Introduction
MicroRNAs (miRNAs or miRs) are small non-coding RNAs
that regulate gene expression at the post-transcriptional level and
can degrade mRNA or/and inhibit mRNA translation by binding to the
3'-untranslated region (UTR) of target mRNAs. For example,
miR-1891b directly binds to the 3'-UTR of cbl proto-oncogene B
(Cbl-b) and degrades Cbl-b mRNA (1). Additionally, miRNAs increase mRNA
translation or stability by binding to the 5'-UTR of target mRNAs.
For example, miR-10a interacts with the 5'-UTR of ribosomal protein
mRNAs to increase their translation (2). miR-483-5p directly binds to the 5'-UTR
of insulin like growth factor 2 (IGF2) and increases IGF2
expression (3). miRNAs have also
been demonstrated to regulate tumor progression in various types of
cancer (4). Liver cancer (LC) is a
malignant tumor with a poor prognosis (5). The molecular mechanism of LC
progression requires further assessment; however, a number of
studies have revealed that miRNAs regulate LC progression. miR-7
has been revealed to inhibit LC growth and metastasis by targeting
phosphatidylinositol-4 5-bisphosphate 3-kinase catalytic subunit Δ,
mTOR and ribosomal protein S6 kinase β-1, and regulate the PI3K/Akt
pathway (6). miR-124-1 inhibits LC
growth by targeting CASC3 exon junction complex subunit to
inactivate the p38/MAPK, JNK and ERK pathways (5).
Protein phosphatase 2 regulatory subunit βα
(PPP2R2A) is a regulatory subunit of tumor suppressor protein
phosphatase 2 (PP2A). PP2A can inhibit PI3K/AKT/mTOR, Wnt/β-catenin
and MAPK signaling to suppress tumor initiation, growth, invasion
and metastasis and induce apoptosis (7,8).
PPP2R2A inhibits the progression of a variety of tumors including
pancreatic (9), bladder and breast
(10) cancer. miR-660 has been
demonstrated to be associated with the development of various
diseases including graves' disease (11), facioscapulohumeral dystrophy
(12) and Alzheimer's disease
(13). miR-660 has also been
revealed to regulate tumor progression. miR-660 has been
demonstrated to be downregulated in lung cancer tissues and
patients with low miR-660 exhibited poor outcomes including
increased mortality (14). The
overexpression of miR-660 has been indicated to inhibit lung cancer
cell proliferation, migration and invasion and induce apoptosis.
Analysis of the mechanisms associated with miR-660 has revealed
that the p53 regulator mouse double minute 2 homolog (MDM2) is a
miR-660 target (14). miR-660
suppresses lung cancer progression by inhibiting MDM2 and
stabilizing p53(14). The role of
miR-660 in LC progression has not, to the best of our knowledge,
been previously assessed. In the present study, the role of
miR-660, also known as miR-660-5p, in LC cell proliferation was
assessed. miR-660 expression was measured in LC cells and tissues,
and the effect of miR-660 overexpression and knockdown on LC cell
proliferation was assessed. Finally, the regulatory mechanism of
miR-660 was investigated.
Materials and methods
Cell culture
A total of six human LC cell lines (MHCC97H,
HCCC-9810, MHCC97L, HepG2, Huh7 and Hep3B) and normal liver cell
line THLE-3 (ATCC® CRL-11233™) were purchased from
American Type Culture Collection and cultured according to
manufacturer's protocol, briefly, DMEM/high glucose (Hyclone; GE
Healthcare Life Sciences) supplemented with 10% FBS (Hyclone; GE
Healthcare Life Sciences) at 37˚C with 5% CO2.
Clinical specimens
LC tissues and adjacent normal liver tissues were
obtained from The Second Affiliated Hospital of Soochow University
(Suzhou, China) between July 2015 and September 2018 according to
Edmondson criteria (5). Adjacent
normal tissues distanced from tumor tissue above 5 cm. The current
study was approved by the Ethics Committee of The Second Affiliated
Hospital of Soochow University. Prior patient consent was obtained
for the obtainment of specimens (Table
S1).
Transfection
The sequence of pre-miR-660 was cloned into a
pMSCV-puro vector (Clontech Laboratories, Inc.). An empty and
miR-660 overexpressing vector (10 µg) were co-transfected with
packaging plasmid pIK (10 µg) into 293T cells (American Type
Culture Collection) using a standard calcium phosphate transfection
method, as previously described (15). The virus supernatants were collected
using 0.45 µm filter (EMD Millipore) 36 h following
co-transfection. HepG2 and HepB3 cells were incubated with virus
supernatants overnight at 37˚C with multiplicity of infection
(MOI)=20. Supernatants were subsequently removed and cells were
cultured using DMEM/high glucose (Hyclone; GE Healthcare Life
Sciences) with 10% FBS (Hyclone; GE Healthcare Life Sciences) and 2
µg/ml puromycin (Sigma-Aldrich; Merck) for 1 week to construct the
stably miR-660 overexpressing cells. The miR-660 mimic (indicated
as miR-660), miR-660 inhibitor (indicated as miR-660-in; cat. no.
