Allergic rhinitis (AR) is a common upper airway
disease, which is characterized by sneezing, rhinorrhea and
obstruction of the nasal passages (1). A previous study revealed that AR was
mediated by immunoglobulin (Ig)E-induced upper airway inflammation,
which is commonly triggered by exposure to allergens (1). AR has been indicated to coexist with
other allergic and/or airway conditions, such as asthma,
rhinosinusitis, nasal polyps and lower airway infection, among
others (2-4).
AR is highly prevalent with an incidence rate of 10-20% worldwide
(1,5), which has rapidly increased along with
industrialization (6). Therefore,
AR is considered a major and emerging public health concern,
especially in developed countries (6,7). The
prevalence of AR in urban China has substantially increased in the
past few years (from 2005 to 2011) (8). An epidemiological study has indicated
that the overall prevalence of AR in adults in major cities across
mainland China has increased from 11.1% in 2005 to 17.6% in 2011,
and patients with AR have been revealed to be more susceptible to
allergic complications compared with the general population
(8). Therefore, subjects diagnosed
with AR have been indicated to experience a reduction in their
quality of life, although not always severe (9). However, AR cannot be completely
treated, which poses a substantial socioeconomic burden (1).
Previous studies have revealed that environmental
stimuli serve pivotal roles in the allergic airway responses that
are essential for the initiation and exacerbation of AR (10,11).
Nevertheless, a potent genetic component has been also associated
with this disease (12). The
heritability of AR has been estimated to be >65% (13,14). A
number of studies have suggested that sequence polymorphisms in
multiple genes and regions are associated with an increased risk of
AR (15-22),
although another study has reported the poor reproducibility of
single-nucleotide polymorphism (SNP) associations with this disease
(23). Nevertheless, certain
genetic variations that have been associated with AR support the
hypothesized pathophysiology of this disease (24). Therefore, genetic and physiological
investigations cross-validate the putative mechanisms of AR,
providing essential insights into its pathogenesis, treatment and
prevention (25).
A previous study has suggested that type 2 T helper
(Th2) cell-mediated inflammatory responses against environmental
stimuli, including pollutants and allergens, contributed in AR
pathophysiology (26). According to
the classical model for allergic airway diseases, including AR and
allergens, which are presented by antigen-presenting cells, result
in the activation and expansion of Th2 cells. These Th2 cells have
been indicated to subsequently produce Th2 cytokines, such as
interleukin (IL)-4, IL-5 and IL-13, which drive IgE antibody
production and promote the proliferation and differentiation of
multiple effector cells, for example mast cells, eosinophils and
basophils (27,28). This results in the secretion of
multiple key inflammatory mediators, such as leukotriene and
histamine (29-31).
In addition to these well-characterized Th2 cytokines, other
mediators that are primarily released by epithelial cells, which
constitute the first line of defense against environmental stimuli,
have also attracted attention owing to their determinant roles in
initiating the inflammatory responses. For example,
epithelium-derived IL-33 has been indicated to induce allergic
airway inflammation (32,33). Previous studies have also reported
that during allergen-driven inflammation, IL-33 was expressed by a
variety of immune cells, including important immune cell types such
as eosinophils (32,33).
Previous studies have examined the potential
association of IL-33 with AR, as IL-33 has been indicated to
exhibit a well-characterized regulatory impact on the initiation
and exacerbation of allergic airway responses (46-48).
In patients with intermittent AR who are sensitive to trees and/or
grass pollens, serum IL-33 has been revealed to be increased
compared with normal controls and positively correlate with disease
severity (49). The serum levels of
IL-33 have also been reported to be higher in patients with
Japanese cedar (JC) pollinosis compared with normal controls
(50), and two IL-33 SNPs
have been associated with the disease (50). Notably, IL-33 in the serum of
patients with AR can be decreased following immunotherapy (51). Intranasal administration of
anti-tumor necrosis factor-α IgY has been indicated to reduce both
IL-33 and ovalbumin-specific IgE antibodies in the serum and nasal
lavage fluids of patients (52).
Although IL-33 is not always detectable in serum, increased levels
of IL-33 in sinus mucosa have also been reported in patients with
JC pollinosis and house dust mite (HDM)-sensitized AR (53). A previous study has demonstrated
that an anti-IL-33 antibody exhibited a therapeutic potential in
experimental AR (54). Moreover,
IL-33 receptor ST2 has been indicated to be highly expressed in the
nasal epithelium of patients with AR or chronic rhinosinusitis
(55-57),
and the concentration of sST2 in the nasal lavage fluids of
patients with AR have been revealed to increase when patients
experience pollen-induced allergic reactions (58). Taken together, these data suggest
that IL-33 likely facilitates the pathogenesis of AR. Therefore,
additional studies on this cytokine may provide novel insights on
potential preventive strategies against AR.
As aforementioned, the prevalence of AR has
increased in recent years in the Chinese population; however little
is known about the susceptibility factors to AR, especially the
genetic basis, in this ethnic group. Therefore, the aim of the
present study was to perform genetic analyses of the IL-33
variations in the Han Chinese population to reveal whether
variations in this gene confer an inheritable risk of AR.
A total of 769 patients with AR (453 men and 316
women) aged between 10 and 65 years (mean ± standard deviation,
37.74±16.52) were recruited between January 2014 and July 2019. The
diagnosis of AR was performed following the criteria of Allergic
Rhinitis and its Impact on Asthma (1). Subjects presenting > two common
symptoms of AR (for example nasal congestion, rhinorrhea, nasal
itching and sneezing) for >4 days/week for at least 3 weeks
during the past 12 months were diagnosed with AR. All patients
received antihistaminic and steroids at The Second Hospital of
Jingzhou and Shishou People's Hospital (Jingzhou, China). Specific
allergens as potential cause of AR were determined for each patient
via skin prick tests (SPT; Allergopharma GmbH & Co. KG) in
accordance with the recommendations by the Subcommittee on Allergen
Standardization and Skin Tests of the European Academy of Allergy
and Clinical Immunology (59). A
total of 18 inhaled allergens, including HDM, grass, tree, mold,
food and weed panel allergens, were examined. The presence of a
wheal ≥ one half of the diameter of the histamine control and >3
mm of the negative control was defined positive in the SPT.
