Introduction
Successful embryo implantation, in which the
blastocyst adheres to and invades the maternal endometrium, is the
first step for establishing natural pregnancy. In the human
endometrium, implantation occurs between 6 and 10 days after
ovulation [the window of implantation (WOI)] (1), during which the human endometrium
undergoes morphological and biochemical changes, including
modulation of estrogen, progesterone, adhesion molecules, growth
factors, cytokines, and chemokines, to facilitate implantation
(2). This process enhances
endometrial receptivity by balancing the expression of adhesion
molecules and inhibitory proteins acting as a barrier to blastocyst
implantation (3). However,
implantation failure (i.e., poor endometrial receptivity) remains a
serious barrier to both spontaneous and assisted pregnancies
(4). To characterize the molecular
mechanisms associated with endometrial receptivity, studies have
identified, using microarrays or RNA sequencing (RNA-seq), hundreds
of differentially expressed genes (DEGs) between prereceptive and
receptive endometria (5-12).
Although these studies have verified many differentially expressed
(DE) transcripts, the overlap between DEGs in different studies is
low, and the molecular mechanisms underlying endometrial
receptivity remain unclear (13).
Only 2% of the human transcriptome is translated
into proteins; the remaining untranslated transcriptome contains
noncoding RNAs (ncRNAs), which have been reported to control
cellular processes and functions by regulating the expression of
target genes (14). Long ncRNAs
(lncRNAs) are over 200 nucleotides in length and display
differential patterns of expression in various tissues, where they
play diverse roles in various physiological processes (15). Additionally, lncRNAs can be used as
biomarkers of implantation failure (16-18).
MicroRNAs (miRNAs) are evolutionarily conserved ncRNAs, measuring
19-22 nucleotides in length, and negatively regulate target gene
expression by binding to the 3'-untranslated region (UTR) of the
target gene (19). Approximately
half of all genes in the genome are miRNA targets regulated at the
post-transcriptional level (20),
and recent studies have linked miRNAs to human endometrial diseases
such as endometriosis and endometrial cancer (21-23).
The competitive endogenous RNA (ceRNA) hypothesis
was first proposed by Salmena et al, who demonstrated that
lncRNAs can regulate other RNA transcripts by competing for target
miRNAs via binding sites known as miRNA response elements (24). Since then, ceRNA networks have been
shown to have roles in the development of various tumors.
In this study, we attempted to identify DE lncRNAs,
miRNAs, and mRNAs associated with endometrial receptivity using
RNA-seq of proliferative and mid-secretory endometrium samples.
Additionally, we performed in silico analysis of genes
previously reported to show differential expression between
prereceptive and receptive endometrium samples. We also constructed
a ceRNA network based on DE miRNA/lncRNA and miRNA/mRNA pairs as
correlations between mRNAs and ncRNAs can indicate complex gene
regulatory events. The hub ncRNAs from this network can be
developed as candidate markers for endometrial receptivity.
Materials and methods
Endometrial tissue collection
Endometrial samples were obtained from 30 fertile
women who attended Konyang University Hospital (Daejeon, Republic
of Korea) and had self-reported regular, normal (21-35 days)
menstrual cycles. Each woman had at least one live birth, fewer
than two spontaneous abortions, and had received no medication,
including hormonal treatments, for at least three months prior to
the day of the biopsy. Endometrial tissues were collected during
the proliferative or secretory phase of the menstrual cycle, which
donors were divided with 2 groups according to the phase of
menstrual period on the day of endometrial biopsy. Before the
biopsy, a gynecologist checked each donor's menstrual period by
checking last menstrual day and endometrial ultrasound
measurements. For endometrial sampling, a sterile speculum was
inserted into the vagina, a betadine dressing was applied, and
endometrial tissue was gently collected using a disposable uterine
sampler (Rampipella, RI.MOS, Mirandola, Italy). Each endometrial
sample was divided into two tubes; one was sent to the pathologist
to identify endometrial pathology and confirm the menstrual cycle
by histological examination using Noyes criteria (25), and the other was used for RNA
extraction. The average age and body index were not different
between 2 groups (37.5±2.7 vs. 36.9±2.4 years, 22.6±3.3 vs.
22.2±3.4 kg/m2). And the number of previous live birth
or abortion history was not also different (Table I). Continuous variables were
compared with the two-sample t-test. And categorical
variables were analyzed with Fisher's exact test. All P-values were
2-sided. P-values less than 0.05 were deemed statistically
significant.
 | Table ICharacteristics of endometrium donors
for RNA sequencing. |
Table I
Characteristics of endometrium donors
for RNA sequencing.
|
Variables/Group | Proliferative phase
(n=15) | Secretory phase
(n=15) | P-value |
|---|
| Age (years) | 37.5±2.7 | 36.9±2.4 | 0.53a |
| BMI
(kg/m2) | 22.6±3.3 | 22.2±3.4 | 0.77a |
| Number of live
births, n (%) | | | 0.09b |
|
1 | 4 (26.6) | 7 (46.6) | |
|
2 | 7 (46.6) | 8 (53.3) | |
|
3 | 4 (26.6) | 0 (0.0) | |
| Number of
abortions, n (%) | | | 0.13b |
|
0 | 13 (86.6%) | 10 (66.6%) | |
|
1 | 1 (6.6%) | 5 (33.3%) | |
|
2 | 1 (6.6%) | 0 (0.0%) | |
Ethical approval
This study was approved by the Bioethics Committee
of Konyang University Hospital (IRB File No. 2018-11-007-005).
Signed informed consent was obtained from each patient.
RNA extraction
Total RNA was isolated from endometrial tissue
immediately after biopsy using TRIzol reagent (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer's instructions.
Total RNA concentrations were calculated using Quant-IT RiboGreen
(Invitrogen), and integrity was assessed using a TapeStation RNA
ScreenTape. Only high-quality RNA preparations with an RNA
integrity number greater than 6.5 were used to construct the RNA
library with total RNA isolated from four proliferative and four
mid-secretory endometrium samples.
RNA library construction
Ten nanograms of RNA isolated from each sample was
used to construct miRNA sequencing libraries using a SMARTer small
noncoding RNA (smRNA)-Seq Kit (Takara Bio Inc.) according to the
manufacturer's instructions. cDNA synthesis was primed using a 3'
smRNA dT primer, which incorporates an adapter sequence at the 5'
end of each RNA template and adds non-template nucleotides bound to
oligo-enhanced SMRT smRNA with locked nucleic acid technology for
greater sensitivity. For template-switching, PrimeScript RT was
used with the SMART smRNA oligo as a template to add a second
adapter sequence to the 3' end of each first-strand cDNA molecule.
Full-length Illumina adapters were added during polymerase chain
reaction (PCR) amplification, and the amplified libraries were
purified to remove the fraction over 171 bp (>18 bp of cDNA plus
153 bp of adaptors).
One microgram total RNA isolated from each sample
was used to prepare a total RNA library using an Illumina TruSeq
Stranded Total RNA Sample Prep Kit (Illumina). rRNA was depleted
using a Ribo-Zero kit (Illumina), and the remaining RNA was
purified, fragmented, and primed for cDNA synthesis. Cleaved RNA
fragments were converted into first-strand cDNA using reverse
transcriptase and random hexamers, followed by second-strand cDNA
synthesis using DNA polymerase I, RNase H, and dUTP. The cDNA
fragments then underwent end repair, single ‘A’ base addition, and
adapter ligation. The products were purified and enriched by PCR to
create the final cDNA library.
Libraries were quantified by quantitative PCR (qPCR)
using KAPA library quantification kits for Illumina sequencing
platforms and qualified using a TapeStation D1000 ScreenTape
(Agilent Technologies).
RNA sequencing
smRNA libraries underwent 51-bp single-end
sequencing using Illumina HiSeq 2500 (Illumina). Indexed cDNA
libraries were subjected to paired-end (2x100 bp) sequencing using
an Illumina Novaseq (Illumina). The reference genome sequence for
Homo sapiens (hg19) and annotation data were downloaded from
the National Center for Biotechnology Information, and known
transcripts were assembled using StringTie v1.3.4d (26). Transcript abundance and gene
expression were calculated as the read count or fragments per
kilobase of exon per million fragments mapped (FPKM) value per
sample. We deposited smRNA sequences derived from sequencing of
smRNA libraries into Gene Expression Omnibus (GEO) of National
Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE167325).
The expression profiles were used to identify DEGs. In groups with
different conditions, DEGs or transcripts were filtered by
statistical hypothesis testing.
Statistical analysis of differential
miRNA and total RNA expression
Raw data were normalized using the relative log
expression method in DESeq2(27).
miRNAs with no count in more than 50% of samples were excluded,
leaving 507 mature miRNAs for further analysis. Various plots were
drawn using normalized log transformation. Two groups were compared
using the ‘nbinomWaldTest’ in DESeq2. Relative gene abundance was
measured in FPKM using StringTie. Statistical analysis was
performed on the estimated abundance of each gene in the samples to
identify DEGs. Genes with more than one ‘zero’ FPKM value in the
samples were excluded. The statistical significance of the
differential expression data was determined using independent
t-tests based on fold changes and the null hypothesis that
no difference existed. The false discovery rate was controlled by
adjusting the P-value using the Benjamini-Hochberg
algorithm.
Hierarchical clustering analysis
Hierarchical clustering analysis was performed using
complete linkage and Euclidean distance as a measure of similarity
to display the expression patterns of DE miRNAs and transcripts
that satisfied fold change (FC) greater than or equal to 1.5 and a
raw P-value of less than 0.05. All data analysis and DEG
visualization were conducted using R v.3.6.0 (www.r-project.org).
Functional enrichment analysis
Gene ontology (GO) and Kyoto Encyclopedia of Genes
and Genomes (KEGG) functional enrichment analyses of DE mRNAs were
performed using the Database for Annotation, Visualization, and
Integrated Discovery (https://david.ncifcrf.gov/). All DEG data analysis and
visualization were conducted using R v.3.6.0 (www.r-project.org).
Reverse transcription (RT)-qPCR
RNA samples isolated from 14 proliferative and nine
mid-secretory phase endometrium samples were subjected to RT-qPCR.
To determine mRNA and lncRNA expression levels, cDNA was
synthesized using M-MLV reverse transcriptase (Promega). qPCR was
performed using a CFX 96 qPCR instrument and iQ SYBR Green Supermix
(Bio-Rad Laboratories) with the following amplification conditions:
Initial denaturation at 95˚C for 3 min; followed by 40 cycles of
denaturation at 95˚C for 10 s, 60˚C for 10 s, and extension at 72˚C
for 15 s. We performed TaqMan microRNA assays for determination of
miRNA expression levels. The primer sequences are presented in
Tables SI and SII. mRNA and lncRNA expression levels
were quantified following normalization to glyceraldehyde
3-phosphate dehydrogenase (GAPDH) expression and miRNA expression
levels were quantified following normalization to RNA, U6 small
nuclear 6, pseudogene (RNU6B) using the 2-∆∆CT method, the fold
change (FC) was evaluated in comparison with the proliferative
phase. Assays were conducted in triplicate. The data are presented
as the mean ± standard error of the mean (SEM). The Student's
t-test or Mann-Whitney U test was used to assess between group
differences.