miR20003338-1-5), miR-660 mimic with seed site mutation
(miR-660-mut) and their negative control were synthesized by
Guangzhou RiboBio Co., Ltd. and transfected into HepG2 and Hep3B
cells using Lipofectamine® 2000 to perform luciferase
assay (Thermo Fisher Scientific, Inc.) according to manufacturers'
protocol. 12 h after transfection, functional experiments could be
performed. The small interfering RNA (siRNA) of PPP2R2A (siPPP2R2A;
Guangzhou RiboBio Co., Ltd.) sequences were as follows:
PPP2R2AsiRNA#1, 5'-GAUCCCAGUAACAGGUCAUUU-3' and PPP2R2AsiRNA#2,
5'-GCAAGUGGCAAGCGAAAGAAA-3'. A total of 10 nM siRNAs were
transfected into cells using Lipofectamine 2000.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was isolated using RNAiso Plus (Takara
Bio, Inc) in LC cells. For miRNA analysis, cDNA was synthesized
using Hiscript Reverse Transcriptase (Vazyme) and a specific
stem-loop primer, the sequence of the primer was:
5'-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAACTC-3'. qPCR was
subsequently performed using AceQ qPCR SYBR Green Master Mix
(Vazyme) and a CFX96 Touch™ Real-time PCR detection system (Bio-Rad
Laboratories, Inc.). miR-660 primer sequences were as follows:
Forward, 5'-GCCCGCTACCCATTGCATATCG-3' and reverse,
5'-GTGCAGGGTCCGAGGT-3'. For mRNA analysis, cDNA was synthesized
using Hiscript High Fidelity One Step RT-PCR kit (Vazyme) and qPCR
was performed using AceQ qPCR SYBR Green Master Mix (Vazyme) and a
CFX96 Touch™ Real-time PCR detection system (Bio-Rad Laboratories,
Inc.) by incubation at 95˚C for 5 min followed by 40 amplification
cycles (10 sec for denaturation at 95˚C and 30 sec of hybridization
and elongation at 60˚C). The specific primer sequences for GAPDH,
p21 and cyclin D1 (CCND1) are as follows: GAPDH forward,
5'-GGTGGTCTCCTCTGACTTC-3' and reverse, 5'-CTCTTCCTCTTGTGCTCTTG-3';
p21 forward, 5'-TGTCCGTCAGAACCCATGC-3' and reverse,
5'-AAAGTCGAAGTTCCATCGCTC-3'; CCND1 forward,
5'-GCTGCGAAGTGGAAACCATC-3' and reverse,
5'-CCTCCTTCTGCACACATTTGAA-3'. U6 was used as an endogenous control
for microRNA data normalization, and GAPDH was used as an
endogenous control for gene normalization.
Western blot analysis
Total protein of HepG2 was isolated using RIPA
buffer [50 mM Tris (pH 7.4); 150 mM NaCl; 1% NP-40; 0.5% sodium
deoxycholate] supplemented with protease inhibitor cocktail (Roche
Applied Science), BCA Protein Assay kit (Thermo Fisher Scientific,
Inc.) was used to determine protein concentration, 20 µg proteins
were separated using 12% SDS-PAGE, transferred onto PVDF membranes,
membranes were blocked using 5% non-fat milk at room temperature
for 2 h, and immunoblotted with the following primary antibodies
for overnight at 4˚C: Anti-p21 (1:1,000; cat. no. ab109520) and
anti-CCND1 (1;1,000; cat. no. ab137875) antibodies were used (all,
Abcam). α-Tubulin (1:1,000; cat. no. ab7291; Abcam) was used as the
loading control. The secondary antibody (HRP-Goat Anti-Mouse IgG
H&L (1:5,000; cat. no. ab205719; Abcam) and HRP-Goat
Anti-Rabbit IgG H&L (1:5,000; cat. no. ab214880; Abcam) for 1 h
at room temperature. Bands were detected using ECL Western Blotting
Detection kit (GE Healthcare).
MTT assay
A total of 3x103 HepG2 cells with miR-660
overexpression were seeded in 96-well plates and stained with 0.05
mg MTT (Sigma-Aldrich; Merck KGaA) at day 0-4 and 5 for 4 h at
37˚C. DMEM medium supplemented with 10% FBS was removed and DMSO
was added (Sigma-Aldrich; Merck KGaA). Absorbance was measured at a
wavelength of 570 nm.
Anchorage-independent growth assay and
bromodeoxyuridine (BrdU) incorporation assay
An anchorage-independent growth assay and a BrdU
incorporation assay were performed according to a previous method
(16).
Cell cycle assay
Cells were trypsined and washed with cold PBS buffer
three times and fixed with 70% cold ethanol at -20˚C for 4 h. Cells
were spun down and resuspended using PBS, and 2 µg/ml RNAase
(Takara Bio, Inc) was added for 30 min at 37˚C, followed by
incubation in 20 µg/ml propidium iodide (Sigma-Aldrich; Merck KGaA)
at 37˚C for 30 min. Finally, cells were analyzed using a flow
cytometer (FACSCalibur; BD Biosciences).
Luciferase reporter assay
The sequence of 3'-UTR of PPP2R2A containing the
binding sites of miR-660 was cloned into a psiCHECK-2 vector
(Promega Corporation). Cells were seeded in 24-well plates and
co-transfected with psiCHECK-2-PPP2R2A-3'-UTR and miR-660 mimic,
miR-660 inhibitor or mutational miR-660 mimic (miR-660-mut) into
HepG2 using Lipofectamine 2000. Luciferase reporter assay was
performed using a Dual-Luciferase Reporter assay kit (Promega
Corporation) at 48 h following co-transfection, according to the
manufacturer's protocol. Renilla luciferase was used to
normal data.