Patients with any other systemic diseases were excluded from the
study.
A total of 769 healthy volunteers (442 men and 327
women) aged between 10 and 65 years (mean ± standard deviation,
37.01±15.94) that tested negative for allergens in the skin prick
tests were recruited as control subjects in the current study. The
controls were also recruited at The Second Hospital of Jingzhou and
Shishou People's Hospital (Jingzhou, China) between January 2014
and July 2019. In addition, the control subjects did not exhibit
any clinical diagnosis or family history of allergy, and did not
experience an upper respiratory tract infection within the last 4
weeks prior to the study. All patients and controls were Han
Chinese.
SNPs were selected based on previous studies and the
genotype data from the 1,000 Genomes Project (60). Based on the high comorbidity rate of
AR and asthma, genome-wide association studies (GWAS) on asthma
were firstly screened to select genome-wide significant variants
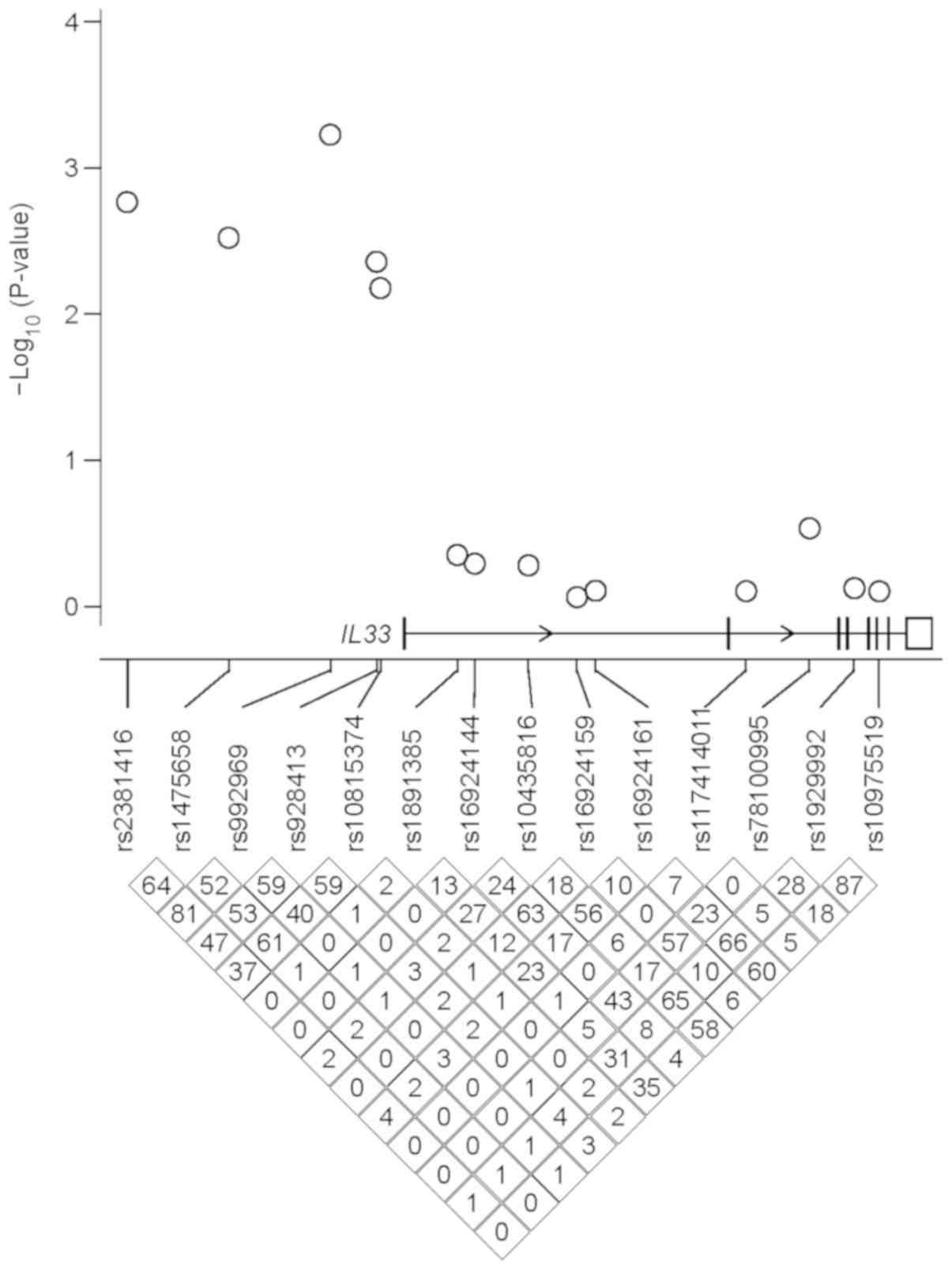
spanning IL-33. Briefly, four SNPs (rs928413, rs9775039,
rs992969 and rs2381416) in IL-33 have been indicated to
exhibit genome-wide significant associations with asthma in
previous GWAS on European and Asian populations (24,61-64).
