notch1 3′-UTR or
pGL3-Notch1-Mut-3′-UTR was co-transfected with miR-145-5p mimic or
inhibitor into 293T cells. When cells were transfected with the
wild-type pGL3-Notch1 3′-UTR, the co-transfection of miR-145-5p
mimic inhibited luciferase activity whereas the co-transfection of
miR-145-5p inhibitor exhibited the opposite effect. By contrast,
co-transfection of miR-145-5p mimic or inhibitor with
pGL3-Notch1-Mut-3′-UTR containing mutations in the predicted
consensus sequences for miR-145-5p exhibited no apparent effect on
luciferase activity (Fig. 4C).
Furthermore, to identify whether miR-145-5p regulated the
expression levels of Notch1 in podocytes, the mRNA levels of Notch1
in different groups were detected. The results confirmed that the
mRNA levels of Notch1 were decreased in podocytes transfected with
miR-145-5p mimics and increased in podocytes transfected with
miR-145-5p inhibitors compared with those in the scrambled control
group (Fig. 4D). These findings
suggest that Notch1 is a direct target of miR-145-5p.
Overexpression of miR-145-5p
suppresses HG-induced activation of the Notch signaling
pathway
The Notch signaling pathway contains numerous
effectors, including NICD, Hes1 and Hey1 (30–32). To
further determine the effect of miR-145-5p on the Notch signaling
pathway, the present study detected the expression levels of NICD,
Hes1 and Hey1, which are important downstream factors of this
signaling pathway. The protein and mRNA expression levels of NICD,
Hes1 and Hey1 were measured by western blotting and RT-qPCR,
respectively. The results demonstrated that in the miR-145-5p mimic
group, the protein and mRNA expression levels of NICD, Hes1 and
Hey1 were significantly decreased compared with those in the
scrambled control group, whereas those in the miR-145-5p inhibitor
group were significantly increased (Fig.
5). Overall, the results suggest that overexpression of
miR-145-5p suppressed the HG-induced activation of the Notch
signaling pathway.
