Introduction
Hepatitis C is one of the most common chronic liver
diseases worldwide, and is caused by hepatitis C virus (HCV)
infection (1). According to World
Health Organization (WHO) guidelines published in 2016, an
increasing number of deaths have been caused by HCV-related
diseases annually (2), and patient
mortality from HCV-associated cirrhosis and hepatic cell cancer
will continue to increase unless more efficient therapies are
applied in the clinic (3). In
addition, a previous study showed that the projected prevalence of
HCV in Asia is 2.8%, which accounts for over 60% of the estimated
cases worldwide (4). Therefore, it
is not surprising that there are >8.9 million people living with
HCV infection in China, and that China has the highest burden of
HCV infection worldwide (5,6). Currently, various direct-acting
antivirals (DAAs) show significant advantages in the treatment of
hepatitis C, including high potency, a higher barrier to
resistance, a favourable tolerability profile, and many other
aspects (7,8). However, for patients infected with
different HCV subtypes, to achieve better antiviral effects, the
use of DAA drugs differs (9).
Therefore, accurate HCV genotyping results directly determine the
therapeutic schedule and treatment effect.
To reduce the global burden of HCV infection and
mortality, more accurate treatment is needed (2). To date, various therapies have been
identified that treat HCV (10).
However, HCV has seven main genotypes as primary divisions
(11), and different subtypes of HCV
infection exhibit different chronic disease progressions, have
different responses to antitoxic therapy, and require different
therapies (12). Therefore, a method
that can offer a rapid and cost-effective HCV diagnosis is greatly
needed. HCV genotyping is of great significance in guiding
antiviral therapy, which not only is an important indicator in the
diagnostic process, but also reflects the treatment effect.
Currently, HCV genotyping methods used in China and
globally vary, such as the Versant HCV genotype assay (LiPA) 2.0,
TaqMan PCR, sequencing, whole-genome deep sequencing (WGS), and the
NS5B-based microarray (13-17).
Sequencing is considered the most accurate method for HCV
genotyping (6). Nevertheless, it has
many drawbacks, as it is time-consuming, expensive, etc. This
coincides with the need for HCV antiviral therapy; therefore, a
more efficient and accurate method for HCV genotyping is
needed.
Based on reverse transcription-quantitative PCR
(RT-qPCR) and regional HCV subtype distribution characteristics, a
method based on a large number of clinical trials to distinguish
five prevalent HCV subtypes in a one-step reaction was designed to
meet clinical needs. For those classic HCV genotyping methods, the
higher requirements for time and capital investment make them more
difficult to promote and broadly apply. According to the
performance validation, this method's favourable reproducibility,
sensitivity, accuracy, specificity and anti-interference shows that
it overcomes these problems. Therefore, this method could be put
into clinical practice and be beneficial for the adjunct diagnosis
and treatment of hepatitis C.
Materials and methods
Samples
The present study included 65 qualified clinical
samples from hepatitis C patients (males, 61.5%) aged 18-76 years
(median age, 49) who were referred to The Second Affiliated
Hospital and Yuying Children's Hospital of Wenzhou Medical
University, and 224 other qualified clinical samples from hepatitis
C patients (males, 60.7%) aged 19-78 years (median age, 50) who
were referred to the Department of First Generation Sequencing,
Hangzhou DiAn Medical Laboratory, Zhejiang, China between January
2018 and January 2019. The concentration of HCV RNA in the above
samples, quantified by the automatic nucleic acid quantitative
detection system (AMPLLY Biotech Co., Ltd.), was above
1x103 IU/ml (range:
1.0x103-5.7x108 IU/ml). The present study was
approved by the Research Ethics Committee of the Second Affiliated
Hospital of Wenzhou Medical University. All patients provided
written informed consent and agreed to the use of their samples in
scientific research.
Materials
The HCV genotype assay kit used in the study was
originally designed by the authors. Aiming at the five most
prevalent HCV subtypes in China, a set of processes was designed
that could detect each of the subtypes above, regardless of whether
an infection was separate or complex. The kit is suitable for
genotyping 1b, 2a, 3a, 3b and 6a HCV subtypes from clinical samples
(plasma or serum). RT-qPCR was performed on the ABI 7500 instrument
as described previously (18).
