With increasing economic development, chronic
non-communicable diseases have emerged as a substantial global
concern due to common risk factors, such as unhealthy diets and
environmental pollution (1,2). Although a range of pharmacological and
surgical interventions are constantly being devised to address the
increased numbers of cases of non-communicable diseases, the side
effects and contraindications of certain medicines or radiotherapy
have limited the number of patients that are able to receive such
treatments (3,4). Furthermore, the unavoidable
postoperative complications such as surgical site infection,
abscess, active bleeding, hematoma and anastomotic leak, resulting
from surgery may worsen a patient's state (5). Therefore, researchers have begun to
consider other possibilities to cope with this global problem
(6,7). The term dysbiosis refers to the major
changes in the gut microbial ecosystems that contribute to a range
of metabolic disorders, including obesity, type 1 and type 2
diabetes and inflammatory bowel disease (8-11).
Numerous other types of disease like autism or allergies have also
been associated with an imbalance in gut microflora composition
(12,13). Several strategies to normalize human
gut microbial ecosystems are available to treat different
syndromes, including fecal microbiota transplantation, which has
demonstrated promising results (14). At present, probiotics are commonly
used to improve the intestinal environment (15-17).
To further determine the relationship between the
gut microflora and a healthy human state, the use of metagenomics
and metatranscriptomic sequencing techniques has been proven to
provide a compositional snapshot of the microbial species in the
gut, as well as to sequence their expressed genes (18,19).
With the elucidated genetic background of different gut microflora,
it also provides a wide range of possibilities to modulate the gut
microflora composition genetically for greater therapy. Advances in
synthetic biology have extended this therapeutic potential, as
selected bacteria can be tailored to deliver drugs or molecules to
act directly on the host (20). An
increasing abundance of human-associated bacteria have been
identified to provide health benefits, including Lactococcus
lactis (L. lactis), Escherichia coli (E.
coli) and Bifidobacterium, making them desirable
engineering targets for therapeutic application (21-27).
Similar efforts may also be applied to Akkermansia
muciniphila (A. muciniphila), a microbial species that
has been proposed as a novel candidate for probiotic therapy
(28). In the present review paper,
the potential of A. muciniphila as an engineered bacterium
was discussed by first reviewing other engineered bacteria. The
review covered its colonization sites in human intestines,
therapeutic effects and probiotic characteristics, prior to
identifying potential avenues for modification.
The most commonly engineered bacteria provide a
platform to develop probiotics as a novel direction in therapeutic
studies. In the following sections, some of these cases are
discussed in further detail. By noting similarities in their
therapeutic effects, characterization status and the strategies
used for their modification, the current review aimed to
demonstrate why A. muciniphila is being considered for
similar engineering approaches.
LAB is a commensal intestinal microbiota species
with widespread use in the production of fermented foods (33,34).
By virtue of its numerous health-promoting effects in humans and
its decoded genetic sequence, LAB has become one of the most
convincing engineered probiotics (35-37).
In 2015, Yang et al (27)
constructed the recombinant strain LAB plantarum (L.
plantarum) NC8, which expresses angiotensin-converting enzyme
inhibitory peptides (ACEIPs) for prolonging antihypertensive
effects; the recombinant expression vector pSIP409-ACEIP was built
by replacing the gusA gene in the pSIP409 plasmid with genes
encoding ACEIPs. Through incubating its DNA with available
methyltransferases in vitro to match the host's DNA
methylation patterns, the transformation efficiency of L.
plantarum was raised to a level comparable with that of E.
coli. This vector was subsequently transformed into L.
plantarum NC8. An antihypertensive effect was noted following
the oral administration of the engineered strain to spontaneously
hypertensive rats, as evidenced by a reduction in abnormal systolic
blood pressure and in triglyceride, endothelin and angiotensin II
levels. For further consideration, the expression levels of the
integrated genes should be monitored and regulated (38).
Different promoter-repressor systems have been
constructed for the induction of recombinant protein expression in
L. plantarum to evaluate their stability and efficiency
(39). An increasing number of
systems have emerged, such as the quorum-sensing system,
chemical-based induction system and temperature-sensitive system,
which enhanced the abilities of microbes to sense, respond to and
record their local environment, as well as improving the ability to
evaluate and control the expression levels of the desired genes,
which are designed to produce the required product (40-42).
Globally, the prevalence of excess weight between
the years 1980 and 2013 has increased to 27.5% in adults and 47% in
children, with 2.1 billion people in the world classifying as
overweight (BMI >25 kg/m2) and over 500 million being
classified as obese (BMI >30 kg/m2) (66). Obesity has become a worldwide health
concern, with current medical and lifestyle interventions largely
failing to offer adequate solutions. Increasing evidence has
indicated that probiotics are involved in gut barrier maintenance
and inflammation normalization, suggesting that their adoption
could eventually result in a long-term treatment for obesity
(67,68).
The prevalence of diabetes has increased in parallel
with the global rise in obesity, with type 2 diabetes accounting
for >90% of all cases of diabetes (73-75).
Both obesity and type 2 diabetes have been associated with changes
in nutrition and more sedentary lifestyles, thus adopting A.
muciniphila interventions for the treatment of diabetes has
been hypothesized to exert similar therapeutic implications
(10,76). Previous studies reported that
prediabetic patients and patients with type 2 diabetes had lower
amounts of A. muciniphila in the gut compared with healthy
individuals (77,78). The relationship between A.
muciniphila and type 2 diabetes was also insinuated following
metformin treatment, which induced high levels of A.
muciniphila in a previous study (79). Notably, Depommier et al
(72) observed more significant
improvements to insulin sensitivity and reductions in insulinemia
following the use of pasteurized, instead of live, A.
muciniphila.
Autism spectrum disorder (ASD) is a complex
neurodevelopmental disorder in which gastrointestinal disturbances
are commonly reported (85).
Through the analysis of fecal samples, Wang et al (86) reported a decreased abundance of
A. muciniphila in children with ASD and their siblings, as
well as a thinner gastrointestinal mucus barrier compared with
control subjects. Other previous studies have also indicated that
intestinal barrier impairment was aggravated in children with ASD
and their immediate relatives, suggesting that A.
muciniphila may guide the implementation of dietary
interventions to reduce gut permeability in individuals with
ASD.
In the majority of the studies discussed, when
supplied in a viable form, therapeutic effects of A.
muciniphila were noted for metabolic disorders. However, such
treatment could also extend to other diseases. For example, in
cancer treatment, A. muciniphila employment was suggested to
enhance the effects of immunotherapy (87,88).
The fecal matter of patients with cancer with positive responses to
immunotherapy has been studied for A. muciniphila, as an
abundance of the bacteria can reflect the state of immunotherapy
(87). In addition, A.
muciniphila was also reported to exhibit protective effects in
immune-mediated diseases, including atopic diseases, IBD and liver
damage (65,89,90).
The association between A. muciniphila and immune-mediated
diseases was explained using whole transcriptome analysis of
intestinal tissue samples, which indicated that A.
muciniphila regulated the expression of the majority of the
genes associated with immune responses (90-94).
The advent of next-generation sequencing and
whole-genome sequencing has provided additional scope for more
bacteria to be genetically modified. Based on this, the prospects
for engineering A. muciniphila are promising.
