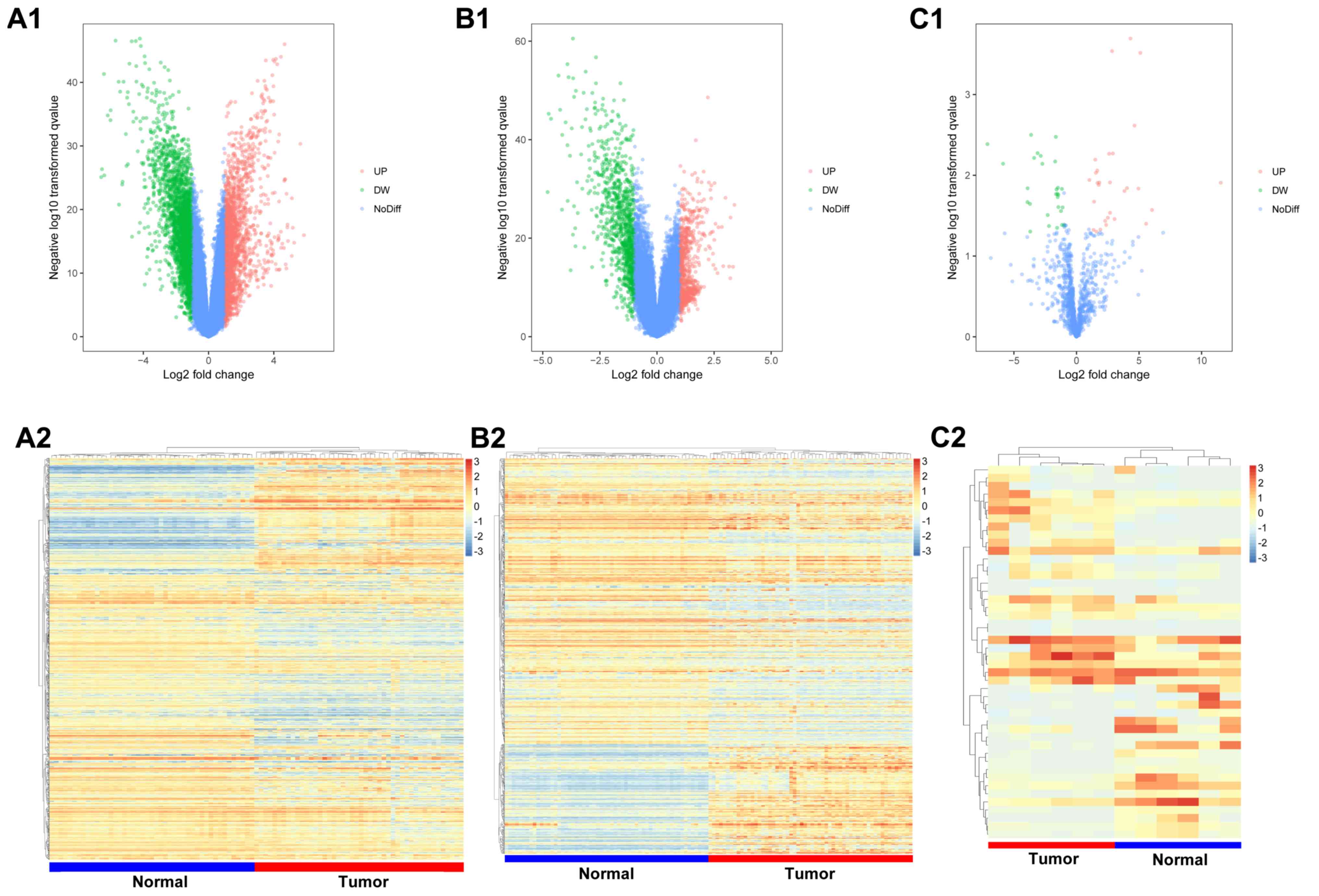
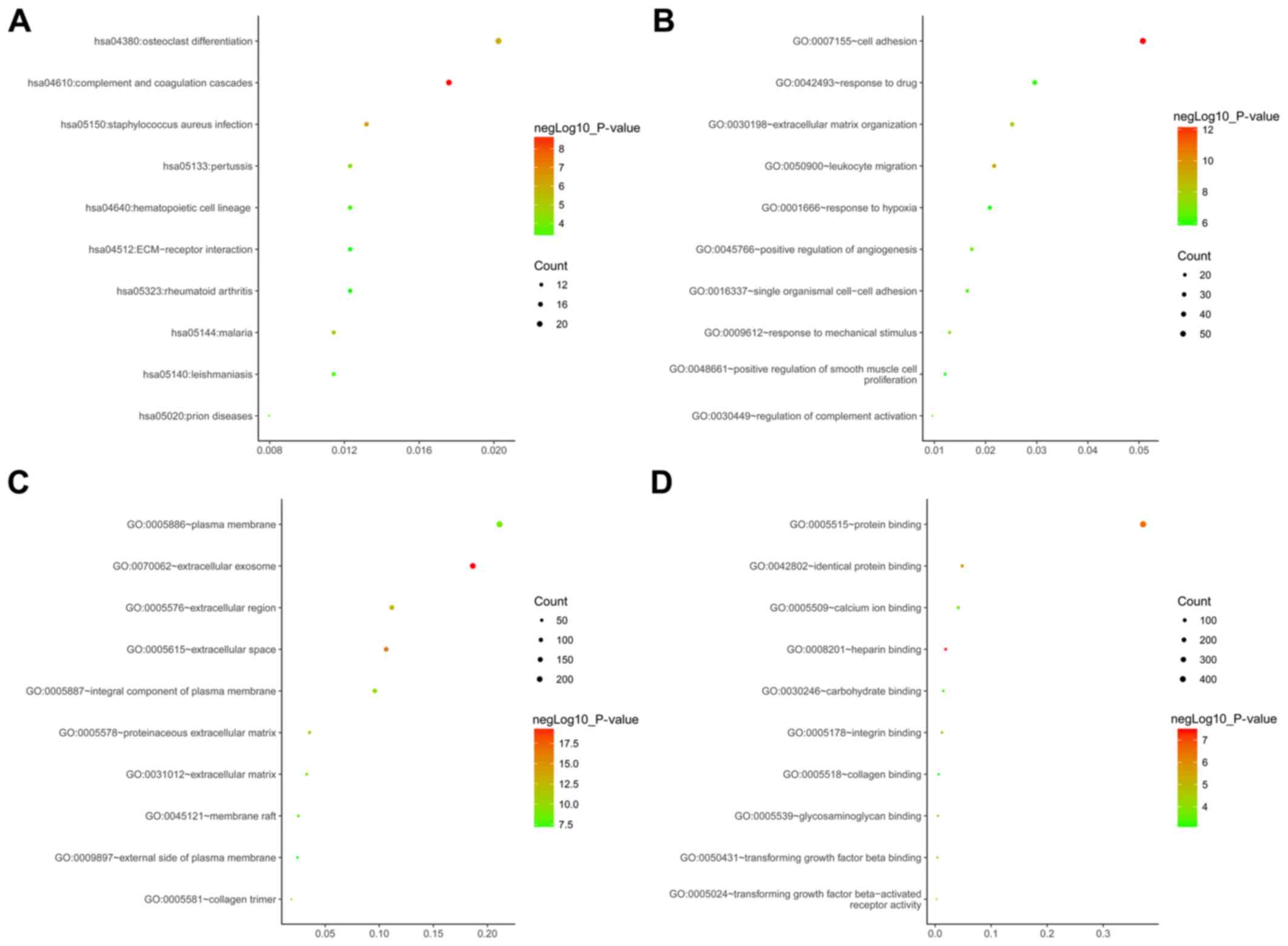
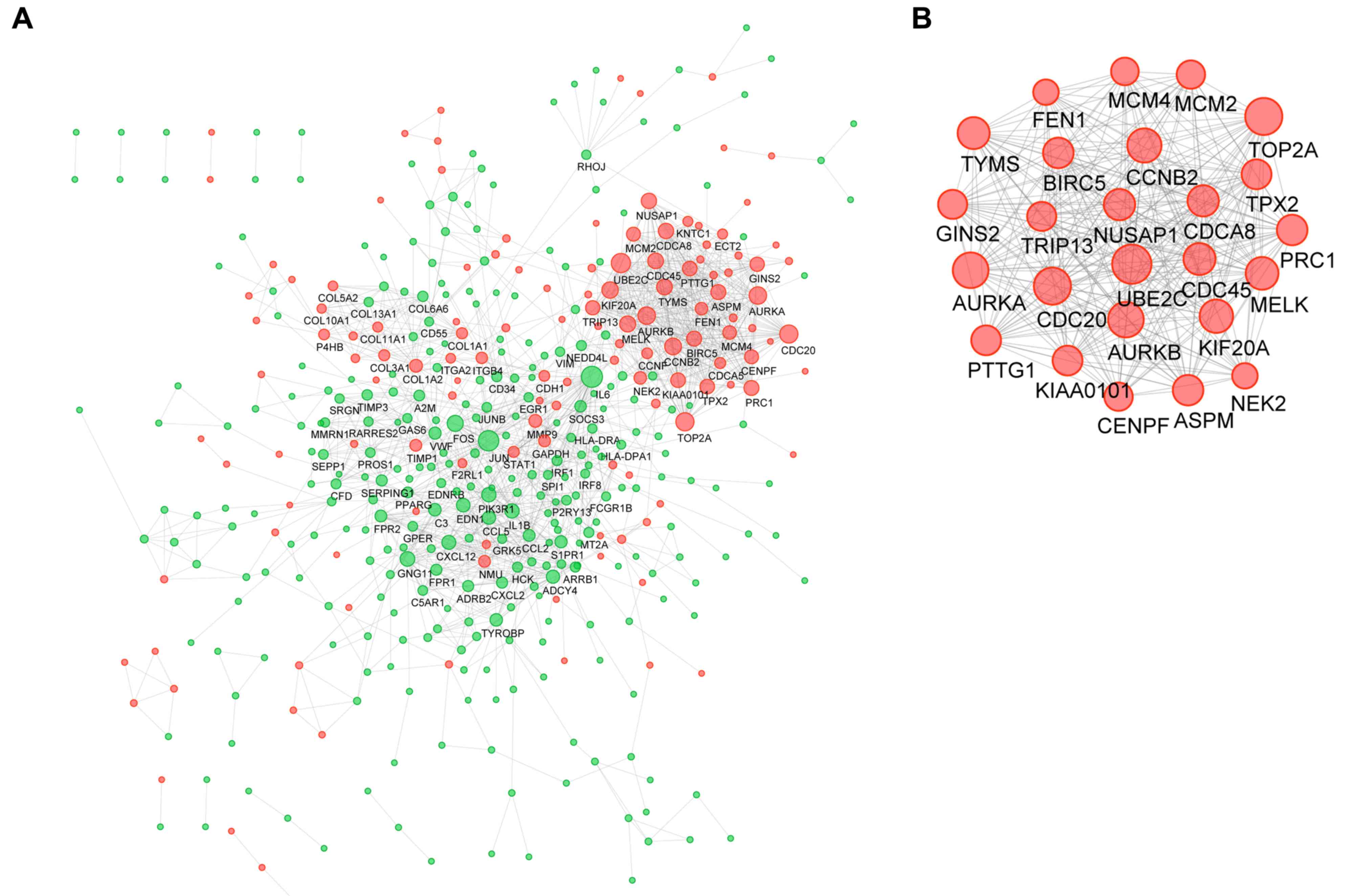
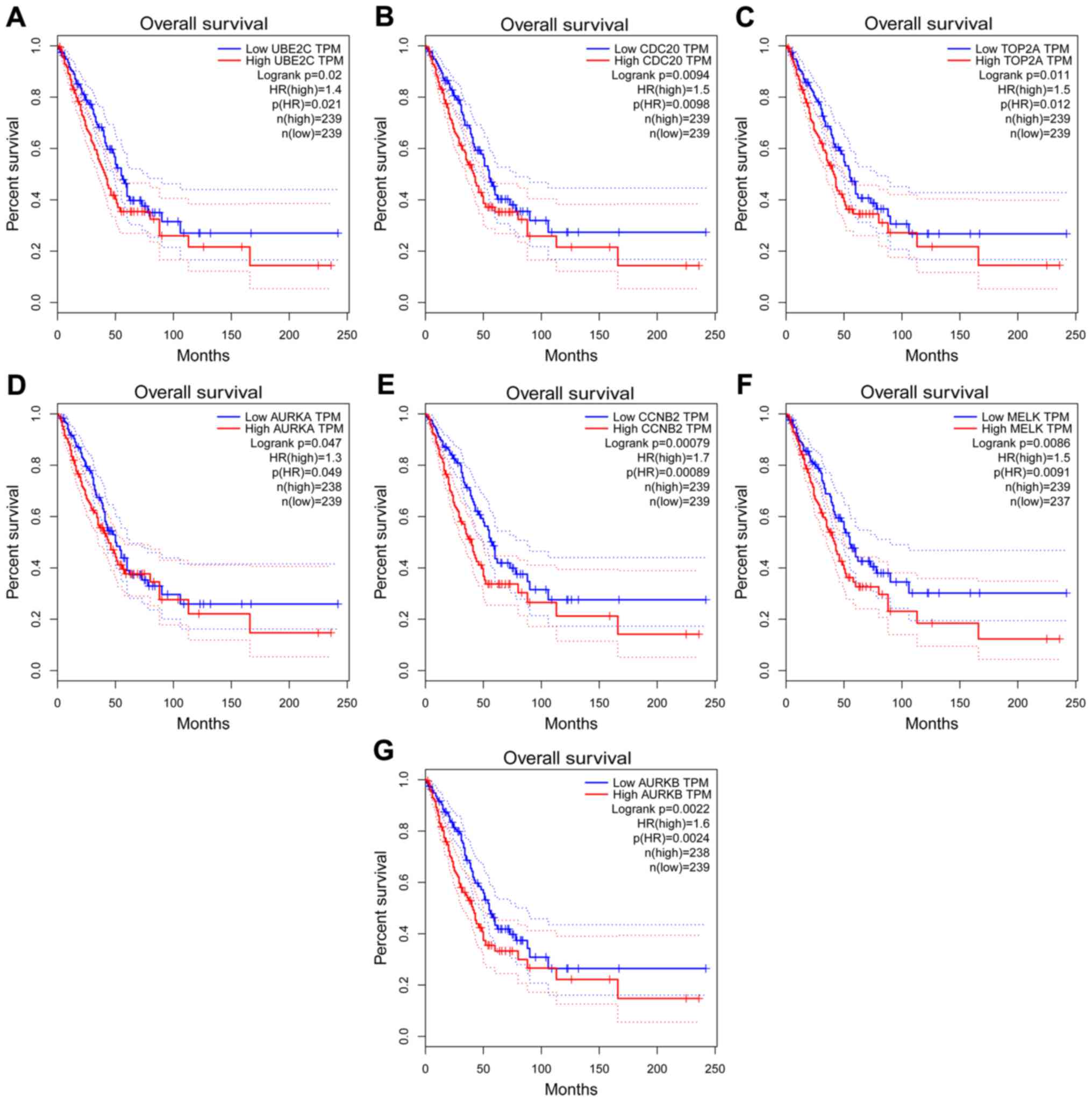
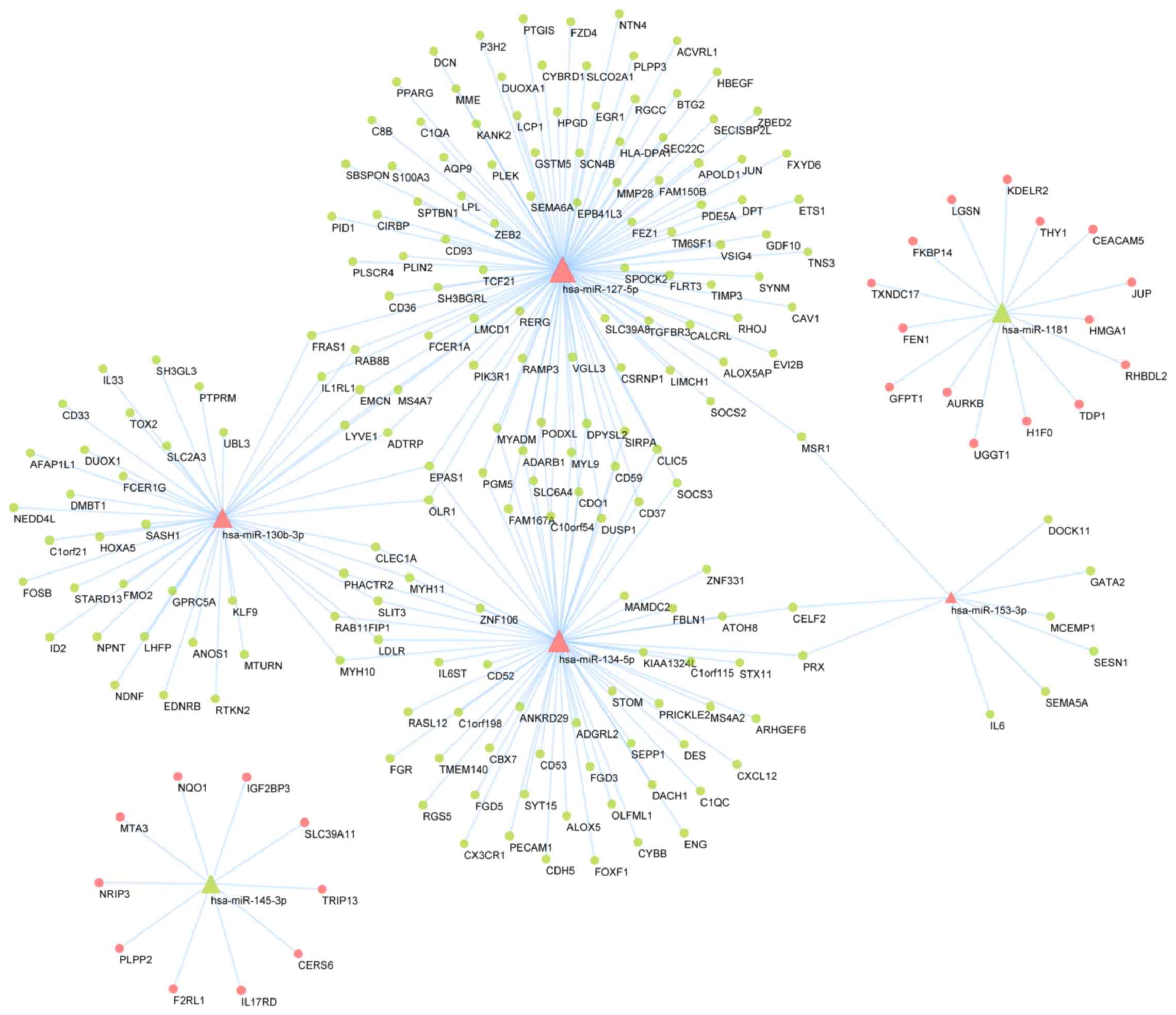