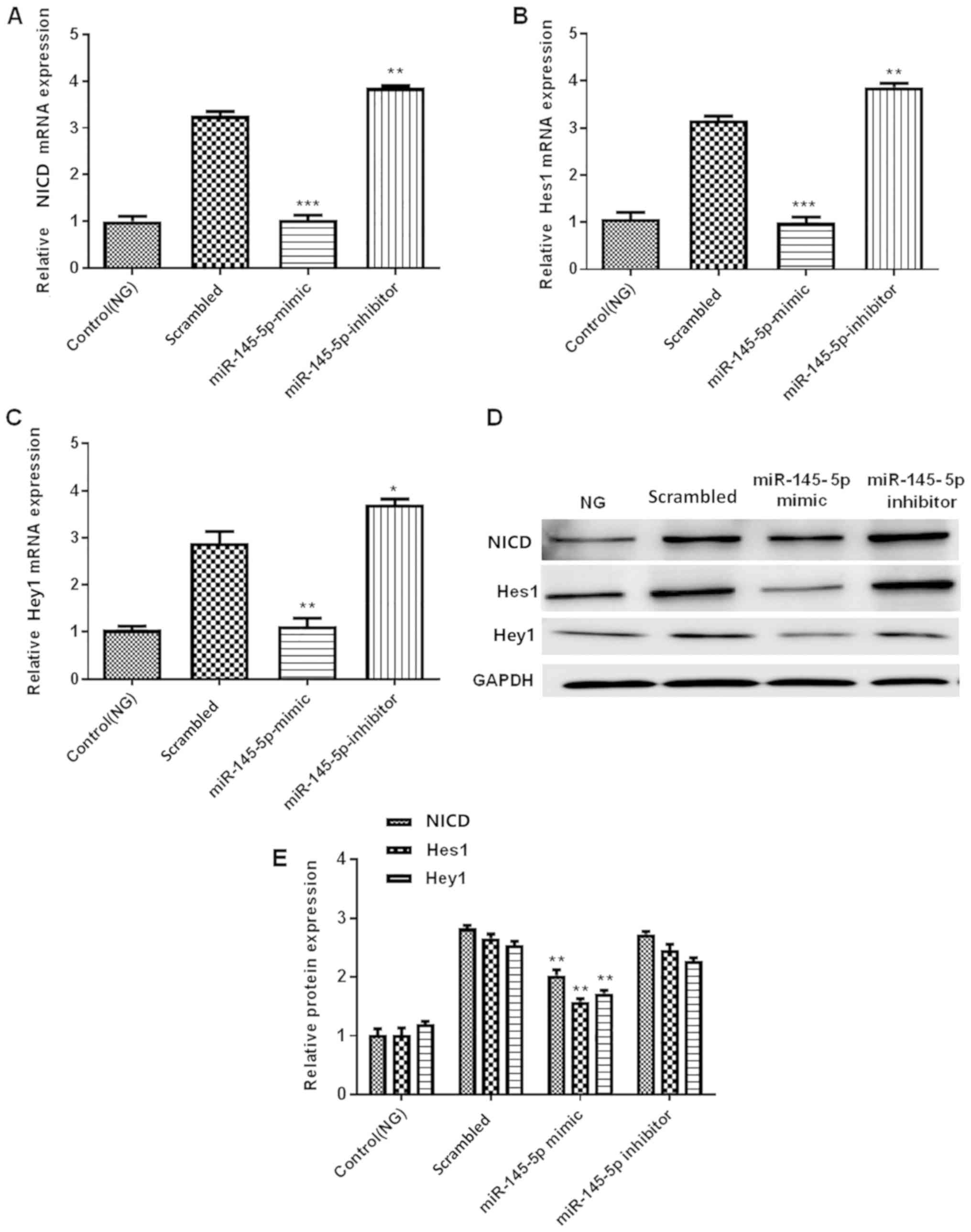 | Figure 5.Overexpression of miR-145-5p
suppresses the HG-induced activation of the Notch signaling
pathway. (A-C) RT-qPCR detection of (A) NICD, (B) Hes1 and (C) Hey1
mRNA expression in control (NG) and transfected podocytes under HG
conditions. (D) Western blotting detection of NICD, Hes1 and Hey1
protein expression in podocytes. (E) Semi-quantification of the
protein levels of NICD, Hes1 and Hey1. Statistical analysis was
performed using one-way ANOVA followed by Tukey's multiple
comparison post hoc test. Data are presented as the mean ± SD, and
shown as the fold change relative to the control group (n=3).
*P<0.05, **P<0.01 and ***P<0.001 vs. the scrambled
control. miR-145-5p, microRNA-145-5p; HG, high glucose; NG, normal
glucose; RT-qPCR, reverse transcription-quantitative PCR; NICD,
intracellular domain of Notch; Hes1, hes family bHLH transcription
factor 1; Hey1, hes related family bHLH transcription factor with
YRPW motif 1. |
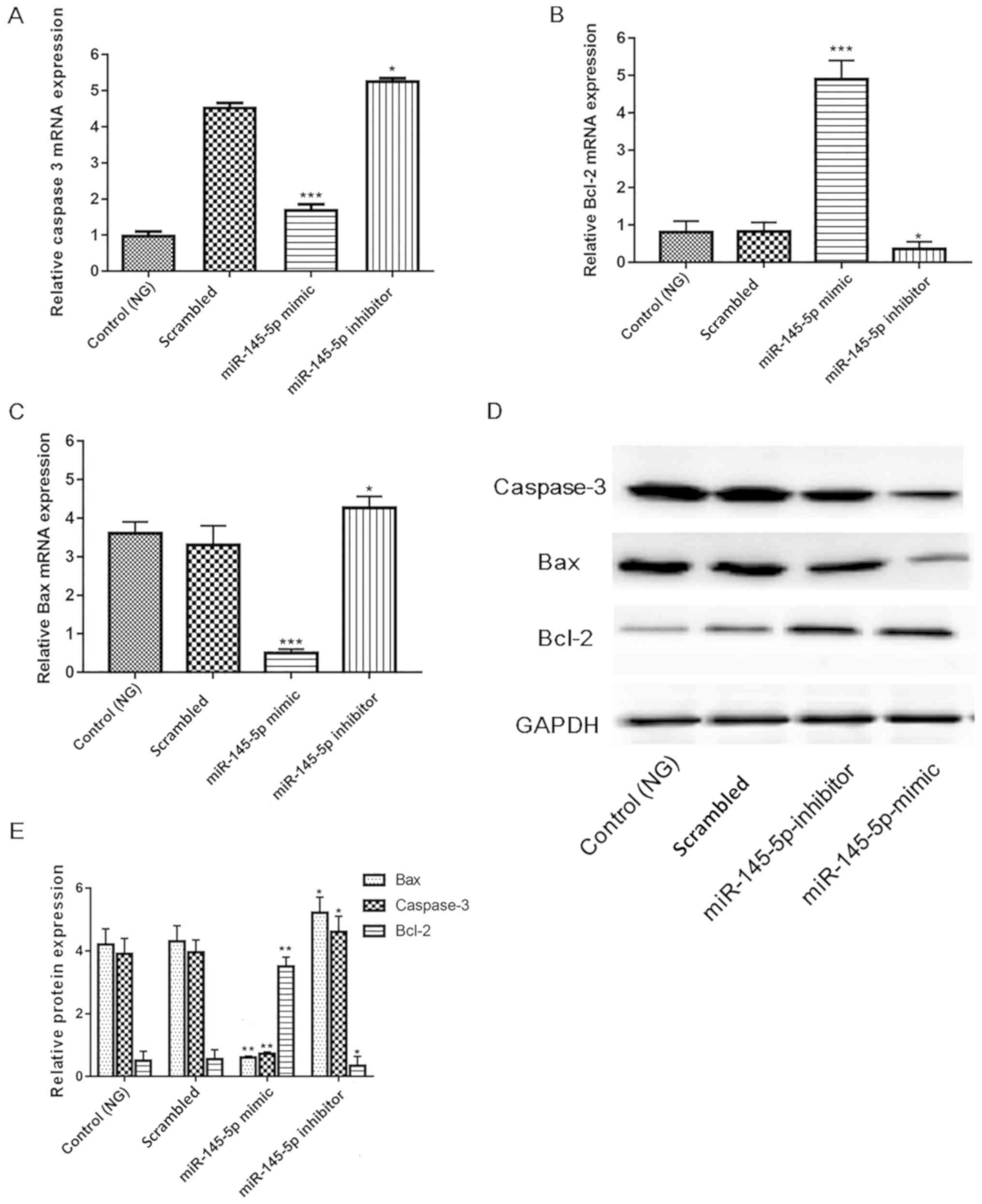
Overexpression of miR-145-5p inhibits
apoptotic pathways
Since numerous key factors, including cleaved
caspase-3, Bcl-2 and Bax serve pivotal roles in apoptosis (33–35), the
present study investigated whether their activity is increased in
HG-induced podocytes. Protein and mRNA expression were measured in
different groups of HG-induced podocytes. Fig. 6 shows that in the miR-145-5p
overexpression group, the mRNA and protein expression levels of
cleaved caspase-3 and Bax were significantly decreased, and those
of Bcl-2 were significantly increased compared with those in the
scrambled control group. Furthermore, opposite results were
observed in the miR-145-5p inhibitor group. These results indicate
that miR-145-5p overexpression suppressed the apoptosis of
podocytes in under HG conditions via the inhibition of apoptotic
signaling.
Discussion
DN is one of the most common complications of
diabetes mellitus and may lead to end-stage renal disease (1). Accumulating evidence has demonstrated