According to the 1,000 Genomes Project, three IL-33 SNPs
(rs928413, rs992969 and rs2381416) have been indicated to be
polymorphic in the Han Chinese population, and therefore were
selected in the present study. The aforementioned study in subjects
with JC pollinosis was also reviewed (50), and two additional SNPs that have
been associated with this disease were selected (rs1929992 and
rs10975519; Fig. 1). The linkage
disequilibrium (LD) pattern of IL-33 in the Han Chinese
population was subsequently examined based on data from the 1,000
Genomes Project, and nine representative tagging SNPs (rs1475658,
rs10815374, rs1891385, rs16924144, rs10435816, rs16924159,
rs16924161, rs117414011 and rs78100995) spanning the IL-33
gene were selected (Fig. 1). In
summary, a total of 14 SNPs (Table
I) were selected for subsequent investigation in the current
study.
Saliva was collected from the participants, and
genomic DNA was extracted according to the manufacturer's protocol
(MagMAX gDNA Saliva Isolation Kit; Thermo Fisher Scientific, Inc.).
The extracted DNA was dissolved in 50 ml Tris-EDTA buffer (10 mM
Tris, pH 7.8; 1 mM EDTA). The concentration of the DNA samples was
quantified via measuring the optical density at 260 nm using
NanoDrop. A multiplex PCR was performed to simultaneously amplify
multiple SNPs in one reaction in 96-well plates, and each well
contained 20 ng genomic DNA, 10 mmol/l primers (the primer
sequences are presented in Table
SI), 2 µl dNTP, 2 µl 10X PCR buffer, 1 µl 25 mmol/l
MgCl2 and 0.4 units Taq polymerase (Takara Biotechnology
Co., Ltd.). The multiplex PCR reaction was conducted on an ABI 9700
cycler (Applied Biosystems; Thermo Fisher Scientific, Inc.) using
the following thermocycling conditions: Denaturation for 5 min at
95˚C; 40 cycles of 30 sec at 95˚C, 30 sec at 60˚C and 30 sec at
72˚C; and extension for 10 min at 72˚C. The PCR products were
subsequently used to genotype candidate SNPs with the SNaPshot
method. In brief, the PCR products were purified via shrimp
alkaline phosphatase (cat. no. M0371L; New England Biolabs) and
exonuclease I (cat. no. M0293L; BioLabs) treatment at 37˚C for 1 h.
Specifically designed SNaPshot primers (the primer sequences are
presented in Table SII) were used
to amplify the SNP target sites for one base extension using
SNaPshot Multiplex Ready Reaction Mix (cat. no. 4323166; Applied
Biosystems; Thermo Fisher Scientific, Inc.), following which the
reaction was terminated and the products were loaded on an ABI 3730
automated sequencer (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The SNP genotype calling results were automatically
retrieved using GeneMarker version 2.2.0 software (SoftGenetics)
and were manually verified. All genotypes were called blind to
sample identity and affection status and were independently
reviewed and confirmed by two authors. A total of 5% of the samples
were randomly selected for re-analysis to ensure 100%
concordance.
Haploview software (version 4.1; Broad Institute of
Massachusetts Institute of Technology and Harvard) (65) and the online program SHEsis
(http://analysis.bio-x.cn/myAnalysis.php) (66,67)
were used to calculate the Hardy-Weinberg equilibrium (HWE) of each
SNP in cases and controls, and the LD between paired SNPs was
defined using the r2 algorithm. SHEsis was also used to
compare the allelic and genotypic distributions of each SNP between
cases and controls, and Pearson's χ2 test was applied to
calculate the P-values, odds ratios (ORs) and 95% confidence
intervals (66,67). Haplotype frequencies and association
signals were estimated with the online program SHEsis using the
χ2 test (66,67). The haplotypes with a frequency
<0.005 in both cases and controls were discarded, and nine
haplotypes remained for subsequent analyses.
No significant differences were observed between
cases and controls in terms of their mean age (P=0.38) and sex
distribution (P=0.29). According to the SPT results of 769 patients
with AR, 352 (45.8%) were sensitized to HDM, 147 (19.1%) were
sensitized to tree pollen and 270 (35.1%) were sensitized to
multiple allergens (data not shown).
The call rate for the SNPs was >99%, and all SNPs
were in HWE in both patients with AR and healthy controls (HWE
P>0.01; Table I). To examine
whether SNPs in the IL-33 gene exhibited an association with
the risk of AR, both the allelic and genotypic frequencies of the
SNPs were compared between cases and controls. The analysis of
allele frequencies revealed that five SNPs in the 5'-flanking
region of IL-33 were significantly associated with the risk
of AR, and the minor alleles of all five SNPs exhibited
significantly increased frequencies in patients with AR compared
with controls (rs2381416, P=0.0017, OR=1.690; rs1475658, P=0.003,
OR=1.50; rs992969, P=0.0006, OR=1.89; rs928413, P=0.0045, OR=1.53;
rs10815374, P=0.0067, OR=1.51; Table
II). These five SNPs exhibited a moderate pairwise LD in the
Han Chinese population (Fig. 1),
and notably, three of them (rs928413, rs992969 and rs2381416) have
previously been indicated to be associated with asthma (61,62,64),
with their asthma risk alleles being the same as their AR risk
alleles, as revealed in the present study. These results verified
the genetic link between AR and asthma in the IL-33 locus.