Statistical analysis
Data analysis was performed using SPSS 20.0 (IBM
Corp.), the results are presented as the mean ± SEM. Comparisons
between two groups were performed using an ANOVA. TCGA data was
downloaded from www.portal.gdc.cancer.gov. TargetScan 7.2 was used to
predict the targets of miR-660. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-660 is upregulated in LC tissues
and cells
The miRNA expression profile of LC tissues (n=372)
and normal liver tissues (n=50) was obtained from The Cancer Genome
Atlas dataset and used to analyze miR-660 expression. miR-660 was
demonstrated to be significantly upregulated in LC tissues (T)
compared with normal liver samples (N) (Fig. 1A). miR-660 expression was also
examined in all LC cell lines and a normal liver cell line, and the
results revealed that miR-660 was significantly upregulated in LC
cells compared with normal liver cells (Fig. 1B). miR-660 expression was further
examined in LC tissues and paired adjacent normal liver tissues and
the results indicated that miR-660 was upregulated in LC tissues
(Fig. 1C), suggesting that miR-660
may promote LC progression.
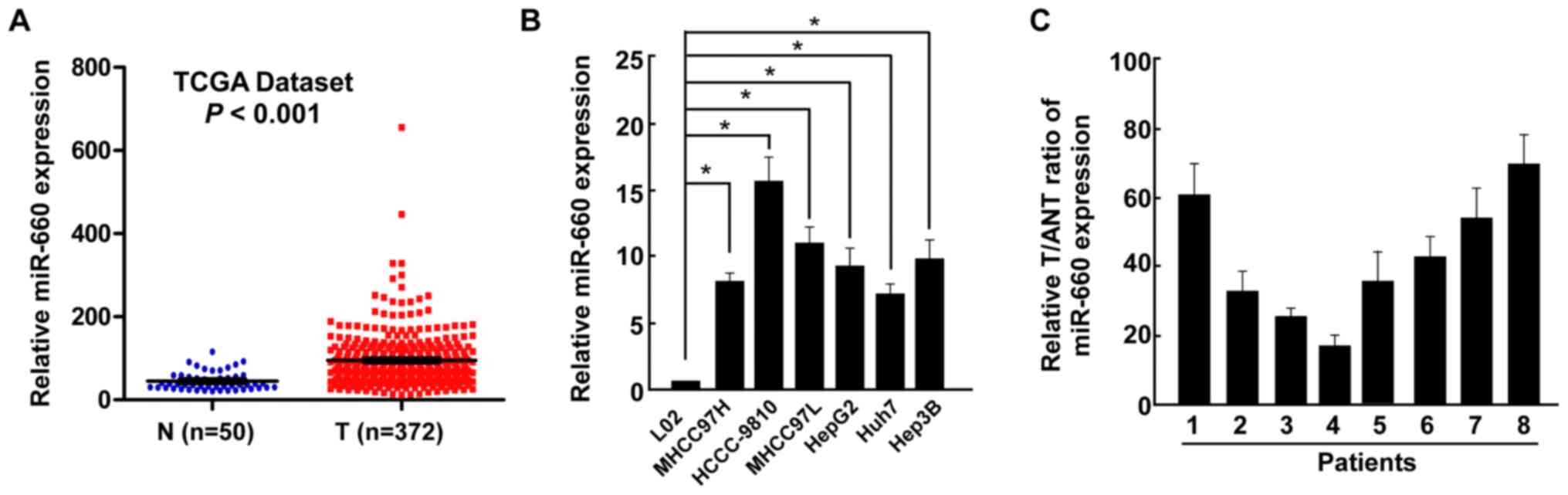 | Figure 1Upregulation of miR-660 in LC tissues.
