Introduction
Glioma is an invasive malignant tumor of the central
nervous system. Surgery is most commonly used for treatment, which
aims to decrease the tumor volume and the number of tumor cells,
relieve symptoms, prolong life and create opportunities for other
treatments (1). However, residual
tumor in the primary site after general surgery will still exist,
which can generate progeny cells that are sufficient to sustain
tumor growth, overcome general cytotoxic therapies, and eventually
result in tumor recurrence (2). The
continuous malignant growth of tumors is the key cause of
recurrence. These types of cells are usually referred to as
tumor-propagating cells, stem cells or persisters, and they are
present in numerous high-grade malignancies, including glioblastoma
(3). Only by overcoming the threat
of recurrence can glioma be cured. In different types of tumor,
there are few cells with self-renewal, unlimited proliferation and
multiple differentiation potential, which exhibit the basic
characteristics of stem cells and are known as cancer stem cells
(CSCs) (4). Glioma stem cells
(GSCs) have been successfully isolated from the brain glioma tissue
in an in-depth study of malignant glioma (5). It has been hypothesized that there is
an association between glioma recurrence and GSCs. For example,
decreased asymmetry in normal stem cells has been associated with
neoplastic transformation, while therapies that increase the rate
of asymmetric division result in decreased numbers of resistant
GSCs (6). In addition, the
frequency of asymmetric division of CSCs is negatively correlated
with their proliferative capacity (7,8). GSCs
exhibit the potential of multidirectional differentiation, and can
differentiate into neurons, oligodendrocytes and astrocytes
(9). At present, it is generally
believed that the way of differentiation of GSCs is very similar to
that of neural stem cells (NSCs) (10). In addition, a number of studies have
also demonstrated that GSCs can be isolated and proliferate in
vitro (11,12). It has also been indicated that NSCs
can proliferate via symmetrical or asymmetric division (13). On the contrary, it was believed that
the main mode of proliferation of GSCs was symmetrical and not
asymmetric cell division (14).
However, when GSCs were inoculated into the subcutaneous layer of
immunodeficient mice, it was observed that they were differentiated
into glioblasts in an asymmetrical manner (15). Numerous features of NSCs have also
been observed in GSCs, including common surface markers, such as
CD133 and Nestin (16,17). Although there is an association
between GSCs and NSCs, it is still essential to determine whether
the induction medium of NSCs can also induce GSCs, and whether
additional stem cell-associated genes in NSCs are also expressed in
GSCs. Based on the aforementioned information, the present study
selected a type of relatively mature culture medium (18,19) of
NSCs to induce GSCs in order to achieve an ideal induction effect
in vitro. Therefore, the aim of the present study was to
establish a glioma cell model in vitro, and further
investigate the pathogenesis, antitumor drug sensitivity and
molecular biological characteristics. The study of GSCs can provide
a new direction to unravel the origin and the mechanism of
development of malignant glioma, and discover novel treatments for
these diseases.
Mitochondria, one of the most important organelles
of the cell, are involved in a dynamic balance process of fission
and fusion, which is important for maintaining the normal
morphology, distribution and function of mitochondria (20) This process determines the
morphology, quantity, function and spatial distribution of
mitochondria, thereby affecting biological processes, such as the
production of ATP, the phagocytosis of mitochondria, apoptosis and
calcium homeostasis (21).
Mitochondria are associated with a number of clinical diseases,
such as cancer, cardiovascular diseases, neurodegenerative diseases
and pulmonary hypertension (22-24).
The occurrence of these diseases is often accompanied by
mitochondrial dysfunction and/or structural disorders. For example,
it has been demonstrated that cancer cells usually exhibit
mitochondrial fragmentation, indicating that the excessive
division, contraction or decreased fusion of mitochondria is
associated with the occurrence of cancer; however the specific
underlying mechanism remains unclear (25). The alterations in the expression
levels of fusion- and fission-associated proteins directly affect
the process of mitochondrial fusion and fission, which indicates
that defects in proteins involved in the mitochondrial dynamics
that regulate mitochondrial fusion and fission can affect cellular
differentiation, proliferation, cellular reprogramming and aging
(26).
Dynamin-related protein 1 (DRP1), which has GTPase
activity, is an essential protein in the process of mitochondrial
division. As a major protein, it is activated and located on the
mitochondrial membrane via non-GTP receptor proteins, such as
mitochondrial fission 1 protein (Fis1), mitofusins (Mfns) and
mitochondrial elongation factor, forming a circular structure,
which contracts and divides the mitochondria. The fission process
may involve DRP1, Mfns and the microregions of pro-apoptotic
proteins (27). Ji et al
(28) suggested that actin induced
the recruitment of DRP1 to the mitochondria to promote
mitochondrial fission, and also demonstrated that actin filaments
were localized close to DRP1 on the mitochondrial membrane to
increase the probability of mitochondrial division before DRP1 is
recruited to the mitochondria. Mitochondrial division is a
multistep process, which is initiated by the recruitment of Fis1 on
the outer mitochondrial membrane, where DRP1 is transposed to and
enriched in potential mitotic sites (29). Several DRP1 molecules form ring
structures around the mitochondria, alter the distance or angle
between molecules by the hydrolysis of GTP, and gradually compress
the mitochondria until they divide, resulting in two independent
mitochondria (30). Similar to
mitochondrial fission, Mfn1 and Mfn2, which are two Mfn isoforms
with a central action in tethering and fusion, are localized in the
outer mitochondrial membrane in mammals (27). However, the GTPase and tethering
actions of Mfn1 are more pronounced than that those of
Mfn2(31). After mitochondrial
division, DRP1 returns to the cytoplasm, and the cycle is repeated.
By using Mdivi-1, which is a selective and transmembrane
mitochondrial mitotic inhibitor, to interfere with the kinetic
equilibrium of mitochondria, the role of DRP1 can be more easily
examined (32). However, to the
best of our knowledge, the interaction mechanism between Mdivi-1
and DRP1 has rarely been reported. Therefore, investigating the
effects on biological characteristics of GSCs by interference with
DRP1 expression is a current field of study. Mdivi-1, bound to an
allosteric site of DRP1 impeding its self-assembly and GTP
hydrolysis, has been indicated to prevent mitochondria fission
resulting in inter-connected, net-like mitochondria (33). It has been indicated that Mdivi-1
restored mitochondrial network organization and energy production
(34). However, the effect of
Mdivi-1 on the induction and differentiation of GSCs has not yet
been investigated. Therefore, the present study aimed to establish
a drug-targeting platform with GSCs as a cell model and DRP1 as a
mitochondrial target. Mdivi-1, a molecular targeting drug, may
further inhibit the general stem cell characteristics of GSCs,
thereby increasing the sensitivity of GSCs to drugs. This method is
likely to provide an important target for the treatment of glioma.
At the same time, it provides a certain experimental basis for a
novel strategy of cancer treatment with tumor stem cells as a
target. Metabolic research has been indicated to be an important
tool in the identification of novel treatments against cancer
(35). However, the role of
mitochondrial metabolism in tumor development remains unclear. If
the current understanding of mitochondrial dynamic regulation and
its intrinsic significance to the maintenance and proliferation of
GSCs is improved, it may become a powerful tool for tumor
treatment.
Materials and methods
Experimental group
When Mdivi-1 (cat no. ab144589; Abcam) interfered
with GSCs, they were divided into 7 groups according to different
treatment conditions at 37˚C in humidified air with 5%
CO2. The 7 groups were as follows: M1-5d (continuous
treatment for 5 days with 1 µM Mdivi-1), M5-2d (continuous
treatment for 2 days with 5 µM Mdivi-1), M5-5d (continuous
treatment for 5 days with 5 µM Mdivi-1), M5-7d (continuous
treatment for 7 days with 5 µM Mdivi-1), M10-2d (continuous
treatment for 2 days with 10 µM Mdivi-1), M10-5d (continuous
treatment for 5 days with 10 µM Mdivi-1) and M10-7d (continuous
treatment for 7 days with 10 µM Mdivi-1). They were compared with
normal control group (untreated GSCs).