However, the other nine SNPs in IL-33 did not exhibit an
association with AR in the current study (Table II). The genotypic analyses of 14
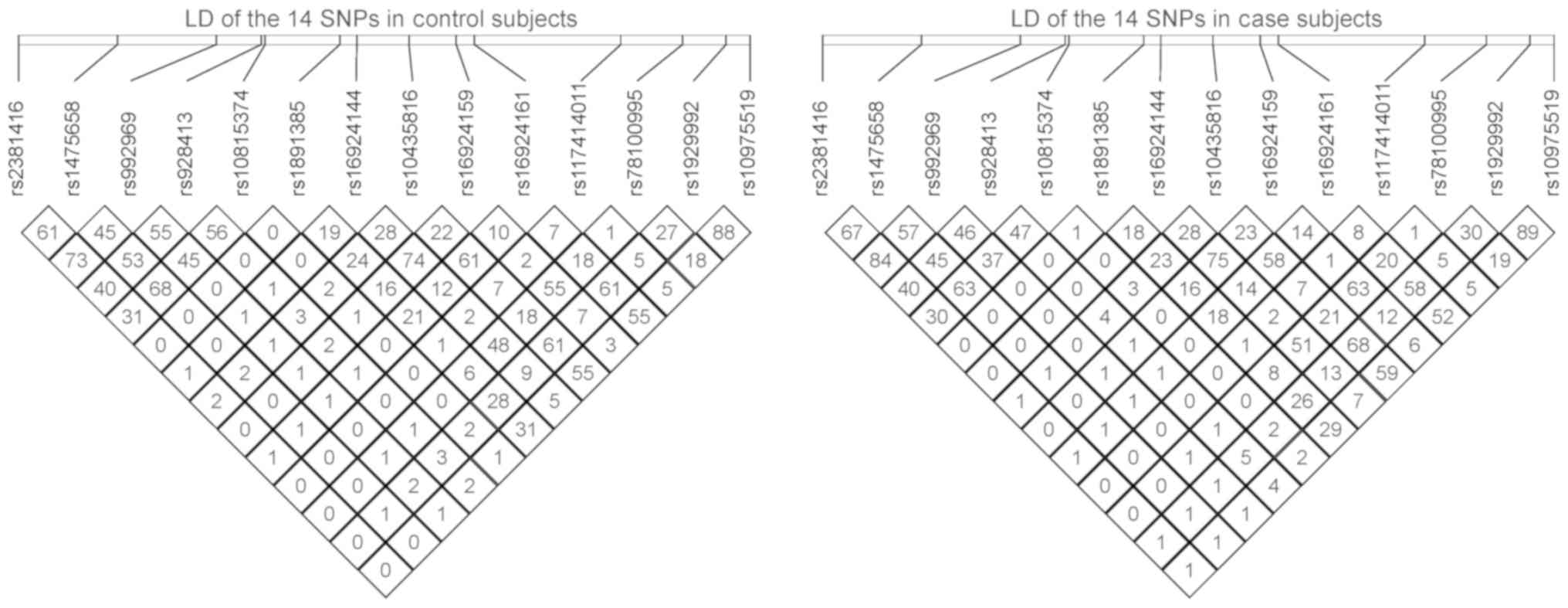
SNPs presented similar results to the allelic analyses (Table I). The LD maps of the 14
IL-33 SNPs in the Han Chinese case and control samples were
constructed, and it was indicated that the LD patterns of cases and
controls were similar (Fig. 2). The
LD map that was constructed using genotype data from the 1,000
Genomes Project was also indicated to be similar to the LD maps of
the cases and controls of the current study (Fig. 1).
Allergic rhinitis (AR) is a common allergic disease
exhibiting important comorbidity with a number of atopic diseases,
for example allergic asthma. Although Th2-mediated allergic
inflammation has been indicated to facilitate AR pathogenesis, the
associated genetic components remain to be fully determined
(1). As >80% of patients with
asthma exhibit rhinitis, and 10-40% of patients with rhinitis
develop asthma, these two diseases are considered to share risk
factors and mechanisms (1).
Therefore, it is of interest to examine the association between the
GWAS risk loci of asthma and AR. IL-33 has been highlighted as an
important initiating factor for allergic asthma, and a recent study
on whole blood gene expression profiles has revealed a consistent
overexpression of the IL1RL1 gene, which encodes IL-33
receptor ST2, in patients diagnosed with multiple morbidities for
asthma, dermatitis and rhinitis compared with normal controls
(68). Therefore, the present study
aimed to explore whether polymorphisms in IL-33 also affect
the susceptibility to AR. Using the SNaPshot method, the current
study indicated that SNPs in the IL-33 gene, which have been
previously associated with asthma (61,62,64),
were also associated with AR. This observation supported the shared
pathological mechanism between asthma and AR, and provided insights
into inflammatory responses associated with AR (69). Previous studies have also reported
that asthma susceptibility loci exhibited an association with AR,
and several other risk variants have also been highlighted
conferring a risk of both asthma and AR (70-73).
The SNPs that were significantly associated with AR
in the present study were localized in the promoter region of
IL-33, and may have potential regulatory impact on this
gene. For example, the A-allele of rs992969, which is associated
with an increased risk of AR in the present study, was also
significantly associated with higher IL-33 expression in
bronchial epithelial cells (P=1.3x10-6) in a previous
study (74). A recent study has
indicated that the A-allele of rs992969 was significantly
associated with a higher mRNA level of IL-33 in bronchial
brushes (P=8.3x10-12) (75). In addition, a functional study has
revealed that the G-allele of rs928413 was associated with higher
IL-33 promoter activity compared with the A-allele in lung
cancer cells, and the G-allele of rs928413 included a binding site
for cyclic AMP-responsive element-binding protein 1, which is
likely an activator of IL-33 transcription in the presence
of this allele (76). These studies
have demonstrated the potential regulatory effects of these SNPs on
the transcription of IL-33.
Although a number of atopic conditions are
considered to arise from altered Th2 immune responses against
environmental allergens (77,78),
the involvement of IL-33 in this process has attracted attention in
recent years. In addition to targeting altered Th2 immune responses
and their downstream effectors, it has been recognized that the
successful prevention of allergic airway diseases is important to
alleviate public health and economic burdens (79-81).
Therefore, pathways mediating the initiation of allergic airway
responses have been extensively studied, and IL-33 has been
indicated to be a potential target molecule for the early control
and prevention of allergic airway inflammation (82-84).
In the current study, the association between IL-33 and AR was
demonstrated, and novel data regarding the genetic basis of AR in
the Han Chinese population were analyzed. Subsequent analyses
validating these findings may reveal further genetic susceptibility
to AR. Moreover, GWAS are still required to determine the shared
and unique mechanisms of AR compared with other atopic
diseases.
