Introduction
IgA nephropathy (IgAN) is one of the most common types of primary glomerular disease worldwide (1). As a chronic progressive disease, 30-50% of patients diagnosed with IgAN ultimately progress to end-stage renal disease (ESRD) (2,3). Therefore, early diagnosis and treatment are crucial to delay or prevent the progression of IgAN to ESRD (4). To date, histopathological diagnosis based on renal biopsy is the only means for the clinical diagnosis and reliable treatment of IgAN (5). However, renal biopsy is an invasive procedure that may give rise to complications, such as pain, fever and perirenal hematoma, and has a number of contraindications, including hypertension, coagulation disorders, atherosclerosis, diabetes and pregnancy (6-9). Furthermore, due to its invasive nature, renal biopsy is difficult to repeat in the same patient during follow-ups; hence, it is not possible to use it to dynamically monitor the effects of treatment. There is therefore a pressing need for minimally invasive, sensitive and specific biomarkers for the early diagnosis of IgAN and the dynamic monitoring of therapeutic effects (10).
Peptidomics is an emerging and potentially promising field for the discovery of clinical biomarkers (11). In the past decade, researchers have shown that patients with IgAN and other glomerular diseases (including acute glomerulonephritis, rapidly progressive glomerulonephritis, chronic glomerulonephritis, asymptomatic hematuria, proteinuria, occult glomerulonephritis and nephrotic syndrome) and healthy controls harbor significantly different peptidome profiles (12-17). They have also discovered certain peptide biomarkers that can distinguish patients with IgAN from healthy controls and patients that have other glomerular diseases with a high degree of sensitivity and specificity. However, all the aforementioned studies were conducted in adult IgAN patients and it has been reported that the peptide profile changes significantly with age (18). Thus, the peptide biomarkers discovered for adult IgAN patients may not be suitable for application to pediatric IgAN patients.
Peptides serve as biomarkers for diagnosis of diseases and some of them are also important bioactive molecules that function as antimicrobials, antioxidants, immunomodulators and angiotensin-converting enzyme inhibitors (ACEIs) (19). The dysregulation of bioactive peptides has been shown to be involved in disease progression (20) and several of these bioactive peptides may prove to be specific targets for novel therapeutics (21).
The present study aimed to characterize the serum peptidome profile of pediatric patients with IgAN. The characterized peptides may serve as potential diagnostic biomarkers or targets of novel drug therapeutics for pediatric IgAN.
Materials and methods
Subject recruitment and sample collection
A total of 17 children (10 males and 7 females; mean age, 8.24±2.44 years; age range, 5-15 years) who were diagnosed with primary IgAN by renal biopsy at the Department of Department of Pediatrics, The First Affiliated Hospital of Sun Yat-sen University, Guangzhou, China from January 2018 to January 2019. Sun Yat-sen University were recruited. Additionally, 18 patients (11 males and 7 females; mean age, 9.28±2.05 years) with other glomerular diseases, including Henoch-Schonlein purpura nephritis, lupus nephritis and minimal change disease were recruited from the First Affiliated Hospital of Sun Yat-sen University as disease controls. An additional 11 healthy subjects were enrolled from the community clinic, Physical Examination Center, Dongguan Eighth People's Hospital, Dongguan, China (5 males and 6 females; mean age 8.27±2.33 years) were recruited as healthy controls. The profiles of the three groups are summarized in Table I.
 |
Table I
Clinical characteristics of the subjects enrolled in the study.
|
Table I
Clinical characteristics of the subjects enrolled in the study.
| A, IgAN group (n=17) |
| No. |
Sex |
Age (years) |
Cr (µmol/l) |
U-P (g/24 h) |
| 1 |
F |
5 |
38 |
1.58 |
| 2 |
M |
5 |
32 |
0.96 |
| 3 |
F |
8 |
27 |
3.14 |
| 4 |
M |
10 |
51 |
0.21 |
| 5 |
F |
13 |
36 |
2.20 |
| 6 |
F |
9 |
37 |
0.15 |
| 7 |
M |
9 |
44 |
0.32 |
| 8 |
F |
7 |
33 |
0.52 |
| 9 |
F |
10 |
40 |
2.32 |
| 10 |
M |
7 |
38 |
1.54 |
| 11 |
F |
11 |
38 |
3.68 |
| 12 |
M |
7 |
27 |
1.16 |
| 13 |
M |
9 |
69 |
1.35 |
| 14 |
M |
6 |
32 |
1.20 |
| 15 |
M |
4 |
32 |
1.12 |
| 16 |
M |
9 |
49 |
3.28 |
| 17 |
M |
11 |
37 |
0.15 |
| B, Healthy control group (n=11) |
|
|
| No. |
Sex |
Age (years) |
Cr (µmol/l) |
U-P (g/24 h) |
| 1 |
M |
6 |
55 |
0 |
| 2 |
F |
5 |
68 |
0 |
| 3 |
F |
6 |
59 |
0 |
| 4 |
F |
10 |
78 |
0 |
| 5 |
M |
9 |
33 |
0 |
| 6 |
M |
10 |
74 |
0 |
| 7 |
F |
8 |
82 |
0 |
| 8 |
M |
11 |
37 |
0 |
| 9 |
M |
8 |
65 |
0 |
| 10 |
F |
6 |
77 |
0 |
| 11 |
F |
12 |
71 |
0 |
| C, Other glomerular disease group (n=18) |
| No. |
Sex |
Age (years) |
Cr (µmol/l) |
U-P (g/24 h) |
| 1 |
M |
8 |
36 |
1.85 |
| 2 |
M |
6 |
42 |
1.23 |
| 3 |
F |
11 |
40 |
0.54 |
| 4 |
M |
9 |
35 |
0.66 |
| 5 |
F |
7 |
49 |
2.24 |
| 6 |
M |
9 |
40 |
0.45 |
| 7 |
F |
9 |
48 |
1.98 |
| 8 |
M |
5 |
33 |
0.44 |
| 9 |
F |
13 |
40 |
1.78 |
| 10 |
M |
12 |
74 |
0.85 |
| 11 |
M |
11 |
45 |
2.11 |
| 12 |
F |
10 |
38 |
0.54 |
| 13 |
M |
8 |
41 |
1.53 |
| 14 |
M |
8 |
40 |
4.72 |
| 15 |
F |
11 |
39 |
0.15 |
| 16 |
F |
10 |
40 |
0.19 |
| 17 |
M |
10 |
31 |
0.20 |
| 18 |
M |
10 |
52 |
0.17 |
Peripheral blood samples were collected in EDTA coagulation-promoting blood collection tubes, allowed to clot at room temperature for 1 h and then centrifuged at 245 x g for 20 min at room temperature. The resulting serum was transferred to 0.5-ml polypropylene microcentrifuge tubes and stored at -80˚C until ready for use.