HCV RNA extraction
The RNA extraction kit was purchased from Taipu
Biosciences (China) Co., Ltd. To extract HCV RNA, 550 µl lysate was
added to several 1.5 ml centrifuge tubes. Next, 100 µl plasma
samples was added to each tube, mixed for 20 sec, and then the
tubes were allowed to stand for 10 min at 50˚C. Afterwards, a
purification column was placed into a 2 ml collection tube. The
mixture was then added to the purification column and centrifuged
at 12,740 x g for 1 min at room temperature before the filtrate was
discarded. The purification column was washed with RNA extraction
buffer I diluted in ethanol and RNA extraction buffer II diluted in
ethanol, successively, and then centrifuged at 12,740 x g for 1 min
at room temperature before the filtrate was discarded. After 2 min
centrifugation at 12,740 x g at room temperature, the purification
column was placed at room temperature for 2-3 min. The purification
column was transferred to a new centrifuge tube, and 50 µl eluant
was added to the centre of the column and allowed to stand for 2
min. After centrifugation at 12,740 x g for 1 min at room
temperature, the RNA solution was collected in a tube. The eluant
was preheated at 65-70˚C, and if the time of elution was prolonged
for 3 min or the eluant was added only once to the centre of the
purification column and eluted again, the extraction efficiency was
improved.
Primer and probe design for HCV
genotyping
The primers and probes were originally designed by
Haifeng Huang and synthesized by Sangon Biotech Co., Ltd. The
fluorescence signal collection of the multi-fluorescence detector
was set to the FAM (494 nm excitation and 522 nm emission
wavelentghs) and JOE (520 nm excitation and 548 nm emission
wavelengths) channels using the ABI 7500 instrument (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The 5'-untranslated
region (UTR) core of the HCV genome region was used to design the
sequences of primers and probes using Primer Premier 5.0 (Premier
Biosoft International) for HCV genotyping, and they were designed
first without three consecutive G or C bases at the end of the
primer, and by avoiding complementarity between themselves or the
primers. The exact sequences of the primers and probes are listed
in Table I. These primers and probes
allowed HCV subtypes 1b, 2a, 3a, 3b and 6a to be distinguished with
only three RT-PCR reaction tubes. The HCV 1b reaction tube
confirmed the presence of HCV 1b subtype infection or a complex
infection with the fluorescein FAM™. The HCV 2a/6a reaction tube
confirmed the presence of HCV 2a subtype infection with fluorescein
FAM™ and HCV 6a subtype infection with the fluorescein JOE™. The
HCV 3a/3b reaction tube confirmed the presence of HCV 3a subtype
infection with fluorescein FAM™ and HCV 3b subtype infection with
fluorescein JOE™. The positive test results were confirmed when one
fluorescein in one tube presented a typical S-type amplification
curve or the cycle threshold (Ct) value was ≤26.5.
 | Table IReverse transcription-quantitative
polymerase chain reaction primers and probes for HCV
genotyping. |
Table I
Reverse transcription-quantitative
polymerase chain reaction primers and probes for HCV
genotyping.
| HCV subtype | Primer/probe | Sequence (5' to
3') |
|---|
| 1b | Upstream primer |
CTCGTAGACCGTGCACCATGA |
| Downstream
primer |
CAGATCGTTGGTGGAGTTTACT |
| Probe |
FAM-GCACGAATCCTAAACCT-MGB |
| 2a | Upstream primer |
CTCGTAGACCGTGCACCATGA |
| Downstream
primer |
CAGATCGTTGGCGGAGTATACT |
| Probe |
FAM-GCACGAATCCTAAACCT-MGB |
| 6a | Upstream primer |
CTCGTAGACCGTGCACCATGA |
| Downstream
primer |
CAGATCGTTGGCGGAGTTTACT |
| Probe |
JOE-GCACTCTTCCAAAACCC-MGB |
| 3a | Upstream primer |
CTCGTAGACCGTGCACCATGA |
| Downstream
primer |
CAGATCGTTGGTGGAGTATACG |
| Probe |
FAM-ACACCATCCGCCGCCCACA-MGB |
| 3b | Upstream primer |
CTCGTAGACCGTGCACCATGA |
| Downstream
primer |
CAGATCGTTGGTGGAGTATATG |
| Probe |
JOE-ACACACCCCGTCGCCCACA-MGB |
Preparation and optimization of the
one-step RT-qPCR system
For RT-qPCR, the essential components of the HCV
RT-PCR reaction are listed as follows: Thermal starter enzyme,
reverse transcriptase, RT-PCR buffer, primer pair, probe and PCR
enhancer. The single component addition optimization was then
performed with a HiScript II One Step RT-PCR kit (Vazyme Biotech
Co., Ltd.), which includes Champagne thermal starter enzyme,
reverse transcriptase and RT-PCR buffer. As the cofactors of
thermally stable DNA polymerase, the concentration of magnesium
ions was carefully set, and a series of magnesium ion concentration
gradients were established to verify the best concentration of
magnesium ions. To better regulate the pH value of the system and
increase the activity of the DNA polymerase, a Tris-based buffering
reagent and a reagent containing potassium ions were added into the
RT-PCR buffer. In addition, the 1% recommended concentrations of
glycerinum and formamide were used as PCR enhancers to promote the
amplification of templates with high GC content. The amounts of
each primer pair and probe added per test were 150 and 50 pmol,
respectively. The final reaction volume was set at 50 µl,
containing 38 µl each subtype of HCV RT-PCR reaction reagent, 2 µl
enzyme, and 10 µl RNA sample. The final reaction conditions were as
follows: 42˚C for 30 min; 95˚C for 3 min; 10 cycles of 94˚C for 20
sec, 55˚C for 20 sec and 72˚C for 30 sec; followed by 30 cycles of
94˚C for 15 sec and 60˚C for 45 sec, with fluorescence signal
collection at 60˚C.