Ingenuity pathway analysis (IPA)
To investigate the associated gene function
networks, IPA was performed (Ingenuity Systems; http://www.ingenuity.com).
ceRNA network construction
To investigate potential interactions among lncRNAs,
miRNAs, and mRNAs, we constructed a ceRNA network. Interactions
between DE lncRNAs and miRNAs were predicted using miRcode
(http://www.mircode.org/). Interactions between DE
miRNAs and mRNAs were predicted using mirDIP (http://ophid.utoronto.ca/mirDIP; ‘bidirectional
search’). The DE lncRNA/miRNA/mRNA ceRNA network was visualized and
constructed using the Cytoscape software (28).
Results
Identification of DE transcripts
To investigate genes associated with endometrial
receptivity, we analyzed DE transcripts between proliferative and
mid-secretory endometrium samples using next-generation sequencing
(NGS). We identified 2,154 DE mRNAs, of which 1,042 (48.37%) were
upregulated and 1,112 (51.63%) were downregulated in the
mid-secretory endometrium (Table
SIII). Variance in the DE mRNAs in the proliferative or
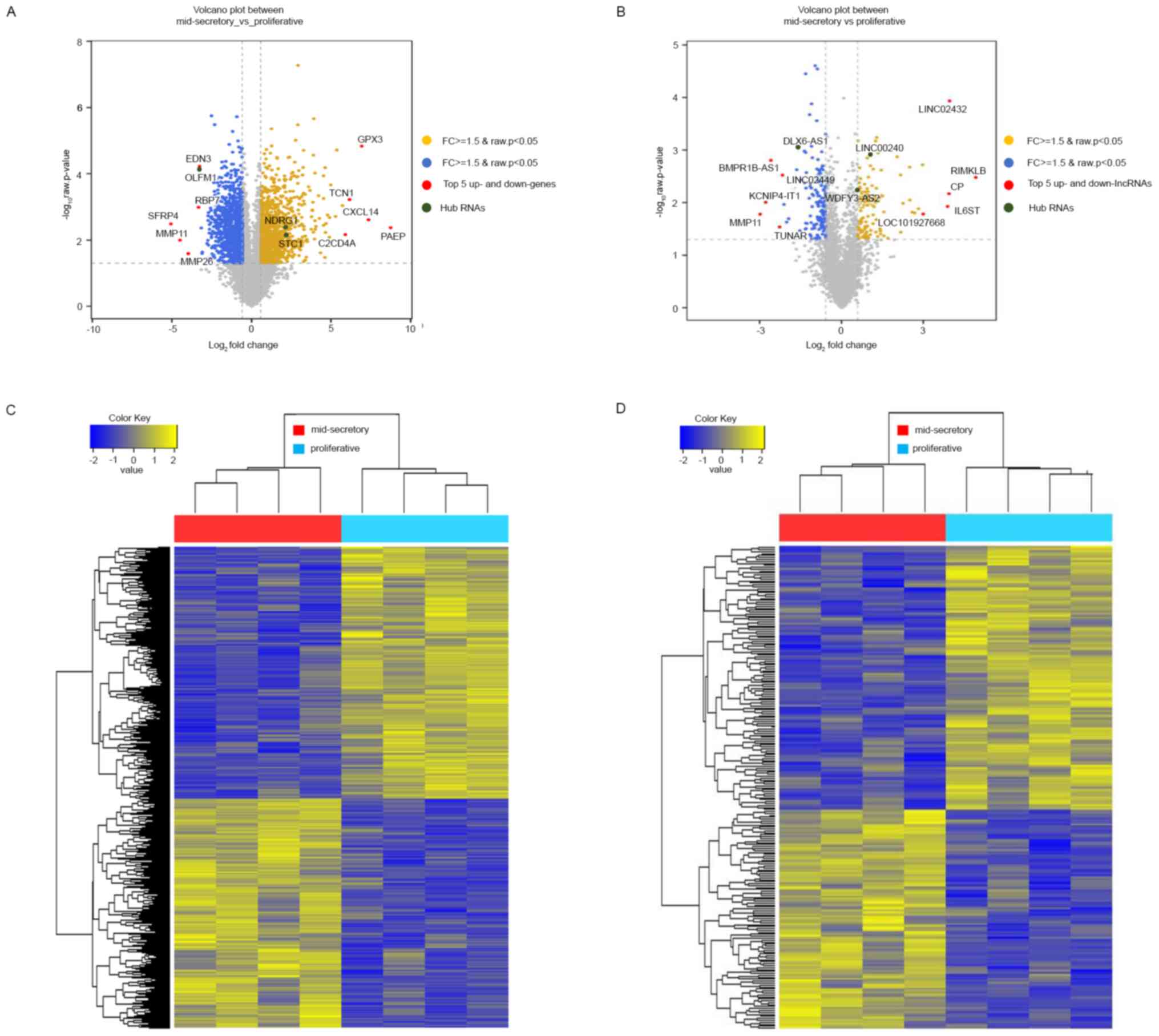
mid-secretory endometrium, visualized using volcano plots,
indicated the top 5 up- and downregulated genes (Fig. 1A). Additionally, we identified 247
significant DE lncRNAs, of which 112 (45.34%) were upregulated and
135 (54.66%) were downregulated in the mid-secretory endometrium
(Table SIV). In Fig. 1B, a volcano plot, displaying the
differences in the expression values of the DE lncRNAs in the
proliferative or mid-secretory endometrium, indicated the top 5 up-
and downregulated lncRNAs. The expression of DE mRNAs and lncRNAs
in the proliferative or mid-secretory endometrium is illustrated in
Fig. 1C and D. Notably, DE mRNAs and lncRNAs were able
to distinguish between the proliferative and mid-secretory
endometria. The top 10 up- and downregulated DE mRNAs and lncRNAs
are indicated in Tables II and
III, respectively.
 | Table IITop 10 differentially expressed
upregulated and downregulated mRNAs with the highest fold
change. |
Table II
Top 10 differentially expressed
upregulated and downregulated mRNAs with the highest fold
change.
| A, Upregulated
mRNAs |
|---|
| Symbol | Gene name | FC | P-value |
|---|
| PAEP | Progestagen
associated endometrial protein | 426.4482 | 0.0043 |
| CXCL14 | C-X-C motif
chemokine ligand 14 | 162.8550 | 0.0024 |
| GPX3 | Glutathione
peroxidase 3 | 121.0577 |
1.45x10-5 |
| TCN1 | Transcobalamin
1 | 71.4916 |
6.03x10-4 |
| C2CD4A | C2
calcium-dependent domain containing 4A | 59.5485 | 0.0069 |
| DPP4 | Dipeptidyl
peptidase 4 | 52.3765 |
9.10x10-4 |
| C4BPA | Complement
component 4 binding protein alpha | 40.8157 |
1.89x10-5 |
| AOX1 | Aldehyde oxidase
1 | 38.7763 | 0.0019 |
| CFD | Complement factor
D | 31.4066 |
2.17x10-4 |
| DKK1 | Dickkopf WNT
signaling pathway inhibitor 1 | 29.9378 | 0.0021 |
| B, Downregulated
mRNAs |
| Symbol | Gene name | FC | P-value |
| SFRP4 | Secreted
frizzled-related protein 4 | -33.4871 | 0.0033 |
| MMP11 | Matrix
metallopeptidase 11 | -22.7452 | 0.0101 |
| MMP26 | Matrix
metallopeptidase 26 | -15.8265 | 0.0255 |
| RBP7 | Retinol binding
protein 7 | -10.1247 | 0.0010 |
| EDN3 | Endothelin 3 | -9.6811 |
5.92x10-5 |
| OLFM1 | Olfactomedin 1 | -9.6540 |
7.57x10-5 |
| SERPINA5 | Serpin family A
member 5 | -8.9736 | 0.0043 |
| POSTN | Periostin | -8.8430 |
7.77x10-4 |
| PKHD1L1 | PKHD1 like 1 | -8.6865 | 0.0254 |
| SLC47A1 | Solute carrier
family 47 member 1 | -8.5516 | 0.0231 |
 | Table IIITop 10 differentially expressed
upregulated and downregulated lncRNAs with the highest fold
change. |
Table III
Top 10 differentially expressed
upregulated and downregulated lncRNAs with the highest fold
change.