Cell lines and culture
The U87 cell line was purchased from The Cell Bank
of Type Culture Collection of Chinese Academy of Sciences, and it
is a glioblastoma cell line but whose origin is unknown. It was
identified by short tandem repeat profiling by Procell Life Science
& Technology Co., Ltd. A maximum number of 5 cell passages were
used before analysis. The cells were routinely cultured in DMEM
complete medium (Gibco; Thermo Fisher Scientific, Inc.) with 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1%
Penicillin/Streptomycin (Thermo Fisher Scientific, Inc.). The N3
medium, included the minimum essential medium/F12 basic medium
(Gibco; Thermo Fisher Scientific, Inc.), 25 µg/ml insulin (cat. no
I6040; Beijing Biotopped Science & Technology Co., Ltd.), 50
µg/ml transferrin (cat. no T6010; Beijing Biotopped Science &
Technology Co., Ltd.), 30 nM sodium selenite (cat. no 214485; Sigma
Aldrich; Merck KGaA), 20 nM progesterone (cat. no IP0400; Beijing
Solarbio Science & Technology Co., Ltd.), 100 nM putrescine
(cat. no D6140; Beijing Biotopped Science & Technology Co.,
Ltd.) and 1% penicillin/streptomycin (Thermo Fisher Scientific,
Inc.). Unlike U87, for the 6 days of induction and maintenance of
GSCs the cells were cultured in N3 medium with 10 ng/ml epidermal
growth factor and 10 ng/ml basic fibroblast growth factor (both
from ProteinTech Group, Inc.), which is also named N3EF. All cells
were cultured at 37˚C in an atmosphere containing 5%
CO2.
Tumorsphere formation assay
U87 cells were cultured in DMEM complete medium in
six-well culture plates with a density of
~1x104/cm2. When the cell density was >60%
the medium was changed to N3EF that is suitable for the growth of
globular cells. After 2 days small clone-like cells appeared and
the medium was refreshed every other day. On the 6th day, the
globular cells were disrupted gently with a pipette tip to prepare
single cells, and the cells were collected after centrifugation at
200 x g for 5 min at room temperature.
Calculation of tumor sphere
proliferation efficiency using the Cell Counting Kit-8 (CCK-8)
assay
Following preparation of cell suspension as
aforementioned, the cells were seeded in a 96-well culture plate
and cultured in a 37˚C incubator with 5% CO2. The cells
were continuously observed for 7 days. Tumorspheres were formed and
proliferation was measured using CCK-8 assay (cat. no BB4202,
BestBio Science). A total of 10 µl CCK-8 reagent was added to 100
µl culture medium per well at 37˚C for 3 h. The absorbance at 450
nm was measured every day for 7 days. The average absorbance values
of the experimental group and the control group were recorded, and
proliferation efficiency was calculated according to the following
equation: Cell proliferation=[(OD-blank OD)/(control cell OD-blank
OD)].
Western blot analysis
U87 cells or GSCs were washed twice in cold PBS, and
subsequently lysed in cold lysis buffer from the Whole Cell Lysis
assay (cat. no KGP250, Nanjing KeyGen Biotech Co., Ltd.) according
to the manufacturer's protocol. The lysate was centrifuged at
10,000 x g for 20 min at 4˚C. The supernatant was collected, and
the protein concentration was determined using the BCA Protein
Quantitation assay (cat. no KGPBCA; Nanjing KeyGen Biotech Co.,
Ltd.), according to the manufacturer's protocol. A total of 30 µg
protein/lane were separated by SDS-PAGE (8-10% gel), and then
transferred to PVDF membranes. Subsequently, the membranes were
blocked with 5% skimmed milk (cat. no. 232100; Difco; BD
Biosciences) for 90 min at room temperature and incubated with
primary antibodies at 4˚C overnight. The antibodies were as
follows: CD133 (1:300; cat. no. bs-0395R; BIOSS), Krueppel-like
factor 4 (Klf4; 1:600; cat. no. 11880-1-AP), Nanog (1:600; cat. no.
14295-1-AP), SOX2 (1:600; cat. no. 20118-1-AP), c-Myc (1:600; cat.
no. 10828-1-AP), OCT4 (1:1,000; cat. no. 60242-1-Ig), Nestin
(1:600; cat. no. 19483-1-AP), DRP1 (1:1,000; cat. no. 12957-1-AP),
Mfn1 (1:1,000; cat. no. 13798-1-AP), Fis1 (1:1,000; cat. no.
0956-1-AP), β-actin (1:5,000; cat. no. 66009-1-Ig), GAPDH (1:5,000;
cat. no. 60004-1-Ig) (all from ProteinTech Group, Inc.), Bax
(1:1,000; cat.no. 2772), Bcl-2 (1:1,000; cat. no. 3498) and cleaved
caspase-3 (1:1,000; cat. no. 14220) (all from Cell Signaling
Technology, Inc.). The membranes were washed with PBST (PBS and
0.1% Tween-20) and incubated with IRDye®680RD Goat
anti-Rabbit IgG (1:5,000; cat. no. 926-68071, LI-COR Biosciences)
and IRDye®680RD Goat anti-Mouse IgG (1:5,000; cat. no.
925-68070, LI-COR Biosciences) secondary antibodies for 1 h at room
temperature. All antibodies were diluted in 5% skimmed milk.
Finally, the PVDF membranes (Millipore, lnc) were quantified using
Odyssey Infrared Imaging System, Image Studio version 4.0 software
(LI-COR Biosciences).
Reverse transcription-quantitative PCR
(RT-qPCR)
RNA was extracted from cells (both U87 and GSCs)
using the AxyPrep Multisource RNA Miniprep Kit (Axygen; Corning,
Inc.). cDNA was performed with the GoScript™ Reverse Transcriptase
System kit (cat. no. A5001, Promega Corporation), according to the
manufacturer's instructions-PCR amplification was performed using
Tip Green qPCR SuperMix (TransGen Biotech Co., Ltd.) in a 20 µl
reaction containing Tip Green qPCR SuperMix buffer, primers and
diluted cDNA. The primer sequences are listed in Table I. The PCR cycling conditions were as
follows: 94˚C for 30 min, followed by 40 cycles of 94˚C for 30 sec,
50-60˚C for 30 sec and 72˚C for 1 min/kb. All relative expression
values were normalized to GAPDH levels using the 2-∆∆Cq
method (36).
 | Table IPrimer sequences. |
Table I
Primer sequences.
| Primer name | Primer
sequences |
|---|
| CD133 | F:
5'-TTCTATGCTGTGTCCTGGGGC-3' |
| | R:
5'-TTGTTGGTGCAAGCTCTTCAAGGT-3' |
| OCT4 | F:
5'-CAGCGACTATGCACAACGAGAGG-3' |
| | R:
5'-CCAGAGTGGTGACGGAGACAGG-3' |
| Nanog | F:
5'-TGCATGCAGTTCCAGCCAAA-3' |
| | R:
5'-ACACGTCTTCAGGTTGCATGT-3' |
| SOX2 | F:
5'-GACCAGCTCGCAGACCTACA-3' |
| | R:
5'-TCGGACTTGACCACCGAAC-3' |
| C-MYC | F:
5'-CTCAGAGAAGCTGGCCTCCTACC-3' |
| | R:
5'-GCGAGCTGCTGTCGTTGAGAG-3' |
| Nestin | F:
5'-TGCCTAGTTCTTCTGCTATCCT-3' |
| | R:
5'-GGGAAGCTCTGATCCTCTTTC-3' |
| Klf4 | F:
5'-CGTCGGTCATCAGCGTCAGC-3' |
| | R:
5'-CCGCCTCCTGCTTGATCTTGG-3' |
| β-actin | F:
5'-CCACACCCGCCACCAGTTCG-3' |
| | R:
5'-TACAGCCCGGGGAGCATCGT-3' |
| GAPDH | F:
5'-CAAGGTCATCCATGACAACTTTG-3' |
| | R:
5'-GTCCACCACCCTGTTGCTGTAG-3' |
Cell immunofluorescence staining
GSCs were fixed using 4% polyformaldehyde at room
temperature for 15 min. After washing 3 times with PBS, the cells
were incubated with 0.5% Triton X-100 in PBS for 20 min at room
temperature. Subsequently, the cells were incubated with 10% goat
serum (OriGene Technologies, Inc.) for 30 min at room temperature.
The liquid was discarded, and the cells were immersed in the
primary antibodies: Rabbit anti-glial fibrillary acidic protein
(GFAP; 1:200; cat. no. 16825-1-AP; ProteinTech Group, Inc.), mouse
anti-O4 (1:200; cat. no. O7139; MilliporeSigma) and mouse
anti-tubulin beta-3 chain (Tuj1; 1:250; cat. no. 480011;
Invitrogen; Thermo Fisher Scientific, Inc.) at 4˚C overnight.