that podocyte apoptosis serves an essential role in the
pathogenesis of proteinuria and deteriorated renal function in DN
(4,36–38).
Previous studies have suggested that the Notch signaling pathway
serves a pivotal role in podocyte apoptosis, which is regulated by
various downstream factors, including NICD, Hes1 and Hey1 (34,36). For
example, the Notch signaling pathway is mediated via the release
and translocation of NICD into the nucleus, where NICD directly
functions as a transcriptional coactivator (39). In the pathological process of DN,
high blood glucose and hemodynamic alterations induce increased
expression of Jagged1 ligand and Notch1 receptor of the Notch
signaling pathway, resulting in a change in the receptor structure
(39). The enzyme g-secretase
induces podocytes to release NICD1, the active form of Notch1, and
activates the downstream genes Hes1 and Hey1 (36,38).
Notably, a previous study found that ubiquitination-dependent
coactivator associated arginine methyltransferase 1 degradation in
podocytes promotes podocyte apoptosis via Notch1 activation in DN
(34).
A series of studies has focused on the crosstalk
between miRNA and the Notch signaling pathway in podocytes
(40,41). For example, a previous study have
demonstrated that miRNA-34 directly interacts with the 3′-UTRs of
Notch1 and Jagged1, which is critical for Notch signaling pathway
activation (41). Liu et al
(42) suggested that miR-34c
overexpression inhibits the Notch signaling pathway by targeting
Notch1 and Jagged1, which are two important factors for Notch
signaling and apoptosis in HG-treated podocytes. Additionally,
Zhang et al (41) provided
evidence that miR-34a overexpression inhibits the Notch signaling
pathway (Notch1, Jagged1, NICD, Hes1 and Hey1 proteins) and
podocyte lesions induced by HG. miR-145 has primarily been defined
by its role in diabetic retinopathy, where it acts as a negative
regulator of toll like receptor 4/NF-κB signaling and attenuates
HG-induced oxidative stress (41).