Validation of RT-qPCR for HCV
genotyping
For the sensitivity validation of RT-qPCR, five
positive HCV subtype (1b, 2a, 3a, 3b and 6a) samples that could be
detected by the kit were taken as the reference. These were diluted
to near the minimum detection limit of 1x103 IU/ml,
which was confirmed by the automatic nucleic acid quantitative
detection system (AMPLLY Biotech Co., Ltd). The detection of each
subtype was repeated 10 times, and the detection rate was
calculated.
To validate the accuracy of this new method,
sequencing, as described by Tong et al (6), was undertaken for genotyping samples
and compared with RT-qPCR. In the test, 11 HCV samples were tested
by sequencing. The nucleic acid was extracted from these samples
and sequenced, and the results compared with NCBI data to determine
the genotype.
For the anti-interference validation of RT-qPCR, a
jaundice sample, a lipid sample and a haemolysis sample were mixed
with high Ct value HCV 3a subtype samples as interfering
substances. The differences in Ct values between samples with
interfering substances (9X the volume of specimens plus 1X the
volume of the interfering substance) and samples without
interfering substances (9X the volume of specimens plus 1X the
volume of normal saline) were recorded.
For the within-run and between-run precision
validation of RT-qPCR, five positive HCV subtype (1b, 2a, 3a, 3b
and 6a) samples were diluted to a low concentration (~1 to
3x103 IU/ml), with five tubes tested repeatedly three
times to a total of 15 repetitions. The Ct value, SD value and
coefficient variable (CV) value of each experiment were
calculated.
RT-qPCR for clinical application
The reaction mixture was prepared in a 1.5 ml
centrifuge tube; three centrifuge tubes were required for the three
different reaction reagents. These centrifuge tubes were
instantaneously mixed and centrifuged at 6,000 x g for 10 sec at
room temperature. Next, 40 µl three reaction reagents were
distributed in three different thin-walled PCR 8-tube strips, to
which 10 µl RNA was subsequently added, including the treated
specimens, negative RNA control samples (extracted from samples
that tested negative from HCV) and positive RNA control samples
(extracted from samples that tested positive for HCV). Furthermore,
the thin-walled PCR tubes were covered, numbered and centrifuged
instantaneously. After that, the reaction tubes were placed on the
ABI 7500 instrument, and the PCR conditions were set as follows:
42˚C for 30 min; 95˚C for 3 min; 94˚C for 20 sec, 55˚C for 20 sec,
and 72˚C for 30 sec for 10 cycles; 94˚C for 15 sec and 60˚C for 45
sec for 30 cycles, with fluorescent signal collection at 60˚C. The
results were analysed after RT-qPCR according to the qPCR
amplification curve and Ct value per sample.
Statistical analysis
The CV was calculated by dividing the SD by the mean
value. Data analysis, including data compilation and percentage
calculation, was performed using SPSS software version 22.0 (IBM
Corp.).