The study was approved by The Clinical Research Ethics Committee of The First Affiliated Hospital, Sun Yat-sen University and all the samples were obtained with informed consent from the subjects' guardians.
Peptide extraction
A 500-µl serum sample from each subject was thawed for further peptide extraction. The samples were successively passed through 300-, 30- and 10-kD ultrafiltration centrifuge tubes (Pall Life Sciences) and the sample (peptide) solutions were collected using filtration membranes by gravity filtration and finally lyophilized. The peptides were dissolved in 600 µl loading buffer. The peptides were then desalted using C18 solid phase extraction cartridges (3 cc/100 mg; Waters Corporation) and lyophilized.
Function study methods of the IgAN-specific peptides
In order to explore the underlying biological functions of IgAN-specific peptides, Gene Ontology (GO) enrichment (www.pathway.com/go/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (www.kegg.jp) was performed on the precursor proteins. To further uncover the functions of the IgAN-specific peptides, previously reported methods were used (22,23). In summary, the IgAN-specific peptides identified were compared with each known functional peptide present in the BIOPEP database (www.uwm.edu.pl/biochemia/biopep/start_biopep.php) using the BLASTP online tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome) (24). The retaining standard used for query sequences was as follows: Identity >50% and E-value <0.5.
Liquid chromatography with tandem mass spectrometry (LC-MS/MS)
The lyophilized samples were dissolved in Nano-RPLC buffer A. Peptide solutions were loaded onto a trap column (C18; size, 100 µm x internal diameter, 3 cm; column length, 3 µm; chromatographic packing particle size, 150 Å; chromatographic packing pore size, 10 umol/l; Suzhou Qiangyao Biological Technology Co., Ltd.) at a flow rate of 2 µl/min and separated on a SPE Supra-Clean® C18 column (size, 150x4.6 mm; Shenzhen Noyadi Chemical Technology Co., Ltd.) on an Eksigent nanoLC-Ultra™ 2D system (AB SCIEX). Mobile phase A was composed of 2% acetonitrile (ACN) and 0.1% formic acid dissolved in H2O. Mobile phase B was composed of 95% ACN and 0.1% formic acid dissolved in H2O. Mobile phase B was increased by a gradient of 5 to 50% within 70 min. An internal standard for the peptide group was not used in the current study; search parameters were strictly controlled to ensure the accuracy of data. Filter parameter setting were as follows: 0≤ retention time ≤50; feature significance, ≥0; feature fold change, ≥1; 1≤ charge ≤10; with peptide ID: True; protein significance, ≥20; protein fold change, ≥2; significance method: PEAKSQ; confident unique supports, ≥1; normalization use Total Ion Chromatography; search parameter settings: Quantification type: Label free quantification; mass error tolerance, 20.0 ppm; retention time shift tolerance, 4.0 min; dependent on PID (25): 20; False Discovery Rate threshold, 1%.
A Triple Time-of-flight (TOF) 5600 system combined with a nanoliter spray III ion source (AB SCIEX) was applied using the following parameters: Spray voltage, 2.3 kV; air curtain pressure, 30 psi; atomization pressure, 5 psi; heater temperature, 150˚C. Mass spectrometry scanning was set on information dependent analysis mode and the first-stage TOF-MS single-sheet scan time was 250 msec.
Bioinformatics
The processing, retrieval and analysis of the original .wiff file data collected by mass spectrometry was conducted with Protein Pilot Software 5.0 (AB SCIEX) using the following parameters: No trypsin digestion and no cysteine modification. Variable modifications included deamidation (NQ), oxidation (M), glutamic acid (Pyro-glu), glutamine (Pyro-gln), acetylation (protein N-terminal), maximum variable modification/peptide set to 3 and no fixed posttranslational modifications. The search method was a thorough search analysis: The mass tolerance of mass spectrometry was 20 ppm, mass spectrometry was 0.1 Da, false positive rate was controlled at 1% false discovery rate and protein search unused score >1.3 was regarded as a reliable result. A peptide with confidence >95% was deemed a reliable sequence.
Statistical analysis
To determine the statistical significance between the IgAN group and the healthy control or disease control groups, one-way ANOVA and a post hoc analysis with Bonferroni correction was applied to compare the peptide peak areas. P<0.05 was considered to indicate a statistically significant difference.
Results
Summary of peptides differentially expressed between the IgAN, healthy control and other glomerular disease groups
The serum peptide profiles of all the 46 participants (characteristics provided in Table I) were analyzed using a Venn diagram (Fig. 1). A total of 4,551 peptides were identified to be differentially expressed between the IgAN group and the healthy control group (Table SI). Among them, 1,173 peptides were significantly differentially expressed (fold change >2; P<0.05; Fig. 1). All the differentially expressed peptides are listed in Table SII. A total of 4,371 peptides were identified to be differentially expressed between the IgAN group and the other glomerular disease group (Table SIII). Among them, 239 peptides were significantly differentially expressed (fold change >2; P<0.05; Fig. 1). All the differentially expressed peptides are listed in Table SIV. Among the 1,173 peptides significantly differentially expressed between the IgAN group and the healthy control group and the 239 peptides significantly differentially expressed between the IgAN group and the other glomerular diseases group, 123 peptides overlapped and were designated as IgAN-specific peptides (Table SV). Of these, 48 peptides had a fold change >5 (P<0.05), including 25 upregulated peptides and 23 downregulated peptides (Table II).