Following incubation of the cells for 30 min at room temperature in
the dark with Alexa Fluor™ 555 goat anti-mouse lgG (1:500; cat. no.
A21422; Invitrogen; Thermo Fisher Scientific, Inc.) or Alexa Flour™
488 goat anti-rabbit lgG (1:500; cat. no. A11008; Invitrogen;
Thermo Fisher Scientific, Inc.), the nuclei were stained using DAPI
at room temperature for 10 min. The stained cells were observed
under a confocal microscope at x20 magnification.
Statistical analysis
Each experiment was repeated three times, and all
data are presented as the mean ± SD. Statistical analyses were
performed using SPSS v22.0 software (IBM Corp.). Comparisons
between two groups were performed using independent samples
Student's t-test, and for comparisons among multiple groups,
one-way ANOVA followed by Bonferroni's post hoc test was used.
P<0.05 was considered to indicate a statistically significant
difference. Quantification analysis of the GSC sphere numbers using
ImageJ v1.47 software (National Institutes of Health). Cell sphere
diameter ≥50 µm was considered to indicate a GSC sphere.
Results
Identification of glioma-like stem
cells in vitro
Following culture of U87 cells (Fig. 1A) at the first day. Morphological
alterations were observed in certain cells on the 2nd day (Fig. 1B) with N3EF medium. It was revealed
that the majority of cells grew in spherical suspension, and the
volume of the sphere increased markedly (Fig. 1C). On the 7th day, it was revealed
that the majority of cells were larger (diameter ≥50 µm; Fig. 1D). After passaging, the cells were
in a good condition the following days, which was considered as the
morphological characteristics of the cells being clear and cell
growth being high (Fig. 1E-G).
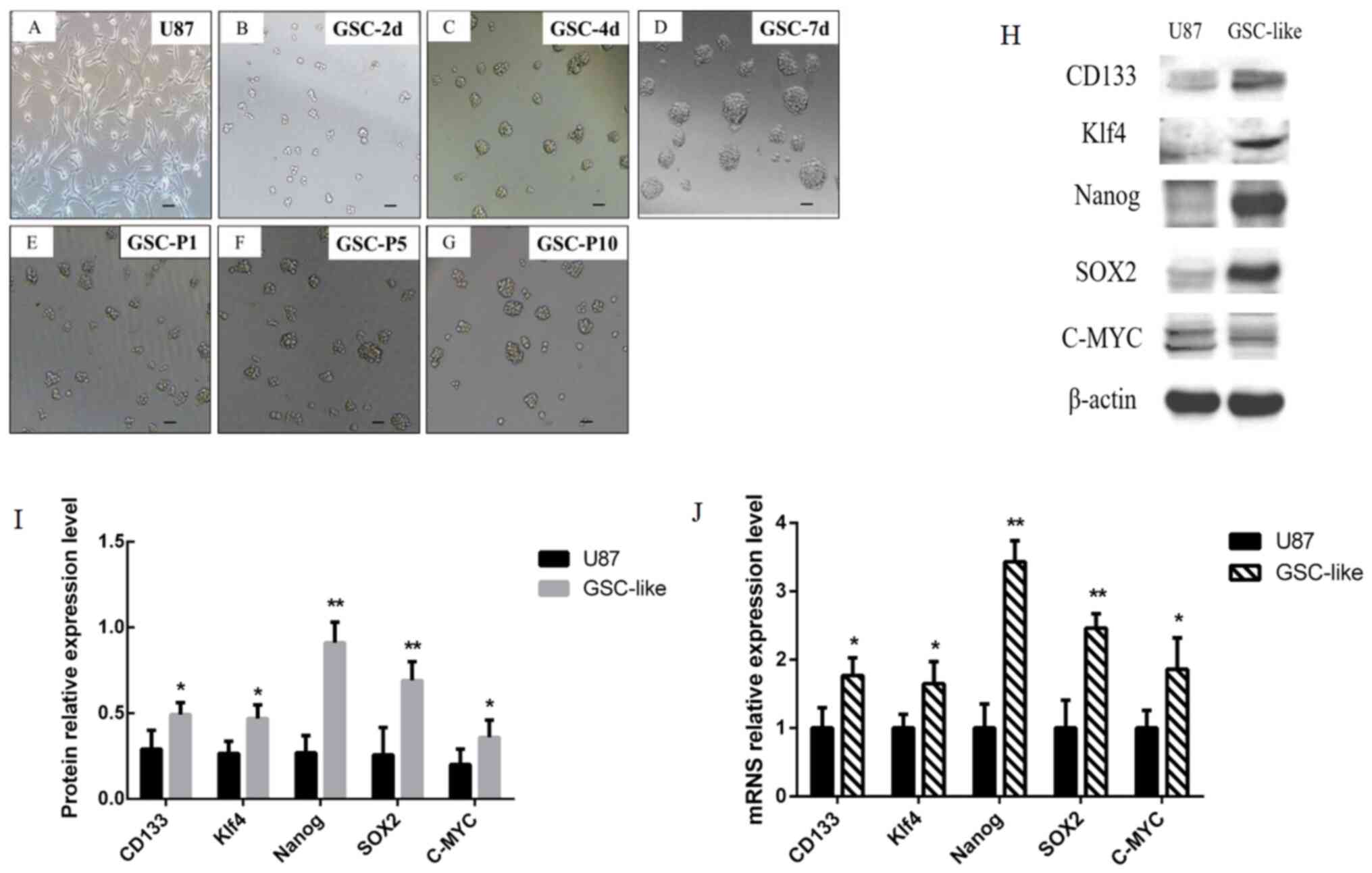 | Figure 1Morphological alterations of GSC-like
cells were observed on different days, and expression of tumor stem
cell-associated genes was detected by western blotting and RT-qPCR.
(A) Glioma cells. (B) GSC-2 days. (C) GSC-3 days. (D) GSC-7 days.
(E-G) GSC-P1, P5 and P10. Scale bars, 50 µm. (H) The expression
level of CD133 (130 kDa), c-Myc (57-70 kDa), Klf4 (65 kDa), SOX2
(35 kDa), Nanog (42 kDa) and β-actin (42 kDa) was examined by
western blotting. (I) Grayscale analysis results of western
blotting. (J) CD133, c-Myc, Klf4, SOX2 and Nanog expression at the
mRNA level was analyzed by RT-qPCR and normalized to β-actin.
*P<0.05 and **P<0.01 vs. U87 as control
group. GSC, glioma stem cells; Klf4, Krueppel-like factor 4; P1,
passage 1; P5, passage 5; P10, passage 10; d, day; RT-qPCR, reverse
transcription-quantitative PCR. |
RT-qPCR data demonstrated that the CD133, C-MYC
andKlf4mRNA expression levels were significantly increased in
GSC-like cells (P<0.05; Fig.
1J). Notably, the changes in mRNA expression levels of SOX2 and
Nanog were the most significant (P<0.01; Fig. 1J). It indicated that the cells
exhibited characteristics of tumor stem cells. The CD133, C-MYC and
Klf4 protein expression levels detected by western blot analysis
were markedly increased in GSC-like cells compared with the control
(P<0.05; Fig. 1H and I). Consistent with the trend of RT-qPCR
results, the expression of SOX2 and Nanog increased more
significantly at the protein level (P<0.01; Fig. 1H and I). These data indicated that the GSC-like
cells exhibited characteristics of stem cells in vitro at
both the mRNA and protein level.