Results
RT-qPCR detection system and
genotyping amplification curve
The HCV RNA genotype was detected with three
reaction tubes. The HCV 1b reaction tube confirmed the presence of
an HCV 1b subtype infection or a complex infection, the HCV 2a/6a
reaction tube confirmed the presence of HCV 2a and 6a subtype
infections, and the HCV 3a/3b reaction tube confirmed the presence
of HCV 3a and 3b subtype infections. The final genotyping
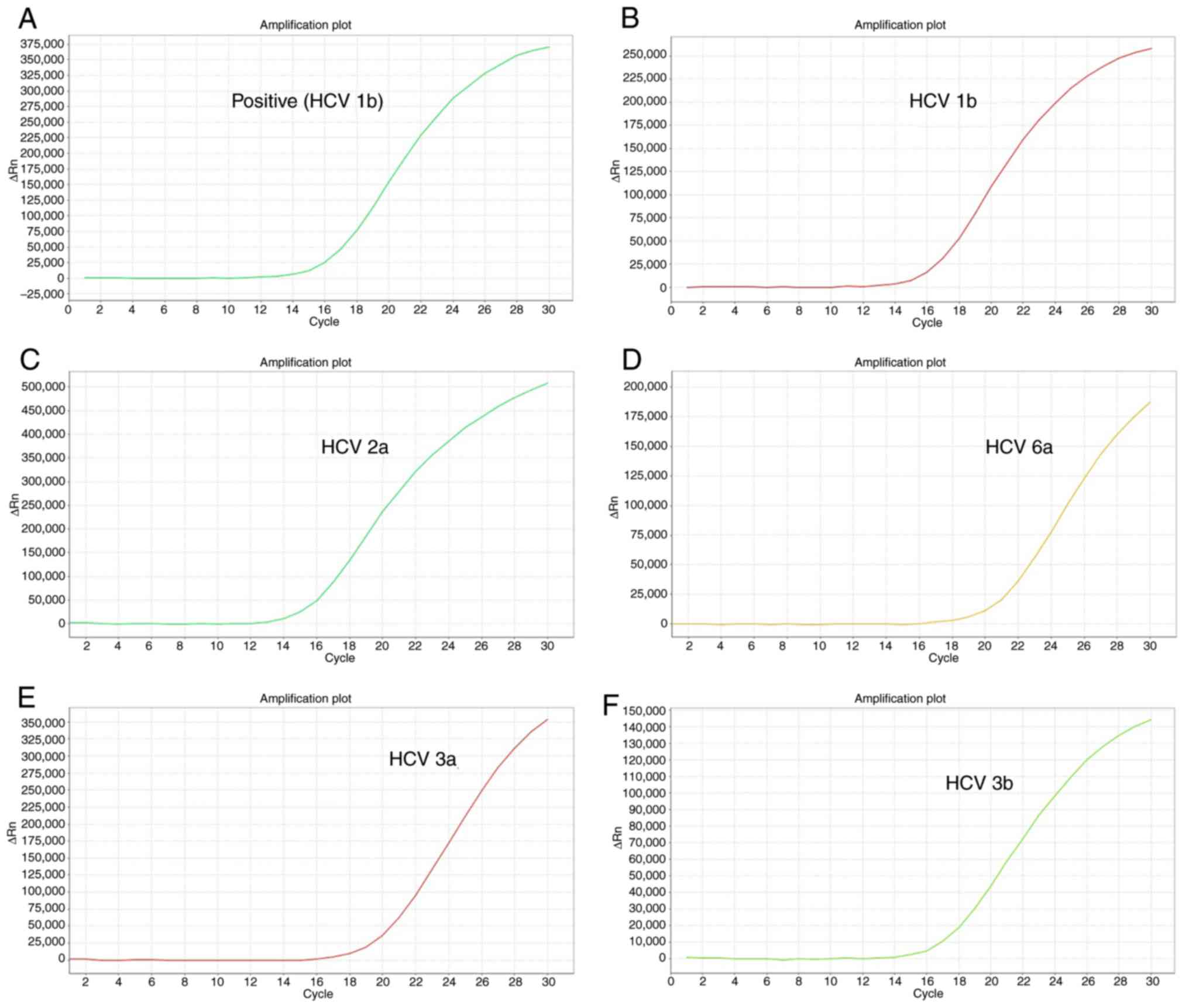
amplification curve of each HCV subtype is shown in Fig. 1. The labeled curves in Fig. 1A-F corresponding to the positive
samples were typical S-amplification curves, which show its good
performance in amplification efficiency.
Sensitivity of RT-qPCR for HCV
genotyping
Regarding the detection rate, five positive HCV
samples with different subtypes were diluted to a confirmed
concentration of 1x103 IU/ml, which was close to the
detection limit, to validate whether the reagent used in RT-qPCR
was sensitive enough. The results showed that the detection rate
were all at 100%, which shows that the sensitivity was sufficient
(Table II).
 | Table IISensitivity of quantitative reverse
transcription PCR for HCV genotyping. |
Table II
Sensitivity of quantitative reverse
transcription PCR for HCV genotyping.