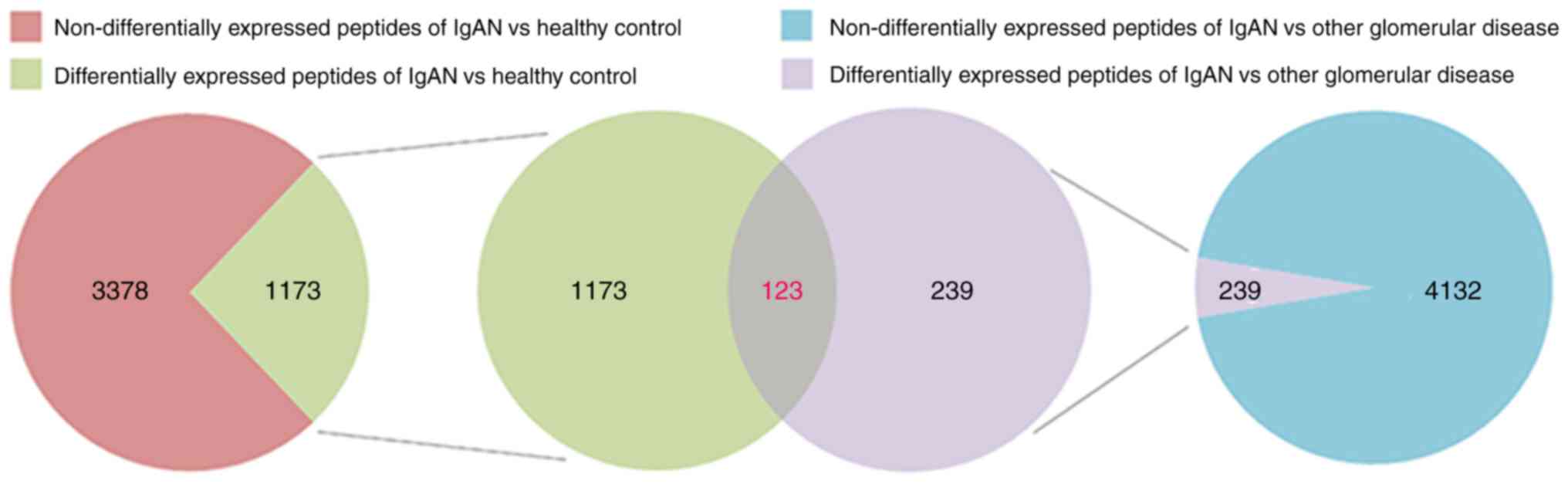 |
Figure 1
Venn diagram illustrating that 123 peptides were exclusively expressed in pediatric patients with IgAN. A total of 4,551 peptides were identified to be differentially expressed between the IgAN group and the healthy control group, and 1,173 of those were significantly differentially expressed (fold change >2; P<0.05). A total of 4,371 peptides were identified to be differentially expressed between the IgAN group and the other glomerular disease group, and 239 of those were significantly differentially expressed (fold change >2; P<0.05). The 123 overlapping peptides were designated as IgAN-specific peptides. IgAN, IgA nephropathy.
|
 |
Table II
Peptides differentially expressed in IgAN patients with a fold change >5.
|
Table II
Peptides differentially expressed in IgAN patients with a fold change >5.
| A, Upregulated in both comparisons |
| |
|
|
IgAN/healthy control |
IgAN/other glomerular diseases |
| Peptide |
MW |
m/z |
Fold change |
P-value |
Fold change |
P-value |
| GKSSSYSKQFTSSTSYNRGDSTFESKSYK MADEAGSEADHEGTHST |
4972.13 |
994.84 |
#DIV/0! |
0.0432 |
#DIV/0! |
0.0098 |
| TSAGLKLILK |
1043.32 |
522.35 |
#DIV/0! |
0.0174 |
#DIV/0! |
0.0025 |
| LLVLITGGK |
913.17 |
457.32 |
#DIV/0! |
0.0099 |
20.83 |
0.0020 |
| CSPDTGSC |
768.81 |
385.13 |
#DIV/0! |
0.0413 |
14.98 |
0.0161 |
| FLPLVAMVLLV |
1214.62 |
1214.74 |
#DIV/0! |
0.0066 |
12.88 |
0.0016 |
| KITHRIHWESASLL |
1690.97 |
564.31 |
#DIV/0! |
0.0306 |
12.63 |
0.0114 |
| VPPNNSNAAEDDLPTVELQGVVPR |
2531.76 |
844.43 |
#DIV/0! |
0.0209 |
10.52 |
0.0086 |
| DSGGQEAN(+.98)N(+.98)PN(+.98)CCNCI |
1641.66 |
547.85 |
#DIV/0! |
0.0000 |
9.99 |
0.0000 |
| IPLDLLLAVPVP |
1259.59 |
630.40 |
#DIV/0! |
0.0032 |
8.64 |
0.0023 |
| QEKNPLPSKETIEQE |
1752.9 |
876.93 |
#DIV/0! |
0.0456 |
8.63 |
0.0294 |
| KKKMKSKKK |
1133.5 |
567.38 |
#DIV/0! |
0.0017 |
7.96 |
0.0008 |
| LVEGEIAEEAAEKATS |
1646.77 |
823.91 |
#DIV/0! |
0.0190 |
7.82 |
0.0109 |
| DDPDAPLQPVTPLQLFEG |
1952.15 |
976.48 |
#DIV/0! |
0.0167 |
7.39 |
0.0085 |
| EFVSETESRGSESGIFTNTKESSSHHP GIAEFPSRG |
3882.09 |
776.96 |
#DIV/0! |
0.0283 |
6.76 |
0.0266 |
| AFKVPAPK |
857.06 |
429.27 |
#DIV/0! |
0.0141 |
6.28 |
0.0120 |
| LAALKKALAAAG |
1097.37 |
549.36 |
#DIV/0! |
0.0074 |
6.09 |
0.0061 |
| MVLKIIAFKPK |
1287.71 |
644.41 |
#DIV/0! |
0.0003 |
5.93 |
0.0002 |
| LLSVLLYAT |
992.22 |
496.81 |
#DIV/0! |
0.0324 |
5.87 |
0.0293 |
| Q(-17.03)AGAAGSRMNFRPGVL |
1614.84 |
807.91 |
#DIV/0! |
0.0161 |
5.81 |
0.0151 |
| Q(-17.03)EKPSSPSPMPSSTPSPSLNLG |
2208.43 |
736.68 |
#DIV/0! |
0.0319 |
5.53 |
0.0333 |
| AEEVHSDPCENNPCLHGGTCNANGT |
2569.69 |
643.00 |
#DIV/0! |
0.0388 |
5.17 |
0.0425 |
| KVMRLRKLAQ(+.98)QIAN |
1670.04 |
835.51 |
14.60 |
0.0121 |
13.13 |
0.0017 |
| GLKQ(+.98)VMAIKSRVVLPYLVPKLT TPPVN(+.98)TRVLA |
3504.32 |