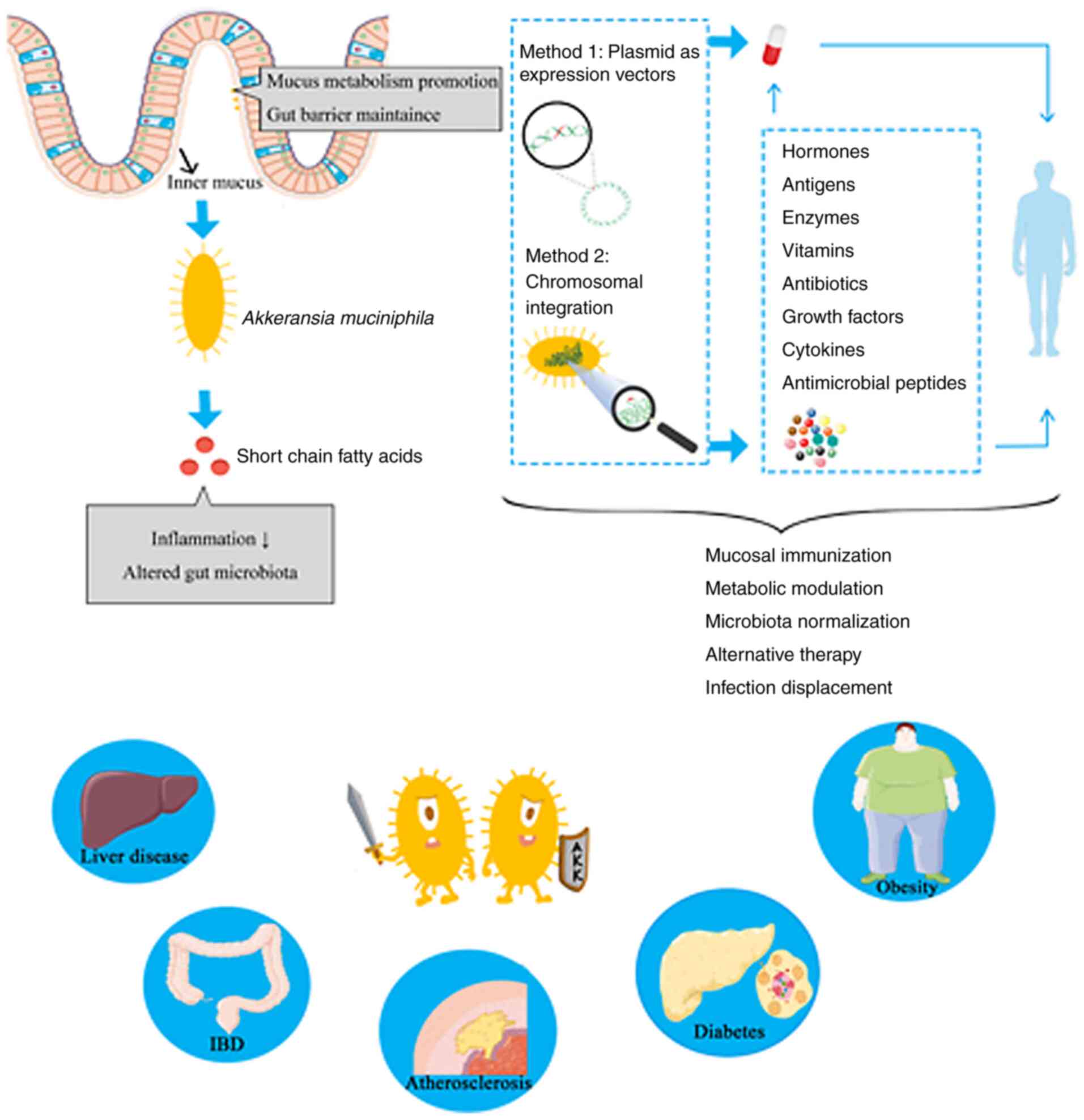
Moreover, an efficient and scalable workflow for the
cultivation and preservation of A. muciniphila cells has
been developed, resulting in viable Akkermansia colonies
with high yields and very high stability, as well as up to
97.9±4.5% survival of >1 year when stored in glycerol-amended
medium at -80˚C (98). The growth
of A. muciniphila can be monitored and controlled by various
quality assessment and control procedures to ensure that viable
cells of A. muciniphila are available. In addition, although
A. muciniphila is an anaerobic bacterium, it has
demonstrated an ability tolerate and even benefit from nanomolar
concentrations of oxygen in liquid medium (99). These properties extend the
possibility of A. muciniphila to be manipulated for
engineering (Fig. 1).
In general, plasmids are the first tool considered
when genome editing is required. Plasmids contain appropriate DNA
as the bacterial origin of replication, an antibiotic resistance
cassette and the gene of interest, which is transcribed from a
prokaryotic promoter (100,101).
Adequate expression of the therapeutic gene or genes is ensured by
using appropriate promoters and other regulatory elements (100,101). In previous years, the genetic
toolbox of plasmids has been greatly expanded by adding sensors,
regulators, memory circuits, delivery devices and kill switches
(102). Once the recombinant
plasmid carrying the desired gene tracks down signal molecules
secreted by target cells or tissues, it releases therapeutics
locally, and is subsequently self-digested as programmed to avoid
any infection (103,104). After construction, plasmids are
converted to the hosts by chemical, mechanical or physical
techniques, with mammalian cell ‘poration’ systems (electroporation
and sonoporation) being the most important and common techniques
used (105-107).
In addition, extra genome integration in a
chromosome of the host cell has been discovered to support the
development of engineered bacteria (108). Normally, a designed homologous
single-stranded DNA donor is provided based on the introduction of
a site-specific double-strand DNA break (DSB) into the locus of
interest (109). Information
encoded on this template can be used to repair the DSB, resulting
in the addition of the desired gene at the site of the break
(109). Recombination systems
carried by helper plasmids are crucial during this process
(109-112).
In the following sections, several mature recombination systems
developed in LAB or E. coli are described, which
could be applied to A. muciniphila once limitations relating
to species differences have been eliminated.
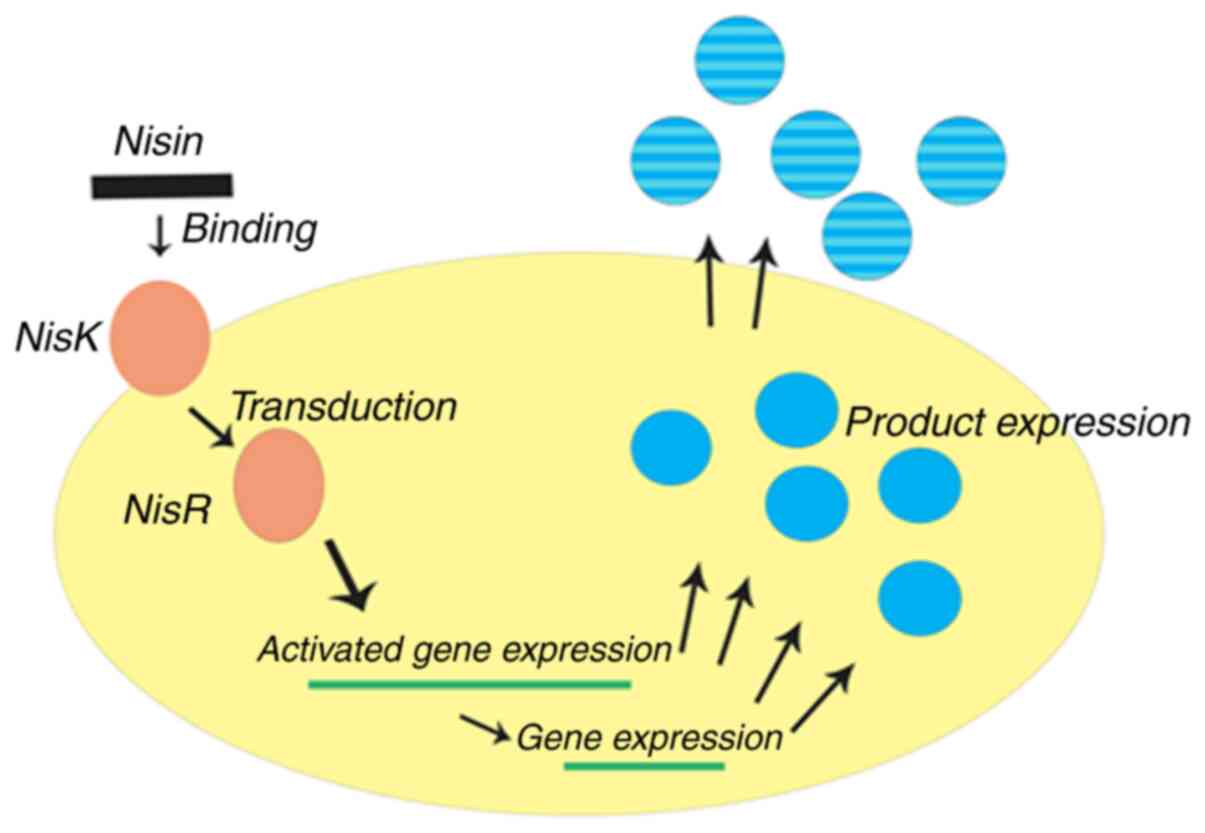
The NICE system is one of the most widely used tools
for chromosomal integration exploited for engineering
Lactobacillus. It is constructed for gene expression based
on nisA and nisF promoters via a two-component regulatory system
consisting of the histidine protein kinase, nisK, and the response
regulator, nisR (113-116).