| Test number | Ct value for
subtype... |
|---|
| 1b | 2a | 3a | 3b | 6a |
|---|
| 1 | 25.32 | 24.99 | 25.08 | 25.37 | 23.12 |
| 2 | 25.72 | 25.63 | 24.75 | 25.32 | 23.66 |
| 3 | 26.50 | 25.82 | 25.19 | 25.70 | 25.68 |
| 4 | 25.27 | 25.94 | 24.63 | 24.52 | 25.42 |
| 5 | 25.43 | 26.27 | 25.85 | 25.90 | 23.66 |
| 6 | 25.58 | 26.10 | 26.37 | 24.91 | 23.66 |
| 7 | 24.17 | 24.23 | 26.42 | 26.02 | 24.49 |
| 8 | 24.30 | 24.28 | 25.34 | 26.42 | 25.93 |
| 9 | 25.08 | 24.83 | 25.83 | 24.48 | 25.33 |
| 10 | 24.92 | 24.11 | 26.21 | 26.33 | 24.64 |
Specificity of RT-qPCR for HCV
genotyping
To verify the specificity of RT-qPCR, two hepatitis
B virus (HBV) samples, nucleic acid from two human cytomegalovirus
(HCMV) samples, and two Mycoplasma pulmonis (MP) samples
were extracted for diagnosis via the HCV genotyping kit. The
results were all negative, showing that none of these samples
cross-reacted with the HCV genotyping reaction system, thereby
verifying its specificity.
Accuracy of RT-qPCR for HCV
genotyping
RT-qPCR and sequencing were used to test 11
different HCV samples. The sequencing results were compared with
data on NCBI to diagnose the subtype of each sample. Among these
samples, the Ct values of three samples could not be detected and
tested negative by the two methods. The genotyping results of the
other samples tested by those two methods were identical (Table III). The RT-qPCR results for the
HCV genotyping method showed concordance with the sequencing
results, the gold standard for HCV genotyping.
 | Table IIIComparison reverse
transcription-quantitative polymerase chain reaction with
sequencing for detection of HCV samples from 11 patients with
hepatitis C. |
Table III
Comparison reverse
transcription-quantitative polymerase chain reaction with
sequencing for detection of HCV samples from 11 patients with
hepatitis C.
| Sample | Ct value | Expected
subtype | Detected
subtype |
|---|
| HCV-1 | 17.20 | 3a | 3a |
| HCV-2 | 17.15 | 1b | 1b |
| HCV-3 | Undetectable | Negative | Negative |
| HCV-4 | 14.16 | 1b | 1b |
| HCV-5 | Undetectable | Negative | Negative |
| HCV-6 | Undetectable | Negative | Negative |
| HCV-7 | 12.76 | 1b | 1b |
| HCV-8 | 15.88 | 2a | 2a |
| HCV-9 | 16.73 | 6a | 6a |
| HCV-10 | 11.29 | 3a | 3a |
| HCV-11 | 12.38 | 6b | 6b |
Anti-interference of RT-qPCR for HCV
genotyping
Regarding anti-interference, the RT-qPCR genotyping
method also performed well. Compared with the high Ct value HCV 3a
subtype samples mixed with saline, the high-value HCV 3a subtype
samples mixed with the jaundice sample, lipid sample and hemolysis
sample reached 2.9% as the maximum difference percentage in Ct
value.
Precision of RT-qPCR for HCV
genotyping
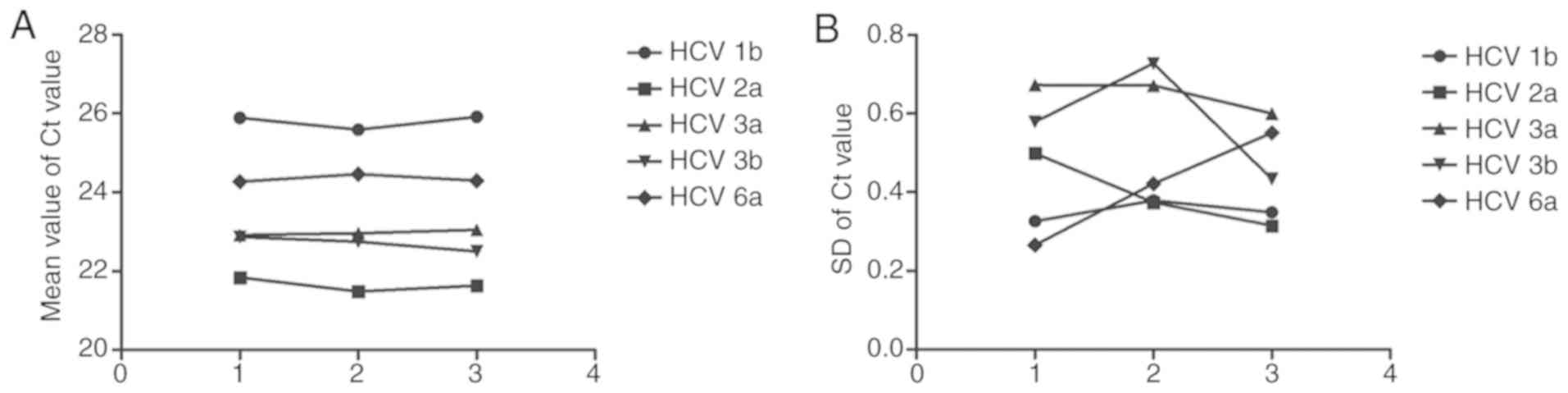
According to the precision test results, the CV
value of every subtype's Ct value in each one of the three parallel
tests of RT-qPCR within-run precision was <5%. Likewise, in the
three tests of the RT-qPCR between-run precision, the CV value of
each subtype's Ct value was also <5%. This shows that both
within-run precision and between-run precision met the requirements
of the clinic for diagnosis (Tables
IV and V, and Fig. 2).