1168.71 |
13.81 |
0.0127 |
7.36 |
0.0040 |
| PGGGYGAA |
648.67 |
649.29 |
7.16 |
0.0460 |
#DIV/0! |
0.0026 |
| RGAASKVKVPM |
1143.41 |
1143.66 |
5.95 |
0.0380 |
11.67 |
0.0032 |
| B, Downregulated in both comparisons |
| |
|
|
IgAN/healthy control |
IgAN/other glomerular diseases |
| Peptide |
MW |
m/z |
Fold change |
P-value |
Fold change |
P-value |
| RTPQGIGLLAKTPLSRPVK |
2032.46 |
1016.63 |
0.15 |
0.0126 |
0.13 |
0.0390 |
| LQVGIPVA |
795.98 |
796.48 |
0.13 |
0.0128 |
0.10 |
0.0256 |
| GRPGPCADVN |
985.08 |
493.23 |
0.12 |
0.0070 |
0.13 |
0.0224 |
| PRVPKYV |
858.05 |
858.53 |
0.08 |
0.0453 |
0.17 |
0.0312 |
| LAETLKREKLKVAN(+.98)KIESIPVKG IIPSKKTKQKEV |
3946.77 |
987.10 |
0.07 |
0.0433 |
0.07 |
0.0390 |
| LRGPHLAKLELIRRLRSQ(+.98) |
2157.59 |
719.78 |
0.07 |
0.0000 |
0.18 |
0.0472 |
| VGGSY |
481.51 |
482.23 |
0.06 |
0.0000 |
0.16 |
0.0043 |
| VKVFSLAVNLIAID |
1501.83 |
751.45 |
0.05 |
0.0470 |
0.10 |
0.0465 |
| AIVGIGGGGGLLLLVIVAVLIAYKRKSR |
2807.51 |
702.43 |
0.04 |
0.0096 |
0.06 |
0.0483 |
| N(+.98)QTILKKGKRENIVNIRKQREKA AILIQ(+.98)AV |
3475.14 |
869.54 |
0.03 |
0.0030 |
0.02 |
0.0094 |
| Peptide |
MW |
m/z |
Fold change |
P-value |
Fold change |
P-value |
| GRIYIQAHIDRDVLIVLVRDAKN(+.98)L |
2792.27 |
931.23 |
0.02 |
0.0253 |
0.07 |
0.0280 |
| Q(-17.03)VKMKPKITRPPINVKII |
2086.38 |
696.10 |
0.02 |
0.0041 |
0.01 |
0.0255 |
| GPRGT |
486.53 |
487.27 |
0.01 |
0.0004 |
0.01 |
0.0043 |
| PPPVLAK |
720.91 |
361.24 |
0.00 |
0.0000 |
0.01 |
0.0026 |
| PVPAL |
495.62 |
496.32 |
0.00 |
0.0000 |
0.00 |
0.0495 |
| LKKFQVT |
863.07 |
863.54 |
0.00 |
0.0004 |
0.00 |
0.0458 |
| PPTPPPLLLLLFPLLLFSRLCGAL |
2602.30 |
867.87 |
0.00 |
0.0016 |
0.00 |
0.0410 |
| LSKHIKT |
826.01 |
826.52 |
0.00 |
0.0244 |
0.00 |
0.0398 |
| LRQLALLLWKNYTLQ(+.98)KRKVLVT |
2699.32 |
675.42 |
0.00 |
0.0238 |
0.00 |
0.0349 |
| HPGMPGGM(+15.99)GT |
957.08 |
957.41 |
0.00 |
0.0015 |
0.00 |
0.0348 |
| EAQGGA |
531.52 |
532.23 |
0.00 |
0.0001 |
0.00 |
0.0265 |
| RLMLTL |
745.98 |
746.47 |
0.00 |
0.0049 |
0.00 |
0.0153 |
| VLLHRGATP |
963.15 |
482.28 |
0.00 |
0.0025 |
0.00 |
0.0006 |
Characteristics of the IgAN-specific peptides
The broad features of the IgAN-specific peptides were analyzed. The molecular weight (MW) of the majority of the peptides ranged from 500-2,000 Da (Fig. 2A) and the isoelectric point (pI) of the majority of peptides ranged from 3.0-6.0 or 8.0-13.0 (Fig. 2B). Fig. 2C demonstrates the distribution of the MW relative to the pI. Of the 123 IgAN-specific peptides, 20 peptides were identified to be proteolytic fragments from 10 precursor proteins (Fig. 2D). The majority of the proteolytic fragments (8 peptides) were derived from fibrinogen α-1 chain (FIBA; P02674) or complement C3 (CO3; P01024; 4 peptides). A total of 8 peptides were derived from FIBA and 4 from CO3 (Fig. 2D).