(A) miR-660 expression in LC tissues and normal liver tissues,
analyzed using The Cancer Genome Atlas data. (B) miR-660 expression
in a normal liver cell line THLE-3 and liver cancer cell lines
MHCC97H, HCCC-9810, MHCC97L, HepG2, Huh7 and Hep3B. (C) miR-660
expression in T and ANT. Error bars indicate SEM.
*P<0.05. miR, microRNA; LC, liver cancer; T, LC
tissues; N, normal liver tissues; ANT, adjacent normal liver
tissues. |
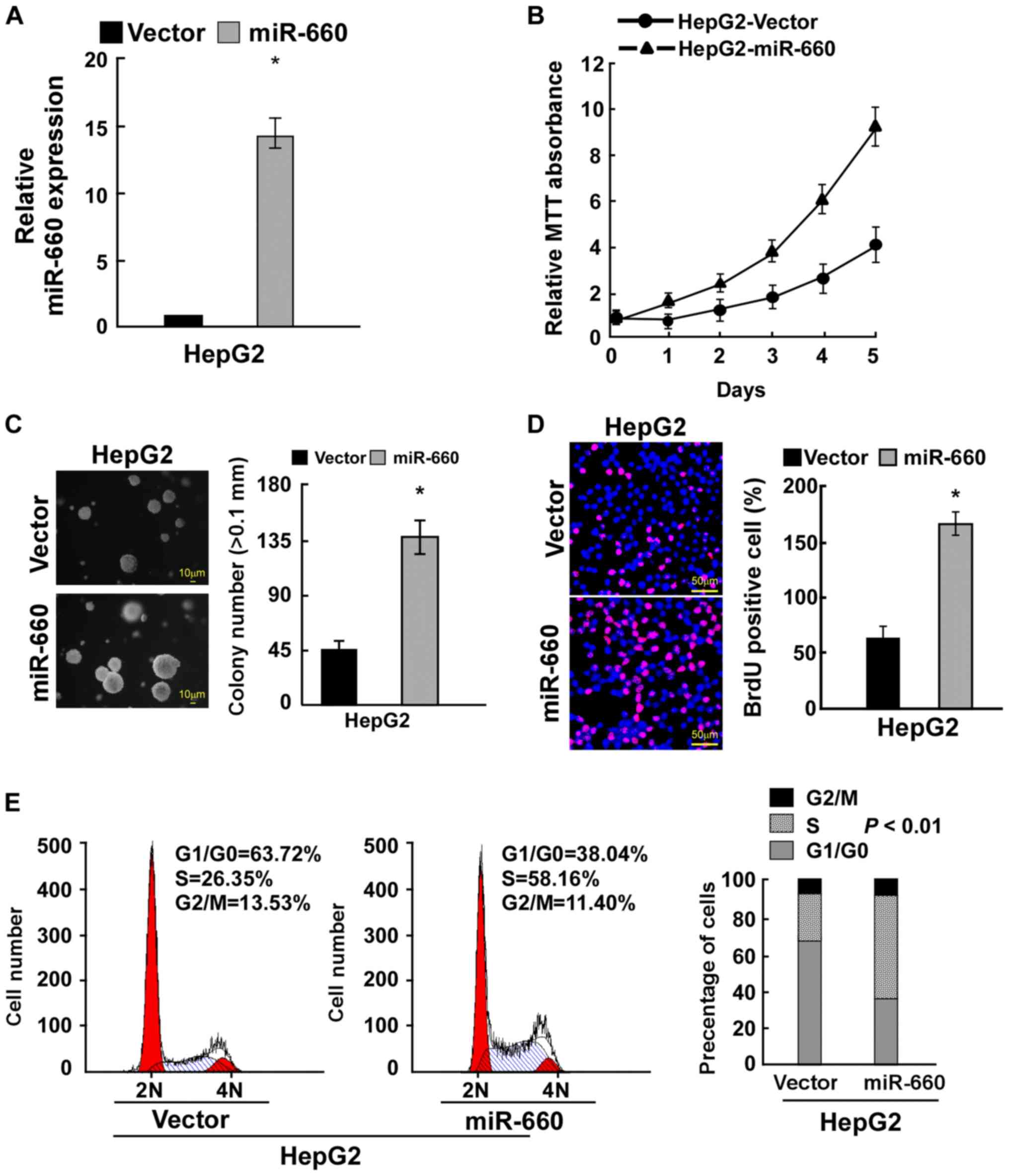
miR-660 promotes LC cell
proliferation
To determine the role of miR-660 in LC cell
proliferation, miR-660 was overexpressed in LC cell HepG2 (Fig. 2A). The MTT assay demonstrated that
miR-660 overexpression promoted LC cell proliferation (Fig. 2B). Anchorage-independent growth
assay revealed that miR-660 overexpression significantly promoted
anchorage-independent growth (Fig.
2C). The BrdU incorporation assay indicated that miR-660
overexpression significantly increased the number of BrdU-positive
cells, suggesting that miR-660 promoted LC cell proliferation
(Fig. 2D). A cell cycle assay was
used to confirm the results observed. miR-660 overexpression