| A, Upregulated
lncRNAs |
|---|
| Transcript ID | Gene symbol | FC | P-value |
|---|
| NR_123740 | RIMKLB,
variant3 | 30.2256 | 0.0033 |
| NR_121625 | LINC02432 | 15.5469 |
1.16x10-4 |
| NR_046371 | CP, variant2 | 15.3309 | 0.0067 |
| NR_120480 | IL6ST,
variant4 | 14.7984 | 0.0118 |
| NR_110114 | LOC101927668 | 7.9406 | 0.0165 |
| NR_126404 | LINC01320 | 7.8229 | 0.0019 |
| NR_045623 | NABP1,
variant4 | 7.0512 | 0.0131 |
| NR_131782 | CCEPR | 6.4217 | 0.0158 |
| NR_026979 | DRAIC | 6.0252 | 0.0151 |
| NR_045580 | RXFP1,
variant9 | 5.6729 | 0.0089 |
| B, Downregulated
lncRNAs |
| Transcript ID | Gene symbol | FC | P-value |
| NR_133013 | MMP11,
variant2 | -7.9496 | 0.0166 |
| NR_002813 | KCNIP4-IT1 | -6.8875 | 0.0098 |
| NR_121610 | BMPR1B-AS1 | -6.0480 | 0.0016 |
| NR_038861 | TUNAR,
variant2 | -4.8339 | 0.0291 |
| NR_120454 | LINC02449 | -4.5053 | 0.0030 |
| NR_027349 | MIR17HG,
variant2 | -4.3154 | 0.0108 |
| NR_072997 | PTK7, variant7 | -4.0389 | 0.0234 |
| NR_120409 | PCSK5,
variant3 | -3.8489 | 0.0202 |
| NR_039982 | LINC00639 | -3.1286 | 0.0024 |
| NR_015448 | DLX6-AS1 | -3.0147 |
8.93x10-4 |
Identification of DE miRNAs
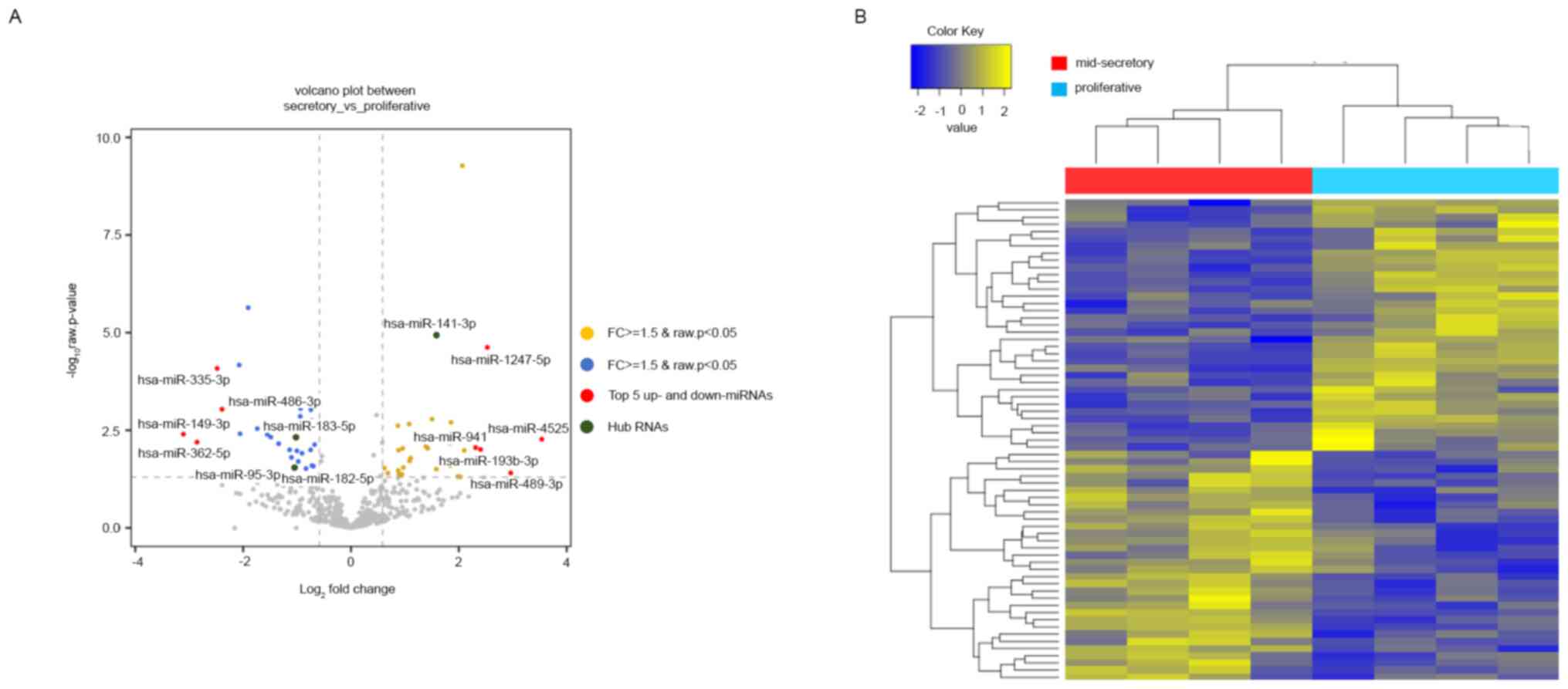
We next checked the expression patterns of miRNAs
from NGS expression data. In total, 507 miRNAs were identified with
small RNA-seq (GSE167325), of which 67 were DE in the mid-secretory
endometrium compared with the proliferative endometrium. Of the 67
DE miRNAs, 32 (47.76%) were upregulated, and 35 (52.24%) were
downregulated in the mid-secretory endometrium (Table SV). The DE miRNAs were visualized
using a volcano plot, which indicated the top 5 up- and
downregulated genes, and a heatmap (Fig. 2A and B). Based on the heatmap of the DE miRNAs,
the mid-secretory endometrium could be clustered separately from
the paired proliferative endometrium. The top ten up- and
downregulated DE miRNAs are summarized in Table IV.
 | Table IVTop 10 differentially expressed
upregulated and downregulated microRNAs with the highest fold
change. |
Table IV
Top 10 differentially expressed
upregulated and downregulated microRNAs with the highest fold
change.
| A, Upregulated
microRNAs |
|---|
| ID | FC | P-value |
|---|
| hsa-miR-4525 | 11.6143 | 0.0054 |
| hsa-miR-489-3p | 7.7917 | 0.0392 |
|
hsa-miR-1247-5p | 5.7667 |
2.39x10-5 |
|
hsa-miR-193b-3p | 5.2862 | 0.0098 |
| hsa-miR-941 | 4.9600 | 0.0088 |
| hsa-miR-876-3p | 4.2922 | 0.0103 |
| hsa-miR-224-5p | 4.1808 |
5.31x10-10 |
|
hsa-miR-376b-3p | 4.0789 | 0.0491 |
| hsa-miR-30d-3p | 4.0318 | 0.0044 |
|
hsa-miR-642a-3p | 3.9458 | 0.0484 |
| B, Downregulated
microRNAs |
| ID | FC | P-value |
| hsa-miR-149-3p | -8.6235 | 0.0477 |
| hsa-miR-362-5p | -7.2379 | 0.0117 |
| hsa-miR-335-3p | -5.5844 | 0.0469 |
| hsa-miR-486-3p | -5.2467 | 0.0075 |
| hsa-miR-95-3p | -5.2448 | 0.0140 |
|
hsa-miR-365b-5p | -5.0526 | 0.0041 |
| hsa-miR-3179 | -4.5559 | 0.0242 |
| hsa-miR-296-3p | -4.2086 | 0.0280 |
|
hsa-miR-135a-5p | -4.1637 | 0.0404 |
|
hsa-miR-1224-5p | -4.1609 | 0.0372 |
GO and KEGG pathway analysis
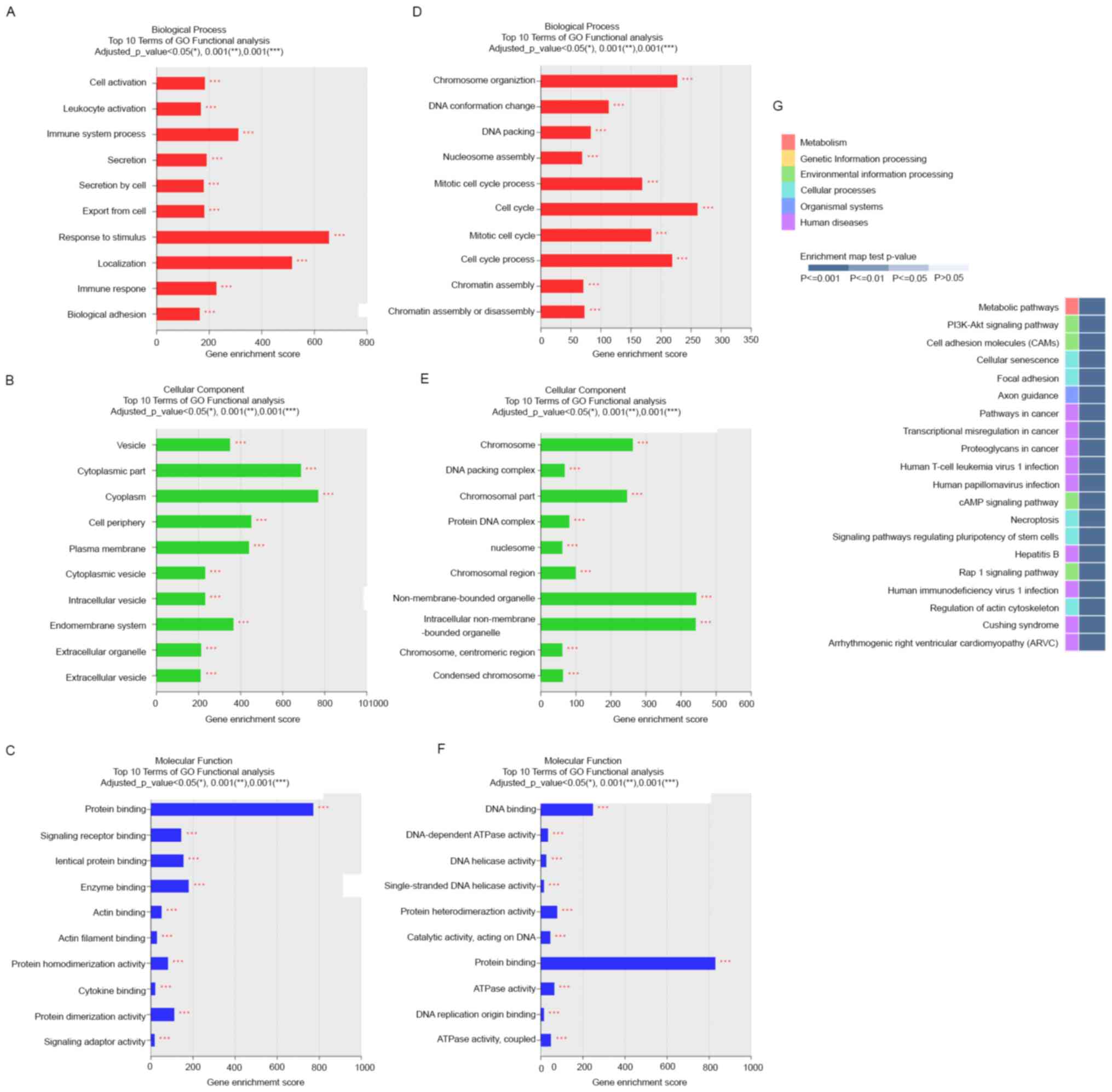
To gain insights into the functions of the DE mRNAs
in the proliferative and mid-secretory endometria, we performed GO
and KEGG analyses of DEGs. GO classes were separated into three
major categories. In the biological process category, DE mRNAs
upregulated in the mid-secretory endometrium were significantly
enriched in response to stimulus, localization, immune system
process, and immune response (Fig.
3A), whereas downregulated DE mRNAs were significantly enriched
in cell cycle and chromosome organization (Fig. 3D). In the cell component category,
upregulated DE mRNAs were mainly enriched in cytoplasm, cytoplasmic
part, cell periphery, and plasma membrane (Fig. 3B), whereas downregulated DE mRNAs
were enriched in nonmembrane-bound organelles, intracellular
nonmembrane-bound organelles, chromosomes, and chromosomal parts
(Fig. 3E). In the molecular
function category, upregulated DE mRNAs were dominantly enriched in
protein binding (Fig. 3C), and
downregulated DE mRNAs were enriched in protein binding and DNA
binding (Fig. 3F).
The top 20 enriched KEGG pathways included
phosphatidylinositol 3-kinase/AKT signaling, cell adhesion
molecules, focal adhesion, cAMP signaling, and Rap1 signaling
pathways (Fig. 3G). These pathways
are associated with the upregulation of essential cell
adhesion-related molecules (integrin, cadherin, selectin, and
laminin) in the mid-secretory endometrium (29). These results suggest that these
pathways mainly contribute to the expression of adhesion molecules
in endometrial epithelial cells during the receptive phase.
