Introduction
Medulloblastoma (MB) is a small round blue cell
tumor of the cerebellum and is one of the most common types of
malignant brain tumors presented during childhood (1). MB is classified based on the
histological features into three major forms of the disease:
Classic, nodular/desmoplastic (ND) and large cell/anaplastic
(2). With recent advances in
genomics, gene expression profiling and DNA methylation analysis,
MB has been further divided into four major subgroups:
WNT/Wingless, Sonic Hedgehog (SHH), Group 3, and Group 4 (3,4). The
overall survival rates range from 60-80% (3). Current treatments for MB include
surgical resection, cranio-spinal radiation (for children older
than 3 years) and chemotherapy; however, long-term survivors of MB
will face significant treatment-related morbidity secondary to the
current treatments (4-6).
The significant complications and reduced quality of life caused by
conventional treatments remain an issue which need to be addressed.
As such, there has been much research interest aimed at dissecting
the molecular genetics underlying the disease.
Cancer is initiated and driven by aberrant genetic
events, such as copy number aberrations (CNAs), also called the
drivers of cancer (7). A number of
cancer types, such as glioblastoma, breast cancer and MB, contain
subtypes, with each subtype possessing distinct molecular profiles
and clinical outcomes (8-10).
In cancer research, driver genes have been defined as genes whose
structural or sequence mutations will confer a selective advantage
to the cancer cell (11,12). A number of driver genes have been
shown to have CNAs or associated changes in the gene expression
profile which may cause oncogenesis (13). A previous study found that driver
genes are essential in carcinogenesis and could be potential
targets for cancer therapy (14).
As such, the identification of tumor driver genes behind the
development of MB is of great interest.
The present study tested for tumor driver genes from
one patient with MB and selected their potential corresponding
targeted drugs. Meanwhile, the MB tissue was successfully implanted
into SCID mice which were used for subsequent drug screening.
Materials and methods
Human tissue sample
For the present study, MB tissue was obtained from a
Chinese patient from the Department of Neurosurgery, Brain and
Nerve Research Laboratory of The First Affiliated Hospital of
Soochow University (Suzhou, China).
The patient was a 2 years and 3 months old boy in
June 2015 when he went through surgery. The patient was treated at
the hospital because of frequent vomiting. The preoperative
examination [CT and MR (magnetic resonance)] revealed a tumor in
the fourth ventricle. The tumor was removed using the suboccipital
midline approach. This was his first operation. Before the
operation, the patient did not receive radiotherapy or
chemotherapy. The tumor specimen obtained during surgery was stored
in DMEM culture medium and transported to laboratory immediately.
The tumor specimen was crushed and used for implantation in SCID
mice.
Statement of informed consent
The human sample was used in accordance with the
policies of the institutional review board of The First Affiliated
Hospital of Soochow University (approval no. 8187101042). The
sample from the MB patient was used with the written consent of
parents.
Intracranial implanted models
A total of 10 female and 10 male SCID mice
(purchased from Shanghai Slack Laboratory Animal Co., Ltd.) which
were 4-6 weeks old were used for experiments. The mice were raised
in specific-pathogen free conditions and the temperature was
maintained between 26 and 28˚C. The mice were exposed to ~10 h
light per day and given food and water following high-temperature
sterilization. The mice were raised separately, but under the same
conditions. The animals had free access to food and water (ad
libitum), animal feed was nutritionally balanced. The weight of
the mouse was 15-20 g when used for implantation. The MB tissue
specimen collected from the surgery was used to make a cell
suspension with a concentration of 106-5x107
cells/ml. Cells were grown in DMEM (Hyclone; GE Healthcare)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.).
The SCID mice were placed on a homemade intracerebral injection
device after anesthesia by diazepam and chloramine intraperitoneal
injection. After head skin disinfection, 0.1 cm right or left of
the midline, 0.3 cm before the coronal suture, the sample trace on
the device was used to penetrate the skin and skull. The depth was
controlled between 1-2 mm. According to the concentration of the
cell suspension, a total of 5-10 µl cell suspension was injected
and the injection time was controlled to be <15 min. The mouse
weight was measured every 2-3 days. If the mice lost 30-35% of
their body weight in 3 days, or if they had a limb movement
disorder, magnetic resonance (MR) examination was performed
immediately to observe the intracranial tumor formation of the SCID
mice. The mice were sacrificed by cervical dislocation. The cell
culture medium of the same volume was injected into 10 control SCID
mice using the aforementioned protocol. The study was approved by
Medical Ethics Committee of Children's Hospital of Soochow
University.