Detection of the differentiation
potential of GSC-like cells
GSCs have the potential of multidirectional
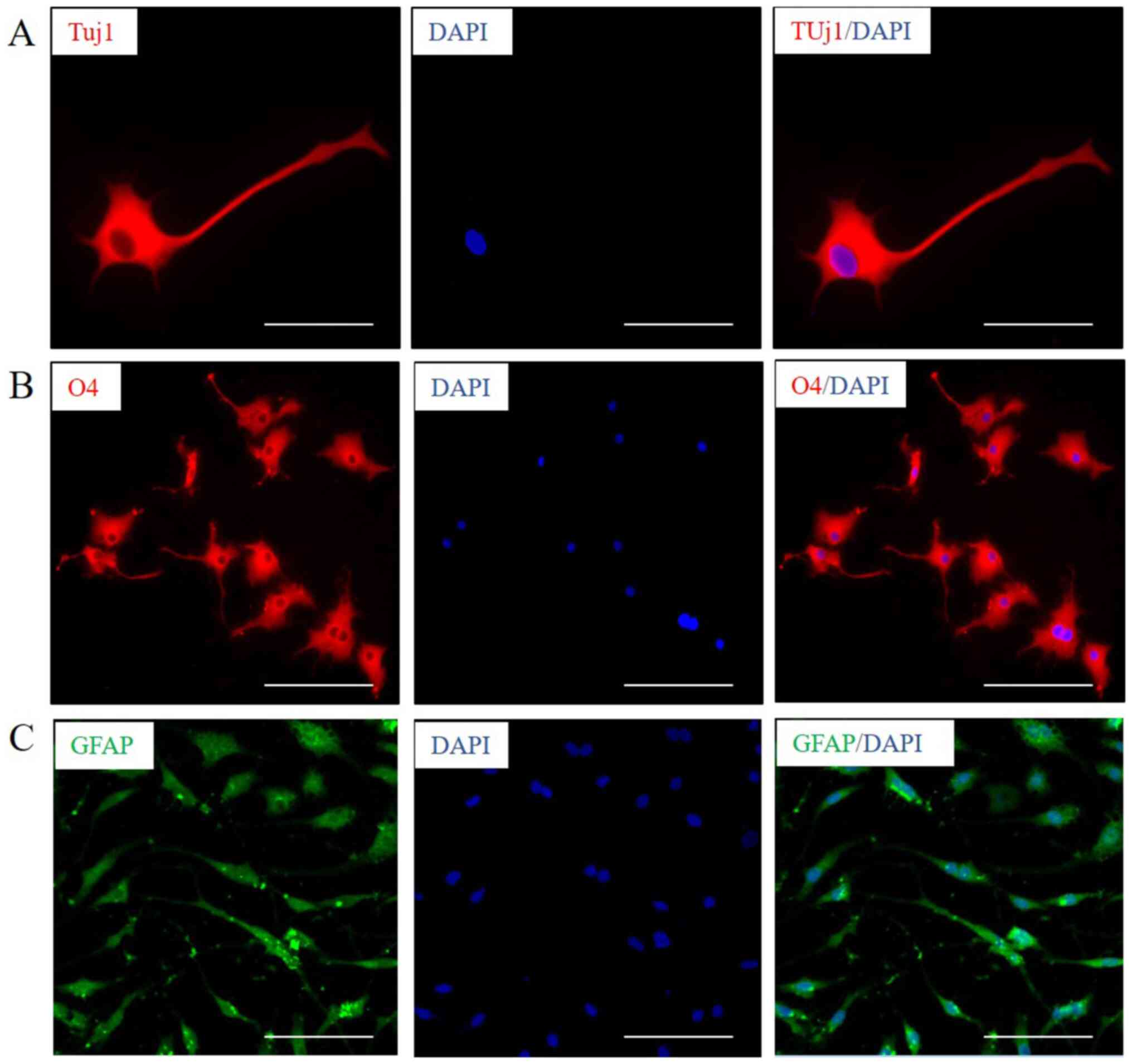
differentiation (37,38). Neuron marker protein Tuj1 (Fig. 2A), oligodendrocyte marker protein O4
(Fig. 2B) and astrocyte marker
protein GFAP (Fig. 2C) were
observed to be expressed after induction of GSC-like cells for 30,
25 and 7 days respectively, by immunofluorescence. The results
revealed that GSC-like cells exhibited a certain multidirectional
differentiation potential. It can be determined that the GSC-like
cells have the characteristics of stem cells, and the present
induction system can be used for the in vitro culture of
GSCs.
Protein expression of
mitochondrial-associated genes in GSCs
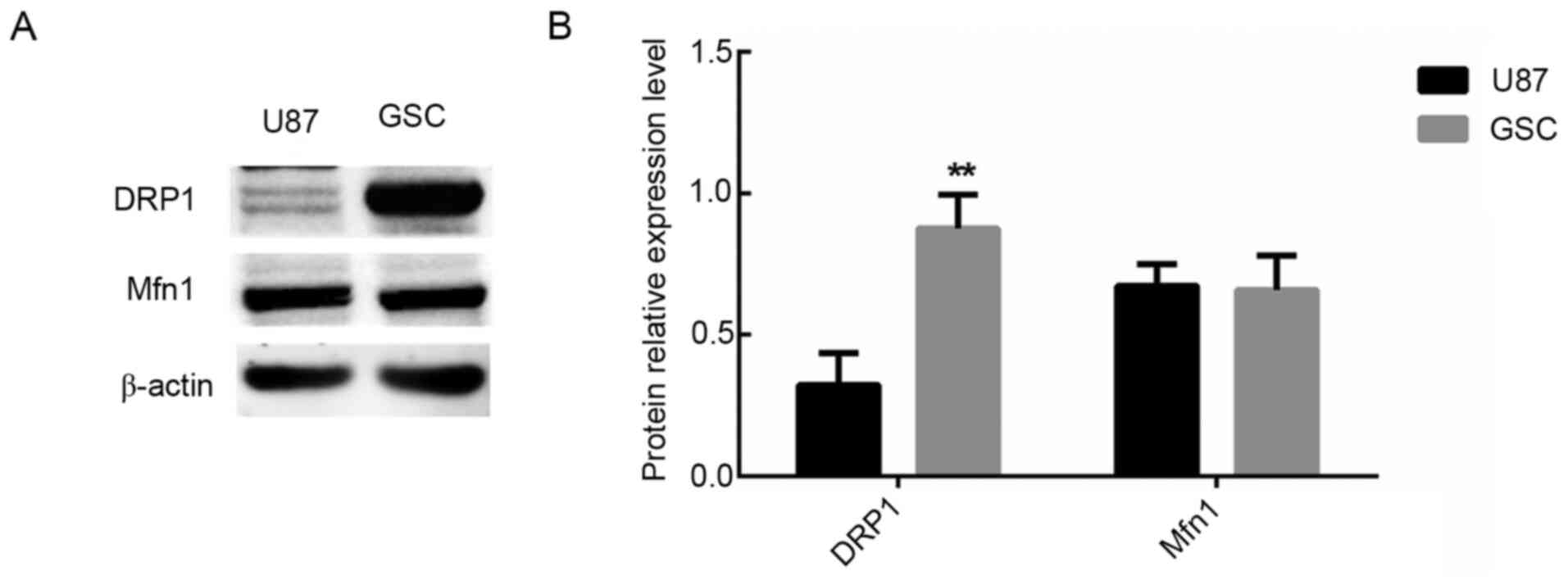
The expression levels of the mitotic protein DRP1
and the fusion protein Mfn1 in the control cell group and GSC cell
group were analyzed. The results revealed that expression of DRP1
was significantly upregulated in the GSC group compared with the
control group (P<0.01; Fig. 3).
However, there was no significant difference in the expression of
Mfn1 (P>0.01; Fig. 3). These
results demonstrated that there was abnormal division of the
mitochondria in GSCs.