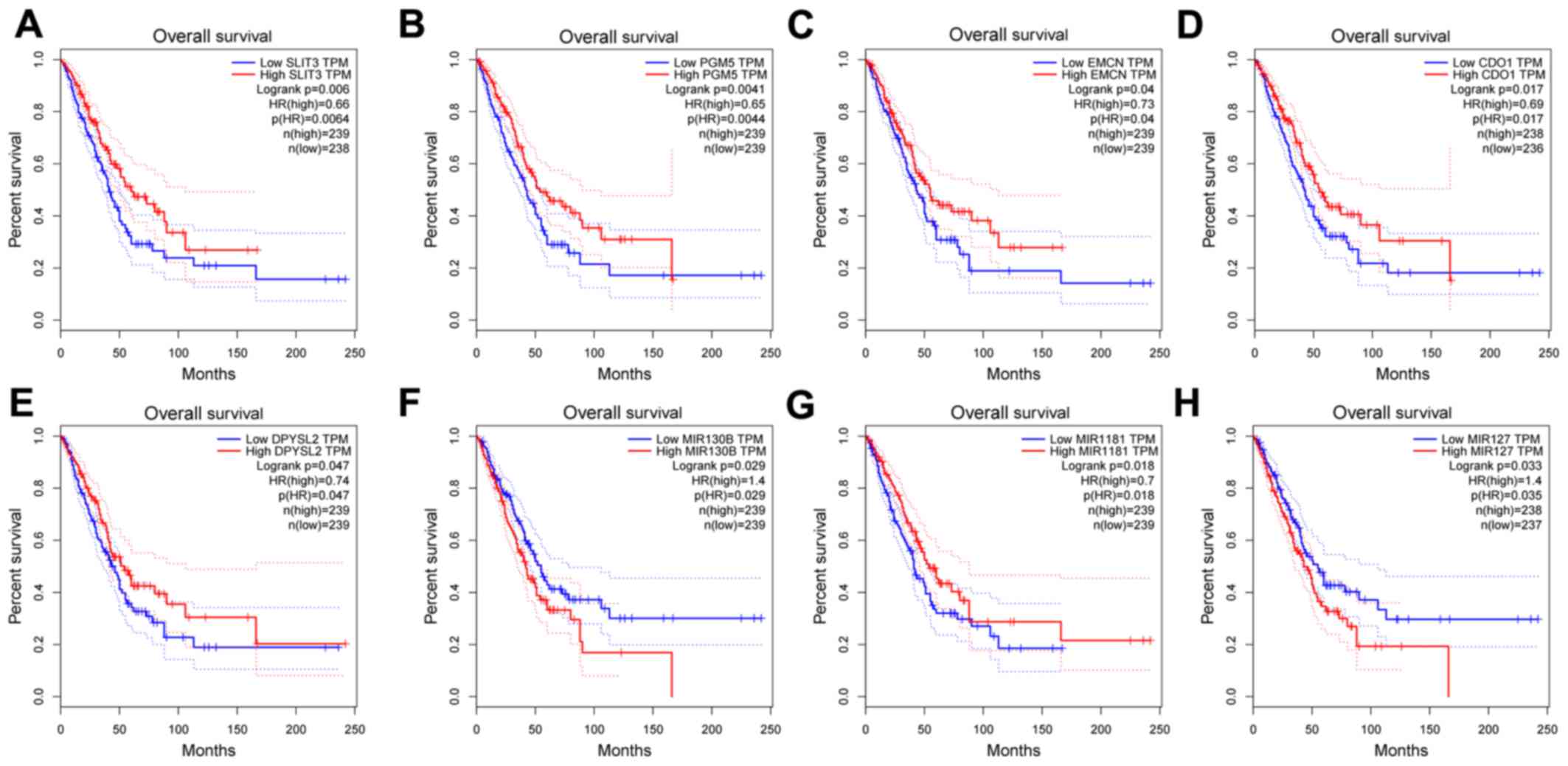
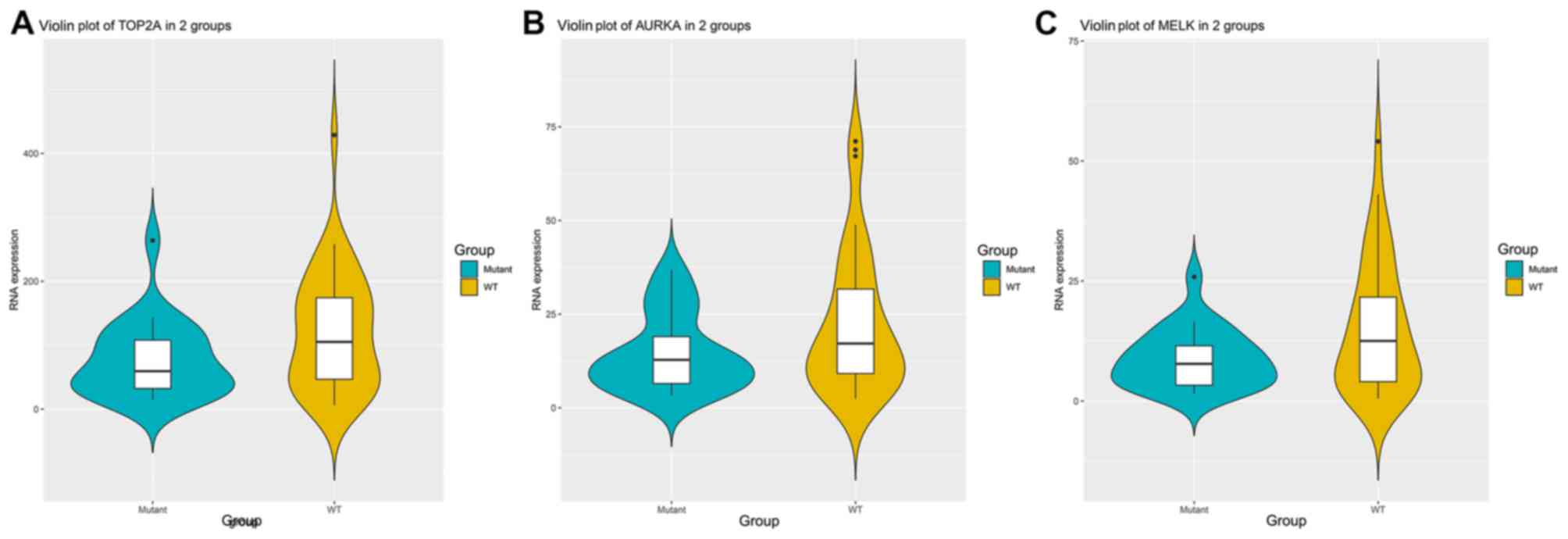
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Osmani L, Askin F, Gabrielson E and Li QK:
Current WHO guidelines and the critical role of immunohistochemical
markers in the subclassification of non-small cell lung carcinoma
(NSCLC): Moving from targeted therapy to immunotherapy. Semin
Cancer Biol. 52:103–109. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Tang J, Kong D, Cui Q, Wang K, Zhang D,
Yuan Q, Liao X, Gong Y and Wu G: Bioinformatic analysis and
identification of potential prognostic microRNAs and mRNAs in
thyroid cancer. PeerJ. 6(e4674)2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297.
2004.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261.