Subcutaneous implanted models
A total of 20 SCID mice, including 10 female mice
and 10 male mice which were 4-6 weeks old were used for
experiments. The weight of the mouse was 15-20 g when used for
implantation. The SCID mice were placed on a homemade injection
device after anesthesia by diazepam and chloramine intraperitoneal
injection. A subcutaneous SCID mouse model was established using
~1x106 MB cells. Tumor cells were injected
subcutaneously into the back of the mice. On day 14
postimplantation, the subcutaneous tumors formed in 6 mice. The
mice were then sacrificed by cervical dislocation. Subcutaneous
tumors were then extracted. Tumor specimens were either preserved
in liquid nitrogen or in 4% paraformaldehyde for 2 h at 37˚C for
further experiments.
Immunohistochemistry (IHC)
Tumor tissues from the patient and from the mice
were prepared and fixed in 4% paraformaldehyde for 2 h at 37˚C.
Formalin-fixed paraffin-embedded tumors were sectioned using a
microtome into 6-µm sections. Antigen retrieval was performed using
10 mM sodium citrate buffer, pH 6, for 16 min at 96-98˚C. Slides
were incubated with primary antibodies including Ki-67 (cat. no.
BA2888; Wuhan Boster Bioengineering Co., Ltd.), GFAP (cat. no.
BA2689; Wuhan Boster Bioengineering Co., Ltd.), P53 (cat. no.
BA2358; Wuhan Boster Bioengineering Co., Ltd.) and Syn
(Synaptophysin; cat. no. ab14692; Abcam). All primary antibodies
were diluted to 1:1000 and incubated for 24 h at 4˚C. Sections were
subsequently incubated with the Cell & Tissue Staining kit
HRP-DAB system (R&D Systems, Inc.). The secondary antibodies
(diluted to 1:200; cat. no. ab6721; Abcam) were added and incubated
for 2 h at 37˚C, according to the manufacturer's instructions.
Immunostaining was performed with known positive and negative tumor
controls and were blindly evaluated by a pathologist. The
percentage of positive cells was calculated by randomly selecting
six sites in the histopathological section of the tumor. - was
defined as no pathological sections being positive for the marker;
+/- was defined as the majority of the pathological sections being
positive for the marker; -/+ was defined as some of the
pathological sections being positive for the marker; and + was
defined as all pathological sections being positive for the
marker.
Detection of the tumor driver
genes
The detection of tumor driver genes was performed by
First Imension. The tumor specimen and peripheral blood from the
patient were used for exon sequencing. The technology used in the
present research was target region capture combined with second
generation high-throughput sequencing. FFPE samples were extracted
and sequenced using the GeneRead™ DNA FFPE kit (cat. no. 180134;
Shanghai YuBo Biotechnology Co., Ltd.). Agarose gel electrophoresis
was used to analyze the degree of DNA degradation and the presence
of RNA and protein contamination. Qubit quantifies DNA
concentrations precisely. Illumina PE150 (Pair end 150 bp)
sequencing was performed according to the effective concentration
and data output requirements of the library. PE150 refers to
high-throughput double-end sequencing, with each end measuring 150
bp. According to the tumor driver gene results, PTCH1 and SMO were
used as the key words to search on the Internet. The web site used
was as follows: https://www.clinicaltrials.gov.