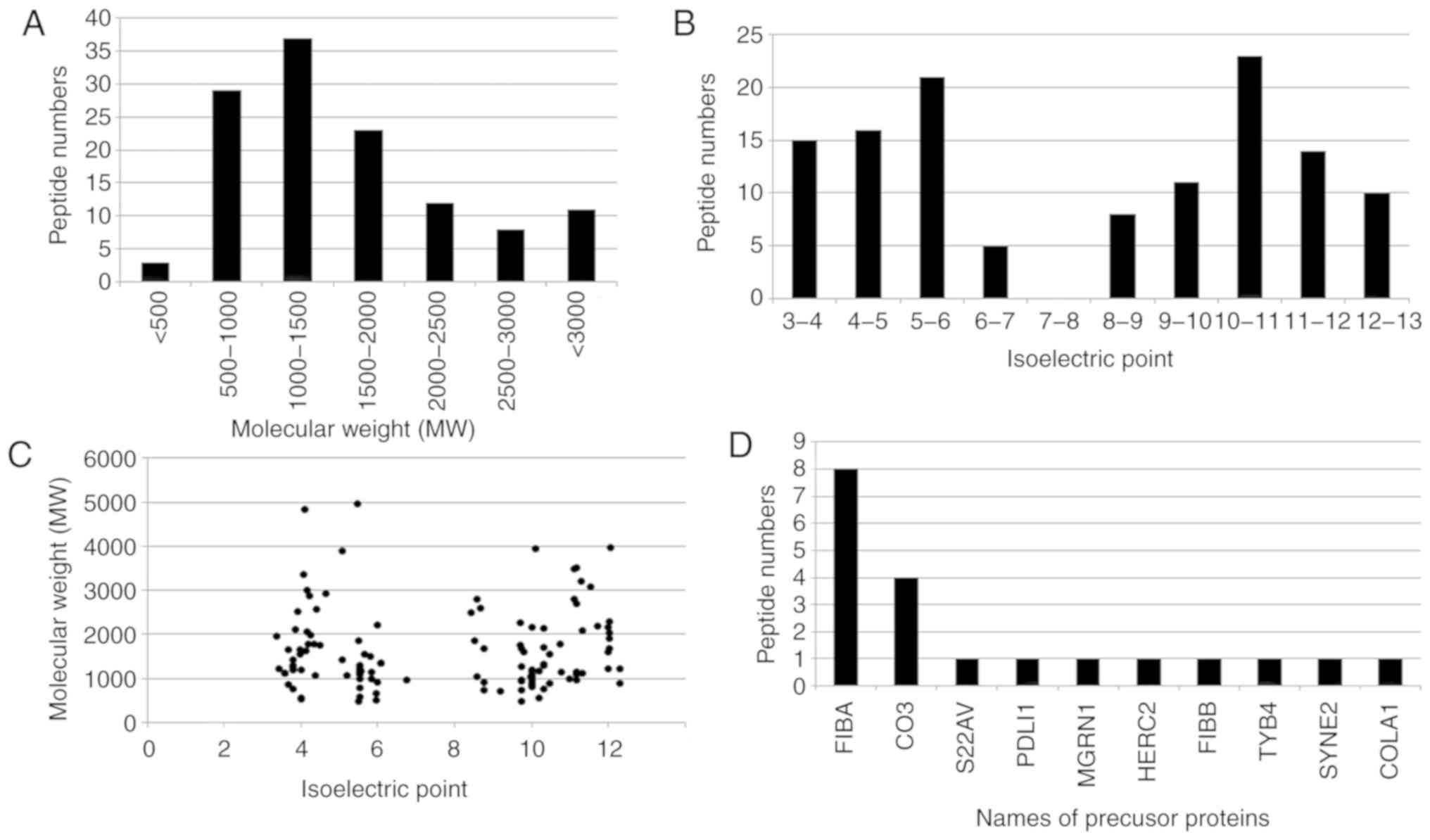 |
Figure 2
Characteristics of the IgAN-specific peptides. (A) Molecular weight distribution of the peptides. (B) Isoelectric point distribution of the peptides. (C) Isoelectric point relative to molecular weight of the peptides. (D) Names of precursor proteins of the peptides. IgAN, IgA nephropathy; FIBA, α-fibrinogen; CO3, cytochrome C oxidase subunit 3; S22AV, solute carrier family 22 member 31; PDLI1, PDZ and LIM domain 1; MGRN1, Mahogunin ring finger 1; HERC2, E3 ubiquitin ligase; FIBB, β-fibrinogen; TYB4, thymosin β4; SYNE2, nesprin 2; COLA1, collagen type 1 α 1.
|
Bioinformatics analysis of the precursor proteins of the IgAN-specific peptides
GO enrichment analysis identified ‘induction of bacterial agglutination’, ‘platelet degranulation’, ‘regulation of response to wounding’, ‘blood coagulation’, ‘hemostasis’, ‘coagulation’, ‘regulated exocytosis’ and ‘regulation of heterotypic cell-cell adhesion’ as biological process-associated terms associated with IgAN-specific peptides (Fig. 3A). With regard to cellular components, ‘secretory granule lumen’, ‘vesicle lumen’, ‘platelet alpha granule lumen’, ‘blood microparticle’, ‘extracellular region’, ‘secretory granule’ and ‘fibrinogen complex’ were among the most highly enriched subcategories (Fig. 3B). Regarding molecular functions, ‘cell adhesion molecule binding’, ‘protein binding’, ‘protein binding, bridging’, ‘binding, bridging’, ‘ubiquitin-like protein ligase activity’, ‘cadherin binding involved in cell-cell adhesion’ and ‘SUMO binding’ were highly enriched subcategories (Fig. 3C). KEGG pathway analysis revealed that the identified peptides were associated with the terms ‘complement and coagulation cascades’, ‘platelet activation’, ‘ubiquitin mediated proteolysis’, ‘legionellosis’, ‘Staphylococcus aureus infection’, ‘leishmaniasis’, ‘pertussis’, ‘protein digestion and absorption’, ‘Chagas disease (American trypanosomiasis)’ and ‘systemic lupus erythematosus’ (Fig. 3D).