2006.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Sempere LF: Integrating contextual miRNA
and protein signatures for diagnostic and treatment decisions in
cancer. Expert Rev Mol Diagn. 11:813–827. 2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Lv K, Yang J, Sun J and Guan J:
Identification of key candidate genes for pancreatic cancer by
bioinformatics analysis. Exp Ther Med. 18:451–458. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Edgar R, Domrachev M and Lash AE: Gene
Expression Omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Sanchez-Palencia A, Gomez-Morales M,
Gomez-Capilla JA, Pedraza V, Boyero L, Rosell R and Fárez-Vidal ME:
Gene expression profiling reveals novel biomarkers in nonsmall cell
lung cancer. Int J Cancer. 129:355–364. 2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Selamat SA, Chung BS, Girard L, Zhang W,
Zhang Y, Campan M, Siegmund KD, Koss MN, Hagen JA, Lam WL, et al:
Genome-scale analysis of DNA methylation in lung adenocarcinoma and
integration with mRNA expression. Genome Res. 22:1197–1211.
2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Ma L, Huang Y, Zhu W, Zhou S, Zhou J, Zeng
F, Liu X, Zhang Y and Yu J: An integrated analysis of miRNA and
mRNA expressions in non-small cell lung cancers. PLoS One.
6(e26502)2011.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7(252)2006.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: The Gene Ontology Consortium: Gene ontology: Tool for the
unification of biology. Nat Genet. 25:25–29. 2000.PubMed/NCBI View
Article : Google Scholar
|
|
14
|
Scardoni G, Petterlini M and Laudanna C:
Analyzing biological network parameters with CentiScaPe.
Bioinformatics. 25:2857–2859. 2009.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Cellai D, Lawlor A, Dawson KA and Gleeson
JP: Tricritical point in heterogeneous k-core percolation. Phys Rev
Lett. 107(175703)2011.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Han JDJ, Bertin N, Hao T, Goldberg DS,
Berriz GF, Zhang LV, Dupuy D, Walhout AJ, Cusick ME, Roth FP, et
al: Evidence for dynamically organized modularity in the yeast
protein-protein interaction network. Nature. 430:88–93.
2004.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43 (D1):D146–D152. 2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The Cancer Genome Atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19 (1A):A68–A77. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Li B and Dewey CN: RSEM: Accurate
transcript quantification from RNA-Seq data with or without a
reference genome. BMC Bioinformatics. 12(323)2011.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Li B, Ruotti V, Stewart RM, Thomson JA and
Dewey CN: RNA-Seq gene expression estimation with read mapping
uncertainty. Bioinformatics. 26:493–500. 2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Carter SL, Eklund AC, Kohane IS, Harris LN
and Szallasi Z: A signature of chromosomal instability inferred
from gene expression profiles predicts clinical outcome in multiple
human cancers. Nat Genet. 38:1043–1048. 2006.PubMed/NCBI View
Article : Google Scholar
|
|
23
|
Choi YL, Park SH, Jang JJ and Park CK:
Expression of the G1-S modulators in hepatitis B virus-related
hepatocellular carcinoma and dysplastic nodule: Association of
cyclin D1 and p53 proteins with the progression of hepatocellular
carcinoma. J Korean Med Sci. 16:424–432. 2001.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Tripathi V, Shen Z, Chakraborty A, Giri S,
Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A, et al:
Long noncoding RNA MALAT1 controls cell cycle progression by
regulating the expression of oncogenic transcription factor B-MYB.
PLoS Genet. 9(e1003368)2013.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Bumbaca B and Li W: Taxane resistance in
castration-resistant prostate cancer: Mechanisms and therapeutic
strategies. Acta Pharm Sin B. 8:518–529. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhong Z, Pannu V, Rosenow M, Stark A and
Spetzler D: KIAA0100 modulates cancer cell aggression behavior of
MDA-MB-231 through microtubule and heat shock proteins. Cancers
(Basel). 10(10)2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Guglietta S and Rescigno M:
Hypercoagulation and complement: Connected players in tumor
development and metastases. Semin Immunol. 28:578–586.
2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
van Ree JH, Jeganathan KB, Malureanu L and
van Deursen JM: Overexpression of the E2 ubiquitin-conjugating
enzyme UbcH10 causes chromosome missegregation and tumor formation.
J Cell Biol. 188:83–100. 2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Berlingieri MT, Pallante P, Guida M, Nappi
C, Masciullo V, Scambia G, Ferraro A, Leone V, Sboner A,
Barbareschi M, et al: UbcH10 expression may be a useful tool in the
prognosis of ovarian carcinomas. Oncogene. 26:2136–2140.
2007.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Parris TZ, Kovács A, Aziz L, Hajizadeh S,
Nemes S, Semaan M, Forssell-Aronsson E, Karlsson P and Helou K:
Additive effect of the AZGP1, PIP, S100A8 and UBE2C molecular
biomarkers improves outcome prediction in breast carcinoma. Int J
Cancer. 134:1617–1629. 2014.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhang HQ, Zhao G, Ke B, Ma G, Liu GL,
Liang H, Liu LR and Hao XS: Overexpression of UBE2C correlates with
poor prognosis in gastric cancer patients. Eur Rev Med Pharmacol
Sci. 22:1665–1671. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Okamoto Y, Ozaki T, Miyazaki K, Aoyama M,
Miyazaki M and Nakagawara A: UbcH10 is the cancer-related E2
ubiquitin-conjugating enzyme. Cancer Res. 63:4167–4173.
2003.PubMed/NCBI
|
|
33
|
Guo J, Wu Y, Du J, Yang L, Chen W, Gong K,
Dai J, Miao S, Jin D and Xi S: Deregulation of UBE2C-mediated
autophagy repression aggravates NSCLC progression. Oncogenesis.