Intervention in GSCs with Mdivi-1
alters the expression of DRP1 and Fis1 at different concentrations
and time points
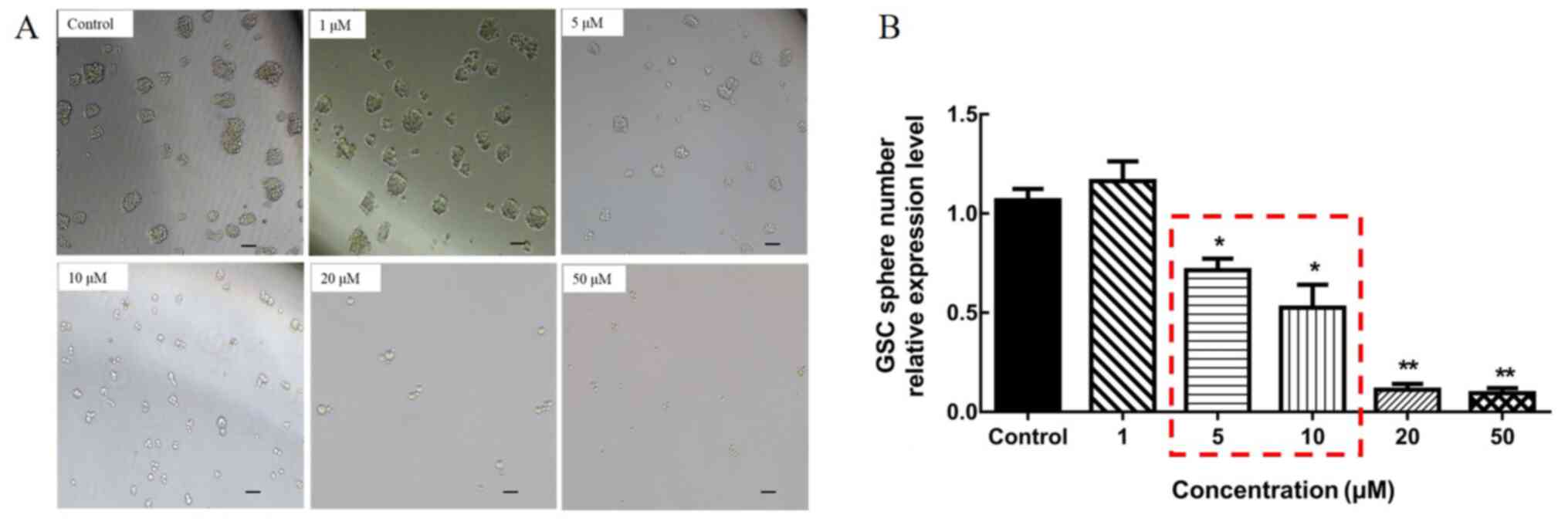
Mdivi-1 was used to intervene in the mitochondrial
division of GSCs, and firstly the optimal intervention
concentration of Mdivi-1 was determined. A total of five
concentrations were selected according to the pharmacological
properties of Mdivi-1(39); 1, 5,
10, 20 and 50 µM. After intervention with different concentrations,
it was preliminarily observed that the cells could not survive at a
concentration of 20 and 50 µM, by observing the morphology and
growth status of the cells (Fig.
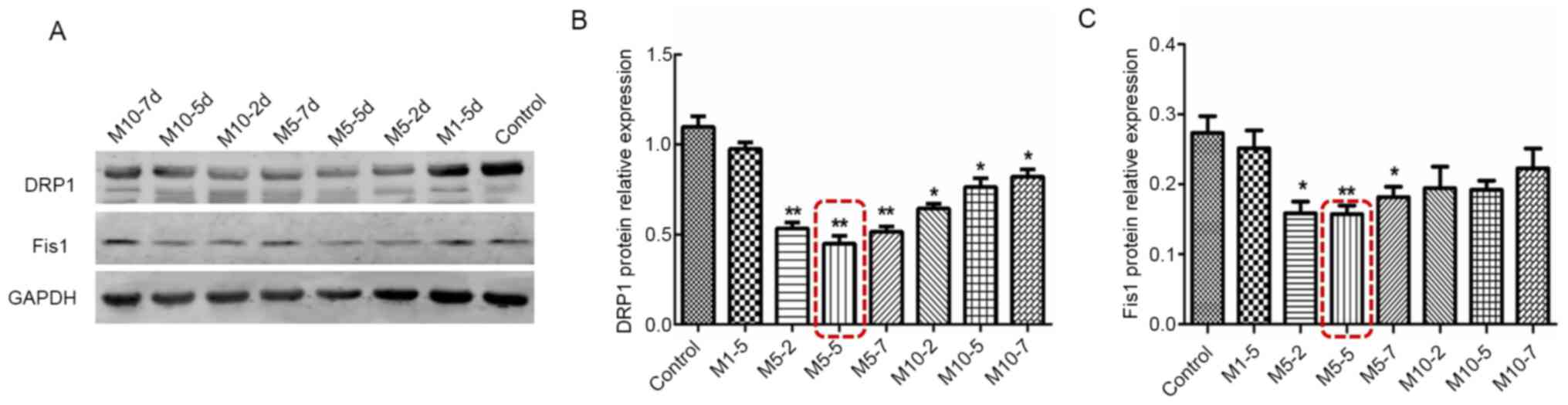
4). In order to identify the most suitable Mdivi-1
concentration, the present study further performed a screening of
the Mdivi-1 intervention concentrations and time points by
detecting the protein expression alterations of DRP1 and Fis. 1.
Compared with the normal control group (untreated GSCs), the
expression levels of DRP1 and Fis1 were more importantly decreased
in the M5-5d group (P<0.01; Fig.
5). It was therefore established that subsequent experiments
could be performed with the M5-5d dose.
Effect on stem cell characteristics of
GSCs with intervention of 5 µM Mdivi-1 for 5 days
The mRNA expression levels of CD133, c-Myc, Klf4,
SOX2, Nestin, Nanog and OCT4 in the control GSC group and the GSC
group treated with a continuous intervention of 5 µM Mdivi-1 for 5
days were analyzed by RT-qPCR assay. The results revealed that the
mRNA expression levels of these genes decreased significantly after
Mdivi-1 intervention (all P<0.001). The CD133 (P<0.01), c-Myc
(P<0.001), Klf4 (P<0.01), SOX2 (P<0.001), Nestin
(P<0.05), Nanog (P<0.01) and OCT4 (P<0.05) protein
expression levels detected via western blot analysis were markedly
decreased in Mdivi-1 treated cells compared with the GSC group
(Fig. 6). These results revealed
that the expression level of the majority of stem cell-associated
genes in GSCs was notably reduced following continuous treatment of
the GSCs with 5 µM Mdivi-1 for 5 days.
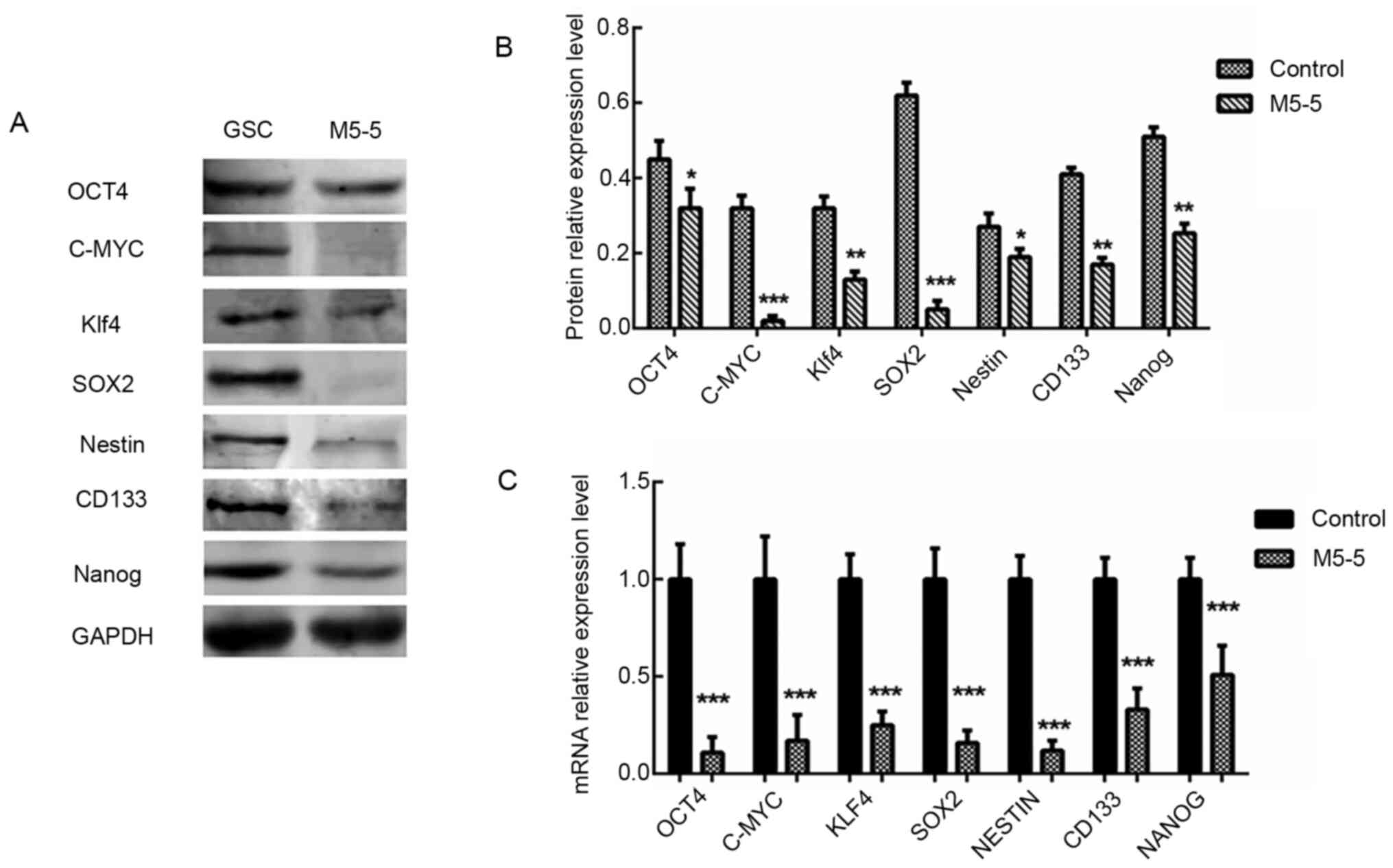 | Figure 6Detection of the effects of M5-5d
intervention on GSC characteristics via western blotting and
reverse transcription-quantitative PCR. (A and B) The protein
expression levels of stem cell-associated genes, CD133 (130 kDa),
c-Myc (57-70 kDa), Klf4 (65 kDa), SOX2 (35 kDa), Nestin (207 kDa),
Nanog (42 kDa) and OCT4 (45 kDa), were analyzed via western
blotting. GAPDH (37 kDa) was used as an internal control. (C) Total
RNA was extracted to quantify CD133, c-Myc, Klf4, SOX2, Nestin,
Nanog and OCT4 at the mRNA level, which was normalized to GAPDH.