Not applicable.
No funding was received.
All data generated or analyzed during this study are
included in this published article.
HR and HX conceived the present study, conducted the
primary experiments and analyses. XZ, LG and SL performed
additional experiments. HR and XZ collected clinical samples. HR
and HX drafted the manuscript. All authors read and approved the
final manuscript.
The present study was approved by the Ethics
Committees of The Second Hospital of Jingzhou, Hubei College of
Chinese Medicine and Shishou People's Hospital. All investigation
procedures were performed in agreement with the Declaration of
Helsinki. Oral informed consent was provided by the participants or
their guardians.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Bousquet J, Khaltaev N, Cruz AA, Denburg
J, Fokkens WJ, Togias A, Zuberbier T, Baena-Cagnani CE, Canonica
GW, van Weel C, et al: Allergic rhinitis and its impact on asthma
(ARIA) 2008 update (in collaboration with the world health
organization, GA(2)LEN and AllerGen). Allergy. 63 (Suppl
86):S8–S160. 2008.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Leynaert B, Neukirch F, Demoly P and
Bousquet J: Epidemiologic evidence for asthma and rhinitis
comorbidity. J Allergy Clin Immunol. 106 (5 Suppl):S201–S205.
2000.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Bousquet J, Van Cauwenberge P and Khaltaev
N: Aria Workshop Group; World Health Organization. Allergic
rhinitis and its impact on asthma. J Allergy Clin Immunol. 108 (5
Suppl):S147–S334. 2001.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Falade AG, Ige OM, Yusuf BO, Onadeko MO
and Onadeko BO: Trends in the prevalence and severity of symptoms
of asthma, allergic rhinoconjunctivitis, and atopic eczema. J Natl
Med Assoc. 101:414–418. 2009.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Nathan RA: The burden of allergic
rhinitis. Allergy Asthma Proc. 28:3–9. 2007.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Worldwide variation in prevalence of
symptoms of asthma, allergic rhinoconjunctivitis, atopic eczema.
ISAAC. The international study of asthma and allergies in childhood
(ISAAC) steering committee. Lancet. 351:1225–1232. 1998.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Haahtela T, Holgate S, Pawankar R, Akdis
CA, Benjaponpitak S, Caraballo L, Demain J, Portnoy J and von
Hertzen L: WAO Special Committee on Climate Change and
Biodiversity. The biodiversity hypothesis and allergic disease:
World allergy organization position statement. World Allergy Organ
J. 6(3)2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Wang XD, Zheng M, Lou HF, Wang CS, Zhang
Y, Bo MY, Ge SQ, Zhang N, Zhang L and Bachert C: An increased
prevalence of self-reported allergic rhinitis in major Chinese
cities from 2005 to 2011. Allergy. 71:1170–1180. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Thompson AK, Juniper E and Meltzer EO:
Quality of life in patients with allergic rhinitis. Ann Allergy
Asthma Immunol. 85:338–347; quiz 347-8. 2000.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Dunlop J, Matsui E and Sharma HP: Allergic
rhinitis: Environmental determinants. Immunol Allergy Clin North
Am. 36:367–377. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Greiner AN, Hellings PW, Rotiroti G and
Scadding GK: Allergic rhinitis. Lancet. 378:2112–2122.
2011.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Portelli MA, Hodge E and Sayers I: Genetic
risk factors for the development of allergic disease identified by
genome-wide association. Clin Exp Allergy. 45:21–31.
2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Willemsen G, van Beijsterveldt TC, van
Baal CG, Postma D and Boomsma DI: Heritability of self-reported
asthma and allergy: A study in adult Dutch twins, siblings and
parents. Twin Res Hum Genet. 11:132–142. 2008.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Fagnani C, Annesi-Maesano I, Brescianini
S, D'Ippolito C, Medda E, Nisticò L, Patriarca V, Rotondi D,
Toccaceli V and Stazi MA: Heritability and shared genetic effects
of asthma and hay fever: An Italian study of young twins. Twin Res
Hum Genet. 11:121–131. 2008.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Bunyavanich S, Schadt EE, Himes BE,
Lasky-Su J, Qiu W, Lazarus R, Ziniti JP, Cohain A, Linderman M,
Torgerson DG, et al: Integrated genome-wide association,
coexpression network, and expression single nucleotide polymorphism
analysis identifies novel pathway in allergic rhinitis. BMC Med
Genomics. 7(48)2014.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Andiappan AK, Wang de Y, Anantharaman R,
Parate PN, Suri BK, Low HQ, Li Y, Zhao W, Castagnoli P, Liu J and
Chew FT: Genome-wide association study for atopy and allergic
rhinitis in a Singapore Chinese population. PLoS One.
6(e19719)2011.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Fujii R, Hishida A, Wu MC, Kondo T,
Hattori Y, Naito M, Endoh K, Nakatochi M, Hamajima N, Kubo M, et
al: Genome-wide association study for pollinosis identified two
novel loci in interleukin (IL)-1B in a Japanese population. Nagoya
J Med Sci. 80:109–120. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Shen Y, Liu Y, Wang XQ, Ke X, Kang HY and
Hong SL: Association between TNFSF4 and BLK gene polymorphisms and
susceptibility to allergic rhinitis. Mol Med Rep. 16:3224–3232.
2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhang Y, Lin X, Desrosiers M, Zhang W,
Meng N, Zhao L, Han D and Zhang L: Association pattern of
interleukin-1 receptor-associated kinase-4 gene polymorphisms with
allergic rhinitis in a Han Chinese population. PLoS One.