When a gene of interest is placed behind the inducible promoter,
PnisA, on a plasmid and transformed into a nisRK strain, the
expression of the cloned gene can be activated by the addition of
nisin (Fig. 2). Using the dual
plasmid system, the classic NICE system can be successfully
introduced into the majority of bacteria. For example, Mohseni
et al (117) genetically
engineered L. lactis using a NICE system with pNZ8148 to
express the native and codon-optimized recombinant E7 [E7 is a good
candidate protein for vaccine development against human
papillomavirus (HPV)-related cervical cancer] oncogenes isolated
from HPV; the results for the overall production of E7 by L.
lactis NZ9000 containing codon-optimized E7 was >2.7-fold
higher compared with NZ9000 containing the native E7 strain. The
findings also indicated that the amount of recombinant E7
oncoprotein accumulation depended on the concentration of nisin
added, with the highest concentration achieved in the presence of
10 ng/ml nisin for both recombinant L. lactis strains.
However, the exposed drawback of the system was that its basal
expression was leaky; therefore, it may not applicable for
production of the desired proteins or for the expression of toxic
proteins (118).
The bacteriophage λ Red homologous recombination
system has been studied over the past 50 years as a model system
for the transfer of chromosomal DNA from species (119). The λ recombination system,
designated ‘Red,’ consists of two proteins; α, an exonuclease that
acts on double-stranded (ds)DNA, and β, a single-stranded (ss)DNA
binding protein capable of annealing complementary ssDNA strands
(120). Red-mediated recombination
is assisted by the γ protein, which increases α and β activity on
linear dsDNA by inhibiting E. coli RecBCD exonuclease
(121,122). In the past, NICE restricted the
integration of molecular weight DNA into the host strain; however,
the new lambda Red recombinase-mediated integration strategy was
found to transform higher molecular weight DNA of variable lengths
into any non-essential locus in the host chromosome (123). Juhas and Ajioka (124) successfully integrated 15 kB DNA
encoding sucrose catabolism and lactose metabolism and transport
operons into the flsu locus of the flagellar region 3b in the E.
coli K12 MG1655 chromosome; this approach preferred the use of
overlapping DNA fragments for integrating the high molecular weight
DNA. Elongation of the integrated DNA sequence is facilitated by
the alternative use of kan and cat-yfp cassettes tagged in
different DNA fragments, which is less time-consuming compared with
the standard lambda Red recombinase-mediated integration (124). Under monitoring, this new strategy
did not reveal any negative effects on the host strain. However,
compared with E. coli, to the best of our knowledge, there
are fewer reports regarding the use of this technique on other
strains.
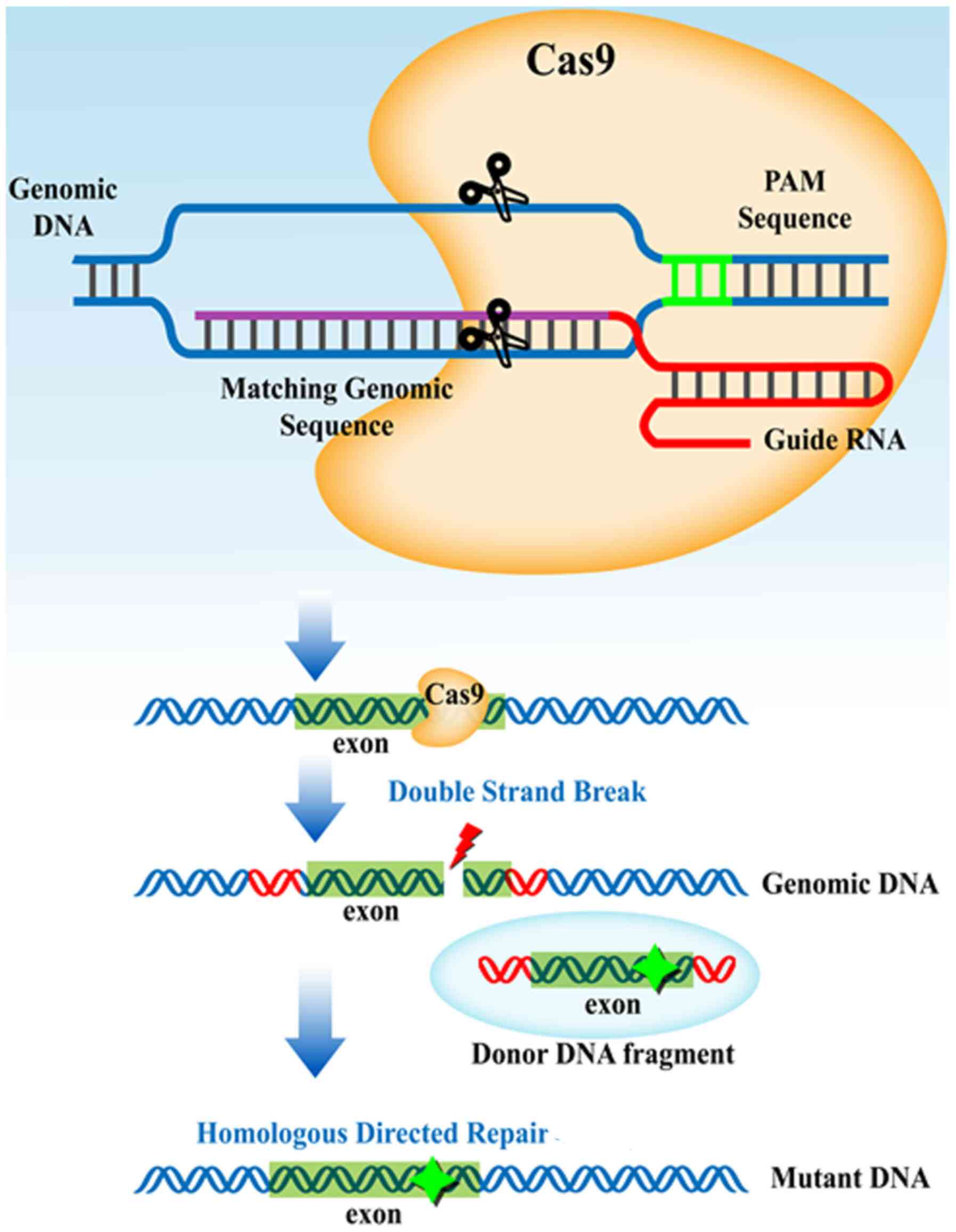
In the CRISPR-Cas system, the small CRISPR RNAs
encoded by CRISPR spacer sequences form a duplex with a
trans-activating CRISPR RNA. The duplex with the Cas9 protein
subsequently searches the presented DNA for a Cas-specific sequence
(Fig. 3) (125-127).