*P<0.05, **P<0.01 and
***P<0.001 vs. untreated GSCs as control group. Klf4,
Krueppel-like factor 4; M, Mdivi-1; GSC, glioma stem cells. |
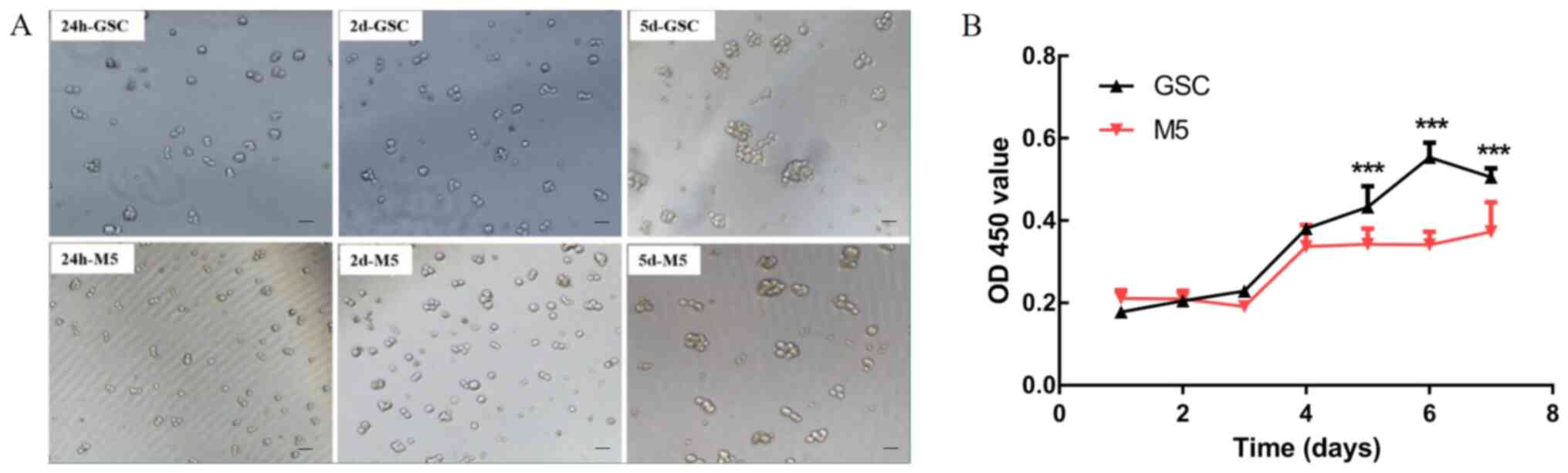
Effects on the proliferation and
apoptosis of GSCs after treatment with 5 µM Mdivi-1 for 5 days
After 5 µM Mdivi-1 continuous intervention for 5
days(M5-5), the number of globular GSCs was reduced compared to
normal untreated GSCs cultured continuously for 5 days (data not
shown). And the treated cells were selected for subsequent
experiments (Fig. 7A). The results
of the CCK-8 assay revealed that the proliferative ability of GSCs
was markedly inhibited following 1 week of Mdivi-1 intervention
(P<0.001; Fig. 7B).
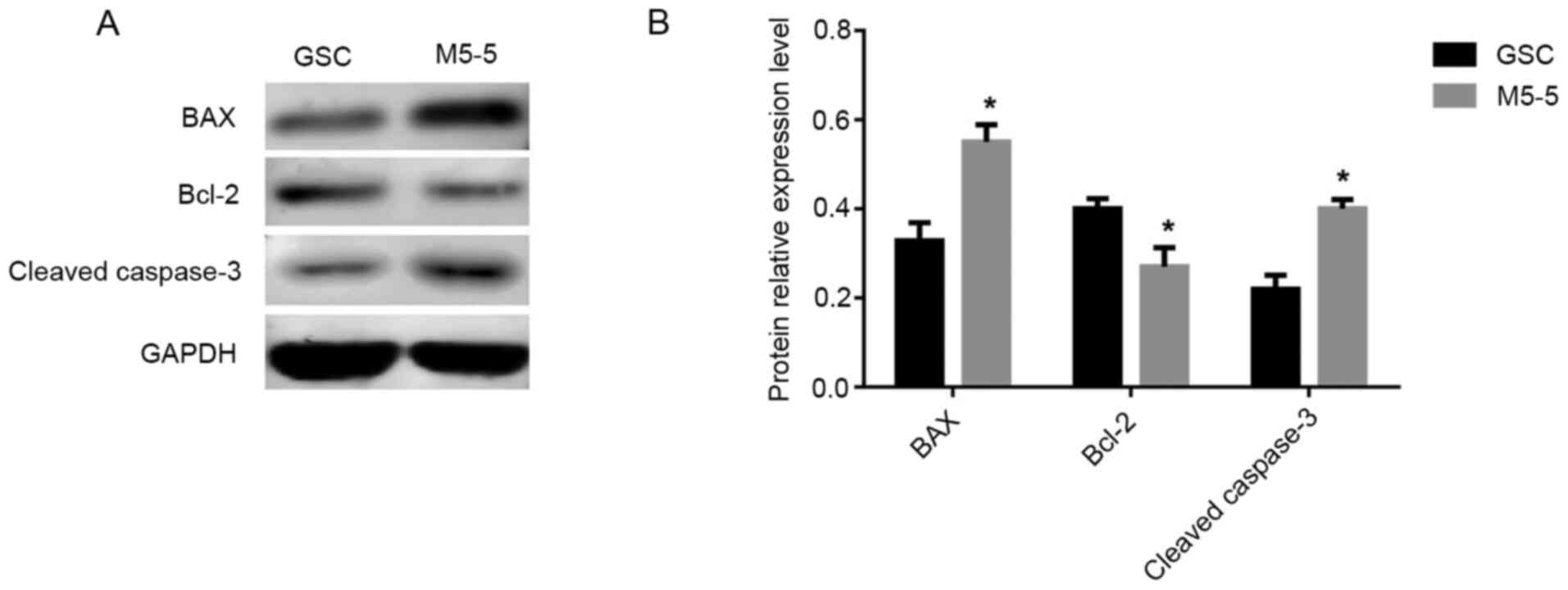
The expression levels of apoptosis-associated
proteins in the control group (GSC group) and the experimental
group (GSCs treated with 5 µM Mdivi-1 for 5 days) was analyzed via
western blotting. The results revealed that compared with the GSC
group, the expression levels of the apoptosis promoting factors Bax
and cleaved caspase-3 were significantly increased and the
expression level of Bcl-2, which is an inhibitor of apoptosis, was
decreased in the GSC group treated with 5 µM Mdivi-1 for 5 days
(all P<0.05; Fig. 8). Therefore,
it was revealed that GSC apoptosis was enhanced following treatment
with Mdivi-1 for 5 days.