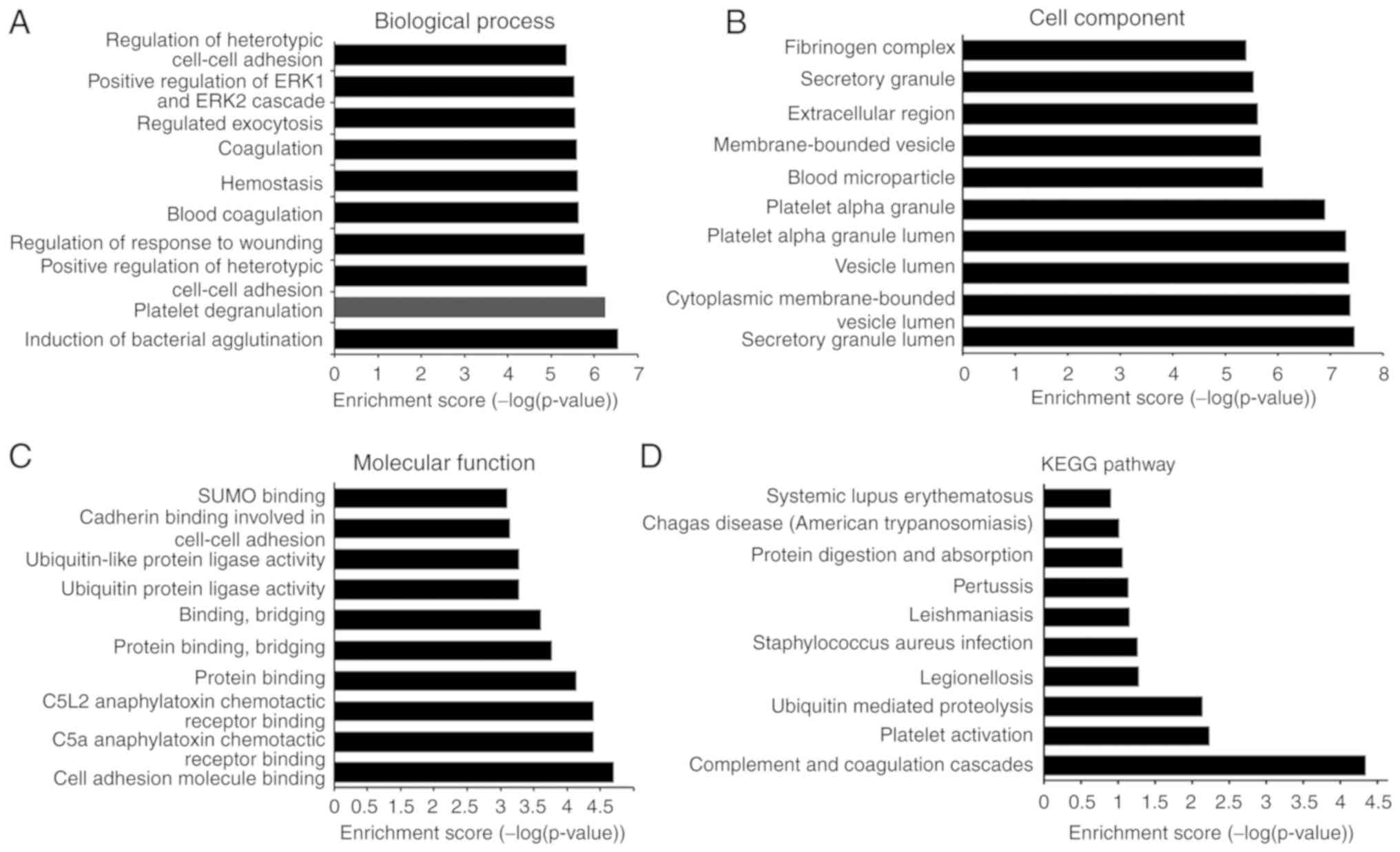 |
Figure 3
Gene Ontology and KEGG pathway analysis of the precursor proteins of the IgAN-specific peptides. (A) Biological processes. (B) Cell components. (C) Molecular function. (D) KEGG pathway analysis. IgAN, immunoglobulin A nephropathy; KEGG, Kyoto Encyclopedia of Genes and Genomes.
|
Identification of potential bioactive peptides associated with IgAN
Consequently, two potential bioactive peptides were identified (Table III). The first peptide was QEKNPLPSKETIEQE, which was upregulated in the IgAN group compared with the BIOPEP database, designated as IgAN-U-P and predicted to have a pro-angiogenic function (26). The second one was PPPVLAK, which was downregulated in the IgAN group compared with the BIOPEP database, designated as IgAN-D-P and predicted to function as an ACEI.
 |
Table III
Potential bioactive peptides associated with IgAN pathogenesis.
|
Table III
Potential bioactive peptides associated with IgAN pathogenesis.
| Designated name |
Sequence |
Expression pattern |
Known functional peptides |
Known activity |
| IgAN-U-P |
QEKNPLPSKETIEQE |
Upregulated |
LKLTETQEKNPLPSKETIEQEKQAGES |
Pro-angiogenic |
| IgAN-D-P |
PPPVLAK |
Downregulated |
PPPVHL |
ACEI |
Discussion
In this preliminary prospective study, a pediatric IgAN group, together with age- and gender-matched healthy control and glomerular disease groups were recruited and peptides from their serum samples were enriched by centrifugal ultrafiltration with an accurate MW cutoff, a method used extensively for peptide enrichment via a size-exclusion mechanism (27,28). The serum of each subject was directly analyzed by LC-MS/MS. The results indicated that the serum peptidome profiles of pediatric patients with IgAN were significantly different from those of the healthy control group and those of the other glomerular diseases group. In total, 123 IgAN-specific peptides with fold change >2 and 48 with fold change >5 were identified.