increased the percentage of S phase cells from 26.35 to 58.16% and
reduced the percentage of G0/G1 phase cells
from 63.72 to 38.04% (Fig. 2E).
These results suggested that miR-660 increased LC cell
proliferation through accelerating cell cycle progression.
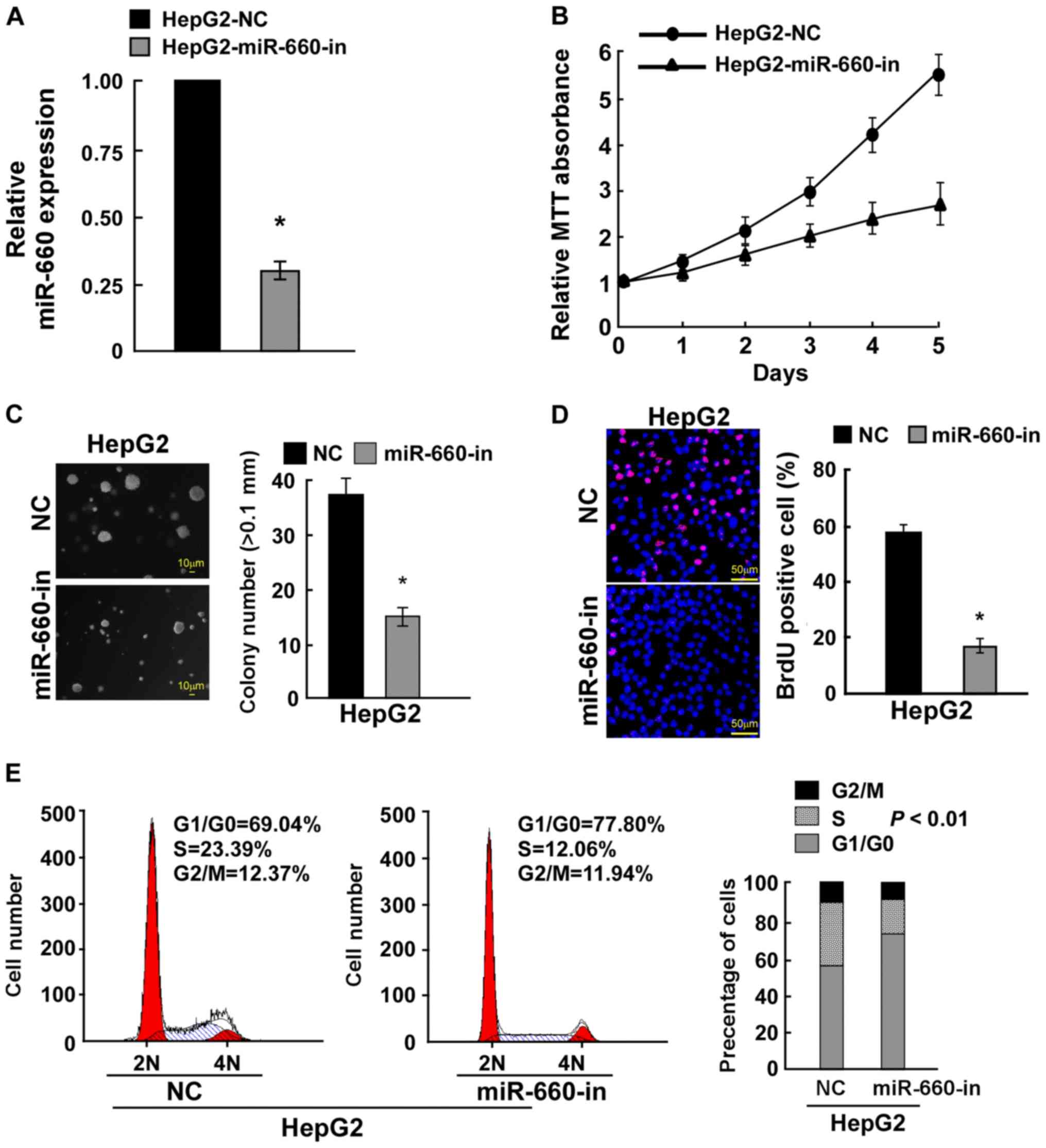
To confirm these results, miR-660 was knocked down
in HepG2 cells (Fig. 3A) The MTT
assay demonstrated that miR-660 knockdown inhibited LC cell
proliferation (Fig. 3B). A
anchorage-independent growth assay indicated that miR-660 knockdown
significantly inhibited cell anchorage-independent growth (Fig. 3C). The BrdU incorporation assay
demonstrated that miR-660 knockdown significantly reduced the
number of BrdU-positive cells (Fig.
3D). The cell cycle assay revealed that miR-660 knockdown
increased the percentage of G0/G1 phase cells
from 69.04 to 77.80% and reduced the percentage of S phase cell
from 23.39 to 12.06% (Fig. 3E).
These findings suggested that miR-660 promoted LC cell
proliferation.
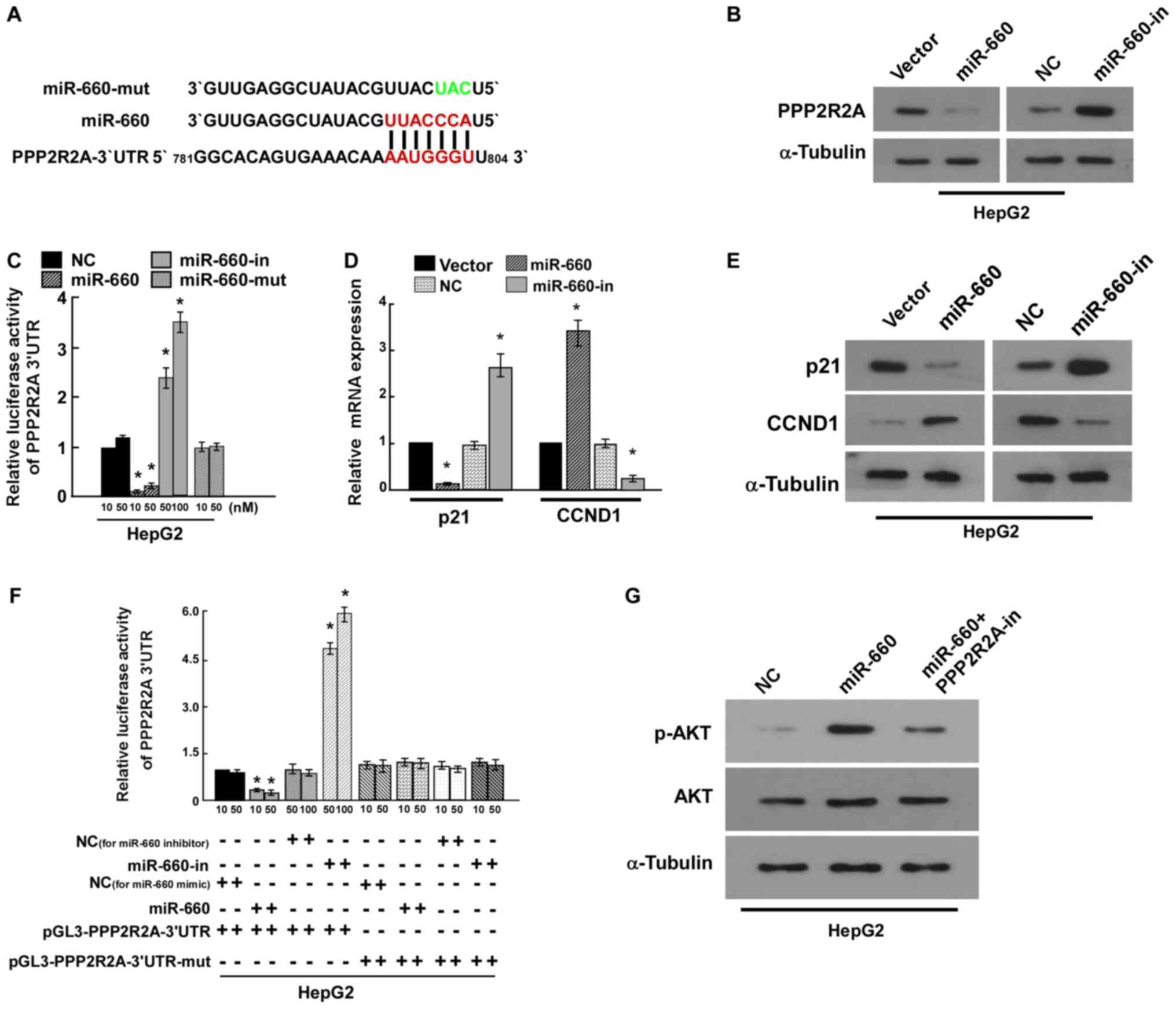
PPP2R2A is the target of miR-660
miRNAs regulate a variety of biological functions
through targeting mRNA. TargetScan 7.2 predicted that PPP2R2A was
the target of miR-660 (Fig. 4A).
