Introduction
Lung cancer is a type of malignant tumor that
continues to be the leading cause of cancer-associated mortality
worldwide (1). The estimated number
of new cases and deaths from lung cancer worldwide in 2020 were
2,206,771 and 1,796,144, respectively (2). As a subtype of lung cancer, lung
adenocarcinoma has the highest incidence of all types of lung
cancer (3). Surgery, chemotherapy,
radiotherapy, targeted therapies and combined therapies are the
therapeutic strategies that are currently available (4). However, poor patient responses to
therapy, individual differences, adverse side effects and
resistance to chemotherapeutic agents mean that clinical
difficulties remain for the treatment of lung adenocarcinoma
(5-7).
Traditional Chinese medicine (TCM) has been reported
to have anti-cancer effects in lung adenocarcinoma, osteosarcoma
and other types of cancer by regulating the tumor microenvironment
and enhancing host immune responses (8-10).
In particular, the red sage Salvia miltiorrhiza (Danshen)
has been reported to improve survival rate for patients with colon
(11) and breast cancer (12). The chemical composition of Salvia
miltiorrhiza includes two categories (13): i) Fat-soluble tanshinone compounds,
which are mainly comprised of Tanshinone I and Tanshinone IIA; and
ii) water-soluble phenolic compounds, consisting mainly of
salvianolate. Tung et al (14) found Tanshinone I to inhibit the
proliferation of lung adenocarcinoma cell lines A549, CL1-0 and
CL1-5, which was in turn more effective compared with Tanshinone
II. However, to the best of our knowledge, no studies have clearly
demonstrated changes in gene expression and pathway enrichment
following Tanshinone I administration on lung adenocarcinoma.
Furthermore, as a major water-soluble component of Salvia
miltiorrhiza extract, it remains currently unclear whether
salvianolate has an effect on lung adenocarcinoma.
ATPase copper transporting α and β (ATP7A and ATP7B)
are heavy metal transporting P-type ATPases that function as copper
efflux transporters to maintain cellular copper homeostasis
(15). It has been previously shown
that ATP7A/7B has effects on tumorigenesis (16), tumor cell differentiation (17) and platinum-based chemotherapy
response (18,19) in breast, lung and ovarian cancer. In
addition, previous studies suggest that ATP7A/7B are considered to
be potential targets for the treatment of non-small cell lung
cancer (NSCLC) (18,20,21).
Therefore, present study investigated the effect of ATP7A/7B
expression following salvianolate treatment.
Numerous profiles in carcinogenesis and cancer
progression have been screened following the development of
microarrays and high-throughput sequencing. The present study
analyzed the gene expression profile matrix file (GSE9315)
(22) using a series of
bioinformatics tools to identify hub genes and key pathways that
are affected by Tanshinone I administration on lung adenocarcinoma.
The effects of salvianolate were then investigated using a
xenograft nude mouse model before the potential underlying
mechanisms of action were assessed. The present study identified
the underlying mechanisms of action and potential targets of the
Salvia miltiorrhiza extract for the treatment of lung
adenocarcinoma.
Materials and methods
Data source
The gene expression profile matrix file (GSE9315)
was downloaded from the Gene Expression Omnibus (GEO) database
(https://www.ncbi.nlm.nih.gov/geo/).
GSE9315 contained five different concentrations of Tanshinone I
(0.00, 0.01, 0.10, 1.00 and 10.00 µg/ml) in macrophage-conditioned
medium (CM) and a control condition without CM in the lung
adenocarcinoma cell line CL1-5. Its platform used was GPL5968
(NCHU_M&A_human 1152 cDNACHIP).
Identification of differentially
expressed genes (DEGs) following different doses of Tanshinone
I
GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) is an
interactive web tool that was used to analyze the raw data and
identify DEGs in the GSE9315 dataset. GSE9315 was divided into the
following three groups: i) Control group (0 µg/ml in CM); ii) low
dosage group (0.01 and 0.1 µg/ml in CM); and iii) medium dosage
group (1 and 10 µg/ml in CM) (22).
In the present study, |log fold change (FC)| >1 was used as the
cut-off criteria to identify DEGs. P<0.05 was not used as a
criterion for screening DEG since there was only one sample in the
control group, which was not suitable for statistical analysis. A
Venn diagram was used to identify overlapping DEGs in the low and
medium dosage groups.
Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of
DEGs
The Database for Annotation, Visualization and
Integrated Discovery (DAVID; version 6.8; https://david.ncifcrf.gov/) (23) was used to perform the GO and KEGG
pathway enrichment analyses. The results were visualized using
ggplot2 package in R Studio (version 1.1.453; The R Foundation)
(24). P<0.05 was considered to
indicate a statistically significant difference.
Protein-protein interaction (PPI)
network construction and module analysis
The Search Tool for the Retrieval of Interacting
Gene (version 11.0; https://string-db.org/cgi/input.pl) database (25) was used to construct a functional PPI
network of the DEGs. An interaction score of 0.4 was regarded as
the cut-off criterion in the present study. The Molecular Complex
Detection (MCODE) in Cytoscape software (version 3.6.0; https://cytoscape.org/) (26) was then used to find closely
connected regions in the PPI network. The selection criteria were
as follows: i) Degree Cut-off, 2; ii) node score cut-off, 0.2; iii)
K-Core, 2; and iv) max depth, 100.
Cell culture
The human lung adenocarcinoma cell line A549
(Changsha Nanke Biotechnology Co., Ltd.; http://www.nkbio.cn) was cultured in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.) at 37˚C with 5%
CO2.