Validation of DE lncRNAs, miRNAs, and
mRNAs
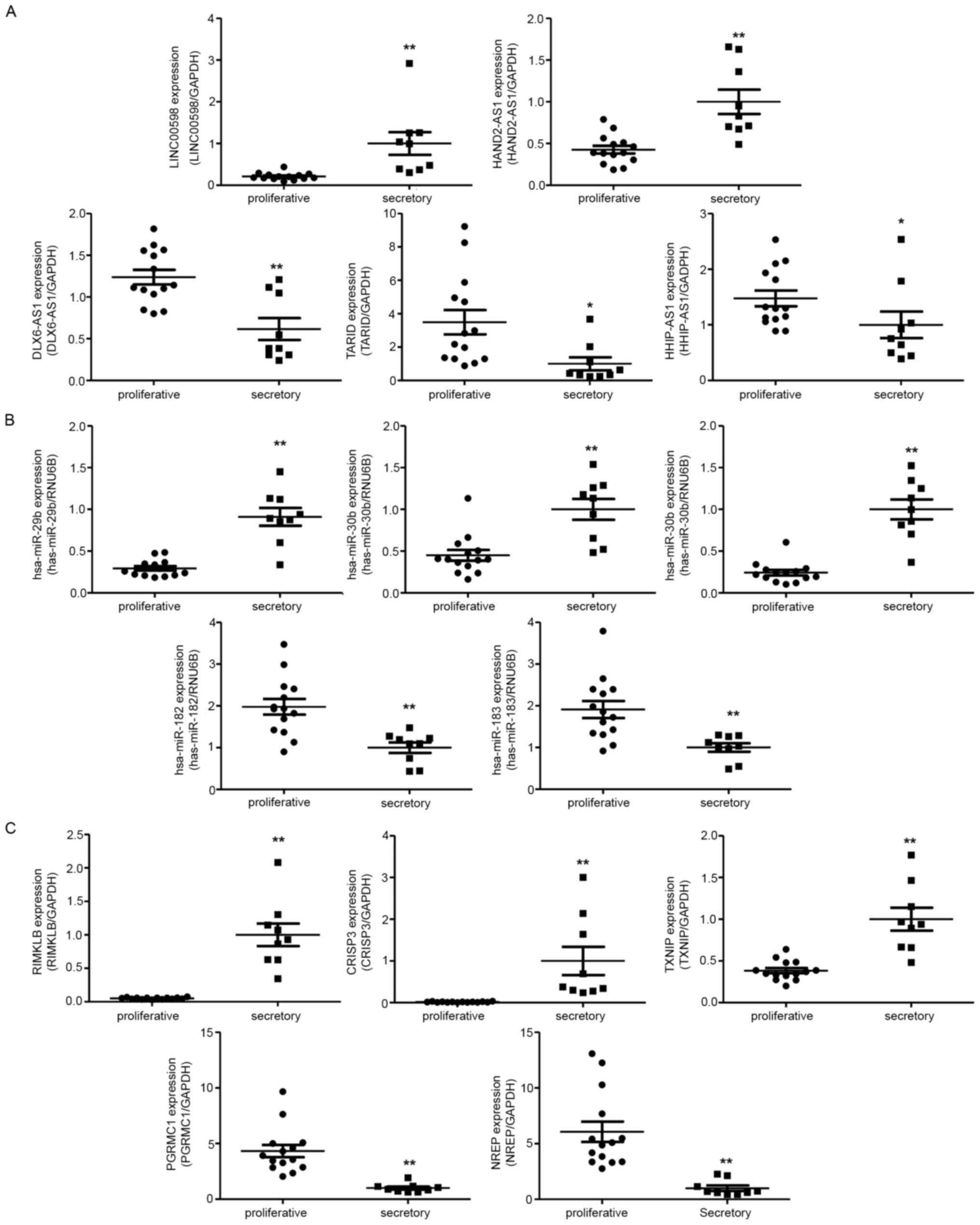
To verify the RNA sequencing results, we
investigated relative ncRNA expression by RT-qPCR. The validated
results of NGS suggested that LINC00598 and HAND2
antisense RNA 1 (HAND2-AS1) were upregulated in the
mid-secretory endometrium, whereas DLX6 antisense RNA1
(DLX6-AS1), TCF21 antisense RNA inducing promoter
demethylation (TARID), and HHIP antisense RNA1
(HHIP-AS1) were downregulated (Fig. 4A). In addition, hsa-miR-29b,
hsa-miR-30b, and hsa-miR-30d were upregulated in the
mid-secretory endometrium, whereas hsa-miR-182 and
hsa-miR-183 were downregulated (Fig. 4B). Of the DE mRNAs, ribosomal
modification protein RimK like family member B (RIMKLB),
cysteine-rich secretory protein 3 (CRISP3), and thioredoxin
interacting protein (TXNIP) were significantly upregulated
in the mid-secretory endometrium, whereas neuronal
regeneration-related protein (NREP) and progesterone
receptor membrane component 1 (PGRMC1) were downregulated
(Fig. 4C).
In silico identification of mRNAs
associated with endometrial receptivity
To investigate the mRNAs associated with endometrial
receptivity, we performed in silico analysis and compared
the DE mRNAs that we identified with those reported by three
previous studies based on RNA-seq (10-12).
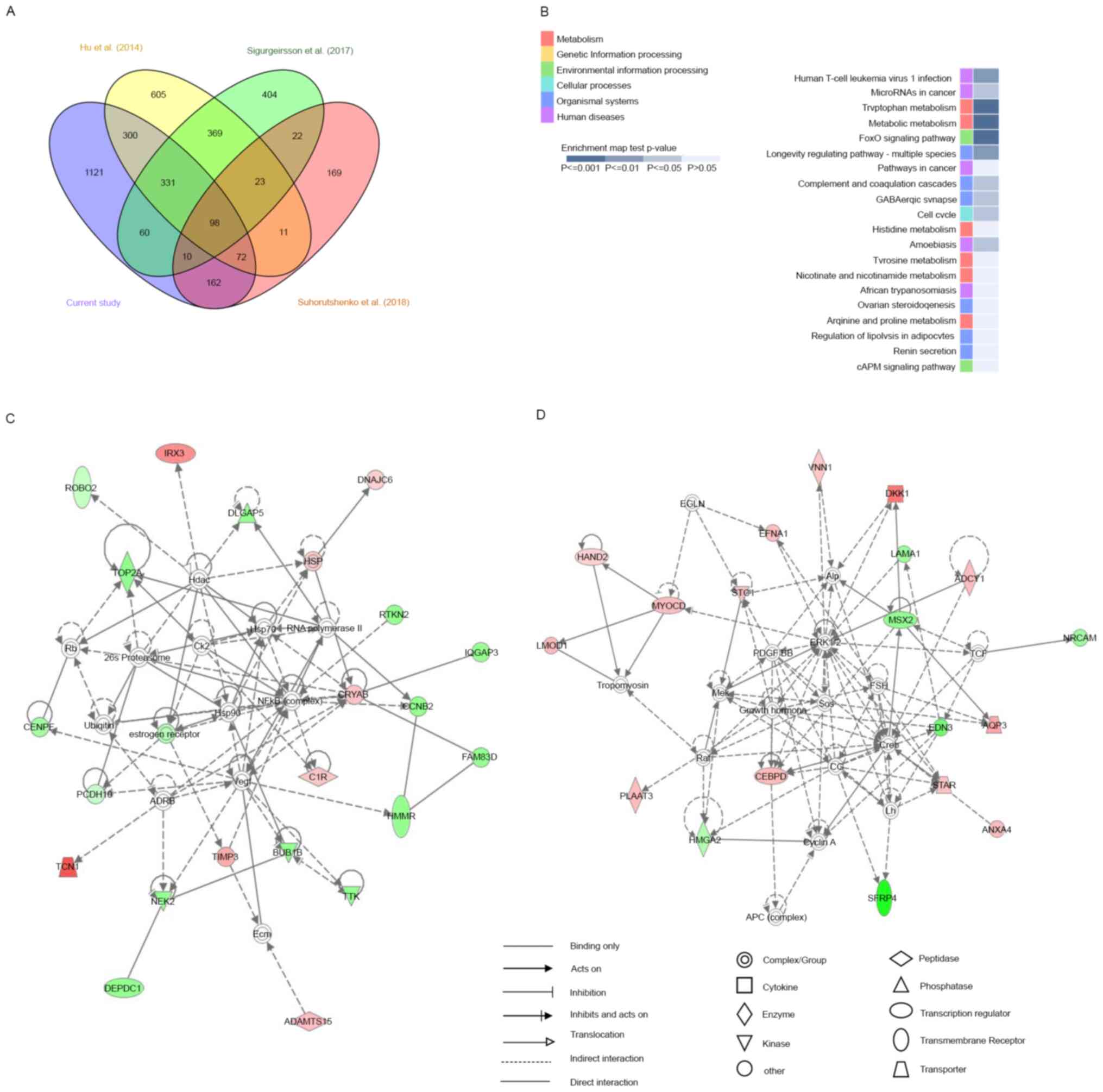
Of the 2,154 DE mRNAs we identified, 501 were identified in at
least two previous studies; 1,121 were uniquely identified in this
study, of which 565 were upregulated and 556 were downregulated.
Moreover, 98 DE mRNAs, of which 60 were upregulated and 38 were
downregulated in the mid-secretory endometrium, were commonly
identified in all four studies (Fig.
5A; Table SVI). Furthermore,
KEGG pathway enrichment analysis revealed that these 98 genes were
significantly involved in tryptophan metabolism, metabolic
pathways, and FoxO signaling (Fig.
5B). In the category of tryptophan metabolism, AOX1,
MAOA, and IDO1 expression levels were highly
increased in the mid-secretory endometrium compared with those in
the proliferative endometrium, whereas in the category of metabolic
pathways, AOX1, MAOA, RIMKLB, IDO1,
NNMT, ARG2, HAL, GALNT13,
PLA2G16, and ADCY1 were upregulated and
GALNT12 was downregulated in the mid-secretory endometrium.
Conversely, in FoxO signaling, SOD2, FBXO32,
GABARAPL1, and BCL6 expression increased in the
mid-secretory endometrium, whereas CCNB2 expression
decreased.
To investigate changes in biological pathways based
on these 98 common DE mRNAs, we performed IPA and identified five
networks, the first of which had a score of 44 and included 23
focus genes (Fig. 5C) whose main
functions were in ‘cell cycle,’ ‘cellular assembly and
organization,’ ‘DNA replication,’ and ‘recombination and repair.’
In this network, ADAMTS15, C1R, CRYAB,
DNAJC6, HSP, IRX3, TCN1, and
TIMP3 mRNAs were upregulated in the mid-secretory
endometrium, whereas BUB1B, CCNB2, CENPF,
DEPDC1, DLGAP5, estrogen receptor, FAM83D,
HMMR, IQGAP3, NEK2, PCDH10,
RTKN2, ROBO1, TOP2A, and TTK mRNAs were
downregulated. The second network had a score of 39 and included 19
focus genes (Fig. 5D), whose main
functions were in ‘cell death and survival,’ ‘cellular
development,’ and ‘connective tissue development and function.’ In
this network, ADCY1, ANXA4, AQP3,
CEBPD, DKK1, EFNA1, HAND2,
LOMD1, MYOCD, PLAAT3, STAR,
STC1, and VNN1 mRNAs were upregulated in the
mid-secretory endometrium, whereas EDN3, HMGA2,
LAMA1, MSX2, NRCAM, and SFRP4 mRNAs
were downregulated. These results suggest that the above networks
were closely related to endometrial receptivity.