PPP2R2A is a regulatory subunit of the PP2A which is a major
Ser/Thr phosphatase (17). Western
blot analysis revealed that miR-660 inhibited PPP2R2A expression
(Fig. 4B). The results of the
luciferase reporter assay indicated that miR-660 overexpression
significantly reduced luciferase activity compared with the vector
control, miR-660 knockdown significantly increased luciferase
activity compared with the negative control, while mutational
miR-660 overexpression luciferase activity did not change (Fig. 4C). These results suggested that
miR-660 directly bound to the 3'-UTR of PPP2R2A and PPP2R2A was the
target of miR-660. p21 and CCND1 are important cell cycle
regulators and have been revealed to regulate cell proliferation
(18,19). The results of the current study
indicated that miR-660 overexpression significantly increased CCND1
expression and decreased p21 mRNA and protein expression. miR-660
knockdown markedly inhibited CCND1 expression and increased p21
expression (Fig. 4D and E). These findings further confirmed that
miR-660 promoted HepG2 proliferation by targeting PPP2R2A.
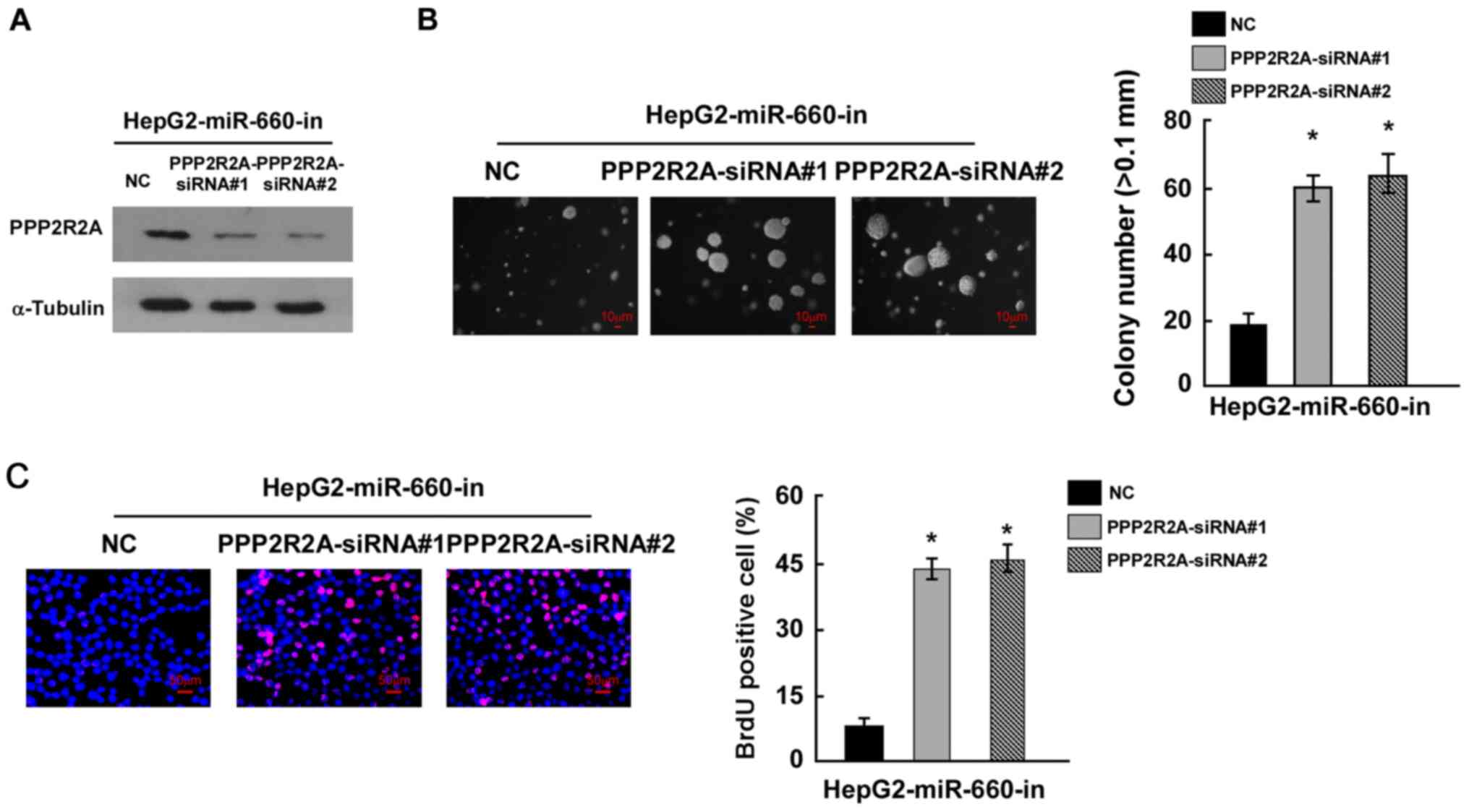
miR-660 promotes LC cell proliferation
through inhibiting PPP2R2A
To confirm whether miR-660 promoted LC cell
proliferation by targeting PPP2R2A, miR-660 and PPP2R2A were
knocked down in HepG2 cells and the results of western blot
analysis indicated that PPP2R2A siRNAs inhibited PPP2R2A expression
(Fig. 5A). The colony formation
assay suggested that miR-660 and PPP2R2A knockdown significantly
promoted HepG2 proliferation compared with miR-660 knockdown alone
(Fig. 5B). The BrdU incorporation
assay demonstrated that miR-660 and PPP2R2A knockdown increased the
number of BrdU positive cells compared with miR-660 knockdown alone
(Fig. 5C). These findings suggested
that miR-660 promoted LC cell proliferation through the inhibition
of PPP2R2A.