Results
Establishment of intracranial and
subcutaneous tumor model in SCID mouse
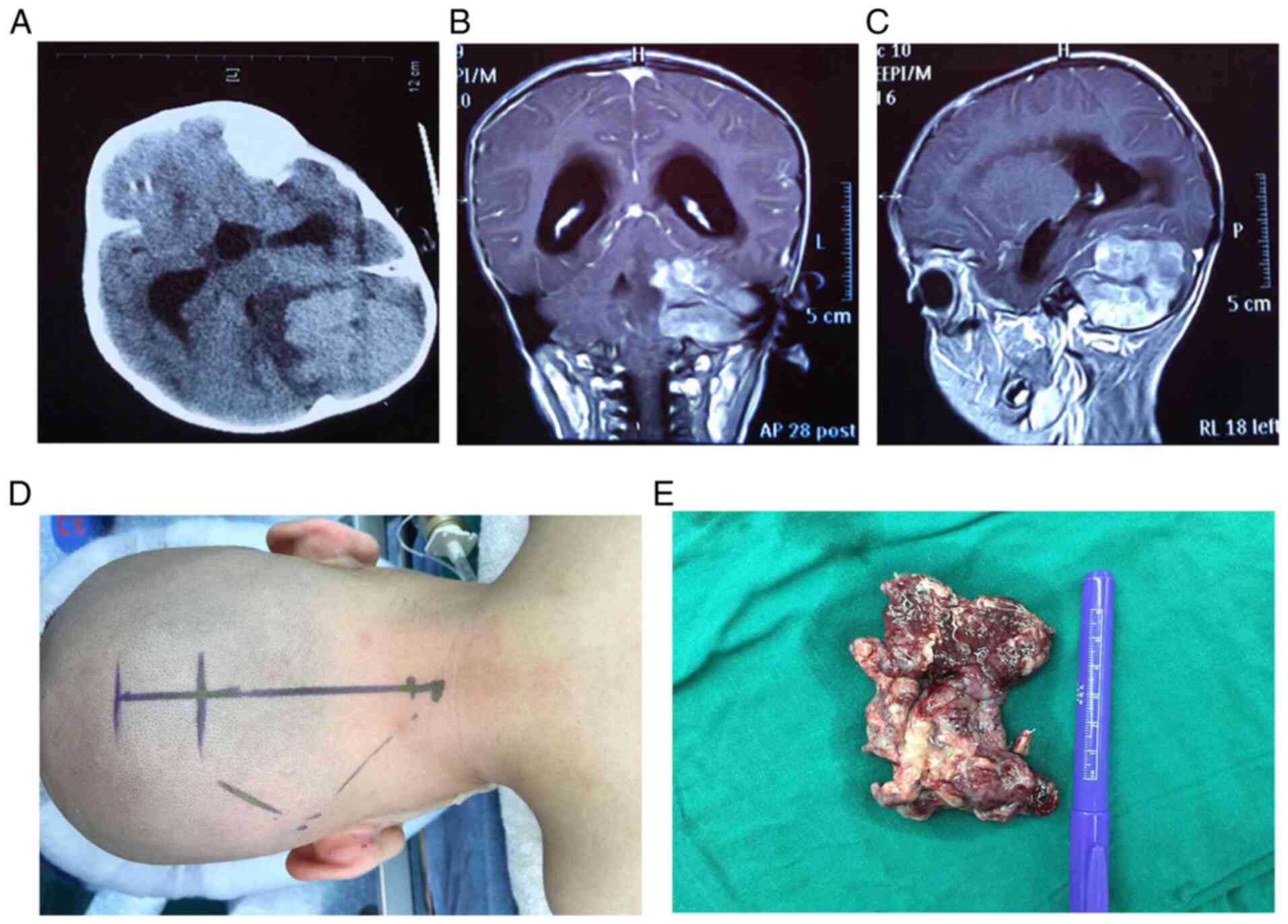
A two-year-old patient was considered to be MB
positive based on imaging data (CT and MR) taken before surgery
(Fig. 1A-C). The tumor was
surgically removed, with a section taken for the present study
(Fig. 1D and E). The remaining section of the tumor
specimen was used for pathological examination to make a definitive
diagnosis. The present study used the tumor specimen for
intracranial and subcutaneous implantation into SCID mice for
subsequent targeted drug screening.