Upon recognition of the specific sequence, Cas9 induces the
targeted DSB, enabling the modification of a target gene sequence
through host bacterial DNA repairing systems (128,129). The presence of a homologous
template ensures the insertion of the addition in the region of the
DSB. CRISPR-based technologies have been implemented for E.
coli, Streptococcus pneumoniae, L. lactis and
probiotic LAB species for the production of pharmaceutical
products and precursors of high industrial significance (130-132).
Various types of CRISPR are currently used for producing desired
strains with therapeutic potentials. Δ-integration CRISPR enables
strains to have multiple loci chromosomal integration, whereas
CRISPR-based homology-directed repair allows site-specific
integration (133). A
catalytically inactive form of Cas9 (dCas9), has been developed to
direct the promoter or coding regions to prevent transcription
rather than cleaving the DNA, known as CRISPR interference
(78). This technique has been used
to control gene expression in Corynebacterium glutamicum, in
which it was employed to downregulate multiple genes by
concatenating single guide RNA sequences encoded on one plasmid
(134). Genomic sequencing of the
A. muciniphila strain determined the CRISPR loci, suggesting
that the A. muciniphila system initiates the CRISPR
defensive mechanism frequently and can be modified using
CRISPR-Cas9(95). An automated
pipeline named CRISPR discovery has since been developed for the
identification of CRISPR repeats and Cas genes in genome
assemblies, to determine the type and subtype and to describe
system completeness (135). With
this knowledge, it is hypothesized that an endogenous CRISPR-Cas9
system can be developed for A. muciniphila, allowing it to
avoid the host's immune system.
Not applicable.
No funding was received.
Not applicable.
TC conceived the idea for the review and designed
its framework. YZ conducted the research and wrote the manuscript.
Both authors edited the manuscript. All authors read and approved
the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
WHO and UNICEF. Health in the post-2015
development agenda report of the Global Thematic Consultation on
Health. World We Want. 31:514–526. 2014.
|
|
2
|
Dora C, Haines A, Balbus J, Fletcher E,
Adair-Rohani H, Alabaster G, Hossain R, de Onis M, Branca F and
Neira M: Indicators linking health and sustainability in the
post-2015 development agenda. Lancet. 385:380–391. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Novelli G, Biancolella M, Latini A,
Spallone A, Borgiani P and Papaluca M: Precision medicine in
non-communicable diseases. High-Throughput. 9(3)2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Imaoka T, Nishimura M, Daino K, Takabatake
M, Moriyama H, Nishimura Y, Morioka T, Shimada Y and Kakinuma S:
Risk of second cancer after ion beam radiotherapy: Insights from
animal carcinogenesis studies. Int J Radiat Biol. 95:1431–1440.
2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
O'Malley RB and Revels JW: Imaging of
abdominal postoperative complications. Radiol Clin North Am.
58:73–91. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Suri S, Kumar V, Kumar S, Goyal A, Tanwar
B and Kaur J and Kaur J: DASH dietary pattern: A treatment for
non-communicable diseases. Curr Hypertens Rev. 16:108–114.
2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zhu J, Yang J and Luo Y: Applications of
engineered intestinal bacteria in disease diagnosis and treatment.
Sheng Wu Gong Cheng Xue Bao. 35:2350–2366. 2019.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
8
|
Parséus A, Sommer N, Sommer F, Caesar R,
Molinaro A, Ståhlman M, Greiner TU, Perkins R and Bäckhed F:
Microbiota-induced obesity requires farnesoid X receptor. Gut.
66:429–437. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Kostic AD, Gevers D, Siljander H, Vatanen
T, Hyötyläinen T, Hämäläinen AM, Peet A, Tillmann V, Pöhö P,
Mattila I, et al: The dynamics of the human infant gut microbiome
in development and in progression toward type 1 diabetes. Cell Host
Microbe. 17:260–273. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F,
Liang S, Zhang W, Guan Y, Shen D, et al: A metagenome-wide
association study of gut microbiota in type 2 diabetes. Nature.
490:55–60. 2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhang M, Sun K, Wu Y, Yang Y, Tso P and Wu
Z: Interactions between intestinal microbiota and host immune
response in inflammatory bowel disease. Front Immunol.
8(942)2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Lu T, Chen Y, Guo Y, Sun J, Shen W, Yuan
M, Zhang S, He P and Jiao X: Altered gut microbiota diversity and
composition in chronic urticaria. Dis Markers.
2019(6417471)2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Cryan JF, O'Riordan KJ, Sandhu K, Peterson
V and Dinan TG: The gut microbiome in neurological disorders.
Lancet Neurol. 19:179–194. 2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Jiménez-Avalos JA, Arrevillaga-Boni G,
González-López L, García-Carvajal ZY and González-Avila M:
Classical methods and perspectives for manipulating the human gut
microbial ecosystem. Crit Rev Food Sci Nutr: Mar 2, 2020 (Epub
ahead of print). doi: 10.1080/10408398.2020.1724075.
|
|
15
|
Szajewska H: What are the indications for
using probiotics in children? Arch Dis Child. 101:398–403.
2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Chua KJ, Kwok WC, Aggarwal N, Sun T and
Chang MW: Designer probiotics for the prevention and treatment of
human diseases. Curr Opin Chem Biol. 40:8–16. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Sanders ME, Akkermans LMA, Haller D,
Hammerman C, Heimbach J, Hörmannsperger G, Huys G, Levy DD,
Lutgendorff F, Mack D, et al: Safety assessment of probiotics for
human use. Gut Microbes. 1:164–185. 2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Khangwal I and Shukla P: Combinatory
biotechnological intervention for gut microbiota. Appl Microbiol
Biotechnol. 103:3615–3625. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yadav M and Shukla P: Recent systems
biology approaches for probiotics use in health aspects: A review.
3 Biotech. 9(448)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Kumar M, Yadav AK, Verma V, Singh B, Mal
G, Nagpal R and Hemalatha R: Bioengineered probiotics as a new hope
for health and diseases: An overview of potential and prospects.
Future Microbiol. 11:585–600. 2016.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yadav R, Singh PK and Shukla P: Metabolic
engineering for probiotics and their genome-wide expression
profiling. Curr Protein Pept Sci. 19:68–74. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Steidler L, Hans W, Schotte L, Neirynck S,
Obermeier F, Falk W, Fiers W and Remaut E: Treatment of murine
colitis by Lactococcus lactis secreting interleukin-10.
Science. 289:1352–1355. 2000.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Braat H, Rottiers P, Hommes DW,
Huyghebaert N, Remaut E, Remon JP, van Deventer SJ, Neirynck S,
Peppelenbosch MP and Steidler L: A phase I trial with transgenic
bacteria expressing interleukin-10 in Crohn's disease. Clin
Gastroenterol Hepatol. 4:754–759. 2006.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Kurtz CB, Millet YA, Puurunen MK,
Perreault M, Charbonneau MR, Isabella VM, Kotula JW, Antipov E,
Dagon Y, Denney WS, et al: An engineered E. Coli Nissle
improves hyperammonemia and survival in mice and shows
dose-dependent exposure in healthy humans. Sci Transl Med.
11(eaau7975)2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Saltzman DA, Katsanis E, Heise CP, Hasz
DE, Vigdorovich V, Kelly SM, Curtiss R III, Leonard AS and Anderson
PM: Antitumor mechanisms of attenuated Salmonella
typhimurium containing the gene for human interleukin-2: A
novel antitumor agent? J Pediatr Surg. 32:301–306. 1997.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Fang X, Tian P, Zhao X, Jiang C and Chen
T: Neuroprotective effects of an engineered commensal bacterium in
the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine Parkinson disease
mouse model via producing glucagon-like peptide-1. J Neurochem.