Chen et al (43) revealed
that miR-145 can suppress the HG-induced proliferation and
migration of vascular smooth muscle cells by targeting Rho
associated coiled-coil containing protein kinase 1, which prevents
the occurrence and progression of atherosclerosis. The present
study first illustrated the transcriptional inhibition of the Notch
signaling pathway by miR-145-5p, which protected podocytes from
HG-induced injury. The present study supported the hypothesis that
miR-145-5p may have a renal protective effect in DN via the
amelioration of renal injury.
A number of factors in the apoptosis signaling
pathway are also involved in the pathological process of DN.
Previous studies indicate that DN can activate cell
apoptosis-related proteins (Bcl-2, p53 and NF-κB) (44–47). Gao
et al (48) reported that the
expression levels of Notch1, Jagged1, NICD, Hes1 and Hey1 were
upregulated in podocytes under HG conditions, and mediate the
apoptosis of podocytes via the Bcl-2 and p53 signaling
pathways.
The present study established an in vitro DN
model by using HG-induced podocytes, and detected the expression
levels of miR-145-5p in different groups, including the HG (25 mM
glucose) and NG (5 mM glucose) groups, at different time points.
The expression levels of miR-145-5p were lower in HG-induced
podocytes than under NG conditions. Functional experiments were
conducted, and the results of flow cytometry and TUNEL staining
assays revealed that the overexpression of miR-145-5p inhibits
HG-induced cell apoptosis. To further clarify possible mechanisms
and signaling pathways, potential target genes of miR-145-5p were
identified using miRanda, TargetScan and miRDB. The results of a
luciferase assay demonstrated that Notch1 was a direct target of
miR-145-5p. In addition, miR-145-5p mimic or inhibitor were
transfected into the HG-induced podocytes. Subsequently, using
western blotting and RT-qPCR, the downstream factors of the Notch
signaling pathway, namely NICD, Hes1 and Hey1 were detected. The
results demonstrated that overexpression of miR-145-5p resulted in
the downregulation of NICD, Hes1 and Hey1 at the protein and mRNA
levels. Finally, the levels of indicators of apoptotic signaling,
including cleaved caspase-3, Bcl-2 and Bax, were measured. The
results revealed that the overexpression of miR-145-5p inhibits
these key factors, which suggests that miR-145-5p inhibits not only
the Notch signaling pathway but also other apoptotic signaling
pathways.
In conclusion, the present study identified that
miR-145-5p overexpression could attenuate the apoptosis of
HG-induced podocytes by inhibiting the activation of the Notch
signaling pathway and other key factors of the apoptotic signaling
pathway. Therefore, miR-145-5p could potentially serve as a novel
and effective therapeutic target for future DN treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and analyzed during the
present study are not publicly available due to further research
being performed, but are available from the corresponding author on
reasonable request.
Authors' contributions
BW and YSL designed the study, read and approved the
final version of the manuscript; BW performed the experiments and
wrote the manuscript. HXG analyzed the data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Tong L and Adler SG: Diabetic kidney
disease. Clin J Am Soc Nephrol. 13:335–338. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ying Q and Wu G: Molecular mechanisms
involved in podocyte EMT and concomitant diabetic kidney diseases:
An update. Ren Fail. 39:474–483. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Asanuma K: The role of podocyte injury in
chronic kidney disease. Nihon Rinsho Meneki Gakkai Kaishi.
38:26–36. 2015.(In Japanese). View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brosius FC and Coward RJ: Podocytes,
signaling pathways and vascular factors in diabetic kidney disease.
Adv Chronic Kidney Dis. 21:304–310. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Armelloni S, Corbelli A, Giardino L, Li M,
Ikehata M, Mattinzoli D, Messa P, Pignatari C, Watanabe S and
Rastaldi MP: Podocytes: Recent biomolecular developments. Biomol
Concepts. 5:319–330. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tetreault N and De Guire V: miRNAs: Their
discovery, biogenesis and mechanism of action. Clin Biochem.
46:842–845. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Duarte FV, Palmeira CM and Rolo AP: The
role of microRNAs in mitochondria: Small players acting wide. Genes
(Basel). 5:865–886. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–354. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li T and Cho WC: MicroRNAs: Mechanisms,
functions and progress. Genomics Proteomics Bioinformatics.