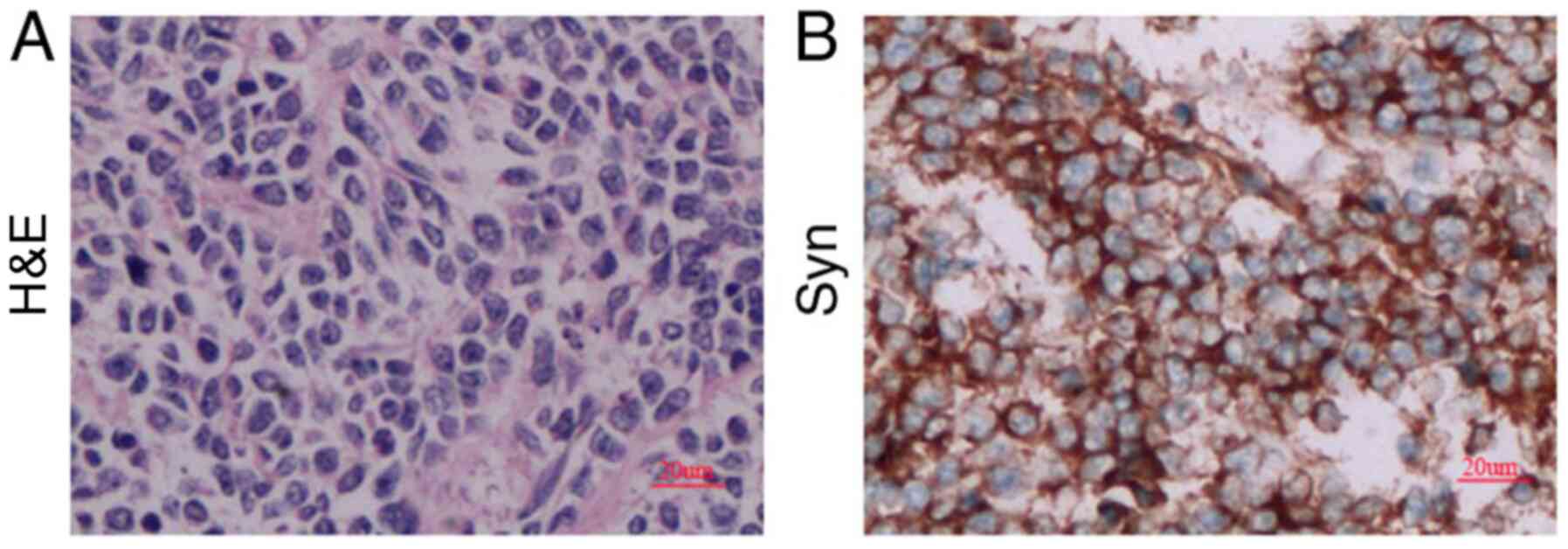
IHC results from the tumor sample taken from the
patient were as follows: Ki-67(+) 80%, GFAP(-/+) and P53(-/+) (data
not shown), as well as Syn(+) (Fig.
2). These markers are used to identify different tumors and
malignancy degree (15). Ki-67 is a
marker of cell proliferation and Syn is a classical marker for MB.