7(49)2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wu Y, Jin D, Wang X, Du J, Di W, An J,
Shao C and Guo J: UBE2C induces cisplatin resistance via
ZEB1/2-dependent upregulation of ABCG2 and ERCC1 in NSCLC cells. J
Oncol. 2019(8607859)2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Wang C, Pan YH, Shan M, Xu M, Bao JL and
Zhao LM: Knockdown of UbcH10 enhances the chemosensitivity of dual
drug resistant breast cancer cells to epirubicin and docetaxel. Int
J Mol Sci. 16:4698–4712. 2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Wang L, Zhang J, Wan L, Zhou X, Wang Z and
Wei W: Targeting Cdc20 as a novel cancer therapeutic strategy.
Pharmacol Ther. 151:141–151. 2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Mondal G, Sengupta S, Panda CK, Gollin SM,
Saunders WS and Roychoudhury S: Overexpression of Cdc20 leads to
impairment of the spindle assembly checkpoint and aneuploidization
in oral cancer. Carcinogenesis. 28:81–92. 2007.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kato T, Daigo Y, Aragaki M, Ishikawa K,
Sato M and Kaji M: Overexpression of CDC20 predicts poor prognosis
in primary non-small cell lung cancer patients. J Surg Oncol.
106:423–430. 2012.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Kidokoro T, Tanikawa C, Furukawa Y,
Katagiri T, Nakamura Y and Matsuda K: CDC20, a potential cancer
therapeutic target, is negatively regulated by p53. Oncogene.
27:1562–1571. 2008.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wu F, Lin Y, Cui P, Li H, Zhang L, Sun Z,
Huang S, Li S, Huang S, Zhao Q, et al: Cdc20/p55 mediates the
resistance to docetaxel in castration-resistant prostate cancer in
a Bim-dependent manner. Cancer Chemother Pharmacol. 81:999–1006.
2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Pommier Y, Sun Y, Huang SN and Nitiss JL:
Roles of eukaryotic topoisomerases in transcription, replication
and genomic stability. Nat Rev Mol Cell Biol. 17:703–721.
2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Liu T, Zhang H, Yi S, Gu L and Zhou M:
Mutual regulation of MDM4 and TOP2A in cancer cell proliferation.
Mol Oncol. 13:1047–1058. 2019.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Chen D, Maruschke M, Hakenberg O,
Zimmermann W, Stief CG and Buchner A: TOP2A, HELLS, ATAD2, and TET3
are novel prognostic markers in renal cell carcinoma. Urology.
102:265.e1–265.e7. 2017.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Lan J, Huang HY, Lee SW, Chen TJ, Tai HC,
Hsu HP, Chang KY and Li CF: TOP2A overexpression as a poor
prognostic factor in patients with nasopharyngeal carcinoma. Tumour
Biol. 35:179–187. 2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Zheng H, Li X, Chen C, Chen J, Sun J, Sun
S, Jin L, Li J, Sun S and Wu X: Quantum dot-based immunofluorescent
imaging and quantitative detection of TOP2A and prognostic value in
triple-negative breast cancer. Int J Nanomedicine. 11:5519–5529.
2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Węsierska-Gądek J and Składanowski A:
Therapeutic intervention by the simultaneous inhibition of DNA
repair and type I or type II DNA topoisomerases: One strategy, many
outcomes. Future Med Chem. 4:51–72. 2012.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Wang TL, Ren YW, Wang HT, Yu H and Zhao
YX: Association of topoisomerase II (TOP2A) and dual-specificity
phosphatase 6 (DUSP6) single nucleotide polymorphisms with
radiation treatment response and prognosis of lung cancer in Han
Chinese. Med Sci Monit. 23:984–993. 2017.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Kovarikova V, Burkus J, Rehak P, Brzakova
A, Solc P and Baran V: Aurora kinase A is essential for correct
chromosome segregation in mouse zygote. Zygote. 24:326–337.
2016.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Xu Z, Ogawa H, Vagnarelli P, Bergmann JH,
Hudson DF, Ruchaud S, Fukagawa T, Earnshaw WC and Samejima K:
INCENP-aurora B interactions modulate kinase activity and
chromosome passenger complex localization. J Cell Biol.
187:637–653. 2009.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Takeshita M, Koga T, Takayama K, Ijichi K,
Yano T, Maehara Y, Nakanishi Y and Sueishi K: Aurora-B
overexpression is correlated with aneuploidy and poor prognosis in
non-small cell lung cancer. Lung Cancer. 80:85–90. 2013.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Schneider MA, Christopoulos P, Muley T,
Warth A, Klingmueller U, Thomas M, Herth FJ, Dienemann H, Mueller
NS, Theis F, et al: AURKA, DLGAP5, TPX2, KIF11 and CKAP5: Five
specific mitosis-associated genes correlate with poor prognosis for
non-small cell lung cancer patients. Int J Oncol. 50:365–372.
2017.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Wu T, Zhang X, Huang X, Yang Y and Hua X:
Regulation of cyclin B2 expression and cell cycle G2/m transition
by menin. J Biol Chem. 285:18291–18300. 2010.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Mo ML, Chen Z, Li J, Li HL, Sheng Q, Ma
HY, Zhang FX, Hua YW, Zhang X, Sun DQ, et al: Use of serum
circulating CCNB2 in cancer surveillance. Int J Biol Markers.