 | Table IVWithin-run precision of reverse
transcription-quantitative polymerase chain reaction for HCV
genotyping. |
Table IV
Within-run precision of reverse
transcription-quantitative polymerase chain reaction for HCV
genotyping.
| Subtype | CV (%) |
|---|
| Test 1 | Test 2 | Test 3 |
|---|
| HCV 1b | 1.26 | 1.48 | 1.34 |
| HCV 2a | 2.28 | 1.74 | 1.45 |
| HCV 3a | 2.94 | 2.93 | 2.60 |
| HCV 3b | 2.53 | 3.19 | 1.92 |
| HCV 6a | 1.09 | 1.72 | 2.26 |
 | Table VBetween-run precision of reverse
transcription-quantitative polymerase chain reaction for HCV
genotyping. |
Table V
Between-run precision of reverse
transcription-quantitative polymerase chain reaction for HCV
genotyping.
| HCV subtype | Mean value | SD | CV (%) |
|---|
| 1b | 25.799 | 0.181 | 0.70 |
| 2a | 21.658 | 0.177 | 0.82 |
| 3a | 22.976 | 0.067 | 0.29 |
| 3b | 22.718 | 0.191 | 0.84 |
| 6a | 24.344 | 0.102 | 0.42 |
Genotyping results for patients with
HCV
In this study, genotyping showed that the HCV 1b
subtype was the most prevalent subtype, accounting for 45% of the
total. This is consistent with other research studying HCV subtype
distribution in China (6). In
addition, the HCV 3b and HCV 6a subtypes accounted for ~18 and 15%,
respectively, while 13% were infected with the HCV 3a subtype. The
population of people infected with the HCV 2a subtype was the
lowest (Table VI).
 | Table VIResults of HCV genotyping for 289
patients with hepatitis C. |
Table VI
Results of HCV genotyping for 289
patients with hepatitis C.
| HCV subtype | No./Percentage
(%) |
|---|
| Wenzhou | Hangzhou | Total |
|---|
| 1b | 23(35) | 107(48) | 130(45) |
| 2a | 4(6) | 22(10) | 26(9) |
| 3a | 10(15) | 28(13) | 38(13) |
| 3b | 15(23) | 38(17) | 53(18) |
| 6a | 13(20) | 29(13) | 42(15) |
| Total | 65 | 224 | 289 |
Discussion
According to WHO guidelines, ~71 million people were
infected with HCV globally in 2015(2). Moreover, a number of clinical practices
have shown that the validation of HCV subtype plays a crucial role
in determining the appropriate treatment for HCV. Thus, various
methods for HCV genotyping have been developed (19). The present study successfully
mitigates some of the problems inherent in HCV genotyping by using
RT-qPCR. To some extent, the method presented here solves the
problem of the expensive and time-consuming nature of HCV
genotyping, and ensures the accuracy of the results.
The method presented here is mainly based on a
one-step RT-PCR and Taqman fluorescence probe technique. Firstly,
to obtain optimally designed primers that could provide the most
accurate genotyping results, the conserved region of five HCV
subtypes (1b, 2a, 3a, 3b and 6a) was focused upon. Probes were
matched with primers to allow the detection of five HCV subtypes in
one step using only three RT-PCR reaction tubes. Nyan and Swinson
(20) reported a method for rapid
detection and genotype identification of HCV 1-6 by one-step
reverse transcription loop-mediated isothermal amplification.
However, the method requires electrophoretic analysis of banding
patterns and visual interpretation of fluorescence intensity in the
reaction tubes, which may result in the contamination of the
non-closed tube test and arbitrary interpretation uncertainty. The
method presented here has the advantage of closed tube detection
and simple analysis. In addition, coupled with the automation of
RT-PCR, the whole procedure requires only a few h to complete and
is easier to operate. This makes it superior to many other classic
approaches for genotyping, and more suitable for widespread
use.