Discussion
It has been revealed that GSCs mediate the
heterogeneity and drug resistance of glioma, and they are also
associated with the prognosis of patients. Therefore, they may
provide a novel direction for the treatment of glioma (40). The results of the present study
revealed that GSCs could be induced to differentiate into neurons,
oligodendrocytes and astrocytes. It was notable that in the process
of GSC-induced differentiation, the results of GFAP identification
demonstrated positive expression, but although the induced cells
were GFAP+ cells, it was difficult to determine whether
they were astrocytes or glioma cells. It was difficult to
distinguish the two cell types using morphology alone; however, the
emergence of the GFAP+ cell population indicated that
the cell model exhibited the potential for redifferentiation.
Subsequently, the alterations in the expression level of stem
cell-associated genes (CD133, C-MYC, Klf4, SOX2, Nestin, Nanog and
OCT4) in the GSC population were determined by western blotting.
These genes were expressed in GSCs. However, the expression level
of these genes in GSCs was significantly decreased after the 5-day
intervention with 5 µM Mdivi-1, thereby altering the GSC
characteristics. Subsequent experimental results revealed that 5 µM
Mdivi-1 significantly inhibited the proliferation of GSCs and
altered the expression level of apoptosis-associated proteins,
thereby potentially promoting apoptosis. At present, the specific
surface markers of GSCs cannot be clearly determined (41). In order to establish a good
therapeutic effect, GSCs must first be distinguished from normal
stem cells or NSCs. Therefore, finding novel GSC-specific markers
will aid follow-up research and clinical treatment.
It has been demonstrated that the proliferation and
survival of tumor stem cells were closely associated with the
function of mitochondria (42). The
dynamic imbalance of the mitochondrial structure has been
demonstrated to be closely associated with tumorigenesis (43). The present study established an
innovative GSC model in vitro. A variety of methods were
used to ensure the effective induction of GSCs. After the
successful establishment of the cell model, the association between
GSCs and mitochondria fission was investigated. Mdivi-1 can act as
a protective agent in non-tumor cells and affect mitochondrial
fusion/fission (33). Therefore, in
the present study Mdivi-1 was selected to inhibit DRP1, thereby
affecting the pathway of mitosis, and observe the role of
mitochondria fission in GSCs. Mdivi-1 exhibits distinct effects in
different cells (44), therefore it
was necessary to examine the optimal intervention concentration and
time of Mdivi-1. A number of different intervention durations and
concentrations were investigated. The results indicated that the
survival of cells treated with M5-5d was enhanced compared with
those treated with M10-5d, which met the requirements of the cell
status for subsequent experiments. In addition, the inhibition
effect of M5-5d was most evident in terms of alterations in the
DRP1 and Fis1 protein expression level among all concentrations and
durations examined. Interestingly, DRP1 expression level was
slightly decreased in M10-5d, but most importantly decreased in
M5-5d. In terms of the survival status of the Mdivi-1 stimulated
cells and the expression level of associated proteins, it was
finally determined that stimulation with 5 µM Mdivi-1 for 5 days
was the most suitable intervention scheme. However, the present
study only used Mdivi-1 as a DRP1 inhibitor to examine its function
in GSCs and did not investigate how Mdivi-1 may interact with DRP1,
which should be thoroughly examined in follow-up experiments.
Mitochondrial dynamics are considered to represent a
novel perspective for understanding complex diseases, and the
association between mitochondrial dynamics and diseases requires
further investigation (45). In
this case, mitochondrial targeting drugs are expected to interfere
with tumor adaptability and promote the elimination of CSCs
(46,47). Although GSCs have gradually become a
popular topic in clinical glioma research, there is still a number
of problems that require further investigation. For example, an
increasing number of studies (48,49)
have found that there is considerable plasticity between non-GSCs
and GSCs subsets in glioblastoma, especially that non-GSCs
differentiated during chemotherapy can transform into GSCs.
However, how plasticity controls the mutual transformation of them
remains unclear (50). At present,
it is not clear what the actual molecular characteristics of GSCs
are, and it is important to identify specific and reliable GSCs
biomarkers (51). This requires
constant investigation and improvement to provide theoretical and
practical basis for basic research and clinical treatment. In
conclusion, the results of the present study demonstrated that GSC
cell models can be cultured in vitro by induction medium.
Furthermore, after the specific inhibitor Mdivi-1 interferes with
GSC, it can significantly reduce the expression of stem
cell-related genes of GSC, inhibit the proliferation of GSC and
promote the expression of apoptosis-related genes of GSC.
Therefore, after using Mdivi-1 to affect the gene expression of
DRP1 may represent a novel strategy of targeting glioma
treatment.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by grants from the Ningxia
Medical University School-level Scientific Research Project (grant
no. XT2018013) and Ningxia High School First-class Disciplines
(West China Top Class Discipline Project in Basic Medical Sciences,
Ningxia Medical University; grant no. NXYLXK2017B07).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LZ and QH designed the study. LZ, HC, HT, JY and WG
performed the research. LZ, HC and QH analyzed the data. LZ, HC and
QH wrote and revised the manuscript. LZ and HC confirm the
authenticity of the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Moini J and Piran P (eds): Chapter 1 -
Histophysiology. In: Functional and Clinical Neuroanatomy. Academic
Press, pp1-49, 2020.
|
|
2
|
Singh SK, Hawkins C, Clarke ID, Squire JA,
Bayani J, Hide T, Henkelman RM, Cusimano MD and Dirks PB:
Identification of human brain tumour initiating cells. Nature.
432:396–401. 2004.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Pattabiraman DR and Weinberg RA: Tackling
the cancer stem cells-what challenges do they pose? Nat Rev Drug
Discov. 13:497–512. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
4
|
Bao B, Ahmad A, Azmi AS, Ali S and Sarkar
FH: Overview of cancer stem cells (CSCs) and mechanisms of their
regulation: implications for cancer therapy. Curr Protoc Pharmacol;
Chapter 14: Unit 14 25, 2013.
|
|
5
|
Yan K, Wu Q, Yan DH, Lee CH, Rahim N,
Tritschler I, DeVecchio J, Kalady MF, Hjelmeland AB, Rich JN, et
al: Glioma cancer stem cells secrete Gremlin1 to promote their
maintenance within the tumor hierarchy. Genes Dev. 28:1085–1100.
2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Sugiarto S, Persson AI, Munoz EG,
Waldhuber M, Lamagna C, Andor N, Hanecker P, Ayers-Ringler J,
Phillips J, Siu J, et al: Asymmetry-defective oligodendrocyte
progenitors are glioma precursors. Cancer Cell. 20:328–340.
2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
O'Brien CA, Kreso A, Ryan P, Hermans KG,
Gibson L, Wang Y, Tsatsanis A, Gallinger S and Dick JE: ID1 and ID3
regulate the self-renewal capacity of human colon cancer-initiating
cells through p21. Cancer Cell. 21:777–792. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Pece S, Tosoni D, Confalonieri S, Mazzarol
G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG and Di Fiore
PP: Biological and molecular heterogeneity of breast cancers
correlates with their cancer stem cell content. Cell. 140:62–73.
2010.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Okano H: Stem cell biology of the central
nervous system. J Neurosci Res. 69:698–707. 2002.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zarco N, Norton E, Quinones-Hinojosa A and
Guerrero-Cazares H: Overlapping migratory mechanisms between neural
progenitor cells and brain tumor stem cells. Cell Mol Life Sci.
76:3553–3570. 2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Yu SC, Ping YF, Yi L, Zhou ZH, Chen JH,
Yao XH, Gao L, Wang JM and Bian XW: Isolation and characterization
of cancer stem cells from a human glioblastoma cell line U87.
Cancer Lett. 265:124–134. 2008.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang R and Liu C: All-trans retinoic acid
therapy induces asymmetric division of glioma stem cells from the
U87MG cell line. Oncol Lett. 18:3646–3654. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Shahriyari L and Komarova NL: Symmetric
vs. asymmetric stem cell divisions: An adaptation against cancer?
PLoS One. 8(e76195)2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Lathia JD, Hitomi M, Gallagher J, Gadani
SP, Adkins J, Vasanji A, Liu L, Eyler CE, Heddleston JM, Wu Q, et
al: Distribution of CD133 reveals glioma stem cells self-renew
through symmetric and asymmetric cell divisions. Cell Death Dis.