According to the aforementioned IHC results, the diagnosis was ND
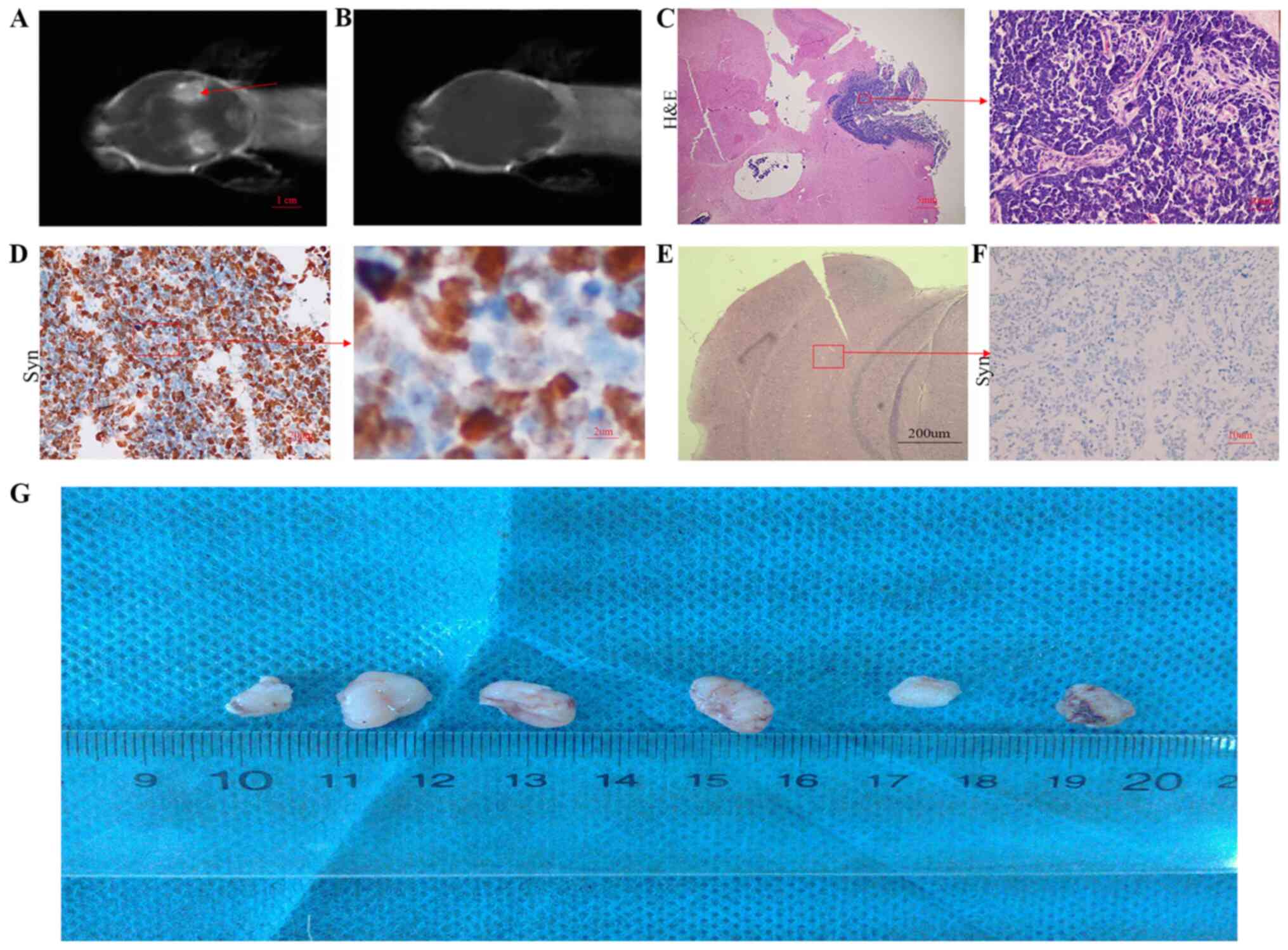
MB with SHH activation. MR examination was used to observe the
intracranial tumors of the SCID mouse. After one month following
implantation, the tumor was found in the brain of the SCID mouse
(Fig. 3A). Tumor formation was not
observed in the brain of the control mouse (Fig. 3B, E
and F). Subsequently, the tumor
tissue from the SCID intracranial tumor mouse was collected and
analyzed. The results were the same as that observed in the
excised, unimplanted tumor tissue (Fig.
3C and D). Tumor cells from the
patient were also used for subcutaneous implantation in SCID mice
(Fig. 3G). Mice with subcutaneously
inoculated cells developed xenograft tumors after 2 weeks, which
may be used for subsequent targeted drug screening experiments.