Furthermore, when genotyping 11 samples, the results
were concordant with sequencing results, showing a high level of
accuracy (Table IV). As accuracy is
always the first concern for genotyping, this suggests that the
method presented here is reliable enough to be used in clinical
practice. Moreover, according to the performance validation, the
RT-qPCR method has good performance in additional aspects. When
samples were diluted to close to the detection limit, their
detection rate still reached 100%, demonstrating a high
sensitivity. None of the HBV, HCMV or MP samples were positive,
showing the high specificity of this method. When HCV samples were
mixed with a jaundice sample, lipid sample and haemolysis sample,
the maximum difference percentage of the Ct value only reached
2.9%, revealing that it also exhibited good anti-interference
qualities. Finally, all CV values of each test for both within-run
precision and between-run precision were <5%, thereby
demonstrating high reproducibility.
No mixed HCV genotypes or subtypes were encountered
in the present study. However, the established method could detect
the mixed positive plasmid of the 5 detectable subtypes (data not
shown), which suggests that the method used in the present study
could detect the 5 detectable subtypes of both single and mixed HCV
samples. Due to the difference between the recombinant DNA strains
and the clinical samples, more studies are needed to confirm this
result.
However, the RT-qPCR method does have limitations.
The most evident one is that it can only detect five HCV subtypes,
and it cannot detect certain low proportion HCV genotypes in China,
such as subtype 1a, Group 5 subtypes and several other Group 6
subtypes. No method for genotyping subtype 1a was used in the
present study, due to the low proportion of this subtype in China.
For example, Tong et al (6)
found only 1.59% of samples are subtype 1a. Other studies have
provided similar data (21,22). Overall, the proportion of subtype 1a
is extremely low in China, and the five subtypes described in the
present study (1b, 2a, 3a, 3b, 6a) are the most common HCV RNA
genotypes in China (21,22). The 289 samples used here fell into
the categories of the 5 detectable types and no other types
(including HCV 1a) were detected. The method presented here could
be useful in regions such as China, where the most prevalent HCV
subtypes are the five described in this study (22). To make up for the deficiency in not
detecting all HCV subtypes, when the genotyping data from HCV
RNA-positive samples showed negative results via the RT-qPCR
detection method, it could be considered as a rare subtype. In this
instance, sequencing should then be performed.
Due to the superiority of DAA drugs, they are now
the main treatment for HCV. The specific treatment of DAA drugs
varies according to the subtype of infecting HCV RNA. Therefore,
HCV genotyping, using methods such as the one presented here,
greatly aids the treatment of HCV. According to the genotyping
results in the present study, patients with HCV were treated
appropriately. However, the small sample size is a limitation of
this study, and future studies with a larger number of clinical
samples are required to verify its findings.
In conclusion, this study presents an accurate,
convenient and cost-effective method for HCV genotyping, which
contributes to the novel use of RT-qPCR techniques and the
elaborate design of primers and probes. Its application in clinical
practice could more accurately, sufficiently and economically
enable the treatment of HCV-infected patients.
Acknowledgements
Not applicable.
Funding
This work was supported by the Lin He's New Medicine
and Clinical Translation Academician Workstation Research Fund
(grant no. 18331203), the PhD Research Launching Fund Project of
The Second Affiliated Hospital of Wenzhou Medical University (grant
no. FEY001), and the Basic Public Welfare Technology Research
Project of Zhejiang Province (grant no. LGF20H200005).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZGC and JY designed and supervised the project. YX
and JY provided the samples, validated them and provided clinical
information about the samples. XY, YX and HH performed the
experiments. HH provided instructions for the experiments. XY and
TD analysed the data and wrote the manuscript. ZC and TD edited the
manuscript. All authors read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of the Second Affiliated Hospital of Wenzhou
Medical University (approval no. L-2018-10). All patients provided
written informed consent, agreed to the use of their samples in
scientific research, and approved the publication of data.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lavanchy D: Evolving epidemiology of
hepatitis C virus. Clin Microbiol Infect. 17:107–115.
2011.PubMed/NCBI View Article : Google Scholar
|
|
2
|
World Health Organization: Guidelines for
the Screening Care and Treatment of Persons with Chronic Hepatitis
C Infection: Updated Version, Geneva, 2016. https://www.who.int/hepatitis/publications/hep-c-guidelines-2016_170.png.
|
|
3
|
Razavi H, Waked I, Sarrazin C, Myers RP,
Idilman R, Calinas F, Vogel W, Mendes Correa MC, Hézode C, Lázaro
P, et al: The present and future disease burden of hepatitis C
virus (HCV) infection with today's treatment paradigm. J Viral
Hepat. 21 (Suppl 1)(S34-S59)2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Petruzziello A, Marigliano S, Loquercio G,
Cozzolino A and Cacciapuoti C: Global epidemiology of hepatitis C
virus infection: An up-date of the distribution and circulation of
hepatitis C virus genotypes. World J Gastroenterol. 22:7824–7840.