Discussion
The role of miRNAs in liver cancer progression has
been extensively studied. For example, miRNA-503 has been indicated
to inhibit the proliferation of liver cancer by targeting cyclin D3
and E2F transcription factor 1(20). miR-122 has also been revealed to
inhibit liver cancer growth by targeting MDM2(21). Numerous miRNAs have been indicated
to serve a role in the progression of liver cancer by targeting a
variety of the same or different genes. In the present study,
miR-660 was revealed to be upregulated in LC cells and tissues, and
cellular function analysis revealed that miR-660 promoted HepG2
proliferation. miR-660 has been previously revealed to be
downregulated in lung cancer cells and tissues, and to inhibit lung
tumorigenesis, while it promotes liver cancer progression. This may
be due to the fact miR-660 targets oncogenes in lung cancer. MDM2
is a target of miR-660 in lung cancer cells and inhibits p53
activity (14,22). In the current study, PPP2R2A was
revealed to be the target of miR-660 in liver cancer. Mechanism
analysis suggested that PPP2R2A was its target, and the double
knockdown of miR-660 and PPP2R2A promoted HepG2 proliferation,
confirming that miR-660 promoted HepG2 proliferation by targeting
PPP2R2A.
A number of miRNAs have been demonstrated to
regulate PPP2R2A expression, including miR-556-5p (23), miR-136, miR-31(24), miR-892a (25) and miR-222(26). These miRNAs promote tumor
progression by inhibiting PPP2R2A, further confirming PPP2R2A as a
tumor suppressor (26). miR-222 is
overexpressed in LC, and promotes LC cell motility by targeting
PPP2R2A (27). In the current
study, miR-660 promoted LC cell proliferation by targeting PPP2R2A,
suggesting that miR-660 may also promote LC metastasis. These
results indicate that miR-660 may be a novel target for LC therapy.
However, studies using animal models are required to support these
findings.
p21 inhibits cell cycle progression and CCND1
promotes cell cycle progression (18,19).
In the current study, it was demonstrated that miR-660 inhibited
p21 expression and upregulated CCND1 expression, which was
consistent with results of the cell cycle assay which indicated
that miR-660 promoted cell cycle progression. These results
revealed that miR-660 promoted HepG2 proliferation by inhibiting
PPP2R2A. In conclusion, miR-660 was upregulated in LC cells and
tissues and contributed to LC cell proliferation by inhibiting
PPP2R2A.
Supplementary Material
Clinicopathological characteristics of
studied patients and expression of miR-660 in LC.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
CYS and YZP designed the study; YZP, LZ, LC and LWC
acquired the data; CYS and YZP wrote and revised the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The current study was approved by the Ethics
Committee of The Second Affiliated Hospital of Soochow University.
Prior patient consent was obtained for the obtainment of
specimens.
Patient consent of publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Dong Q, Li C, Che X, Qu J, Fan Y, Li X, Li
Y, Wang Q, Liu Y, Yang X and Qu X: MicroRNA-891b is an independent
prognostic factor of pancreatic cancer by targeting Cbl-b to
suppress the growth of pancreatic cancer cells. Oncotarget.
7:82338–82353. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Ørom UA, Nielsen FC and Lund AH:
MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and
enhances their translation. Mol Cell. 30:460–471. 2008.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Liu M, Roth A, Yu M, Morris R, Bersani F,
Rivera MN, Lu J, Shioda T, Vasudevan S, Ramaswamy S, et al: The
IGF2 intronic miR-483 selectively enhances transcription from IGF2
fetal promoters and enhances tumorigenesis. Genes Dev.
27:2543–2548. 2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Lin Q, Ma L, Liu Z, Yang Z, Wang J, Liu J
and Jiang G: Targeting microRNAs: A new action mechanism of natural
compounds. Oncotarget. 8:15961–15970. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Xu L, Dai W, Li J, He L, Wang F, Xia Y,
Chen K, Li S, Liu T, Lu J, et al: Methylation-regulated miR-124-1
suppresses tumorigenesis in hepatocellular carcinoma by targeting
CASC3. Oncotarget. 7:26027–26041. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Fang Y, Xue JL, Shen Q, Chen J and Tian L:
MicroRNA-7 inhibits tumor growth and metastasis by targeting the
phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma.