10:237–238. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Graves P and Zeng Y: Biogenesis of
mammalian microRNAs: A global view. Genomics Proteomics
Bioinformatics. 10:239–245. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fu Y, Wang C, Zhang D, Chu X, Zhang Y and
Li J: miR-15b-5p ameliorated high glucose-induced podocyte injury
through repressing apoptosis, oxidative stress, and inflammatory
responses by targeting Sema3A. J Cell Physiol. 234:20869–20878.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ma J, Li YT, Zhang SX, Fu SZ and Ye XZ:
miR-590-3p attenuates acute kidney injury by inhibiting tumor
necrosis factor receptor-associated factor 6 in septic mice.
Inflammation. 42:637–649. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li H, Zhu X, Zhang J and Shi J:
MicroRNA-25 inhibits high glucose-induced apoptosis in renal
tubular epithelial cells via PTEN/AKT pathway. Biomed Pharmacother.
96:471–479. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Peng J, Wu Y, Deng Z, Zhou Y, Song T, Yang
Y, Zhang X, Xu T, Xia M, Cai A, et al: miR-377 promotes white
adipose tissue inflammation and decreases insulin sensitivity in
obesity via suppression of sirtuin-1 (SIRT1). Oncotarget.
8:70550–70563. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Barutta F, Tricarico M, Corbelli A,
Annaratone L, Pinach S, Grimaldi S, Bruno G, Cimino D, Taverna D,
Deregibus MC, et al: Urinary exosomal microRNAs in incipient
diabetic nephropathy. PLoS One. 8:e737982013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yamamoto S, Schulze KL and Bellen HJ:
Introduction to Notch signaling. Methods Mol Biol. 1187:1–14. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hori K, Sen A and Artavanis-Tsakonas S:
Notch signaling at a glance. J Cell Sci. 126:2135–2140. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Penton AL, Leonard LD and Spinner NB:
Notch signaling in human development and disease. Semin Cell Dev
Biol. 23:450–457. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Braune EB and Lendahl U: Notch-a
goldilocks signaling pathway in disease and cancer therapy. Discov
Med. 21:189–196. 2016.PubMed/NCBI
|
|
21
|
Voelkel JE, Harvey JA, Adams JS, Lassiter
RN and Stark MR: FGF and Notch signaling in sensory neuron
formation: A multifactorial approach to understanding signaling
pathway hierarchy. Mech Dev. 134:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Saleem MA, O'Hare MJ, Reiser J, Coward RJ,
Inward CD, Farren T, Xing CY, Ni L, Mathieson PW and Mundel P: A
conditionally immortalized human podocyte cell line demonstrating
nephrin and podocin expression. J Am Soc Nephrol. 13:630–638.
2002.PubMed/NCBI
|
|
23
|
Shepard BD, Natarajan N, Protzko RJ, Acres
OW and Pluznick JL: A cleavable N-terminal signal peptide promotes
widespread olfactory receptor surface expression in HEK293T cells.
PLoS One. 8:e687582013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Loo DT: In situ detection of apoptosis by
the TUNEL assay: An overview of techniques. Methods Mol Biol.
682:3–13. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shan H, Zhang Y, Lu Y, Zhang Y, Pan Z, Cai
B, Wang N, Li X, Feng T, Hong Y and Yang B: Downregulation of
miR-133 and miR-590 contributes to nicotine-induced atrial
remodelling in canines. Cardiovasc Res. 83:465–472. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pockley AG, Foulds GA, Oughton JA,
Kerkvliet NI and Multhoff G: Immune cell phenotyping using flow
cytometry. Curr Protoc Toxicol. 66:18.8.1–34. 2015. View Article : Google Scholar
|
|
28
|
Agarwal V, Bell GW, Nam J and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
eLife. 4:e050052015. View Article : Google Scholar
|
|
29
|
Liu W and Wang X: Prediction of functional
microRNA targets by integrative modeling of microRNA binding and
target expression data. Genome Biol. 20:182019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zheng R, Pan L, Gao J, Ye X, Chen L, Zhang
X, Tang W and Zheng W: Prognostic value of miR-106b expression in
breast cancer patients. J Surg Res. 195:158–165. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu ZH, Dai XM and Du B: Hes1: A key role
in stemness, metastasis and multidrug resistance. Cancer Biol Ther.