6(e21769)2011.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ke X, Song S, Wang X, Shen Y, Kang H and
Hong S: Associations of single nucleotide polymorphisms of PTPN22
and Ctla4 genes with the risk of allergic rhinitis in a Chinese Han
population. Hum Immunol. 78:227–231. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Kanazawa J, Masuko H, Yatagai Y, Sakamoto
T, Yamada H, Kitazawa H, Iijima H, Naito T, Saito T, Noguchi E, et
al: Association analyses of eQTLs of the TYRO3 gene and allergic
diseases in Japanese populations. Allergol Int. 68:77–81.
2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Ke X, Yang Y, Shen Y, Wang X and Hong S:
Association between TNFAIP3 gene polymorphisms and risk of allergic
rhinitis in a Chinese Han population. Iran J Allergy Asthma
Immunol. 15:46–52. 2016.PubMed/NCBI
|
|
23
|
Nilsson D, Andiappan AK, Hallden C, Tim
CF, Säll T, Wang de Y and Cardell LO: Poor reproducibility of
allergic rhinitis SNP associations. PLoS One.
8(e53975)2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Waage J, Standl M, Curtin JA, Jessen LE,
Thorsen J, Tian C and Schoettler N: 23andMe Research Team; AAGC
collaborators, Flores C, et al. Genome-wide association and
HLA fine-mapping studies identify risk loci and genetic pathways
underlying allergic rhinitis. Nat Genet. 50:1072–1080.
2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Li J, Zhang Y and Zhang L: Discovering
susceptibility genes for allergic rhinitis and allergy using a
genome-wide association study strategy. Curr Opin Allergy Clin
Immunol. 15:33–40. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Romagnani S: The increased prevalence of
allergy and the hygiene hypothesis: Missing immune deviation,
reduced immune suppression, or both? Immunology. 112:352–363.
2004.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Romagnani S: Lymphokine production by
human T cells in disease states. Annu Rev Immunol. 12:227–257.
1994.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Abbas AK, Murphy KM and Sher A: Functional
diversity of helper T lymphocytes. Nature. 383:787–793.
1996.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Li L, Xia Y, Nguyen A, Lai YH, Feng L,
Mosmann TR and Lo D: Effects of Th2 cytokines on chemokine
expression in the lung: IL-13 potently induces eotaxin expression
by airway epithelial cells. J Immunol. 162:2477–2487.
1999.PubMed/NCBI
|
|
30
|
Durham SR, Ying S, Varney VA, Jacobson MR,
Sudderick RM, Mackay IS, Kay AB and Hamid QA: Cytokine messenger
RNA expression for IL-3, IL-4, IL-5, and
granulocyte/macrophage-colony-stimulating factor in the nasal
mucosa after local allergen provocation: Relationship to tissue
eosinophilia. J Immunol. 148:2390–2394. 1992.PubMed/NCBI
|
|
31
|
Bischoff SC, Sellge G, Lorentz A, Sebald
W, Raab R and Manns MP: IL-4 enhances proliferation and mediator
release in mature human mast cells. Proc Natl Acad Sci USA.
96:8080–8085. 1999.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Hardman CS, Panova V and McKenzie AN:
IL-33 citrine reporter mice reveal the temporal and spatial
expression of IL-33 during allergic lung inflammation. Eur J
Immunol. 43:488–498. 2013.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Bystrom J, Patel SY, Amin K and
Bishop-Bailey D: Dissecting the role of eosinophil cationic protein
in upper airway disease. Curr Opin Allergy Clin Immunol. 12:18–23.
2012.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Prefontaine D, Nadigel J, Chouiali F,
Audusseau S, Semlali A, Chakir J, Martin JG and Hamid Q: Increased
IL-33 expression by epithelial cells in bronchial asthma. J Allergy
Clin Immunol. 125:752–754. 2010.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Lloyd CM: IL-33 family members and
asthma-bridging innate and adaptive immune responses. Curr Opin
Immunol. 22:800–806. 2010.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Shimizu M, Matsuda A, Yanagisawa K, Hirota
T, Akahoshi M, Inomata N, Ebe K, Tanaka K, Sugiura H, Nakashima K,
et al: Functional SNPs in the distal promoter of the ST2 gene are
associated with atopic dermatitis. Hum Mol Genet. 14:2919–2927.
2005.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Grotenboer NS, Ketelaar ME, Koppelman GH
and Nawijn MC: Decoding asthma: Translating genetic variation in
IL-33 and IL1RL1 into disease pathophysiology. J Allergy Clin
Immunol. 131:856–865. 2013.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Watanabe M, Nakamoto K, Inui T, Sada M,
Honda K, Tamura M, Ogawa Y, Yokoyama T, Saraya T, Kurai D, et al:
Serum sST2 levels predict severe exacerbation of asthma. Respir
Res. 19(169)2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Li R, Yang G, Yang R, Peng X and Li J:
Interleukin-33 and receptor ST2 as indicators in patients with
asthma: A meta-analysis. Int J Clin Exp Med. 8:14935–14943.
2015.PubMed/NCBI
|
|
40
|
Schmitz J, Owyang A, Oldham E, Song Y,
Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X, et
al: IL-33, an interleukin-1-like cytokine that signals via the IL-1
receptor-related protein ST2 and induces T helper type 2-associated
cytokines. Immunity. 23:479–490. 2005.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Oshikawa K, Yanagisawa K, Tominaga S and
Sugiyama Y: Expression and function of the ST2 gene in a murine
model of allergic airway inflammation. Clin Exp Allergy.
32:1520–1526. 2002.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Lohning M, Stroehmann A, Coyle AJ, Grogan
JL, Lin S, Gutierrez-Ramos JC, Levinson D, Radbruch A and Kamradt
T: T1/ST2 is preferentially expressed on murine Th2 cells,
independent of interleukin 4, interleukin 5, and interleukin 10,
and important for Th2 effector function. Proc Natl Acad Sci USA.