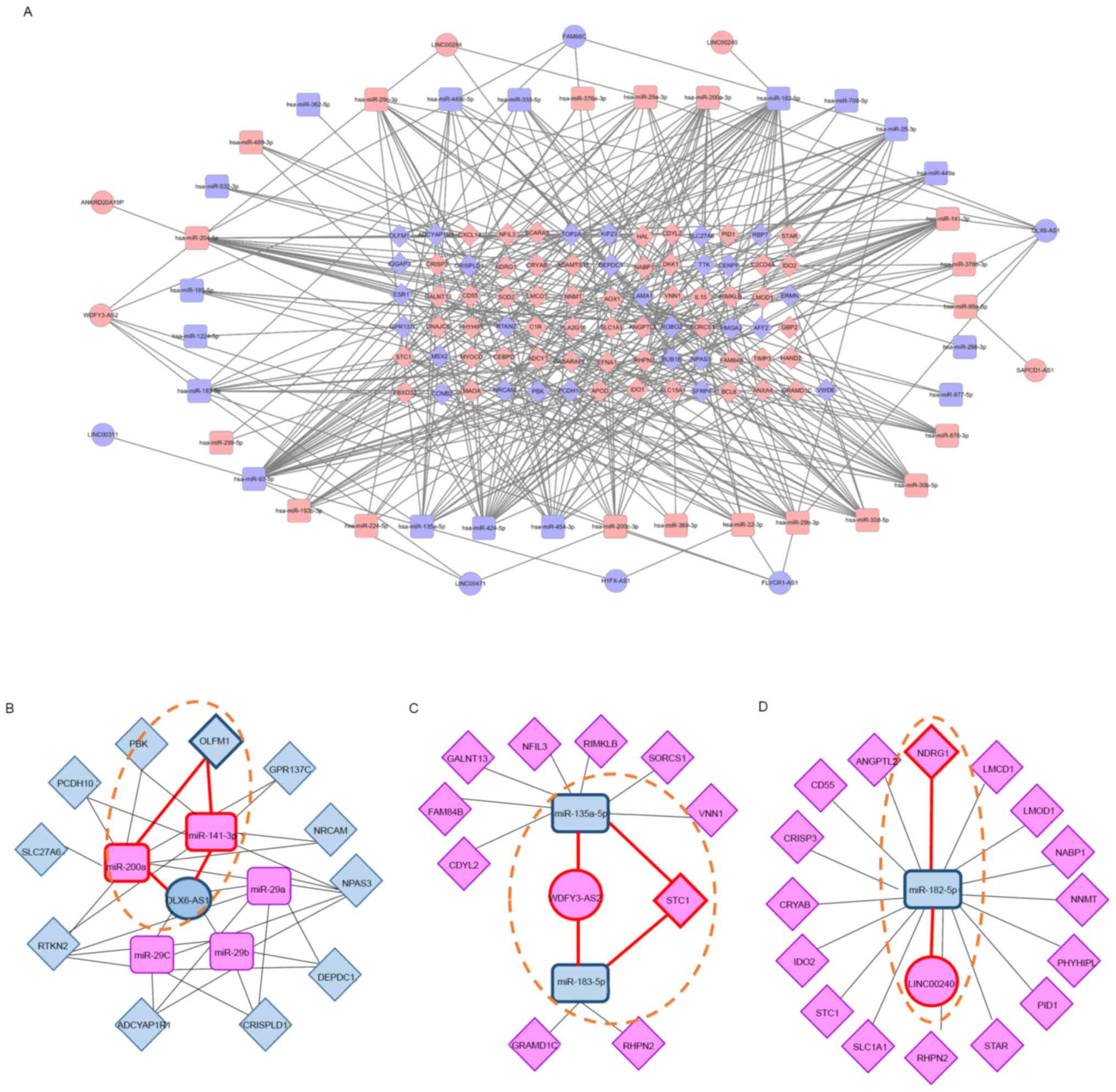
ceRNA network construction
To better understand the pivotal combined roles of
DE lncRNAs, miRNAs, and mRNAs, we constructed and visualized a
ceRNA network (Fig. 6A). Eleven DE
lncRNAs (five upregulated and six downregulated) interacted with 13
DE miRNAs from the miRcode database, and 36 DE miRNAs (19
upregulated and 17 downregulated) interacted with 79 DE mRNAs (51
upregulated and 28 downregulated) retrieved from the mirDIP
database. These 11 DE lncRNAs, 36 DE miRNAs, and 79 DE mRNAs were
then used to establish a ceRNA network that consisted of 126 nodes
and 370 edges, where the number of edges derived from nodes
indicates regulatory interactions between RNAs and the importance
of biological functions. The top hub RNAs included two lncRNAs
(WDFY3-AS2, upregulated; DLX6-AS1, downregulated) and
six miRNAs (hsa-miR-141, hsa-miR-200a, and
hsa-miR-204: Upregulated; hsa-miR-93,
hsa-miR-182, and hsa-miR-424: Downregulated). Of the
hub lncRNAs, DLX6-AS1 formed a connecting network with two
DE miRNAs and 10 DE mRNAs (Fig.
6B). WDFY3-AS2 was also associated with five DE miRNAs
and 11 DE mRNAs (Fig. 6C). Finally,
LINC00240 was associated with one DE miRNA and 16 DE mRNAs
(Fig. 6D).
We also identified hub RNAs forming three axes: The
DLX6-AS1/miR-141 or miR-200a/OLFM1 axis,
WDFY3-AS2/miR-135a or miR-183/STC1 axis, and
LINC00240/miR-182/NDRG1 axis, which may be related to
endometrial receptivity (Fig. 6B,
C and D; dotted lines). To evaluate the RNA
expression correlation between the tree axes, we examined the
relative RNA expression by RT-qPCR in proliferative and
mid-secretory phase endometrial tissues. As shown in Fig. 7, the miRNA expression patterns were
negatively correlated with lncRNAs in both proliferative and
secretory tissues. The mRNA expression patterns were contrary to
those of miRNA. In addition, three hub RNAs showed different
expression patterns between secretory and proliferative tissues.
Taken together, these results indicate that the ceRNA network might
play crucial roles in the regulation of endometrial
receptivity.
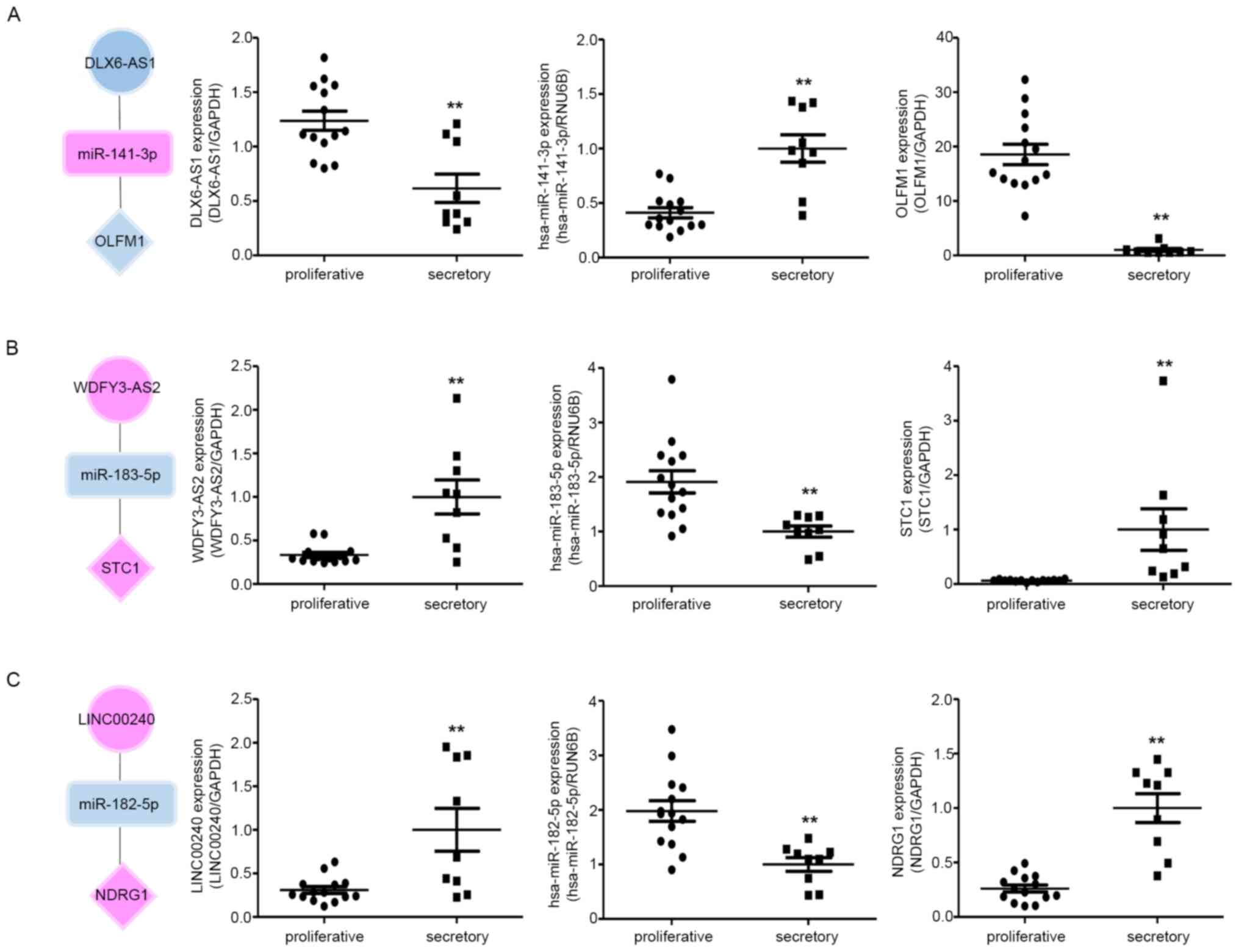 | Figure 7RNA expression correlation between
the three ceRNA subnetworks in proliferative and secretory
endometria. The expression of (A) DLX6-AS1, hsa-miR-141-3p and
OLFM; (B) WDFY3-AS2, hsa-miR-183-5p and STC1; and (C) LINC00240,
hsa-miR-182-5p and NDRG1 (C) was detected by quantitative PCR in
proliferative and secretory endometria. The circles, squares and
diamonds represent lncRNAs, miRNAs and genes, respectively.
Upregulated genes are indicated in red and downregulated genes are
indicated in blue. All data are presented as the mean ± SEM, and
experiments were repeated in triplicate. **P<0.01.
ceRNA, competitive endogenous RNA; lncRNA, long non-coding RNA;
miRNA, microRNA>=. |
Discussion
Many studies have investigated the gene expression
profiles of the prereceptive and receptive endometria to identify
the transcriptome related to endometrial receptivity; however, the
interactions between genes and ncRNAs associated with endometrial
receptivity have remained unclear. In this study, we investigated
whether the interplay among genes, miRNAs, and lncRNAs affect
molecular signaling associated with endometrial receptivity. Of the
top 10 up- and downregulated DE mRNAs identified in our study
(Table II), eight upregulated
genes and three downregulated genes belonged to 57 genes that were
previously proposed as biomarkers of human endometrial receptivity
(30). Of the DEGs, CXC motif
chemokine ligand (CXCL14), transcobalamin 1 (TCN1),
complement component 4 binding protein alpha (C4BPA),
aldehyde oxidase 1 (AOX1), Dickkopf Wnt signaling pathway
inhibitor 1 (DKK1), secreted frizzled-related protein
(SFRP4), endothelin 3 (EDN3), and olfactomedin 1
(OLFM1) were related to 238 endometrial receptivity analysis
array genes (31). The PAEP
gene showed the greatest upregulation in the mid-secretory
endometrium in this study. This gene has been reported to display
negative or low expression in the proliferative endometrium
(32) and may serve as a candidate
biomarker for endometrial receptivity (33,34).