160:441–452. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Yang G, Jiang Y, Yang W, Du F, Yao Y, Shi
C and Wang C: Effective treatment of hypertension by recombinant
Lactobacillus plantarum expressing angiotensin converting
enzyme inhibitory peptide. Microb Cell Fact. 14(202)2015.
|
|
28
|
Cani PD and de Vos WM: Next-generation
beneficial microbes: The case of Akkermansia muciniphila.
Front Microbiol. 8(1765)2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Rodríguez V, Asenjo JA and Andrews BA:
Design and implementation of a high yield production system for
recombinant expression of peptides. Microb Cell Fact.
13(65)2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Sahdev S, Khattar SK and Saini KS:
Production of active eukaryotic proteins through bacterial
expression systems: A review of the existing biotechnology
strategies. Mol Cell Biochem. 307:249–264. 2008.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Chance RE and Frank BH: Research,
development, production, and safety of biosynthetic human insulin.
Diabetes Care. 16 (Suppl 3):S133–S142. 1993.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Whelan RA, Rausch S, Ebner F, Günzel D,
Richter JF, Hering NA, Schulzke JD, Kühl AA, Keles A, Janczyk P, et
al: A transgenic probiotic secreting a parasite immunomodulator for
site-directed treatment of gut inflammation. Mol Ther.
22:1730–1740. 2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zoetendal EG, Vaughan EE and De Vos WM: A
microbial world within us. Mol Microbiol. 59:1639–1650.
2006.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Coeuret V, Dubernet S, Bernardeau M,
Gueguen M and Vernoux JP: Isolation, characterisation and
identification of lactobacilli focusing mainly on cheeses and other
dairy products. Lait. 83:269–306. 2003.
|
|
35
|
Kikuchi Y, Kunitoh-Asari A, Hayakawa K,
Imai S, Kasuya K, Abe K, Adachi Y, Fukudome S, Takahashi Y and
Hachimura S: Oral administration of Lactobacillus plantarum
strain AYA enhances IgA secretion and provides survival protection
against influenza virus infection in mice. PLoS One.
9(e86416)2014.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Sazawal S, Hiremath G, Dhingra U, Malik P,
Deb S and Black RE: Efficacy of probiotics in prevention of acute
diarrhoea: A meta-analysis of masked, randomised,
placebo-controlled trials. Lancet Infect Dis. 6:374–382.
2006.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Wolvers D, Antoine JM, Myllyluoma E,
Schrezenmeir J, Szajewska H and Rijkers GT: Guidance for
substantiating the evidence for beneficial effects of probiotics:
Prevention and management of infections by probiotics. J Nutr.
140:S698–S712. 2010.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Spath K, Heinl S and Grabherr R: ‘Direct
cloning in Lactobacillus plantarum: Electroporation with
non-methylated plasmid DNA enhances transformation efficiency and
makes shuttle vectors obsolete’. Microb Cell Fact.
11(141)2012.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Heiss S, Hörmann A, Tauer C, Sonnleitner
M, Egger E, Grabherr R and Heinl S: Evaluation of novel inducible
promoter/repressor systems for recombinant protein expression in
Lactobacillus plantarum. Microb Cell Fact.
15(50)2016.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Yadav R and Shukla P: An overview of
advanced technologies for selection of probiotics and their
expediency: A review. Crit Rev Food Sci Nutr. 57:3233–3242.
2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Allain T, Mansour NM, Bahr MMA, Martin R,
Florent I, Langella P and Bermúdez-Humarán LG: A new lactobacilli
in vivo expression system for the production and delivery of
heterologous proteins at mucosal surfaces. FEMS Microbiol Lett.
363(fnw117)2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Rong G, Corrie SR and Clark HA: In vivo
biosensing: Progress and perspectives. ACS Sens. 2:327–338.
2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Rowland IR, Rumney CJ, Coutts JT and
Lievense LC: Effect of Bifidobacterium longum and inulin on
gut bacterial metabolism and carcinogen-induced aberrant crypt foci
in rats. Carcinogenesis. 19:281–285. 1998.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Rafter J, Bennett M, Caderni G, Clune Y,
Hughes R, Karlsson PC, Klinder A, O'Riordan M, O'Sullivan GC,
Pool-Zobel B, et al: Dietary synbiotics reduce cancer risk factors
in polypectomized and colon cancer patients. Am J Clin Nutr.
85:488–496. 2007.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Le Leu RK, Hu Y, Brown IL, Woodman RJ and
Young GP: Synbiotic intervention of Bifidobacterium lactis
and resistant starch protects against colorectal cancer development
in rats. Carcinogenesis. 31:246–251. 2010.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Bae EA, Han MJ, Song MJ and Kim DH:
Purification of Rotavirus infection-inhibitory protein from
Bifidobacterium breve K-110. J Microbiol Biotechnol.
12:553–556. 2002.
|
|
47
|
Patole SK, Rao SC, Keil AD, Nathan EA,
Doherty DA and Simmer KN: Benefits of Bifidobacterium breve
M-16V Supplementation in preterm neonates-A retrospective cohort
study. PLoS One. 11(e0150775)2016.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Venturi A, Gionchetti P, Rizzello F,
Johansson R, Zucconi E, Brigidi P, Matteuzzi D and Campieri M:
Impact on the composition of the faecal flora by a new probiotic
preparation: Preliminary data on maintenance treatment of patients
with ulcerative colitis. Aliment Pharmacol Ther. 13:1103–1108.
1999.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Yadav R, Kumar V, Baweja M and Shukla P:
Gene editing and genetic engineering approaches for advanced
probiotics: A review. Crit Rev Food Sci Nutr. 58:1735–1746.
2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Wei C, Xun AY, Wei XX, Yao J, Wang JY, Shi
RY, Yang GH, Li YX, Xu ZL, Lai MG, et al: Bifidobacteria expressing
tumstatin protein for antitumor therapy in tumor-bearing mice.
Technol Cancer Res Treat. 15:498–508. 2015.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Xu YF, Zhu LP, Hu B, Fu GF, Zhang HY, Wang
JJ and Xu GX: A new expression plasmid in Bifidobacterium
longum as a delivery system of endostatin for cancer gene
therapy. Cancer Gene Ther. 14:151–157. 2007.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Zhu LP, Yin Y, Xing J, Li C, Kou L, Hu B,
Wu ZW, Wang JJ and Xu GX: Therapeutic efficacy of
Bifidobacterium longum-mediated human granulocyte
colony-stimulating factor and/or endostatin combined with
cyclophosphamide in mouse-transplanted tumors. Cancer Sci.
100:1986–1990. 2009.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Hu B, Kou L, Li C, Zhu LP, Fan YR, Wu ZW,
Wang JJ and Xu GX: Bifidobacterium longum as a delivery
system of TRAIL and endostatin cooperates with chemotherapeutic
drugs to inhibit hypoxic tumor growth. Cancer Gene Ther.
16:655–663. 2009.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Bolhassani A and Zahedifard F: Therapeutic
live vaccines as a potential anticancer strategy. Int J Cancer.
131:1733–1743. 2012.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Chen T, Zhao X, Ren Y, Wang Y, Tang X,
Tian P, Wang H and Xin H: Triptolide modulates tumour-colonisation
and anti-tumour effect of attenuated Salmonella encoding DNase I.