2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Polaris Observatory HCV Collaborators;
Blach S, Zeuzem S, Manns M, Altraif I, Duberg AS, Muljono DH, Waked
I, Alavian SM, Lee MH, et al: Global prevalence and genotype
distribution of hepatitis C virus infection in 2015: A modelling
study. Lancet Gastroenterol Hepatol. 2:161–176. 2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Tong YQ, Liu B, Liu H, Zheng HY, Gu J, Liu
H, Song EJ, Song C and Li Y: Accurate genotyping of hepatitis C
virus through nucleotide sequencing and identification of new HCV
subtypes in China population. Clin Microbiol Infect.
21(874.e9-e847.e21)2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Grebely J and Dore GJ: What is killing
people with hepatitis C virus infection? Semin Liver Dis.
31:331–339. 2011.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Schinazi R, Halfon P, Marcellin P and
Asselah T: HCV direct-acting antiviral agents: The best
interferon-free combinations. Liver Int. 34 (Suppl
1)(S69-S78)2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
De Clercq E: Current race in the
development of DAAs (direct-acting antivirals) against HCV. Biochem
Pharmacol. 89:441–452. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
European Association for the Study of the
Liver. EASL recommendations on treatment of hepatitis C 2018. J
Hepatol. 69:461–511. 2018.
|
|
11
|
Manns MP, Buti M, Gane E, Pawlotsky JM,
Razavi H, Terrault N and Younossi Z: Hepatitis C virus infection.
Nat Rev Dis Primers. 3(17006)2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
European Association for the Study of the
Liver. EASL recommendations on treatment of Hepatitis C 2016. J
Hepatol. 66:153–194. 2017.
|
|
13
|
Yang R and Wei L: Profile of the VERSANT
HCV genotype 2.0 assay. Expert Rev Mol Diagn. 18:995–1004.
2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Verbeeck J, Stanley MJ, Shieh J, Celis L,
Huyck E, Wollants E, Morimoto J, Farrior A, Sablon E,
Jankowski-Hennig M, et al: Evaluation of versant Hepatitis C virus
genotype assay (LiPA) 2.0. J Clin Microbiol. 46:1901–1906.
2008.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Gryadunov D, Nicot F, Dubois M,
Mikhailovich V, Zasedatelev A and Izopet J: Hepatitis C virus
genotyping using an oligonucleotide microarray based on the NS5B
sequence. J Clin Microbiol. 48:3910–3917. 2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Rodriguez-Frias F, Nieto-Aponte L, Gregori
J, Garcia-Cehic D, Casillas R, Tabernero D, Homs M, Blasi M, Vila
M, Chen Q, et al: High HCV subtype heterogeneity in a chronically
infected general population revealed by high-resolution hepatitis C
virus subtyping. Clin Microbiol Infect.
23(775.e1-775.e6)2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Soria ME, Gregori J, Chen Q, García-Cehic
D, Llorens M, de Ávila AI, Beach NM, Domingo E, Rodríguez-Frías F,
Buti M, et al: Pipeline for specific subtype amplification and drug
resistance detection in hepatitis C virus. BMC Infect Dis.
18(446)2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Roy F, Mendoza L, Hiebert J, McNall RJ,
Bankamp B, Connolly S, Lüdde A, Friedrich N, Mankertz A, Rota PA
and Severini A: Rapid identification of measles virus vaccine
genotype by real-time PCR. J Clin Microbiol. 55:735–743.
2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Warkad SD, Nimse SB, Song KS and Kim T:
HCV detection, discrimination, and genotyping technologies. Sensors
(Basel). 18(pii: E3423)2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Nyan DC and Swinson KL: A method for rapid
detection and genotype identification of hepatitis C virus 1-6 by
one-step reverse transcription loop-mediated isothermal
amplification. Int J Infect Dis. 43:30–36. 2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhang Y, Chen LM and He M: Hepatitis C
Virus in mainland China with an emphasis on genotype and subtype
distribution. Virol J. 14(41)2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chen Y, Yu C, Yin X, Guo X, Wu S and Hou
J: Hepatitis C virus genotypes and subtypes circulating in Mainland
China. Emerg Microbes Infect. 6(e95)2017.PubMed/NCBI View Article : Google Scholar
|