SFRP4 modulates Wnt signaling and is more highly expressed in
proliferative endometrium compared with that in secretory
endometrium (35); upregulation of
SFRP4 in the placenta is associated with severe preeclampsia
(36). OLFM1 is an extracellular
matrix protein that is expressed at significantly lower levels in
the secretory endometrium than in the proliferative endometrium and
negatively regulates spheroid attachment between choriocarcinoma
JAr cells and Ishikawa cells (37).
Moreover, human chorionic gonadotropin secreted by pre-implantation
embryos significantly downregulates OLFM1 expression (38). Thus, the genes identified in this
study may be closely related to endometrial receptivity.
Of the 247 DE lncRNAs identified in this study,
RIMKLB transcript variant 3 showed the highest upregulation,
whereas MMP11 transcript variant 2 showed the greatest
downregulation. Among the top 10 up- and downregulated lncRNAs
(Table III), LINC01320 was
upregulated in the endometrium during the WOI, consistent with
findings of previous studies (11,12).
KCNIP4-IT1 and DLX6-AS1 are also downregulated in the
secretory endometrium (11),
whereas DLX6-AS1 is overexpressed in the placenta of
patients with preeclampsia and is associated with reduced
trophoblast proliferation, migration, and invasion (39). However, the correlations between
lncRNAs and implantation are not clear. Of the 67 DE miRNAs
identified in our study, hsa-miR-4525 showed the highest
upregulation, and hsa-miR-149-3p showed the most
downregulation. Mmu-miR-193 influences embryo implantation
by modulating growth factor receptor-bound protein 7 expression in
mice (40). Similarly,
miR-30d knockout reduces the implantation rate in mice
(41), and miR-135a and
miR-135b are downregulated in receptive endometrium
(11). FOXO1 is a decidualization
marker that is regulated post-transcriptionally by miR-135a
in human melanoma cells (42);
however, the roles of miRNAs in the endometrium remain unclear.
Of top 20 enriched KEGG pathways, adhesion molecules
are known to be expressed in human endometrial epithelial cells and
can be used as markers of endometrial receptivity (43). Focal adhesion, cAMP signaling, and
RAP1 signaling pathways were significantly enhanced in the
mid-secretory endometrium. Consistent with this, Kusama et
al demonstrated that RAP1 is crucial for cAMP-mediated
decidualization in rat and human endometrial stromal cells
(44). Thus, focal adhesion, cAMP
signaling, and RAP1 signaling may be important for decidualization
during the receptive phase.
Of the 2,154 DEGs identified herein, 1,121 mRNAs
were uniquely identified in this study. Recently, 18,210 structural
variations have been identified in the Korean human genome compared
with the human reference genome GRCh38, and most of the insertions
among structural variants are causes of the variance in the
transcriptome (45). Therefore, as
compared with previously published data, the mRNAs that were
uniquely expressed in our study were probably derived from
differences between human populations. Moreover, the study
endometrium donors (about 37.5 years of age) were about 10 years
older than donors in previous studies (about 28 years of age).
Aging regulates complex biological processes in humans (46). Therefore, differences in the age of
volunteers may have contributed to the uniquely expressed mRNA. By
analyzing previously published RNA-seq expression profiling
studies, we identified 98 common genes involved in mid-secretory
endometrial function, of which 34 were shared with the endometrial
receptivity analysis array, a commercial tool used for endometrial
receptivity diagnosis (31).
Moreover, 50 of these 98 common genes were identical to 57 genes
proposed as putative receptivity markers through meta-analysis by
Altmäe et al (30).
Therefore, we propose that the 98 common genes identified in this
study may be related to endometrial receptivity. In the KEGG
pathways of these 98 common genes, previous studies have reported
the expression patterns of AOX1, MAOA, IDO1,
NNMT, ARG2, HAL, GALNT12, SOD2,
GABARAPL1, BCL6, and CCNB2 in the endometrium
(30,31). In particular, the localization and
involvement of AOX1, MAOA, IDO1, ARG2, NNMT, and BCL6 in the
mid-secretory endometrium have been well described (30). MAOA is known to affect human
endometrial receptivity, and its expression may be altered by
inadequate decidualization (47);
indeed, patients who experience implantation failure display
decreased MAOA expression (48,49).
IDO1 inhibits the expression of the decidualization marker genes
Prl and Igfbp1 in mice under in vitro
decidualization (50). FOXO1 is an
important cAMP-dependent transcription factor in decidualizing
human endometrial stromal cells and exerts antioxidant properties
by targeting and regulating SOD2 expression (51-55).
SOD2 and FoxO1 expression is induced during the differentiation of
the stromal compartment in the mid- to late-secretory phase of the
cycle and these proteins are expressed in decidualizing endometrial
stromal cells in culture (52). In
addition, decidualization is associated with the induction of
various free radical scavengers, including SOD2(56). Although MAOA, IDO1, and SOD2 are
known to participate in decidualization and endometrial
receptivity, further studies are required to determine the effects
of the other genes on endometrial receptivity.
In this study, we also demonstrated that networks
related to the cell cycle, DNA replication, DNA recombination, and
DNA repair for cell proliferation were mostly downregulated in the
mid-secretory endometrium, whereas networks involved in cell
survival, cellular development, and connective tissue development
were upregulated. Within these networks, the DKK1 protein has been
shown to affect spheroid attachment on endometrial epithelial
cells, and MSX2 knockout directly affects endometrial receptivity
and embryo implantation in mice (57,58).
Therefore, these networks and the genes involved in them may play
important roles in endometrial receptivity and implantation.
LncRNA/miRNA/mRNA interactions are known to form a
network of ceRNAs with key roles in biological networks. Until
recently, most studies of ceRNAs have been related to cancer,
including tumor diagnosis, prognosis, and targeted treatments, with
few studies examining endometrial receptivity by constructing ceRNA
networks. Recently, in patients with and without endometriosis,
Wang and Yu identified four ceRNA networks as biomarkers for
endometrial receptivity (59).
Similarly, Xu et al identified potential novel biomarkers
for repeated implantation failure from a ceRNA network constructed
from DE RNAs (60). In this study,
we successfully constructed ceRNA networks to identify the RNA
interactions that affect endometrial receptivity and discovered
that the top hub RNAs includ two lncRNAs and six miRNAs. Of the hub
lncRNAs, DLX6-AS1 acts as a sponge for many miRNAs and is
significantly overexpressed in various cancers, including cervical
cancer (61,62). DLX6-AS1 is also upregulated
in the placenta of patients with preeclampsia and negatively
regulates the proliferation, migration, and invasion of
trophoblasts (39). Of the hub
miRNAs, miR-141-3p can directly sponge DLX6-AS1
(63), and miR-141 is
upregulated in endometriomas compared with the eutopic endometrium
of patients with endometriosis (64). Interestingly, mmu-miR-141
affects the proliferation of endometrial cells and the number of
embryo implantation sites in mice, suggesting essential roles in
embryo implantation (65).
miR-141 belongs to the miR-200 family, which also
includes miR-200a, miR-200b, miR-200c, and
miR-429. miR-200a/b/c are upregulated during
endometrial stromal cell decidualization in vitro (66), whereas OLFM1 has been reported as a
target of miR-141 and miR-200a in human gastric
cancer cells (67). In a
trophoblastic spheroid (JAr)-endometrial epithelial cell (Ishikawa)
co-culture model, recombinant OLFM1 protein treatment suppressed
the attachment of JAr spheroids onto the Ishikawa cell monolayer,
suggesting that OLFM1 inhibits endometrial receptivity (68). Therefore, the
DLX6-AS1/miR-141 or miR-200a/OLFM1 axes may be
important regulators of endometrial receptivity.
Endometrial cell proliferation decreased between
the proliferative to mid-secretory phases; however, uncontrolled
endometrial epithelial cell proliferation can lead to implantation
failure and has been observed in the eutopic secretory endometrium
of patients with endometriosis (69). Therefore, endometrial cell
proliferation is closely related to pregnancy. WDFY3-AS2,
which was identified from our ceRNA network, has no known
endometrial function but has been reported to be related to cancer
progression. For example, WDFY3-AS2 inhibits cancer cell
proliferation and invasion (70,71),
and miR-135a, which is a potential target of WDFY3-AS
in diffuse glioma (72), promotes
cancer cell proliferation, migration, and invasion (73) and directly suppresses FOXO1 in
hepatocellular carcinoma cells (74). Other pregnancy-related studies
showed strong expression of stanniocalcin (STC) 1 and
STC2 mRNA in decidualized rat cells, suggesting that STC1
and STC2 play important roles in implantation and decidualization
(75). In humans, STC1 is
upregulated in the secretory endometrium compared with that in the
proliferative endometrium, and is dysregulated in the eutopic
endometrium of patients with endometriosis, suggesting its roles in
the pathogenesis of decidualization defects (76). Therefore, accumulating evidence
indicates that the WDFY3-AS2/miR-135a/STC1 axis may be
involved in endometrial stromal cell decidualization. The ceRNA
network in this study revealed a potential, previously unknown
interaction between WDFY3-AS2 and miR-183, which is
decreased in vitro during human endometrial stromal cell
decidualization (77). In addition,
miR-183 suppresses FOXO1 in non-small cell lung cancer
(NSCLC), significantly increasing NSCLC growth in vitro and
in vivo (78). Recently,
Akbar et al suggested that miR-183-5p may be a
potential biomarker for endometrial receptivity (79), and STC1 has been reported as a
potential target of miR-183-5p in bladder cancer (80). Thus, our study suggests that the
WDFY3-AS2/miR-183-5p/STC1 axis could be a biomarker for
endometrial receptivity.
Although LINC00240 was more highly expressed
in the secretory endometrium than in the proliferative endometrium
in this study, no interaction between LINC00240 and
miR-182 has been reported. LINC00240 promotes cell
proliferation, invasion, and migration in gastric and cervical
cancer (81,82), and miR-182 suppresses FOXO1
expression in endometrial cancer cells (83). In addition, N-myc downregulated gene
1 (NDRG1) directly affects pregnancy in mice, and reduced NDRG1
expression has been reported in decidual samples from patients with
recurrent miscarriage (84). Thus,
the LINC00240/miR-182/NDRG1 axis may also play important
roles in endometrial receptivity.
There were some limitations to this study. First,
the sample size was small, and tissue heterogeneity could have
limited the generalization of the results. Additionally, sample
collection was based on menstrual cycle history, ultrasound
finding, and histological confirmation rather than serum
luteinizing hormone concentrations, which are generally used for
cycle dating. In fact, endometrial receptivity is the receptive
status of the endometrium that allows the embryo to implant and is
usually compared between endometria of fertile and infertile
patients in the WOI. Therefore, we are currently collecting
endometrium samples from infertile patients to further explore
genes associated with endometrial receptivity.
In conclusion, the ceRNA networks constructed in
this study may partially explain the regulatory mechanisms
underlying endometrial receptivity; however, further studies are
required to define the relationships between these ceRNA networks
and endometrial receptivity.
Supplementary Material
Primer sequences for the validation of
next-generation sequencing results.