Appl Microbiol Biotechnol. 103:929–939. 2019.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Shahabi V, Reyes-Reyes M, Wallecha A,
Rivera S, Paterson Y and MacIag P: Development of a Listeria
monocytogenes based vaccine against prostate cancer. Cancer
Immunol Immunother. 57:1301–1313. 2008.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Chen Y, Yang D, Li S, Gao Y, Jiang R, Deng
L, Frankel FR and Sun B: Development of a Listeria
monocytogenes-based vaccine against hepatocellular carcinoma.
Oncogene. 31:2140–2152. 2012.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Chan CT, Lee JW, Cameron DE, Bashor CJ and
Collins JJ: ‘Deadman’ and ‘Passcode’ microbial kill switches for
bacterial containment. Nat Chem Biol. 12:82–86. 2016.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Derrien M, Vaughan EE, Plugge CM and de
Vos WM: Akkermansia municiphila gen. nov., sp. nov., a human
intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol.
54:1469–1476. 2004.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Reunanen J, Kainulainen V, Huuskonen L,
Ottman N, Belzer C, Huhtinen H, de Vos WM and Satokari R:
Akkermansia muciniphila adheres to enterocytes and
strengthens the integrity of the epithelial cell layer. Appl
Environ Microbiol. 81:3655–3662. 2015.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Ottman N, Geerlings SY, Aalvink S, de Vos
WM and Belzer C: Action and function of Akkermansia
muciniphila in microbiome ecology, health and disease. Best
Pract Res Clin Gastroenterol. 31:637–642. 2017.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Thibault R, Blachier F, Darcy-Vrillon B,
De Coppet P, Bourreille A and Segain JP: Butyrate utilization by
the colonic mucosa in inflammatory bowel diseases: A transport
deficiency. Inflamm Bowel Dis. 16:684–695. 2010.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Puertollano E, Kolida S and Yaqoob P:
Biological significance of short-chain fatty acid metabolism by the
intestinal microbiome. Curr Opin Clin Nutr Metab Care. 17:139–144.
2014.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Ottman N, Reunanen J, Meijerink M, Pietilä
TE, Kainulainen V, Klievink J, Huuskonen L, Aalvink S, Skurnik M,
Boeren S, et al: Pili-like proteins of Akkermansia
muciniphila modulate host immune responses and gut barrier
function. PLoS One. 12(e0173004)2017.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Plovier H, Everard A, Druart C, Depommier
C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T,
Lichtenstein L, et al: A purified membrane protein from
Akkermansia muciniphila or the pasteurized bacterium
improves metabolism in obese and diabetic mice. Nat Med.
23:107–113. 2017.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Ng M, Fleming T, Robinson M, Thomson B,
Graetz N, Margono C, Mullany EC, Biryukov S, Abbafati C, Abera SF,
et al: Global, regional, and national prevalence of overweight and
obesity in children and adults during 1980-2013: A systematic
analysis for the Global Burden of Disease Study 2013. Lancet.
384:766–781. 2014.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Cani PD, Neyrinck AM, Fava F, Knauf C,
Burcelin RG, Tuohy KM, Gibson GR and Delzenne NM: Selective
increases of bifidobacteria in gut microflora improve
high-fat-diet-induced diabetes in mice through a mechanism
associated with endotoxaemia. Diabetologia. 50:2374–2383.
2007.PubMed/NCBI View Article : Google Scholar
|
|
68
|
GBD 2015 Obesity Collaborators. Afshin A,
Forouzanfar MH, Reitsma MB, Sur P, Estep K, Lee A, Marczak L,
Mokdad AH, Moradi-Lakeh M, et al: Health effects of overweight and
obesity in 195 countries over 25 years. N Engl J Med. 377:13–27.
2017.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Santacruz A, Collado MC, García-Valdés L,
Segura MT, Martín-Lagos JA, Anjos T, Martí-Romero M, Lopez RM,
Florido J, Campoy C and Sanz Y: Gut microbiota composition is
associated with body weight, weight gain and biochemical parameters
in pregnant women. Br J Nutr. 104:83–92. 2010.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Karlsson CL, Önnerfält J, Xu J, Molin G,
Ahrné S and Thorngren-Jerneck K: The microbiota of the gut in
preschool children with normal and excessive body weight. Obesity
(Silver Spring). 20:2257–2261. 2012.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Everard A, Belzer C, Geurts L, Ouwerkerk
JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne
NM, et al: Cross-talk between Akkermansia muciniphila and
intestinal epithelium controls diet-induced obesity. Proc Natl Acad
Sci USA. 110:9066–9071. 2013.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Depommier C, Everard A, Druart C, Plovier
H, Van Hul M, Vieira-Silva S, Falony G, Raes J, Maiter D, Delzenne
NM, et al: Supplementation with Akkermansia muciniphila in
overweight and obese human volunteers: A proof-of-concept
exploratory study. Nat Med. 25:1096–1103. 2019.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Hu FB, Manson JAE and Willett WC: Types of
dietary fat and risk of coronary heart disease: A critical review.
J Am Coll Nutr. 20:5–19. 2001.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Almdal T, Scharling H, Jensen JS and
Vestergaard H: The independent effect of type 2 diabetes mellitus
on ischemic heart disease, stroke, and death: A population-based
study of 13,000 men and women with 20 years of follow-up. Arch
Intern Med. 164:1422–1426. 2004.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Maleckas A, Venclauskas L, Wallenius V and
Fändriks HL: Metabolic surgery in the treatment of type 2 diabetes
mellitus. Oxford Textb Endocrinol Diabetes. 61:257–264. 2011.
|
|
76
|
Cani PD, Amar J, Iglesias MA, Poggi M,
Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, et
al: Metabolic endotoxemia initiates obesity and insulin resistance.
Diabetes. 56:1761–1772. 2007.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Zhang X, Shen D, Fang Z, Jie Z, Qiu X,
Zhang C, Chen Y and Ji L: Human gut microbiota changes reveal the
progression of glucose intolerance. PLoS One.
8(e711108)2013.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Hansen CH, Krych L, Nielsen DS, Vogensen
FK, Hansen LH, Sørensen SJ, Buschard K and Hansen AK: Early life
treatment with vancomycin propagates Akkermansia muciniphila
and reduces diabetes incidence in the NOD mouse. Diabetologia.
55:2285–2294. 2012.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Forslund K, Hildebrand F, Nielsen T,
Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-Silva S,
Gudmundsdottir V, Pedersen HK, et al: Disentangling type 2 diabetes
and metformin treatment signatures in the human gut microbiota.
Nature. 528:262–266. 2015.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Tedgui A and Mallat Z: Cytokines in
atherosclerosis: Pathogenic and regulatory pathways. Physiol Rev.
86:515–581. 2006.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Li J, Zhao F, Wang Y, Chen J, Tao J, Tian
G, Wu S, Liu W, Cui Q, Geng B, et al: Gut microbiota dysbiosis
contributes to the development of hypertension. Microbiome.
5(14)2017.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Jonsson AL and Bäckhed F: Role of gut
microbiota in atherosclerosis. Nat Rev Cardiol. 14:79–87.
2017.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Barrington WT and Lusis AJ:
Atherosclerosis: Association between the gut microbiome and
atherosclerosis. Nat Rev Cardiol. 14:699–700. 2017.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Li J, Lin S, Vanhoutte PM, Woo CW and Xu
A: Akkermansia muciniphila protects against atherosclerosis
by preventing metabolic endotoxemia-induced inflammation in
Apoe-/- Mice. Circulation. 133:2434–2446.
2016.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Campion D, Ponzo P, Alessandria C, Saracco
GM and Balzola F: The role of microbiota in autism spectrum
disorders. Minerva Gastroenterol Dietol. 64:333–350.