2(e200)2011.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Berika M, Elgayyar ME and El-Hashash AH:
Asymmetric cell division of stem cells in the lung and other
systems. Front Cell Dev Biol. 2(33)2014.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Najbauer J, Kraljik N and Nemeth P: Glioma
stem cells: Markers, hallmarks and therapeutic targeting by
metformin. Pathol Oncol Res. 20:789–797. 2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Jin X, Jin X, Jung JE, Beck S and Kim H:
Cell surface Nestin is a biomarker for glioma stem cells. Biochem
Biophys Res Commun. 433:496–501. 2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zou Q, Yan Q, Zhong J, Wang K, Sun H, Yi X
and Lai L: Direct conversion of human fibroblasts into neuronal
restricted progenitors. J Biol Chem. 289:5250–5260. 2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wernig M, Tucker KL, Gornik V, Schneiders
A, Buschwald R, Wiestler OD, Barde YA and Brüstle O: Tau EGFP
embryonic stem cells: An efficient tool for neuronal lineage
selection and transplantation. J Neurosci Res. 69:918–924.
2002.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Liu YJ, McIntyre RL, Janssens GE and
Houtkooper RH: Mitochondrial fission and fusion: A dynamic role in
aging and potential target for age-related disease. Mech Ageing
Dev. 186(111212)2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Vyas S, Zaganjor E and Haigis MC:
Mitochondria and cancer. Cell. 166:555–566. 2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Taguchi N, Ishihara N, Jofuku A, Oka T and
Mihara K: Mitotic phosphorylation of dynamin-related GTPase Drp1
participates in mitochondrial fission. J Biol Chem.
282:11521–11529. 2007.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Song M and Dorn GW II: Mitoconfusion:
Noncanonical functioning of dynamism factors in static mitochondria
of the heart. Cell Metab. 21:195–205. 2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Ryan JJ, Marsboom G, Fang YH, Toth PT,
Morrow E, Luo N, Piao L, Hong Z, Ericson K, Zhang HJ, et al:
PGC1α-mediated mitofusin-2 deficiency in female rats and humans
with pulmonary arterial hypertension. Am J Respir Crit Care Med.
187:865–878. 2013.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Senft D and Ronai ZA: Regulators of
mitochondrial dynamics in cancer. Curr Opin Cell Biol. 39:43–52.
2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Seo BJ, Yoon SH and Do JT: Mitochondrial
dynamics in stem cells and differentiation. Int J Mol Sci.
19(3893)2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Kim D, Sankaramoorthy A and Roy S:
Downregulation of Drp1 and Fis1 inhibits mitochondrial fission and
prevents high glucose-induced apoptosis in retinal endothelial
cells. Cells. 9(1662)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ji WK, Hatch AL, Merrill RA, Strack S and
Higgs HN: Actin filaments target the oligomeric maturation of the
dynamin GTPase Drp1 to mitochondrial fission sites. Elife.
4(e11553)2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Yu R, Jin SB, Lendahl U, Nister M and Zhao
J: Human Fis1 regulates mitochondrial dynamics through inhibition
of the fusion machinery. EMBO J. 38(e99748)2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Chen H, Detmer SA, Ewald AJ, Griffin EE,
Fraser SE and Chan DC: Mitofusins Mfn1 and Mfn2 coordinately
regulate mitochondrial fusion and are essential for embryonic
development. J Cell Biol. 160:189–200. 2003.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Allegra A, Innao V, Allegra AG and
Musolino C: Relationship between mitofusin 2 and cancer. Adv
Protein Chem Struct Biol. 116:209–236. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ruiz A, Alberdi E and Matute C:
Mitochondrial division inhibitor 1 (mdivi-1) protects neurons
against excitotoxicity through the modulation of mitochondrial
function and intracellular Ca2+ signaling. Front Mol
Neurosci. 11(3)2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Cassidy-Stone A, Chipuk JE, Ingerman E,
Song C, Yoo C, Kuwana T, Kurth MJ, Shaw JT, Hinshaw JE, Green DR
and Nunnari J: Chemical inhibition of the mitochondrial division
dynamin reveals its role in Bax/Bak-dependent mitochondrial outer
membrane permeabilization. Dev Cell. 14:193–204. 2008.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Valenti D, Rossi L, Marzulli D, Bellomo F,
De Rasmo D, Signorile A and Vacca RA: Inhibition of Drp1-mediated
mitochondrial fission improves mitochondrial dynamics and
bioenergetics stimulating neurogenesis in hippocampal progenitor
cells from a Down syndrome mouse model. Biochim Biophys Acta Mol
Basis Dis. 1863:3117–3127. 2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Martinez-Outschoorn UE, Peiris-Pages M,
Pestell RG, Sotgia F and Lisanti MP: Cancer metabolism: A
therapeutic perspective. Nat Rev Clin Oncol. 14(113)2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Caren H, Stricker SH, Bulstrode H, Gagrica
S, Johnstone E, Bartlett TE, Feber A, Wilson G, Teschendorff AE,
Bertone P, et al: Glioblastoma stem cells respond to
differentiation cues but fail to undergo commitment and terminal
cell-cycle arrest. Stem Cell Reports. 5:829–842. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Srikanth M, Kim J, Das S and Kessler JA:
BMP signaling induces astrocytic differentiation of clinically
derived oligodendroglioma propagating cells. Mol Cancer Res.
12:283–294. 2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Smith G and Gallo G: To mdivi-1 or not to
mdivi-1: Is that the question? Dev Neurobiol. 77:1260–1268.
2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Takashima Y, Kawaguchi A and Yamanaka R:
Promising Prognosis Marker Candidates on the Status of
Epithelial-Mesenchymal Transition and Glioma Stem Cells in
Glioblastoma. Cells. 8:2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Kim WT and Ryu CJ: Cancer stem cell
surface markers on normal stem cells. BMB Rep. 50:285–298.
2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Fiorillo M, Lamb R, Tanowitz HB, Cappello
AR, Martinez-Outschoorn UE, Sotgia F and Lisanti MP: Bedaquiline,
an FDA-approved antibiotic, inhibits mitochondrial function and
potently blocks the proliferative expansion of stem-like cancer
cells (CSCs). Aging (Albany NY). 8:1593–1607. 2016.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Roberts ER and Thomas KJ: The role of
mitochondria in the development and progression of lung cancer.
Comput Struct Biotechnol J. 6(e201303019)2013.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Bordt EA, Clerc P, Roelofs BA, Saladino
AJ, Tretter L, Adam-Vizi V, Cherok E, Khalil A, Yadava N, Ge SX, et
al: The Putative Drp1 Inhibitor mdivi-1 is a reversible
mitochondrial complex I inhibitor that modulates reactive oxygen
species. Dev Cell. 40:583–594.e6. 2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Tilokani L, Nagashima S, Paupe V and
Prudent J: Mitochondrial dynamics: Overview of molecular
mechanisms. Essays Biochem. 62:341–360. 2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Semenza GL: Hypoxia-inducible factors:
Coupling glucose metabolism and redox regulation with induction of
the breast cancer stem cell phenotype. EMBO J. 36:252–259.
2017.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Peiris-Pages M, Martinez-Outschoorn UE,
Pestell RG, Sotgia F and Lisanti MP: Cancer stem cell metabolism.
Breast Cancer Res. 18(55)2016.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Ohka F, Natsume A and Kondo Y: Chapter 15
- Clinical significance of epigenetic alterations in glioblastoma.
In: Epigenetic Cancer Therapy. Gray SG (ed). Academic Press,
Boston, pp339-350, 2015.
|
|
49
|
Garnier D, Renoult O, Alves-Guerra MC,
Paris F and Pecqueur C: Glioblastoma stem-like cells,
metabolic strategy to kill a challenging target. Front Oncol.
9(118)2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Safa AR, Saadatzadeh MR, Cohen-Gadol AA,
Pollok KE and Bijangi-Vishehsaraei K: Glioblastoma stem cells
(GSCs) epigenetic plasticity and interconversion between
differentiated non-GSCs and GSCs. Genes Dis. 2:152–163.
2015.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Yuan Y, Yan Z, Miao J, Cai R, Zhang M,
Wang Y, Wang L, Dang W, Wang D, Xiang D, et al: Autofluorescence of
NADH is a new biomarker for sorting and characterizing cancer stem
cells in human glioma. Stem Cell Res Ther. 10(330)2019.PubMed/NCBI View Article : Google Scholar
|