Primers information for the validation
of microRNA.
Differentially expressed mRNAs with
1.5 fold change between mid-secretory phase and proliferative phase
endometrium.
Differentially expressed lncRNAs with
1.5 fold change between mid-secretory phase and proliferative phase
endometrium. Sorted fold change from highly expressed in secretory
phase to proliferative phase.
Differentially expressed miRNAs with
1.5 FC between the mid-secretory phase and proliferative phase
endometrium.
Commonly expressed mRNA with 1.5 fold
change between mid-secretory phase and proliferative phase
endometrium.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Basic Science
Research Program through the National Research Foundation of the
Ministry of Education, Korea (grant no.
NRF-2017R1A6A1A03015713).
Availability of data and materials
The datasets used an/or analyzed are available from
the corresponding author on reasonable request. The smRNA
sequencing data that support the findings of this study are openly
available in the GEO database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE167325).
Authors' contributions
JK and SRP conceived the current study and designed
the experiments. THK provided resources. SLY, THK and YHH designed
the methods. SLY, YHH, YK and DUJ constructed the figures. DCL
performed ingenuity pathway analysis. SLY, YHH, YK and DUJ
validated the genes, lncRNAs and miRNAs identified from RNA-seq.
SLY, THK, YHH and SRP wrote the original draft of the manuscript.
SLY, THK, JK and SRP wrote and reviewed the manuscript. JK and SRP
supervised the current study. JK and SRP acquired funding. All
authors have read and approved the final manuscript, and agree to
the published version of the manuscript. JK and SRP confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
The current study was approved by the Bioethics
Committee of KYU (Institutional Review Board File No.
2018-11-007-005). All patients provided written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Psychoyos A: Hormonal control of
ovoimplantation. Vitam Horm. 31:201–256. 1973.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Simon C, Martin JC and Pellicer A:
Paracrine regulators of implantation. Best Pract Res Clin Obstet
Gynaecol. 14:815–826. 2000.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Aplin JD: The cell biological basis of
human implantation. Best Pract Res Clin Obstet Gynaecol.
14:757–764. 2000.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Cha J, Sun X and Dey SK: Mechanisms of
implantation: Strategies for successful pregnancy. Nat Med.
18:1754–1767. 2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Carson DD, Lagow E, Thathiah A, Al-Shami
R, Farach-Carson MC, Vernon M, Yuan L, Fritz MA and Lessey B:
Changes in gene expression during the early to mid-luteal
(receptive phase) transition in human endometrium detected by
high-density microarray screening. Mol Hum Reprod. 8:871–879.
2002.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kao LC, Tulac S, Lobo S, Imani B, Yang JP,
Germeyer A, Osteen K, Taylor RN, Lessey BA and Guidice LC: Global
gene profiling in human endometrium during the window of
implantation. Endocrinology. 143:2119–2138. 2002.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Borthwick JM, Charnock-Jones DS, Tom BD,
Hull ML, Teirney R, Phillips SC and Smith SK: Determination of the
transcript profile of human endometrium. Mol Hum Reprod. 9:19–33.
2003.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Riesewijk A, Martin J, van Os R,
Horcajadas JA, Polman J, Pellicer A, Mosselman S and Simón C: Gene
expression profiling of human endometrial receptivity on days LH+2
versus LH+7 by microarray technology. Mol Hum Reprod. 9:253–264.
2003.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Mirkin S, Arslan M, Churikov D, Corica A,
Diaz JI, Williams S, Bocca S and Oehninger S: In search of
candidate genes critically expressed in the human endometrium
during the window of implantation. Hum Reprod. 20:2104–2117.
2005.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Hu S, Yao G, Wang Y, Xu H, Ji X, He Y, Zhu
Q, Chen Z and Sun Y: Transcriptomic changes during the
pre-receptive to receptive transition in human endometrium detected
by RNA-Seq. J Clin Endocrinol Metab. 99:E2744–E2753.
2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Sigurgeirsson B, Amark H, Jemt A, Ujvari
D, Westgren M, Lundeberg J and Gidlöf S: Comprehensive RNA
sequencing of healthy human endometrium at two time points of the
menstrual cycle. Biol Reprod. 96:24–33. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Suhorutshenko M, Kukushkina V,
Velthut-Meikas A, Altmäe S, Peters M, Mägi R, Krjutškov K, Koel M,
Codoñer FM, Martinez-Blanch JF, et al: Endometrial receptivity
revisited: Endometrial transcriptome adjusted for tissue cellular
heterogeneity. Hum Reprod. 33:2074–2086. 2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Gomez E, Ruiz-Alonso M, Miravet J and
Simon C: Human endometrial transcriptomics: Implications for
embryonic implantation. Cold Spring Harb Perspect Med.
5(a022996)2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yamamura S, Imai-Sumida M, Tanaka Y and
Dahiya R: Interaction and cross-talk between non-coding RNAs. Cell
Mol Life Sci. 75:467–484. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Fan LJ, Han HJ, Guan J, Zhang XW, Cui QH,
Shen H and Shi C: Aberrantly expressed long noncoding RNAs in
recurrent implantation failure: A microarray related study. Syst
Biol Reprod Med. 63:269–278. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Feng C, Shen JM, Lv PP, Jin M, Wang LQ,
Rao JP and Feng L: Construction of implantation failure related
lncRNA-mRNA network and identification of lncRNA biomarkers for
predicting endometrial receptivity. Int J Biol Sci. 14:1361–1377.
2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Chen MY, Liao GD, Zhou B, Kang LN, He YM
and Li SW: Genome-wide profiling of long noncoding RNA expression
patterns in women with repeated implantation failure by RNA
sequencing. Reprod Sci. 26:18–25. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Lim LP, Lau NC, Garrett-Engele P, Grimson
A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Rižner TL: Discovery of biomarkers for
endometrial cancer: Current status and prospects. Expert Rev Mol
Diagn. 16:1315–1336. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Wilczynski M, Danielska J, Dzieniecka M,
Szymanska B, Wojciechowski M and Malinowski A: Prognostic and
clinical significance of miRNA-205 in endometrioid endometrial
cancer. PLoS One. 11(e0164687)2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Bashti O, Noruzinia M, Garshasbi M and
Abtahi M: miR-31 and miR-145 as potential non-invasive regulatory
biomarkers in patients with endometriosis. Cell J.
20(293)2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Noyes RW, Hertig AT and Rock J: Reprint
of: Dating the endometrial biopsy. Fertil Steril. 112 (Suppl
1):e93–e115. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Pertea M, Kim D, Pertea GM, Leek JT and
Salzberg SL: Transcript-level expression analysis of RNA-seq
experiments with HISAT, StringTie and Ballgown. Nat Protoc.
11:1650–1667. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15(550)2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Re. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Achache H and Revel A: Endometrial
receptivity markers, the journey to successful embryo implantation.
Hum Reprod Update. 12:731–746. 2006.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Altmäe S, Koel M, Võsa U, Adler P,
Suhorutšenko M, Laisk-Podar T, Kukushkina V, Saare M,
Velthut-Meikas A, Krjutškov K, et al: Meta-signature of human
endometrial receptivity: A meta-analysis and validation study of
transcriptomic biomarkers. Sci Rep. 7(10077)2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Díaz-Gimeno P, Horcajadas JA,
Martínez-Conejero JA, Esteban FJ, Alamá P, Pellicer A and Simón C:
A genomic diagnostic tool for human endometrial receptivity based
on the transcriptomic signature. Fertil Steril. 95:50–60, 60 e1-15.
2011.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Julkunen M, Koistinen R, Sjöberg J,
Rutanen EM, Wahlström T and Seppälä M: Secretory endometrium
synthesizes placental protein 14. Endocrinology. 118:1782–1786.
1986.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Chan C, Virtanen C, Winegarden NA, Colgan
TJ, Brown TJ and Greenblatt EM: Discovery of biomarkers of
endometrial receptivity through a minimally invasive approach: A
validation study with implications for assisted reproduction.
Fertil Steril. 100:810–817. 2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Burmenskaya OV, Bozhenko VK, Smolnikova
VY, Kalinina EA, Korneeva IE, Donnikov AE, Beyk EP, Naumov VA,
Aleksandrova NV, Borovikov PI and Trofimov DY: Transcription
profile analysis of the endometrium revealed molecular markers of
the personalized ‘window of implantation’ during in vitro
fertilization. Gynecol Endocrinol. 33 (Supp1):S22–S27.
2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhang D, Sun C, Ma C, Dai H and Zhang W:
Data mining of spatial-temporal expression of genes in the human
endometrium during the window of implantation. Reprod Sci.
19:1085–1098. 2012.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhang Z, Zhang L, Zhang L, Jia L, Wang P
and Gao Y: Association of Wnt2 and sFRP4 expression in the third
trimester placenta in women with severe preeclampsia. Reprod Sci.
20:981–989. 2013.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Kodithuwakku SP, Ng PY, Liu Y, Ng EH,
Yeung WS, Ho PC and Lee KF: Hormonal regulation of endometrial
olfactomedin expression and its suppressive effect on spheroid
attachment onto endometrial epithelial cells. Hum Reprod.
26:167–175. 2011.PubMed/NCBI View Article : Google Scholar
|
|
38
|
So KH, Kodithuwakku SP, Kottawatta KS, Li
RH, Chiu PC, Cheung AN, Ng EH, Yeung WS and Lee KF: Human chorionic
gonadotropin stimulates spheroid attachment on fallopian tube
epithelial cells through the mitogen-activated protein kinase
pathway and down-regulation of olfactomedin-1. Fertil Steril.
104:474–482. 2015.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Tan Y, Xiao D, Xu Y and Wang C: Long
non-coding RNA DLX6-AS1 is upregulated in preeclampsia and
modulates migration and invasion of trophoblasts through the
miR-376c/GADD45A axis. Exp Cell Res. 370:718–724. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Li R, He J, Chen X, Ding Y, Wang Y, Long
C, Shen L and Liu X: Mmu-miR-193 is involved in embryo implantation
in mouse uterus by regulating GRB7 gene expression. Reprod Sci.
21:733–742. 2014.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Balaguer N, Moreno I, Herrero M,
Gonzáléz-Monfort M, Vilella F and Simón C: MicroRNA-30d deficiency
during preconception affects endometrial receptivity by decreasing
implantation rates and impairing fetal growth. Am J Obstet Gynecol.
221:46 e1–46 e16. 2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Ren JW, Li ZJ and Tu C: MiR-135
post-transcriptionally regulates FOXO1 expression and promotes cell
proliferation in human malignant melanoma cells. Int J Clin Exp
Pathol. 8:6356–6366. 2015.PubMed/NCBI
|
|
43
|
Singh H and Aplin JD: Adhesion molecules
in endometrial epithelium: Tissue integrity and embryo
implantation. J Anat. 215:3–13. 2009.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Kusama K, Yoshie M, Tamura K, Daikoku T,
Takarada T and Tachikawa E: Possible roles of the cAMP-mediators
EPAC and RAP1 in decidualization of rat uterus. Reproduction.
147:897–906. 2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Seo JS, Rhie A, Kim J, Lee S, Sohn MH, Kim
CU, Hastie A, Cao H, Yun JY, Kim J, et al: De novo assembly and
phasing of a Korean human genome. Nature. 538:243–247.