2018.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Wang L, Christophersen CT, Sorich MJ,
Gerber JP, Angley MT and Conlon MA: Low relative abundances of the
mucolytic bacterium Akkermansia muciniphila and
Bifidobacterium spp. in feces of children with autism. Appl
Environ Microbiol. 77:6718–6721. 2011.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Naito Y, Uchiyama K and Takagi T: A
next-generation beneficial microbe: Akkermansia muciniphila.
J Clin Biochem Nutr. 63:33–35. 2018.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Routy B, Le Chatelier E, Derosa L, Duong
CPM, Alou MT, Daillère R, Fluckiger A, Messaoudene M, Rauber C,
Roberti MP, et al: Gut microbiome influences efficacy of PD-1-based
immunotherapy against epithelial tumors. Science. 359:91–97.
2018.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Zheng H, Liang H, Wang Y, Miao M, Shi T,
Yang F, Liu E, Yuan W, Ji ZS and Li DK: Altered gut microbiota
composition associated with eczema in infants. PLoS One.
11(e0166026)2016.PubMed/NCBI View Article : Google Scholar
|
|
90
|
Wu W, Lv L, Shi D, Ye J, Fang D, Guo F, Li
Y, He X and Li L: Protective effect of Akkermansia
muciniphila against immune-mediated liver injury in a mouse
model. Front Microbiol. 8(1804)2017.PubMed/NCBI View Article : Google Scholar
|
|
91
|
Png CW, Lindén SK, Gilshenan KS, Zoetendal
EG, McSweeney CS, Sly LI, McGuckin MA and Florin TH: Mucolytic
bacteria with increased prevalence in IBD mucosa augment in vitro
utilization of mucin by other bacteria. Am J Gastroenterol.
105:2420–2428. 2010.PubMed/NCBI View Article : Google Scholar
|
|
92
|
Wang HX, Liu M, Weng SY, Li JJ, Xie C, He
HL, Guan W, Yuan YS and Gao J: Immune mechanisms of Concanavalin a
model of autoimmune hepatitis. World J Gastroenterol. 18:119–125.
2012.PubMed/NCBI View Article : Google Scholar
|
|
93
|
Drell T, Larionova A, Voor T, Simm J,
Julge K, Heilman K, Tillmann V, Štšepetova J and Sepp E:
Differences in gut microbiota between atopic and healthy children.
Curr Microbiol. 71:177–183. 2015.PubMed/NCBI View Article : Google Scholar
|
|
94
|
Lukovac S, Belzer C, Pellis L, Keijser BJ,
de Vos WM, Montijn RC and Roeselers G: Differential modulation by
Akkermansia muciniphila and Faecalibacterium
prausnitzii of host peripheral lipid metabolism and histone
acetylation in mouse gut organoids. mBio. 5:e01438–14.
2014.PubMed/NCBI View Article : Google Scholar
|
|
95
|
van Passel MWJ, Kant R, Zoetendal EG,
Plugge CM, Derrien M, Malfatti SA, Chain PS, Woyke T, Palva A, de
Vos WM and Smidt H: The genome of Akkermansia muciniphila, a
dedicated intestinal mucin degrader, and its use in exploring
intestinal metagenomes. PLoS One. 6(e16876)2011.PubMed/NCBI View Article : Google Scholar
|
|
96
|
Caputo A, Dubourg G, Croce O, Gupta S,
Robert C, Papazian L, Rolain JM and Raoult D: Whole-genome assembly
of Akkermansia muciniphila sequenced directly from human
stool. Biol Direct. 10(5)2015.PubMed/NCBI View Article : Google Scholar
|
|
97
|
Guo X, Li S, Zhang J, Wu F, Li X, Wu D,
Zhang M, Ou Z, Jie Z, Yan Q, et al: Genome sequencing of 39
Akkermansia muciniphila isolates reveals its population
structure, genomic and functional diverisity, and global
distribution in mammalian gut microbiotas. BMC Genomics.
18(800)2017.PubMed/NCBI View Article : Google Scholar
|
|
98
|
Ouwerkerk JP, Aalvink S, Belzer C and De
Vos WM: Preparation and preservation of viable Akkermansia
muciniphila cells for therapeutic interventions. Benef
Microbes. 8:163–169. 2017.PubMed/NCBI View Article : Google Scholar
|
|
99
|
Ouwerkerk JP, van der Ark KCH, Davids M,
Claassens NJ, Finestra TR, de Vos WM and Belzer C: Adaptation of
Akkermansia muciniphila to the oxic-anoxic interface of the
mucus layer. Appl Environ Microbiol. 82:6983–6993. 2016.PubMed/NCBI View Article : Google Scholar
|
|
100
|
Vectors DP: Suicide gene therapy of
cancer. Mol Ther. 3:S98–S115. 2001.
|
|
101
|
Baban CK, Cronin M, O'Hanlon D, O'Sullivan
GC and Tangney M: Bacteria as vectors for gene therapy of cancer.
Bioeng Bugs. 1:385–394. 2010.PubMed/NCBI View Article : Google Scholar
|
|
102
|
Pedrolli DB, Ribeiro NV, Squizato PN, de
Jesus VN and Cozetto DA: Team AQA Unesp at iGEM 2017. Engineering
microbial living therapeutics: The synthetic biology toolbox.
Trends Biotechnol. 37:100–115. 2019.PubMed/NCBI View Article : Google Scholar
|
|
103
|
Waller MC, Bober JR, Nair NU and Beisel
CL: Toward a genetic tool development pipeline for host-associated
bacteria. Curr Opin Microbiol. 38:156–164. 2017.PubMed/NCBI View Article : Google Scholar
|
|
104
|
Riglar DT and Silver PA: Engineering
bacteria for diagnostic and therapeutic applications. Nat Rev
Microbiol. 16:214–225. 2018.PubMed/NCBI View Article : Google Scholar
|
|
105
|
Welker DL, Hughes JE, Steele JL and
Broadbent R: High efficiency electrotransformation of
Lactobacillus casei. FEMS Microbiol Lett. 362:1–6.
2015.PubMed/NCBI View Article : Google Scholar
|
|
106
|
Walsh M, Tangney M, O'Neill MJ, Larkin JO,
Soden DM, McKenna SL, Darcy R, O'Sullivan GC and O'Driscoll CM:
Evaluation of cellular uptake and gene transfer efficiency of
pegylated poly-L-lysine compacted DNA: Implications for cancer gene
therapy. Mol Pharm. 3:644–653. 2006.PubMed/NCBI View Article : Google Scholar
|
|
107
|
Ahmad S, Casey G, Sweeney P, Tangney M and
O'Sullivan GC: Optimised electroporation mediated DNA vaccination
for treatment of prostate cancer. Genet Vaccines Ther.
8(1)2010.PubMed/NCBI View Article : Google Scholar
|
|
108
|
Kado CI: Historical events that spawned
the field of plasmid biology. Microbiol Spectr 2, 2014.
|
|
109
|
St-Pierre F, Cui L, Priest DG, Endy D,
Dodd IB and Shearwin KE: One-step cloning and chromosomal
integration of DNA. ACS Synth Biol. 2:537–541. 2013.PubMed/NCBI View Article : Google Scholar
|
|
110
|
Urnov FD, Rebar EJ, Holmes MC, Zhang HS
and Gregory PD: Genome editing with engineered zinc finger
nucleases. Nat Rev Genet. 11:636–646. 2010.PubMed/NCBI View Article : Google Scholar
|
|
111
|
Miller JC, Tan S, Qiao G, Barlow KA, Wang
J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al: A TALE
nuclease architecture for efficient genome editing. Nat Biotechnol.
29:143–148. 2011.PubMed/NCBI View Article : Google Scholar
|
|
112
|
Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G,
Forest CR and Church GM: Programming cells by multiplex genome
engineering and accelerated evolution. Nature. 460:894–898.