16:353–359. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lopez-Mateo I, Arruabarrena-Aristorena A,
Artaza-Irigaray C, Lopez JA, Calvo E and Belandia B: HEY1 functions
are regulated by its phosphorylation at Ser-68. Biosci Rep.
36(pii): e003432016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Choudhary GS, Al-Harbi S and Almasan A:
Caspase-3 activation is a critical determinant of genotoxic
stress-induced apoptosis. Methods Mol Biol. 1219:1–9. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Laulier C and Lopez BS: The secret life of
Bcl-2: Apoptosis- independent inhibition of DNA repair by Bcl-2
family members. Mutat Res. 751:247–257. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhu S, Li T, Tan J, Yan X, Zhang D, Zheng
C, Chen Y, Xiang Z and Cui H: Bax is essential for death
receptor-mediated apoptosis in human colon cancer cells. Cancer
Biother Radiopharm. 27:577–581. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim D, Lim S, Park M, Choi J, Kim J, Han
H, Yoon K, Kim K, Lim J and Park S: Ubiquitination-dependent CARM1
degradation facilitates Notch1-mediated podocyte apoptosis in
diabetic nephropathy. Cell Signal. 26:1774–1782. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Matoba K, Kawanami D, Nagai Y, Takeda Y,
Akamine T, Ishizawa S, Kanazawa Y, Yokota T and Utsunomiya K:
Rho-kinase blockade attenuates podocyte apoptosis by inhibiting the
notch signaling pathway in diabetic nephropathy. Int J Mol Sci.
18(pii): E17952017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gao F, Yao M, Cao Y, Liu S, Liu Q and Duan
H: Valsartan ameliorates podocyte loss in diabetic mice through the
Notch pathway. Int J Mol Med. 37:1328–1336. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yamamoto S, Schulze KL and Bellen HJ:
Introduction to Notch signaling. Notch Signaling: Methods and
Protocols. Bellen HJ and Yamamoto S: Springer; New York, NY: pp.
1–14. 2014
|
|
40
|
Sun J, Zhao F, Zhang W, Lv J, Lv J and Yin
A: BMSCs and miR-124a ameliorated diabetic nephropathy via
inhibiting notch signalling pathway. J Cell Mol Med. 22:4840–4855.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang X, Song S and Luo H: Regulation of
podocyte lesions in diabetic nephropathy via miR-34a in the Notch
signaling pathway. Medicine (Baltimore). 95:e50502016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu XD, Zhang LY, Zhu TC, Zhang RF, Wang
SL and Bao Y: Overexpression of miR-34c inhibits high
glucose-induced apoptosis in podocytes by targeting Notch signaling
pathways. Int J Clin Exp Pathol. 8:4525–4534. 2015.PubMed/NCBI
|
|
43
|
Chen M, Zhang Y, Li W and Yang J:
MicroRNA-145 alleviates high glucose-induced proliferation and
migration of vascular smooth muscle cells through targeting ROCK1.
Biomed Pharmacother. 99:81–86. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hui Y and Yin Y: MicroRNA-145 attenuates
high glucose-induced oxidative stress and inflammation in retinal
endothelial cells through regulating TLR4/NF-κB signaling. Life
Sci. 207:212–218. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lim JH, Youn DY, Yoo HJ, Yoon HH, Kim MY,
Chung S, Kim YS, Chang YS, Park CW and Lee JH: Aggravation of
diabetic nephropathy in BCL-2 interacting cell death suppressor
(BIS)-haploinsufficient mice together with impaired induction of
superoxide dismutase (SOD) activity. Diabetologia. 57:214–223.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Deshpande SD, Putta S, Wang M, Lai JY,
Bitzer M, Nelson RG, Lanting LL, Kato M and Natarajan R:
Transforming growth factor-β-induced cross talk between p53 and a
microRNA in the pathogenesis of diabetic nephropathy. Diabetes.
62:3151–3162. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kolati SR, Kasala ER, Bodduluru LN,
Mahareddy JR, Uppulapu SK, Gogoi R, Barua CC and Lahkar M: BAY
11–7082 ameliorates diabetic nephropathy by attenuating
hyperglycemia-mediated oxidative stress and renal inflammation via
NF-κB pathway. Environ Toxicol Pharmacol. 39:690–699. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gao F, Yao M, Shi Y, Hao J, Ren Y, Liu Q,
Wang X and Duan H: Notch pathway is involved in high
glucose-induced apoptosis in podocytes via Bcl-2 and p53 pathways.
J Cell Biochem. 114:1029–1038. 2013. View Article : Google Scholar : PubMed/NCBI
|