25:236–242. 2010.PubMed/NCBI
|
|
54
|
Qian X, Song X, He Y, Yang Z, Sun T, Wang
J, Zhu G, Xing W and You C: CCNB2 overexpression is a poor
prognostic biomarker in Chinese NSCLC patients. Biomed
Pharmacother. 74:222–227. 2015.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Takashima S, Saito H, Takahashi N, Imai K,
Kudo S, Atari M, Saito Y, Motoyama S and Minamiya Y: Strong
expression of cyclin B2 mRNA correlates with a poor prognosis in
patients with non-small cell lung cancer. Tumour Biol.
35:4257–4265. 2014.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Ganguly R, Mohyeldin A, Thiel J, Kornblum
HI, Beullens M and Nakano I: MELK-a conserved kinase: Functions,
signaling, cancer, and controversy. Clin Transl Med.
4(11)2015.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Wang J, Wang Y, Shen F, Xu Y, Zhang Y, Zou
X, Zhou J and Chen Y: Maternal embryonic leucine zipper kinase: A
novel biomarker and a potential therapeutic target of cervical
cancer. Cancer Med. 7:5665–5678. 2018.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Speers C, Zhao SG, Kothari V, Santola A,
Liu M, Wilder-Romans K, Evans J, Batra N, Bartelink H, Hayes DF, et
al: Maternal embryonic leucine zipper kinase (MELK) as a novel
mediator and biomarker of radioresistance in human breast cancer.
Clin Cancer Res. 22:5864–5875. 2016.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Li S, Li Z, Guo T, Xing XF, Cheng X, Du H,
Wen XZ and Ji JF: Maternal embryonic leucine zipper kinase serves
as a poor prognosis marker and therapeutic target in gastric
cancer. Oncotarget. 7:6266–6280. 2016.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Lou W, Ding B, Xu L and Fan W:
Construction of Potential Glioblastoma Multiforme-Related
miRNA-mRNA Regulatory Network. Front Mol Neurosci.
12(66)2019.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Gao X, Wang X, Cai K, Wang W, Ju Q, Yang
X, Wang H and Wu H: MicroRNA-127 is a tumor suppressor in human
esophageal squamous cell carcinoma through the regulation of
oncogene FMNL3. Eur J Pharmacol. 791:603–610. 2016.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Goswami RS, Atenafu EG, Xuan Y, Waldron L,
Reis PP, Sun T, Datti A, Xu W, Kuruvilla J, Good DJ, et al:
MicroRNA signature obtained from the comparison of aggressive with
indolent non-Hodgkin lymphomas: Potential prognostic value in
mantle-cell lymphoma. J Clin Oncol. 31:2903–2911. 2013.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Shi L, Wang Y, Lu Z, Zhang H, Zhuang N,
Wang B, Song Z, Chen G, Huang C, Xu D, et al: miR-127 promotes EMT
and stem-like traits in lung cancer through a feed-forward
regulatory loop. Oncogene. 36:1631–1643. 2017.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Fort RS, Mathó C, Oliveira-Rizzo C, Garat
B, Sotelo-Silveira JR and Duhagon MA: An integrated view of the
role of miR-130b/301b miRNA cluster in prostate cancer. Exp Hematol
Oncol. 7(10)2018.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Dettmer MS, Perren A, Moch H, Komminoth P,
Nikiforov YE and Nikiforova MN: MicroRNA profile of poorly
differentiated thyroid carcinomas: New diagnostic and prognostic
insights. J Mol Endocrinol. 52:181–189. 2014.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Ruiz-Martinez M, Navarro A, Marrades RM,
Viñolas N, Santasusagna S, Muñoz C, Ramírez J, Molins L and Monzo
M: YKT6 expression, exosome release, and survival in non-small cell
lung cancer. Oncotarget. 7:51515–51524. 2016.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Mirzadeh Azad F, Naeli P, Malakootian M,
Baradaran A, Tavallaei M, Ghanei M and Mowla SJ: Two lung
development-related microRNAs, miR-134 and miR-187, are
differentially expressed in lung tumors. Gene. 577:221–226.
2016.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Sun CC, Li SJ and Li DJ: Hsa-miR-134
suppresses non-small cell lung cancer (NSCLC) development through
down-regulation of CCND1. Oncotarget. 7:35960–35978.
2016.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Qin Q, Wei F, Zhang J, Wang X and Li B:
miR-134 inhibits non-small cell lung cancer growth by targeting the
epidermal growth factor receptor. J Cell Mol Med. 20:1974–1983.
2016.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Pan JY, Zhang F, Sun CC, Li SJ, Li G, Gong
FY, Bo T, He J, Hua RX, Hu WD, et al: miR-134: A human cancer
suppressor? Mol Ther Nucleic Acids. 6:140–149. 2017.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Hua X and Fan KC: Down-regulation of
miR-1181 indicates a dismal prognosis for nasopharyngeal carcinoma
and promoted cell proliferation and metastasis by modulating
Wnt/β-catenin signaling. Eur Rev Med Pharmacol Sci. 23:1077–1086.
2019.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Nam EJ, Lee M, Yim GW, Kim JH, Kim S, Kim
SW and Kim YT: MicroRNA profiling of a CD133(+) spheroid-forming
subpopulation of the OVCAR3 human ovarian cancer cell line. BMC Med
Genomics. 5(18)2012.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Wang J, Guo XJ, Ding YM and Jiang JX:
miR-1181 inhibits invasion and proliferation via STAT3 in
pancreatic cancer. World J Gastroenterol. 23:1594–1601.
2017.PubMed/NCBI View Article : Google Scholar
|