2009.PubMed/NCBI View Article : Google Scholar
|
|
113
|
Kuipers OP, Beerthuyzen MM, de Ruyter PG,
Luesink EJ and de Vos WM: Autoregulation of nisin biosynthesis in
lactococcus lactis by signal transduction. J Biol Chem.
270:27299–27304. 1995.PubMed/NCBI View Article : Google Scholar
|
|
114
|
Van der Meer JR, Polman J, Beerthuyzen MM,
Siezen RJ, Kuipers OP and De Vos WM: Characterization of the
Lactococcus lactis nisin A operon genes nisP, encoding a
subtilisin-like serine protease involved in precursor processing,
and nisR, encoding a regulatory protein involved in nisin
biosynthesis. J Bacteriol. 175:2578–2588. 1993.PubMed/NCBI View Article : Google Scholar
|
|
115
|
Engelke G, Gutowski-Eckel Z, Kiesau P,
Siegers K, Hammelmann M and Entian KD: Regulation of nisin
biosynthesis and immunity in Lactococcus lactis 6F3. Appl
Environ Microbiol. 60:814–825. 1994.PubMed/NCBI View Article : Google Scholar
|
|
116
|
Kleerebezem M, Bongers R, Rutten G, Vos
WMD and Kuipers OP: Autoregulation of subtilin biosynthesis in
Bacillus subtilis: The role of the spa-box in
subtilin-responsive promoters. Peptides. 25:1415–1424.
2004.PubMed/NCBI View Article : Google Scholar
|
|
117
|
Mohseni AH, Razavilar V, Keyvani H, Razavi
MR and Khavari-Nejad RA: Efficient production and optimization of
E7 oncoprotein from Iranian human papillomavirus type 16 in
Lactococcus lactis using nisin-controlled gene expression
(NICE) system. Microb Pathog. 110:554–560. 2017.PubMed/NCBI View Article : Google Scholar
|
|
118
|
Van Hoang V, Ochi T, Kurata K, Arita Y,
Ogasahara Y and Enomoto K: Nisin-induced expression of recombinant
T cell epitopes of major Japanese cedar pollen allergens in
Lactococcus lactis. Appl Microbiol Biotechnol. 102:261–268.
2018.PubMed/NCBI View Article : Google Scholar
|
|
119
|
Kaiser AD: A genetic study of the
temperate coliphage λ. Virology. 1:424–443. 1955.PubMed/NCBI View Article : Google Scholar
|
|
120
|
Carter DM and Radding CM: The role of
exonuclease and beta protein of phage lambda in genetic
recombination. II. Substrate specificity and the mode of action of
lambda exonuclease. J Biol Chem. 246:2502–2512. 1971.PubMed/NCBI
|
|
121
|
Murphy KC: Lambda Gam protein inhibits the
helicase and chi-stimulated recombination activities of
Escherichia coli RecBCD enzyme. J Bacteriol. 173:5808–5821.
1991.PubMed/NCBI View Article : Google Scholar
|
|
122
|
Karu AE, Sakaki Y, Echols H and Linn S:
The gamma protein specified by bacteriophage gamma. Structure and
inhibitory activity for the recBC enzyme of Escherichia
coli. J Biol Chem. 250:7377–7387. 1975.PubMed/NCBI
|
|
123
|
Murphy KC: λ recombination and
recombineering. EcoSal Plus 7, 2016.
|
|
124
|
Juhas M and Ajioka JW: Lambda Red
recombinase-mediated integration of the high molecular weight DNA
into the Escherichia coli chromosome. Microb Cell Fact.
15(172)2016.PubMed/NCBI View Article : Google Scholar
|
|
125
|
Deltcheva E, Chylinski K, Sharma CM,
Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J and Charpentier
E: CRISPR RNA maturation by trans-encoded small RNA and host factor
RNase III. Nature. 471:602–607. 2011.PubMed/NCBI View Article : Google Scholar
|
|
126
|
Bolotin A, Quinquis B, Sorokin A and Dusko
Ehrlich S: Clustered regularly interspaced short palindrome repeats
(CRISPRs) have spacers of extrachromosomal origin. Microbiology
(Reading). 151:2551–2561. 2005.PubMed/NCBI View Article : Google Scholar
|
|
127
|
Deveau H, Barrangou R, Garneau JE, Labonté
J, Fremaux C, Boyaval P, Romero DA, Horvath P and Moineau S: Phage
response to CRISPR-encoded resistance in Streptococcus
thermophilus. J Bacteriol. 190:1390–1400. 2008.PubMed/NCBI View Article : Google Scholar
|
|
128
|
Tong Y, Charusanti P, Zhang L, Weber T and
Lee SY: CRISPR-Cas9 based engineering of actinomycetal genomes. ACS
Synth Biol. 4:1020–1029. 2015.PubMed/NCBI View Article : Google Scholar
|
|
129
|
Garneau JE, Dupuis MÈ, Villion M, Romero
DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH and
Moineau S: The CRISPR/Cas bacterial immune system cleaves
bacteriophage and plasmid DNA. Nature. 468:67–71. 2010.PubMed/NCBI View Article : Google Scholar
|
|
130
|
Jiang W, Bikard D, Cox D, Zhang F and
Marraffini LA: RNA-guided editing of bacterial genomes using
CRISPR-Cas systems. Nat Biotechnol. 31:233–239. 2013.PubMed/NCBI View Article : Google Scholar
|
|
131
|
Oh JH and Van Pijkeren JP:
CRISPR-Cas9-assisted recombineering in Lactobacillus
reuteri. Nucleic Acids Res. 42(e131)2014.PubMed/NCBI View Article : Google Scholar
|
|
132
|
van der Els S, James JK, Kleerebezem M and
Bron PA: Versatile Cas9-driven subpopulation selection toolbox for
Lactococcus lactis. Appl Environ Microbiol. 84:e02752–17.
2018.PubMed/NCBI View Article : Google Scholar
|
|
133
|
Manghwar H, Lindsey K, Zhang X and Jin S:
CRISPR/Cas system: Recent advances and future prospects for genome
editing. Trends Plant Sci. 24:1102–1125. 2019.PubMed/NCBI View Article : Google Scholar
|
|
134
|
Gauttam R, Seibold GM, Mueller P, Weil T,
Weiß T, Handrick R and Eikmanns BJ: A simple dual-inducible CRISPR
interference system for multiple gene targeting in
Corynebacterium glutamicum. Plasmid. 103:25–35.
2019.PubMed/NCBI View Article : Google Scholar
|
|
135
|
Crawley AB, Henriksen JR and Barrangou R:
CRISPRdisco: An automated pipeline for the discovery and analysis
of CRISPR-Cas systems. CRISPR J. 1:171–181. 2018.PubMed/NCBI View Article : Google Scholar
|
|
136
|
Takei S, Omoto C, Kitagawa K, Morishita N,
Katayama T, Shigemura K, Fujisawa M, Kawabata M, Hotta H and
Shirakawa T: Oral administration of genetically modified
Bifidobacterium displaying HCV-NS3 multi-epitope fusion
protein could induce an HCV-NS3-specific systemic immune response
in mice. Vaccine. 32:3066–3074. 2014.PubMed/NCBI View Article : Google Scholar
|
|
137
|
Pathmakanthan S, Meance S and Edwards CA:
Probiotics: A review of human studies to date and methodological
approaches. Microb Ecol Health Dis. 12:10–30. 2000.
|
|
138
|
Lawenius L, Scheffler JM, Gustafsson KL,
Henning P, Nilsson KH, Colldén H, Islander U, Plovier H, Cani PD,
de Vos WM, et al: Pasteurized Akkermansia muciniphila
protects from fat mass gain but not from bone loss. Am J Physiol
Endocrinol Metab. 318:E480–E491. 2020.PubMed/NCBI View Article : Google Scholar
|