95:6930–6935. 1998.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Coyle AJ, Lloyd C, Tian J, Nguyen T,
Erikkson C, Wang L, Ottoson P, Persson P, Delaney T, Lehar S, et
al: Crucial role of the interleukin 1 receptor family member T1/ST2
in T helper cell type 2-mediated lung mucosal immune responses. J
Exp Med. 190:895–902. 1999.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Xu D, Chan WL, Leung BP, Huang FP, Wheeler
R, Piedrafita D, Robinson JH and Liew FY: Selective expression of a
stable cell surface molecule on type 2 but not type 1 helper T
cells. J Exp Med. 187:787–794. 1998.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Lecart S, Lecointe N, Subramaniam A, Alkan
S, Ni D, Chen R, Boulay V, Pène J, Kuroiwa K, Tominaga S and Yssel
H: Activated, but not resting human Th2 cells, in contrast to Th1
and T regulatory cells, produce soluble ST2 and express low levels
of ST2L at the cell surface. Eur J Immunol. 32:2979–2987.
2002.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Trajkovic V, Sweet MJ and Xu D: T1/ST2-an
IL-1 receptor-like modulator of immune responses. Cytokine Growth
Factor Rev. 15:87–95. 2004.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Ding W, Zou GL, Zhang W, Lai XN, Chen HW
and Xiong LX: Interleukin-33: Its emerging role in allergic
diseases. Molecules. 23(1665)2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Haenuki Y, Matsushita K,
Futatsugi-Yumikura S, Ishii KJ, Kawagoe T, Imoto Y, Fujieda S,
Yasuda M, Hisa Y, Akira S, et al: A critical role of IL-33 in
experimental allergic rhinitis. J Allergy Clin Immunol. 130:184–194
e11. 2012.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Gluck J, Rymarczyk B and Rogala B: Serum
IL-33 but not ST2 level is elevated in intermittent allergic
rhinitis and is a marker of the disease severity. Inflamm Res.
61:547–550. 2012.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Sakashita M, Yoshimoto T, Hirota T, Harada
M, Okubo K, Osawa Y, Fujieda S, Nakamura Y, Yasuda K, Nakanishi K
and Tamari M: Association of serum interleukin-33 level and the
interleukin-33 genetic variant with Japanese cedar pollinosis. Clin
Exp Allergy. 38:1875–1881. 2008.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Du Y, Luo Y, Yang C, Liu J, Wan J and Wang
K: Discussion IL-33 and its receptor ST2 associated with the
pathogenesis of allergic rhinitis. Lin Chuang Er Bi Yan Hou Tou
Jing Wai Ke Za Zhi. 29:811–814. 2015.PubMed/NCBI(In Chinese).
|
|
52
|
Guo-Zhu H, Xi-Ling Z, Zhu W, Li-Hua W, Dan
H, Xiao-Mu W, Wen-Yun Z and Wei-Xu H: Therapeutic potential of
combined anti-IL-1β IgY and anti-TNF-α IgY in guinea pigs with
allergic rhinitis induced by ovalbumin. Int Immunopharmacol.
25:155–161. 2015.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Asaka D, Yoshikawa M, Nakayama T,
Yoshimura T, Moriyama H and Otori N: Elevated levels of
interleukin-33 in the nasal secretions of patients with allergic
rhinitis. Int Arch Allergy Immunol. 158 (Suppl 1):S47–S50.
2012.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Kim YH, Yang TY, Park CS, Ahn SH, Son BK,
Kim JH, Lim DH and Jang TY: Anti-IL-33 antibody has a therapeutic
effect in a murine model of allergic rhinitis. Allergy. 67:183–190.
2012.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Kamekura R, Kojima T, Takano K, Go M,
Sawada N and Himi T: The role of IL-33 and its receptor ST2 in
human nasal epithelium with allergic rhinitis. Clin Exp Allergy.
42:218–228. 2012.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Shaw JL, Fakhri S, Citardi MJ, Porter PC,
Corry DB, Kheradmand F, Liu YJ and Luong A: IL-33-responsive innate
lymphoid cells are an important source of IL-13 in chronic
rhinosinusitis with nasal polyps. Am J Respir Crit Care Med.
188:432–439. 2013.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Soyka MB, Holzmann D, Basinski TM,
Wawrzyniak M, Bannert C, Bürgler S, Akkoc T, Treis A, Rückert B,
Akdis M, et al: The induction of IL-33 in the sinus epithelium and
its influence on T-helper cell responses. PLoS One.
10(e0123163)2015.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Baumann R, Rabaszowski M, Stenin I,
Tilgner L, Gaertner-Akerboom M, Scheckenbach K, Wiltfang J, Chaker
A, Schipper J and Wagenmann M: Nasal levels of soluble IL-33R ST2
and IL-16 in allergic rhinitis: Inverse correlation trends with
disease severity. Clin Exp Allergy. 43:1134–1143. 2013.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Position paper: Allergen standardization
and skin tests The European academy of allergology and clinical
immunology. Allergy. 48:48–82. 1993.PubMed/NCBI
|
|
60
|
1000 Genomes Project Consortium. Auton A,
Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL,
McCarthy S, McVean GA and Abecasis GR. A global reference for human
genetic variation. Nature. 526:68–74. 2015.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Torgerson DG, Ampleford EJ, Chiu GY,
Gauderman WJ, Gignoux CR, Graves PE, Himes BE, Levin AM, Mathias
RA, Hancock DB, et al: Meta-analysis of genome-wide association
studies of asthma in ethnically diverse North American populations.
Nat Genet. 43:887–892. 2011.PubMed/NCBI View
Article : Google Scholar
|
|
62
|
Demenais F, Margaritte-Jeannin P, Barnes
KC, Cookson WOC, Altmüller J, Ang W, Barr RG, Beaty TH, Becker AB,
Beilby J, et al: Multiancestry association study identifies new
asthma risk loci that colocalize with immune-cell enhancer marks.