Although there is an urgent need for a minimally invasive and reliable biomarker for the diagnosis of IgAN, the identification of such specific peptides is just the first step in a long process before they can be used in the clinic, which will first require their successful validation. There are a substantial number of urinary peptides that show promise as biomarkers of IgAN; however, all of them require further, rigorous validation in well-planned studies (29). A previous study found 5 peptides that may be candidate serum markers for IgAN and may be associated with the pathogenesis of IgA (30). One of the critical steps in progressing this research into the clinic is the evaluation of these peptides in independent test sets, collected using a multicentric approach (31,32). Additionally, in order to improve accuracy, the mass spectrometry results will be verified in other IgAN patients in a future study.
Although ~60 years have passed since the first description of IgAN in 1968(33), its exact pathogenesis is largely unknown. A large number of studies have shown that some peptides function as bioactive molecules and are not simply protein degradation products (34). The results of the present study indicated that the upregulated IgAN-specific peptide, QEKNPLPSKETIEQE, may have a pro-angiogenic function. Capillary endothelial cell proliferation is consistently shown to be one of the indicators of the pathological activity of IgAN. Clinically, it is more characteristic of the acute phase and is associated with greater numbers of crescent cells (obtained by performing pathological analyses) compared to cases with no endothelial cell proliferation (35). Hence, capillary endothelial cell proliferation could be considered as a predictor of early disease activity (36). The results of the present study also indicated that the downregulated IgAN-specific peptide, PPPVLAK, may function as an ACEI. Consistently, ACEI agents have been revealed to be a promising therapy for IgAN due to their significant effects on reducing proteinuria (37). Therefore, based on the results of the present study, it is proposed that absence of the pro-angiogenic peptide combined with deficiency of ACEI peptide may be involved in the pathogenesis of IgAN in pediatric patients.
Supplementary Material
Table SI: Total peptides expressed in between the IgAN group and the healthy control group
Table SII: Differentially expressed peptides between the IgAN group and the healthy control group
Table SIII: Total peptides expressed in between the IgAN group and the other glomerular disease group
Table SIV: Differentially expressed peptides between the IgAN group and the other glomerular disease group
Table SV: IgAN?specifc peptides
Acknowledgements
Not applicable.
Funding
The current study was supported by the Dongguan Bureau of Science and Technology for the City Key Program of Science and Technology (grant no. 2016108101038).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
ZL conceived and designed the current study and interpreted experimental results. SC, XJ and CR contributed to the design of the study and interpretation of experimental results. XL and FY performed experiments, analyzed data, prepared figures and drafted the manuscript. SC and XJ approved the final version of manuscript. XL and XJ edited and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The current study was approved by The Clinical Research Ethics Committee of The First Affiliated Hospital, Sun Yat-sen University (approval no. 20180706) and all samples were obtained with informed consent.
Patient consent for publication
All study participants had given their written informed consent before participating in the study.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Diniz H, Bandeira M, Teresa A, Besteiro B, Coimbra J, Gomes F and Sampaio S: IgA nephropathy with thrombotic microangiopathy: Is this secondary thrombotic microangiopathy or IgA nephropathy-triggered atypical Hemolytic Uremic Syndrome? Portuguese J Nephrol Hypertension. 33:239–243. 2019.
|
|
2
|
Barratt J and Feehally J: IgA nephropathy. J Am Soc Nephrol. 16:2088–2097. 2005.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Smith AC and Feehally J: New insights into the pathogenesis of IgA nephropathy. Pathogenesis of IgA nephropathy. Springer Semin Immunopathol. 24:477–493. 2003.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Galla JH: IgA nephropathy. Kidney Int. 47:377–387. 1995.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Schwartz D and Pullman J: Renal biopsy in pregnancy: Risks, benefits, pathologic findings, and illustrative examples. In: Obstetric and Gynecologic Nephrology. Springer, pp87-99, 2020.
|
|
6
|
Bandari J, Fuller TW, Ii RM and D'Agostino LA: Renal biopsy for medical renal disease: Indications and contraindications. Can J Urol. 23:8121–8126. 2016.PubMed/NCBI
|
|
7
|
Sumnu A, Gursu M and Ozturk S: Primary glomerular diseases in the elderly. World J Nephrol. 4:263–270. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Xu DM, Chen M, Zhou FD and Zhao MH: Risk factors for severe bleeding complications in percutaneous renal biopsy. Am J Med Sci. 353:230–235. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhu MS, Chen JZ and Xu AP: Factors that can minimize bleeding complications after renal biopsy. Int Urol Nephrol. 46:1969–1975. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Hollis EM: Diffusion-Weighted magnetic resonance imaging in diagnosing graft dysfunction: A non-invasive alternative to renal biopsy. Electronic Theses and Dissertations, 2017 https://doi.org/10.18297/etd/2661.