Detection of tumor driver genes in
MB
Peripheral blood and tumor specimens from the
patient were used for the detection of tumor driver genes. Sequence
capture technology was used to test all exons using high-throughput
sequencing. The results found point mutation, deletion, insertion,
copy number and structural variations. In the present study, three
genetic variants were found from the samples taken from the patient
with MB, including PTCH1p.F573S, PTCH1p.S574P, PTCH1p.L575P
(Tables I and II).
 | Table ISignificant mutations of germ
cells. |
Table I
Significant mutations of germ
cells.
| Gene | Reference
sequence | Nucleotide
mutation | Amino acid
mutation | Cover depth | Cover depth of
mutation | Frequency of
mutation |
|---|
| BRCA2 | NM_000059 | c.T943A | p.C315S | 330 | 160 | 48% |
| BRCA2 | NM_000059 | c.A10234G | p.I3412V | 241 | 122 | 51% |
| PRDM1 | NM_182907 | c.C998T | p.P333L | 812 | 412 | 51% |
| HEY1 | NM_001282851 | c.C50T | p.T17M | 303 | 175 | 58% |
| MN1 | NM_002430 | c.T1682C | p.M561T | 289 | 138 | 48% |
 | Table IISignificant mutations of somatic
cells. |
Table II
Significant mutations of somatic
cells.
| Gene | Reference
sequence | Mutation type | Nucleotide
mutation | Amino acid
mutation | Cover depth | Cover depth of
mutation | Frequency of
mutation |
|---|
| PTCH1 | NM_000264 | Nonsynonymous
mutation | c.T1718C | p.F573S | 44 | 40 | 91% |
| PTCH1 | NM_000264 | Nonsynonymous
mutation | c.T1720C | p.S574P | 44 | 40 | 91% |
| PTCH1 | NM_000264 | Nonsynonymous
mutation | T1724C | p.L575P | 44 | 42 | 95% |
| ROS1 | NM_002944 | Increase of | | | | | |
| Copy number | - | - | CN=6 | - | - | | |
Screening for tumor driver
gene-related targeted drugs
It was found that the drugs Vismodegib and
Sonidegib, which target mutations in PTCH1, have been approved by
the FDA for the therapy of other types of tumors (Table III). Currently, there are no drugs
for the treatment of mutations to PTCH1 in MB. The drug BMS-833923,
which targets PTCH1 mutations in solid and blood tumors is still in
clinical trials (Table III).
 | Table IIIInformation of the targeted drugs
which may be of use for treating the MB. |
Table III
Information of the targeted drugs
which may be of use for treating the MB.
| Drugs | Clinical trial
title | Testing stage | Gene mutation type
and tumor type | NCT ID | State |
|---|
| Sonidegib | Molecular phase II
study to link targeted therapy to patients with pathway activated
tumors: Module-5 LDE225 for patients with PTCH1 or SMO mutated
tumors | II | Solid tumor and blood
tumor with PTCH1 or SMO mutation | NCT02002689 | Completed |
| Vismodegib and
temozolomide | An international,
Randomized open label phase I/II study of Vismodegib in combination
with temozolomide versus temozolomide alone in adult patients with
recurrent or refractory medulloblastomas presenting an activation
of the SHH pathway | II | MB with SHH pathway
activation | NCT01601184 | In the
recruitment |
| Vismodegib | A phase II clinical
trial evaluating the efficacy and safety of GDC-0499 in children
with recurrent or refractory medulloblastomas | II | Recurrent MB with SHH
pathway activated or inactivated | NCT01239316 | Completed |
| BMS-833923 | A phase Ib multiple
ascending dose study of BMS-833923 alone or in combination with
lenalidomide plus dexamethasone or in combination with
bortezomib | Ib | Multiple progressive
tumors | NCT00884546 | Completed |
Discussion
Previously, clinicians have developed comprehensive
treatment plans and predicted recurrence and metastasis risk for
patients with MB according to the pathological type (16). Despite these classifications,
patients from the high risk group who received identical treatment
regimens have presented with varied prognoses. Therefore, the use
of only the pathological classification does not meet the current
treatment request. With the progress of genomics, MB has been found
to be a type of brain tumor which present with varied molecular
characteristics (17). It is now
clear that MB is not a single disease entity, but instead consists
of at least four distinct molecular subgroups: WNT/Wingless, SHH,
Group 3 and Group 4 (18,19). The heterogeneity within the same
molecular subgroup has led to the identification of 12 subtypes
within the current molecular subgroups, demonstrating the
requirement to better characterize the specific driving factors
that contribute to the subgroup heterogeneity (19). With the appearance of molecular
classification, molecular targeted therapy has been expected to be
more effective for the treatment of MB. Although there may be