2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Galkin F, Mamoshina P, Aliper A, de
Magalhaes JP, Gladyshev VN and Zhavoronkov A: Biohorology and
biomarkers of aging: Current state-of-the-art, challenges and
opportunities. Ageing Res Rev. 60(101050)2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Gibson DA, Simitsidellis I, Cousins FL,
Critchley HO and Saunders PT: Intracrine androgens enhance
decidualization and modulate expression of human endometrial
receptivity genes. Sci Rep. 6(19970)2016.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Henriquez S, Tapia A, Quezada M, Vargas M,
Cardenas H, Rios M, Salvatierra AM, Croxatto H, Orihuela P,
Zegers-Hochschild F, et al: Deficient expression of monoamine
oxidase A in the endometrium is associated with implantation
failure in women participating as recipients in oocyte donation.
Mol Hum Reprod. 12:749–754. 2006.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Tapia A, Gangi LM, Zegers-Hochschild F,
Balmaceda J, Pommer R, Trejo L, Pacheco IM, Salvatierra AM,
Henríquez S, Quezada M, et al: Differences in the endometrial
transcript profile during the receptive period between women who
were refractory to implantation and those who achieved pregnancy.
Hum Reprod. 23:340–351. 2008.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Li DD, Yin YH, Wu JY, Yang ZQ, Cao H,
Zhang QL, Guo B and Yue ZP: Effects of Ido1 on mouse
decidualization. Mol Biol (Mosk). 49:649–657. 2015.PubMed/NCBI View Article : Google Scholar : (In Russian).
|
|
51
|
Christian M, Zhang X, Schneider-Merck T,
Unterman TG, Gellersen B, White JO and Brosens JJ: Cyclic
AMP-induced forkhead transcription factor, FKHR, cooperates with
CCAAT/enhancer-binding protein beta in differentiating human
endometrial stromal cells. J Biol Chem. 277:20825–20832.
2002.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Kajihara T, Jones M, Fusi L, Takano M,
Feroze-Zaidi F, Pirianov G, Mehmet H, Ishihara O, Higham JM, Lam EW
and Brosens JJ: Differential expression of FOXO1 and FOXO3a confers
resistance to oxidative cell death upon endometrial
decidualization. Mol Endocrinol. 20:2444–2455. 2006.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Labied S, Kajihara T, Madureira PA, Fusi
L, Jones MC, Higham JM, Varshochi R, Francis JM, Zoumpoulidou G,
Essafi A, et al: Progestins regulate the expression and activity of
the forkhead transcription factor FOXO1 in differentiating human
endometrium. Mol Endocrinol. 20:35–44. 2006.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Lehtinen MK, Yuan Z, Boag PR, Yang Y,
Villén J, Becker EB, DiBacco S, de la Iglesia N, Gygi S, Blackwell
TK and Bonni A: A conserved MST-FOXO signaling pathway mediates
oxidative-stress responses and extends life Span. Cell.
125:987–1001. 2006.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Rached MT, Kode A, Xu L, Yoshikawa Y, Paik
JH, Depinho RA and Kousteni S: FoxO1 is a positive regulator of
bone formation by favoring protein synthesis and resistance to
oxidative stress in osteoblasts. Cell Metab. 11:147–160.
2010.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Gellersen B, Brosens IA and Brosens JJ:
Decidualization of the human endometrium: Mechanisms, functions,
and clinical perspectives. Semin Reprod Med. 25:445–453.
2007.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Liu Y, Kodithuwakku SP, Ng PY, Chai J, Ng
EH, Yeung WS, Ho PC and Lee KF: Excessive ovarian stimulation
up-regulates the Wnt-signaling molecule DKK1 in human endometrium
and may affect implantation: An in vitro co-culture study. Hum
Reprod. 25:479–490. 2010.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Daikoku T, Cha J, Sun X, Tranguch S, Xie
H, Fujita T, Hirota Y, Lydon J, DeMayo F, Maxson R and Dey SK:
Conditional deletion of Msx homeobox genes in the uterus inhibits
blastocyst implantation by altering uterine receptivity. Dev Cell.
21:1014–1025. 2011.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Wang X and Yu Q: Endometriosis-related
ceRNA network to identify predictive biomarkers of endometrial
receptivity. Epigenomics. 11:147–167. 2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Xu H, Zhou M, Cao Y, Zhang D, Han M, Gao
X, Xu B and Zhang A: Genome-wide analysis of long noncoding RNAs,
microRNAs, and mRNAs forming a competing endogenous RNA network in
repeated implantation failure. Gene. 720(144056)2019.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Fang C, Xu L, He W, Dai J and Sun F: Long
noncoding RNA DLX6-AS1 promotes cell growth and invasiveness in
bladder cancer via modulating the miR-223-HSP90B1 axis. Cell Cycle.
18:3288–3299. 2019.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Wang X, Lin Y and Liu J: Long noncoding
RNA DLX6AS1 promotes proliferation by acting as a ceRNA targeting
miR199a in cervical cancer. Mol Med Rep. 19:1248–1255.
2019.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Guo Q, Sun H, Zheng K, Yin S and Niu J:
Long non-coding RNA DLX6-AS1/miR-141-3p axis regulates osteosarcoma
proliferation, migration and invasion through regulating Rab10. RSC
Adv. 9:33823–33833. 2019.
|
|
64
|
Hawkins SM, Creighton CJ, Han DY, Zariff
A, Anderson ML, Gunaratne PH and Matzuk MM: Functional microRNA
involved in endometriosis. Mol Endocrinol. 25:821–832.
2011.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Liu X, Gao R, Chen X, Zhang H, Zheng A,
Yang D, Ding Y, Wang Y and He J: Possible roles of mmu-miR-141 in
the endometrium of mice in early pregnancy following embryo
implantation. PLoS One. 8(e67382)2013.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Jimenez PT, Mainigi MA, Word RA, Kraus WL
and Mendelson CR: miR-200 regulates endometrial development during
early pregnancy. Mol Endocrinol. 30:977–987. 2016.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Wu XM, Shao XQ, Meng XX, Zhang XN, Zhu L,
Liu SX, Lin J and Xiao HS: Genome-wide analysis of microRNA and
mRNA expression signatures in hydroxycamptothecin-resistant gastric
cancer cells. Acta Pharmacol Sin. 32:259–269. 2011.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Kottawatta KS, So KH, Kodithuwakku SP, Ng
EH, Yeung WS and Lee KF: MicroRNA-212 regulates the expression of
olfactomedin 1 and C-terminal binding protein 1 in human
endometrial epithelial cells to enhance spheroid attachment in
vitro. Biol Reprod. 93(109)2015.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Franco-Murillo Y, Miranda-Rodríguez JA,
Rendón-Huerta E, Montaño LF, Cornejo GV, Gómez LP, Valdez-Morales
FJ, Gonzalez-Sanchez I and Cerbón M: Unremitting cell proliferation
in the secretory phase of eutopic endometriosis: Involvement of
pAkt and pGSK3β. Reprod Sci. 22:502–510. 2015.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Li W, Ma S, Bai X, Pan W, Ai L and Tan W:
Long noncoding RNA WDFY3-AS2 suppresses tumor progression by acting
as a competing endogenous RNA of microRNA-18a in ovarian cancer. J
Cell Physiol. 235:1141–1154. 2020.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Zhang Q, Guan F, Fan T, Li S, Ma S, Zhang
Y, Guo W and Liu H: LncRNA WDFY3-AS2 suppresses proliferation and
invasion in oesophageal squamous cell carcinoma by regulating
miR-2355-5p/SOCS2 axis. J Cell Mol Med. 24:8206–8220.
2020.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Wu F, Zhao Z, Chai R, Liu Y, Wang K, Wang
Z, Li G, Huang R, Jiang H and Zhang K: Expression profile analysis
of antisense long non-coding RNA identifies WDFY3-AS2 as a
prognostic biomarker in diffuse glioma. Cancer Cell Int.
18(107)2018.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Wang J, Zhang L, Jiang W, Zhang R, Zhang
B, Silayiding A and Duan X: MicroRNA-135a promotes proliferation,
migration, invasion and induces chemoresistance of endometrial
cancer cells. Eur J Obstet Gynecol Reprod Biol X.
5(100103)2019.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Zeng YB, Liang XH, Zhang GX, Jiang N,
Zhang T, Huang JY, Zhang L and Zeng XC: miRNA-135a promotes
hepatocellular carcinoma cell migration and invasion by targeting
forkhead box O1. Cancer Cell Int. 16(63)2016.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Xiao LJ, Yuan JX, Song XX, Li YC, Hu ZY
and Liu YX: Expression and regulation of stanniocalcin 1 and 2 in
rat uterus during embryo implantation and decidualization.
Reproduction. 131:1137–1149. 2006.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Aghajanova L, Altmae S, Kasvandik S,
Salumets A, Stavreus-Evers A and Giudice LC: Stanniocalcin-1
expression in normal human endometrium and dysregulation in
endometriosis. Fertil Steril. 106:681–691 e1. 2016.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Estella C, Herrer I, Moreno-Moya JM,
Quiñonero A, Martínez S, Pellicer A and Simón C: miRNA signature
and Dicer requirement during human endometrial stromal
decidualization in vitro. PLoS One. 7(e41080)2012.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Zhang L, Quan H, Wang S, Li X and Che X:
MiR-183 promotes growth of non-small cell lung cancer cells through
FoxO1 inhibition. Tumour Biol. 36:8121–8126. 2015.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Akbar R, Ullah K, Rahman TU, Cheng Y, Pang
HY, Jin LY, Wang QJ, Huang HF and Sheng JZ: miR-183-5p regulates
uterine receptivity and enhances embryo implantation. J Mol
Endocrinol. 64:43–52. 2020.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Gao JM, Huang LZ, Huang ZG and He RQ:
Clinical value and potential pathways of miR-183-5p in bladder
cancer: A study based on miRNA-seq data and bioinformatics
analysis. Oncol Lett. 15:5056–5070. 2018.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Li Y, Yan J, Wang Y, Wang C, Zhang C and
Li G: LINC00240 promotes gastric cancer cell proliferation,
migration and EMT via the miR-124-3p/DNMT3B axis. Cell Biochem
Funct. 38:1079–1088. 2020.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Zhang Y, Li X, Zhang J and Liang H:
Natural killer T cell cytotoxic activity in cervical cancer is
facilitated by the LINC00240/microRNA-124-3p/STAT3/MICA axis.
Cancer Lett. 474:63–73. 2020.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Myatt SS, Wang J, Monteiro LJ, Christian
M, Ho KK, Fusi L, Dina RE, Brosens JJ, Ghaem-Maghami S and Lam EW:
Definition of microRNAs that repress expression of the tumor
suppressor gene FOXO1 in endometrial cancer. Cancer Res.
70:367–377. 2010.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Meng N, Yang Q, He Y, Gu WW, Gu Y, Zhen XX
and Wang J, Zhang X, Sun ZG and Wang J: Decreased NDRG1 expression
is associated with pregnancy loss in mice and attenuates the in
vitro decidualization of endometrial stromal cells. Mol Reprod Dev.
86:1210–1223. 2019.PubMed/NCBI View Article : Google Scholar
|