|
|
11
|
Silberring J and Ciborowski P: Biomarker discovery and clinical proteomics. Trends Analyt Chem. 29(128)2010.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Graterol F, Navarro-Muñoz M, Ibernon M, López D, Troya MI, Pérez V, Bonet J and Romero R: Poor histological lesions in IgA nephropathy may be reflected in blood and urine peptide profiling. BMC Nephrol. 14(82)2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Haubitz M, Wittke S, Weissinger EM, Walden M, Rupprecht HD, Floege J, Haller H and Mischak H: Urine protein patterns can serve as diagnostic tools in patients with IgA nephropathy. Kidney Int. 67:2313–2320. 2005.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Julian BA, Wittke S, Novak J, Good DM, Coon JJ, Kellmann M, Zürbig P, Schiffer E, Haubitz M, Moldoveanu Z, et al: Electrophoretic methods for analysis of urinary polypeptides in IgA-associated renal diseases. Electrophoresis. 28:4469–4483. 2007.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kaneshiro N, Xiang Y, Nagai K, Kurokawa MS, Okamoto K, Arito M, Masuko K, Yudoh K, Yasuda T, Suematsu N, et al: Comprehensive analysis of short peptides in sera from patients with IgA nephropathy. Rapid Commun Mass Spectrom. 23:3720–3728. 2009.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Rocchetti MT, Papale M, d'Apollo AM, Suriano IV, Di Palma AM, Vocino G, Montemurno E, Varraso L, Grandaliano G, Di Paolo S and Gesualdo L: Association of urinary laminin G-like 3 and free K light chains with disease activity and histological injury in IgA nephropathy. Clin J Am Soc Nephrol. 8:1115–1125. 2013.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wu J, Wang N, Wang J, Xie Y, Li Y, Liang T, Wang J, Yin Z, He K and Chen X: Identification of a uromodulin fragment for diagnosis of IgA nephropathy. Rapid Commun Mass Spectrom. 24:1971–1978. 2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zurbig P, Decramer S, Dakna M, Jantos J, Good DM, Coon JJ, Bandin F, Mischak H, Bascands JL and Schanstra JP: The human urinary proteome reveals high similarity between kidney aging and chronic kidney disease. Proteomics. 9:2108–2117. 2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Baum F, Fedorova M, Ebner J, Hoffmann R and Pischetsrieder M: Analysis of the endogenous peptide profile of milk: Identification of 248 mainly casein-derived peptides. J Proteome Res. 12:5447–5462. 2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjöberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, et al: Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 11:76–83. 2010.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Buse JB, Sesti G, Schmidt WE, Montanya E, Chang CT, Xu Y, Blonde L and Rosenstock J: Liraglutide Effect Action in Diabetes-6 Study Group: Switching to once-daily liraglutide from twice-daily exenatide further improves glycemic control in patients with type 2 diabetes using oral agents. Diabetes Care. 33:1300–1303. 2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Cui X, Li Y, Yang L, You L, Wang X, Shi C, Ji C and Guo X: Peptidome analysis of human milk from women delivering macrosomic fetuses reveals multiple means of protection for infants. Oncotarget. 7:63514–63525. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Dallas DC, Guerrero A, Khaldi N, Castillo PA, Martin WF, Smilowitz JT, Bevins CL, Barile D, German JB and Lebrilla CB: Extensive in vivo human milk peptidomics reveals specific proteolysis yielding protective antimicrobial peptides. J Proteome Res. 12:2295–2304. 2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Minkiewicz P, Dziuba J, Iwaniak A, Dziuba M and Darewicz M: BIOPEP database and other programs for processing bioactive peptide sequences. J AOAC Int. 91:965–980. 2008.PubMed/NCBI
|
|
25
|
Wang Z, Wang H, Peng Y, Chen F, Zhao L, Li X, Qin J, Li Q, Wang B, Pan B and Guo W: A liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based assay to profile 20 plasma steroids in endocrine disorders. Clin Chem Lab Med. 21(0869)2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Dettin M, Ghezzo F, Conconi MT, Urbani L, D'Auria G, Falcigno L, Guidolin D, Nico B, Ribatti D, Di Bello C and Parnigotto PP: In vitro and in vivo pro-angiogenic effects of thymosin-beta4-derived peptides. Cell Immunol. 271:299–307. 2011.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Soloviev M and Finch P: Peptidomics: Bridging the gap between proteome and metabolome. Proteomics. 6:744–747. 2006.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Srinivasan M and Patel MS: Metabolic programming in the immediate postnatal period. Trends Endocrinol Metab. 19:146–152. 2008.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Marek-Bukowiec K, Konieczny A, Ratajczyk K and Witkiewicz W: Candidate urine peptide biomarkers for IgA nephropathy: Where are we now? Dis Markers. 21(5205831)2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ishii E, Okamura J, Tsuchida M, Kobayashi M, Akiyama Y, Nakahata T, Kojima S, Hanada R, Horibe K, Sato T, et al: Infant leukemia in Japan: Clinical and biological analysis of 48 cases. Med Pediatr Oncol. 19:28–32. 1991.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Alkhalaf A, Zürbig P, Bakker SJ, Bilo HJ, Cerna M, Fischer C, Fuchs S, Janssen B, Medek K, Mischak H, et al: Multicentric validation of proteomic biomarkers in urine specific for diabetic nephropathy. PLoS One. 5(e13421)2010.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Knepper MA: Common sense approaches to urinary biomarker study design. J Am Soc Nephrol. 20:1175–1178. 2009.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Berger J and Hinglais N: Intercapillary deposits of IgA-IgG. J Urol Nephrol (Paris). 74:694–695. 1968.(In French). PubMed/NCBI
|
|
34
|
Pessione E and Cirrincione S: Bioactive molecules released in food by lactic acid bacteria: Encrypted peptides and biogenic amines. Front Microbiol. 7(876)2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Lai KN, Tang SC, Schena FP, Novak J, Tomino Y, Fogo AB and Glassock RJ: IgA nephropathy. Nat Rev Dis Primers. 2(16001)2016.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Woo KT, Lim CC, Foo MW, Loh HL, Jin AZ, Chin YM, Choo JC, Tan PH Chow KY, Choong LH, et al: 30-Year follow-up study of IgA nephritis in a southeast Asian population: An evaluation of the oxford histological classification. Clin Nephrol. 86:270–278. 2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Tan J, Dong L, Ye D, Tang Y, Hu T, Zhong Z, Tarun P, Xu Y and Qin W: The efficacy and safety of immunosuppressive therapies in the treatment of IgA nephropathy: A network meta-analysis. Sci Rep. 10(6062)2020.PubMed/NCBI View Article : Google Scholar
|

