treatments that work on multiple forms of MB, the diversity of the
genetic and epigenetic events even within a particular subgroup,
makes it likely that each patient will be responsive to distinct
therapies and combinations thereof (20). Identifying appropriate therapies for
each patient may require detailed molecular and cellular analysis
of tumor tissues (20). At present,
drugs which target the signaling pathways behind MB, such as the
AKT pathway, SHH pathway and NOTCH pathway are already in clinical
trials (21). Some of these
targeted drugs have been proven to be ineffective in the clinical
trials, while others have presented a significant influence on the
prognosis, even within the same molecular subtype of MB (22). These results indicate that there are
still significant deficiencies in the selection of targeted drugs
based on the present MB molecular classification, making the
selection of the appropriate treatment for patients with MB,
unclear. In a previous study which investigated other types of
malignant human tumors, it was proposed that individualized
targeted therapies could be developed based on tumor driver genes
which was considered as the main reference index for treatment
(11). This may provide a new
method to improve the effect of targeted therapies for the
treatment for MB. A new study performed by the Children's Hospital
of Philadelphia (USA) found that PF-06463922 targets the tumor
driver gene, anaplastic lymphoma (ALK), in lung cancer and is
effective for treating Neuroblastomas with ALK mutations (21). These results provide confidence that
by analyzing the tumor driver genes from various patients with MB,
it will be possible to the best targeted drugs which have been used
for the treatment of other tumor types.
In conclusion, the present study analyzed tumor
driver genes from the tumor tissue collected from one patient with
MB and screened for targeted drugs of the tumor driver genes. An
intracranial and subcutaneous implanted models of MB were also
developed in mice. The subcutaneous implanted model of MB may be of
use for the preliminary screening of drugs for the treatment of MB.
Due to the differences in the microenvironment between subcutaneous
and intracranial tumors, further drug screening requires
intracranial tumor models. Subsequent experiments could then
validate the findings on the effects of these targeted drugs in an
animal model of MB. Further experiments should investigate the
characteristics of tumor driver genes in patients with different
molecular subtypes.
Acknowledgements
Not applicable.
Funding
The study was supported by Science and education
program of Suzhou (grant no. KJXW2017023).
Availability of data and materials
The datasets used and/or anlayzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HW designed the study. YH performed the experiments.
MC analyzed and interpreted the data and edited the manuscript. All
authors read and approved the manuscript.
Ethics approval and consent to
participate
All methods were carried out in accordance with
guidelines and regulations of Soochow University. The human sample
was used in accordance with the policies of the institutional
review board of Children's Hospital of Soochow University. The use
of the sample from the MB patient was approved by his parents. The
study was approved by medical ethics committee of Children's
Hospital of Soochow University.
Patient consent for publication
Written informed consent obtained from patient's
parents prior to publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kumar R, Liu AP and Northcott PA:
Medulloblastoma genomics in the modern molecular era. Brain Pathol.
30:679–690. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Dhall G, O'Neil SH, Ji L, Haley K,
Whitaker AM, Nelson MD, Gilles F, Gardner SL, Allen JC, Cornelius
AS, et al: Excellent outcome of young children with nodular
desmoplastic medulloblastoma treated on ‘Head Start’ III: A
multi-institutional, prospective clinical trial. Neuro Oncol: Apr
18, 2020 (Epub ahead of print). doi: 10.1093/neuonc/noaa102.
|
|
3
|
Batora NV, Sturm D, Jones DT, Kool M,
Pfister SM and Northcott PA: Transitioning from genotypes to
epigenotypes: Why the time has come for medulloblastoma
epigenomics. Neuroscience. 264:171–185. 2014.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ris MD, Packer R, Goldwein J,
Jones-Wallace D and Boyett JM: Intellectual outcome after
reduced-dose radiation therapy plus adjuvant chemotherapy for
medulloblastoma: A Children's Cancer Group study. J Clin Oncol.