Hepatology. 55:1852–1862. 2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Liu CC, Lin SP, Hsu HS, Yang SH, Lin CH,
Yang MH, Hung MC and Hung SC: Suspension survival mediated by
PP2A-STAT3-Col XVII determines tumour initiation and metastasis in
cancer stem cells. Nat Commun. 7(11798)2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Cristóbal I, Manso R, Rincón R, Caramés C,
Senin C, Borrero A, Martínez-Useros J, Rodriguez M, Zazo S,
Aguilera O, et al: PP2A inhibition is a common event in colorectal
cancer and its restoration using FTY720 shows promising therapeutic
potential. Mol Cancer Ther. 13:938–947. 2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wang Q, Li J, Wu W, Shen R, Jiang H, Qian
Y, Tang Y, Bai T, Wu S, Wei L, et al: Smad4-dependent suppressor
pituitary homeobox 2 promotes PPP2R2A-mediated inhibition of Akt
pathway in pancreatic cancer. Oncotarget. 7:11208–11222.
2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Li X, Zhang J and Ma D: PPP2R2A binds and
dephosphorylates GFPT2 in breast cancer cells. Sheng Wu Gong Cheng
Xue Bao. 34:956–963. 2018.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
11
|
Qin Q, Wang X, Yan N, Song RH, Cai TT,
Zhang W, Guan LJ, Muhali FS and Zhang JA: Aberrant expression of
miRNA and mRNAs in lesioned tissues of Graves' disease. Cell
Physiol Biochem. 35:1934–1942. 2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Dmitriev P, Stankevicins L, Ansseau E,
Petrov A, Barat A, Dessen P, Robert T, Turki A, Lazar V, Labourer
E, et al: Defective regulation of microRNA target genes in
myoblasts from facioscapulohumeral dystrophy patients. J Biol Chem.
288:34989–35002. 2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Satoh J, Kino Y and Niida S: MicroRNA-seq
data analysis pipeline to identify blood biomarkers for Alzheimer's
disease from public data. Biomarker insights. 10:21–31.
2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Fortunato O, Boeri M, Moro M, Verri C,
Mensah M, Conte D, Caleca L, Roz L, Pastorino U and Sozzi G:
Mir-660 is downregulated in lung cancer patients and its
replacement inhibits lung tumorigenesis by targeting MDM2-p53
interaction. Cell Death Dis. 5(e1564)2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Cribbs AP, Kennedy A, Gregory B and
Brennan FM: Simplified production and concentration of lentiviral
vectors to achieve high transduction in primary human T cells. BMC
Biotechnol. 13(98)2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lin H, Dai T, Xiong H, Zhao X, Chen X, Yu
C, Li J, Wang X and Song L: Unregulated miR-96 induces cell
proliferation in human breast cancer by downregulating
transcriptional factor FOXO3a. PLoS One. 5(e15797)2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Alvarez-Fernández M, Halim VA, Aprelia M,
Laoukili J, Mohammed S and Medema RH: Protein phosphatase 2A (B55α)
prevents premature activation of forkhead transcription factor
FoxM1 by antagonizing cyclin A/cyclin-dependent kinase-mediated
phosphorylation. J Biol Chem. 286:33029–33036. 2011.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Forte D, Salvestrini V, Corradi G, Rossi
L, Catani L, Lemoli RM, Cavo M and Curti A: The tissue inhibitor of
metalloproteinases-1 (TIMP-1) promotes survival and migration of
acute myeloid leukemia cells through CD63/PI3K/Akt/p21 signaling.
Oncotarget. 8:2261–2274. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Pestell RG: New roles of cyclin D1. Am J
Pathol. 183:3–9. 2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Xiao F, Zhang W, Chen L, Chen F, Xie H,
Xing C, Yu X, Ding S, Chen K, Guo H, et al: MicroRNA-503 inhibits
the G1/S transition by downregulating cyclin D3 and E2F3 in
hepatocellular carcinoma. J Transl Med. 11(195)2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Simerzin A, Zorde-Khvalevsky E, Rivkin M,
Adar R, Zucman-Rossi J, Couchy G, Roskams T, Govaere O, Oren M,
Giladi H and Galun E: The liver-specific microRNA-122*, the
complementary strand of microRNA-122, acts as a tumor suppressor by
modulating the p53/mouse double minute 2 homolog circuitry.
Hepatology. 64:1623–1636. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Fei L and Xu H: Role of MCM2-7 protein
phosphorylation in human cancer cells. Cell Biosci.
8(43)2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhao W, Cao L, Zeng S, Qin H and Men T:
Upregulation of miR-556-5p promoted prostate cancer cell
proliferation by suppressing PPP2R2A expression. Biomed
Pharmacother. 75:142–147. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Shen S, Yue H, Li Y, Qin J, Li K, Liu Y
and Wang J: Upregulation of miR-136 in human non-small cell lung
cancer cells promotes Erk1/2 activation by targeting PPP2R2A.
Tumour Biol. 35:631–640. 2014.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Liang WL, Cao J, Xu B, Yang P, Shen F, Sun
Z, Li WL, Wang Q and Liu F: miR-892a regulated PPP2R2A expression
and promoted cell proliferation of human colorectal cancer cells.
Biomed Pharmacother. 72:119–124. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zeng LP, Hu ZM, Li K and Xia K: miR-222
attenuates cisplatin-induced cell death by targeting the
PPP2R2A/Akt/mTOR Axis in bladder cancer cells. J Cell Mol Med.
20:559–567. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wong QW, Ching AK, Chan AW, Choy KW, To
KF, Lai PB and Wong N: MiR-222 overexpression confers cell
migratory advantages in hepatocellular carcinoma through enhancing
AKT signaling. Clin Cancer Res. 16:867–875. 2010.PubMed/NCBI View Article : Google Scholar
|