Nat Genet. 50:42–53. 2018.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Zhu Z, Lee PH, Chaffin MD, Chung W, Loh
PR, Lu Q, Christiani DC and Liang L: A genome-wide cross-trait
analysis from UK Biobank highlights the shared genetic architecture
of asthma and allergic diseases. Nat Genet. 50:857–864.
2018.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Bonnelykke K, Sleiman P, Nielsen K,
Kreiner-Møller E, Mercader JM, Belgrave D, den Dekker HT, Husby A,
Sevelsted A, Faura-Tellez G, et al: A genome-wide association study
identifies CDHR3 as a susceptibility locus for early childhood
asthma with severe exacerbations. Nat Genet. 46:51–55.
2014.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Barrett JC, Fry B, Maller J and Daly MJ:
Haploview: Analysis and visualization of LD and haplotype maps.
Bioinformatics. 21:263–265. 2005.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Shi YY and He L: SHEsis, a powerful
software platform for analyses of linkage disequilibrium, haplotype
construction, and genetic association at polymorphism loci. Cell
Res. 15:97–98. 2005.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z,
He L and Shi Y: A partition-ligation-combination-subdivision EM
algorithm for haplotype inference with multiallelic markers: Update
of the SHEsis. (http://analysis.bio-x.cn).
Cell Res. 19:519–523. 2009.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Lemonnier N, Melen E, Jiang Y, Joly S,
Ménard C, Aguilar D, Acosta-Perez E, Bergström A, Boutaoui N,
Bustamante M, et al: A novel whole blood gene expression signature
for asthma, dermatitis, and rhinitis multimorbidity in children and
adolescents. Allergy 2020 (Epub ahead of print).
|
|
69
|
Rogala B and Gluck J: The role of
interleukin-33 in rhinitis. Curr Allergy Asthma Rep. 13:196–202.
2013.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Tomita K, Sakashita M, Hirota T, Tanaka S,
Masuyama K, Yamada T, Fujieda S, Miyatake A, Hizawa N, Kubo M, et
al: Variants in the 17q21 asthma susceptibility locus are
associated with allergic rhinitis in the Japanese population.
Allergy. 68:92–100. 2013.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Li Y, Chen J, Rui X, Li N, Jiang F and
Shen J: The association between sixteen genome-wide association
studies-related allergic diseases loci and childhood allergic
rhinitis in a Chinese Han population. Cytokine. 111:162–170.
2018.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Yoon D, Ban HJ, Kim YJ, Kim EJ, Kim HC,
Han BG, Park JW, Hong SJ, Cho SH, Park K and Lee JS: Replication of
genome-wide association studies on asthma and allergic diseases in
Korean adult population. BMB Rep. 45:305–310. 2012.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Amarin JZ, Naffa RG, Suradi HH, Alsaket
YM, Obeidat NM, Mahafza TM and Zihlif MA: An intronic
single-nucleotide polymorphism (rs13217795) in FOXO3 is associated
with asthma and allergic rhinitis: A case-case-control study. BMC
Med Genet. 18(132)2017.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Li X, Hastie AT, Hawkins GA, Moore WC,
Ampleford EJ, Milosevic J, Li H, Busse WW, Erzurum SC, Kaminski N,
et al: eQTL of bronchial epithelial cells and bronchial alveolar
lavage deciphers GWAS-identified asthma genes. Allergy.
70:1309–1318. 2015.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Ketelaar ME, Portelli MA, Dijk FN, Shrine
N, Faiz A, Vermeulen CJ, Xu CJ, Hankinson J, Bhaker S, Henry AP, et
al: Phenotypic and functional translation of IL-33 genetics in
asthma. J Allergy Clin Immunol 2020 (Epub ahead of print).
|
|
76
|
Gorbacheva AM, Korneev KV, Kuprash DV and
Mitkin NA: The risk G allele of the single-nucleotide polymorphism
rs928413 creates a CREB1-Binding site that activates IL-33 promoter
in lung epithelial cells. Int J Mol Sci. 19(2911)2018.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Paller AS, Spergel JM, Mina-Osorio P and
Irvine AD: The atopic march and atopic multimorbidity: Many
trajectories, many pathways. J Allergy Clin Immunol. 143:46–55.
2019.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Akdis CA, Arkwright PD, Bruggen MC, Busse
W, Gadina M, Guttman-Yassky E, Kabashima K, Mitamura Y, Vian L, Wu
J and Palomares O: Type 2 immunity in the skin and lungs. Allergy.
75:1582–1605. 2020.PubMed/NCBI View Article : Google Scholar
|
|
79
|
D'Amato G, Ortega OPM, Annesi-Maesano I
and D'Amato M: Prevention of allergic asthma with allergen
avoidance measures and the role of exposome. Curr Allergy Asthma
Rep. 20(8)2020.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Liccardi G, Cazzola M, Walter Canonica G,
Passalacqua G and D'Amato G: New insights in allergen avoidance
measures for mite and pet sensitized patients. A critical
appraisal. Respir Med. 99:1363–1376. 2005.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Halken S: Prevention of allergic disease
in childhood: Clinical and epidemiological aspects of primary and
secondary allergy prevention. Pediatr Allergy Immunol. 15 (Suppl
16):4–5, 9-32. 2004.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Cook J and Saglani S: Pathogenesis and
prevention strategies of severe asthma exacerbations in children.
Curr Opin Pulm Med. 22:25–31. 2016.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Chen WY, Tsai TH, Yang JL and Li LC:
Therapeutic strategies for targeting IL-33/ST2 signalling for the
treatment of inflammatory diseases. Cell Physiol Biochem.
49:349–358. 2018.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Takatori H, Makita S, Ito T, Matsuki A and
Nakajima H: Regulatory mechanisms of IL-33-ST2-Mediated allergic
inflammation. Front Immunol. 9(2004)2018.PubMed/NCBI View Article : Google Scholar
|