19:3470–3476. 2001.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Xu W, Janss A, Packer RJ, Phillips P,
Goldwein J and Moshang T Jr: Endocrine outcome in children with
medulloblastoma treated with 18 Gy of craniospinal radiation
therapy. Neuro Oncol. 6:113–118. 2004.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Hoppe-Hirsch E, Renier D, Lellouch-Tubiana
A, Sainte-Rose C, Pierre-Kahn A and Hirsch JF: Medulloblastoma in
childhood: Progressive intellectual deterioration. Childs Nerv
Syst. 6:60–65. 1990.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Chen P, Fan Y, Man TK, Hung YS, Lau CC and
Wong ST: A gene signature based method for identifying subtypes and
subtype-specific drivers in cancer with an application to
medulloblastoma. BMC Bioinformatics. 14 (Suppl
18)(S1)2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Sanai N: Integrated genomic analysis
identifies clinically relevant subtypes of glioblastoma. World
Neurosurg. 74:4–5. 2010.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kool M, Koster J, Bunt J, Hasselt NE,
Lakeman A, van Sluis P, Troost D, Meeteren NS, Caron HN, Cloos J,
et al: Integrated genomics identifies five medulloblastoma subtypes
with distinct genetic profiles, pathway signatures and
clinicopathological features. PLoS One. 3(e3088)2008.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yang J, Wang X, Kim M, Xie Y and Xiao G:
Detection of candidate tumor driver genes using a fully integrated
Bayesian approach. Stat Med. 33:1784–1800. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Vogelstein B, Papadopoulos N, Velculescu
VE, Zhou S, Diaz LA Jr and Kinzler KW: Cancer genome landscapes.
Science. 339:1546–1558. 2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ohshima K, Hatakeyama K, Nagashima T,
Watanabe Y, Kanto K, Doi Y, Ide T, Shimoda Y, Tanabe T, Ohnami S,
et al: Integrated analysis of gene expression and copy number
identified potential cancer driver genes with
amplification-dependent overexpression in 1,454 solid tumors. Sci
Rep. 7(641)2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Akavia UD, Litvin O, Kim J, Sanchez-Garcia
F, Kotliar D, Causton HC, Pochanard P, Mozes E, Garraway LA and
Pe'er D: An integrated approach to uncover drivers of cancer. Cell.
143:1005–1017. 2010.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Son EI, Kim IM, Kim DW, Yim MB, Kang YN,
Lee SS, Kwon KY, Suh SI, Kwon TK, Lee JJ, et al:
Immunohistochemical analysis for histopathological subtypes in
pediatric medulloblastomas. Pathol Int. 53:67–73. 2003.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Aref D and Croul S: Medulloblastoma:
Recurrence and metastasis. CNS Oncol. 2:377–385. 2013.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kijima N and Kanemura Y: Molecular
Classification of Medulloblastoma. Neurol Med Chir (Tokyo).
56:687–697. 2016.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Gottardo NG, Hansford JR, McGlade JP,
Alvaro F, Ashley DM, Bailey S, Baker DL, Bourdeaut F, Cho YJ, Clay
M, et al: Medulloblastoma Down Under 2013: A report from the third
annual meeting of the International Medulloblastoma Working Group.
Acta Neuropathol. 127:189–201. 2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ferrucci V, de Antonellis P, Pennino FP,
Asadzadeh F, Virgilio A, Montanaro D, Galeone A, Boffa I, Pisano I,
Scognamiglio I, et al: Metastatic group 3 medulloblastoma is driven
by PRUNE1 targeting NME1-TGF-β-OTX2-SNAIL via PTEN inhibition.
Brain. 141:1300–1319. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang J, Garancher A, Ramaswamy V and
Wechsler-Reya RJ: Medulloblastoma: From molecular subgroups to
molecular targeted therapies. Annu Rev Neurosci. 41:207–232.
2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Mackall CL: In search of targeted
therapies for childhood cancer. Front Oncol. 1(18)2011.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kumar V, Kumar V, McGuire T, Coulter DW,
Sharp JG and Mahato RI: Challenges and recent advances in
medulloblastoma therapy. Trends Pharmacol Sci. 38:1061–1084.
2017.PubMed/NCBI View Article : Google Scholar
|