Establishment and drug administration
in animal models
The present study was performed according to the
guidelines of the Ethics Committee of Institutional Animal Care and
Use in Central South University (27) and approved by the Medical Ethics
Committee of Xiangya Hospital, Central South University (approval
no. 2017121170; Changsha, China). A total of 30, 6-week-old nude
mice (weight range, 16-18 g; sex, 1:1 ratio of males and females)
purchased from Hunan SJA Laboratory Animal Co., Ltd, were kept
under specific-pathogen free conditions (sterile laminar flow
chamber, room temperature 21-25˚C; humidity of 60-70%; 12 h
light/dark cycle) with access to food and drinking water ad
libitum at the Department of Laboratory Animals, Central South
University. A549 cells in the logarithmic growth phase were
harvested and resuspended in PBS at a density of
1x106/ml. After being anesthetized by isoflurane
inhalation (1.5%), the nude mice were injected subcutaneously into
the left armpit in a cell suspension of 0.2 ml (2x105
cells per mouse). The next day, the tumor volume of nude mice was
measured, which was regarded as the first day after injection (day
1). The tumor volume was measured once every 3 days for 16 days
(day 1, 4, 7, 10, 13 and 16). When the tumors became visible (day
4), 10, 20 and 50 mg/kg salvianolate (Green Valley, Inc.) and
normal saline (Sichuan Kelun Pharmaceutical Co., Ltd.) were
injected into the tumor-adjacent tissue once every 3 days (day 4,
7, 10 and 13). Mice were sacrificed by cervical dislocation on day
16 following the final treatment of salvianolate and normal saline
before the tumor tissues were collected for further research. The
tumor volume was measured using the following equation: V
(mm3) = a x b2 x0.52, where ‘a’ and ‘b’ were
the longest and shortest diameters of the tumor. The tumor
inhibition rate was calculated using the following equation:
Inhibition rate (%)=
(1-Vsalvianolate/Vmean-control) x100%. Lack
of breathing or no basic vital signs were evaluated to ensure death
after cervical dislocation. In the present study, ‘The animal
exhibiting depression and hypothermia without anesthesia or
sedation (dying)’ was regarded as the criterion for euthanizing the
animals.
Hematoxylin and eosin (H&E)
staining
The tumor tissues were fixed in 4% paraformaldehyde
for 24 h at room temperature and embedded in paraffin. Tissue
samples were then cut into 3-µm-thick slices and the slices were
heated at 60˚C for 1.5 h. The slices were deparaffinized in xylene
and rehydrated in graded ethanol solutions (100, 95, 80 and 70%
ethanol for 5 min at each concentration) at room temperature. After
washing with water, slices were stained with hematoxylin for 2 min,
rinsed with water, incubated in 0.5% HCl ethanol for 1-3 sec,
rinsed with water and stained with eosin for 3 min, all at room
temperature. Then the slices were dehydrated using 70, 80, 95 and
100% gradient ethanol and xylene for 5 min at room temperature, and
sealed with neutral gum. The nuclei were blue-violet in color,
whilst the cytoplasm and intercellular matrix were red under light
microscopy when the histological changes were assessed
(magnification, x400).
Immunohistochemistry (IHC)
Paraffin-embedded tumor tissues, as aforementioned,
were cut into 4-µm-thick slices. Slices were dewaxed using xylene,
rehydrated in an descending ethanol gradient and treated with 3%
hydrogen peroxide for 10 min at room temperature to remove
endogenous oxidases. The slices were then repaired using a citrate
repairing solution (0.01 mol/l; pH 6.0; cat. no. C1010; Beijing
Solarbio Science & Technology Co., Ltd.), heated at 100˚C for
15 min for antigen retrieval and washed for 3 min with PBS (0.01
mol/l, pH 7.2) after air cooling. Subsequently, each slice was
blocked with 5% goat serum (cat. no. SL038; Beijing Solarbio
Science & Technology Co., Ltd.) for 10 min at room temperature
and incubated with primary antibodies against ATP7A (1:100; cat.
no. NBP2-59376; Novus Biologicals, LLC.) or ATP7B (1:100; cat. no.
ab133731; Abcam) overnight at 4˚C. After three washes with PBS, the
slices were incubated with rabbit anti-mouse and goat anti-rabbit
IgG horseradish peroxidase-conjugated secondary antibodies (both
1:1,000; cat. nos. ab6721 and ab6728, respectively; Abcam) for 2 h
at room temperature. The sections were washed three times with PBS
and incubated with streptavidin-peroxidase solution for 10 min at
room temperature. Finally, DAB (Fuzhou Maixin Biotech Co., Ltd.)
was used for visualization and hematoxylin for counterstaining,
followed by dehydration with ethanol gradient (70, 80, 90, 95 and
100%) followed by xylene and mounting with neutral gum. After
visualizing, images of sections were captured using a light
microscope (magnification, x400) and analyzed using Image-Pro Plus
6.0 software (Media Cybernetics, Inc.)
Statistical analysis
All data were processed using SPSS 19.0 statistical
software (IBM Corp.). One-way ANOVA with Bonferroni's correction
was used for the comparison between the various groups. Data are
presented as the mean ± SD. P<0.05 was considered to indicate a
statistically significant difference. The number of repeats in each
group is ≥3.
Results
Identification of DEGs after low or
medium dosage treatment of Tanshinone I
Based on the aforementioned threshold (|log FC|
>1), 28 and 102 DEGs were identified in the low dosage group and
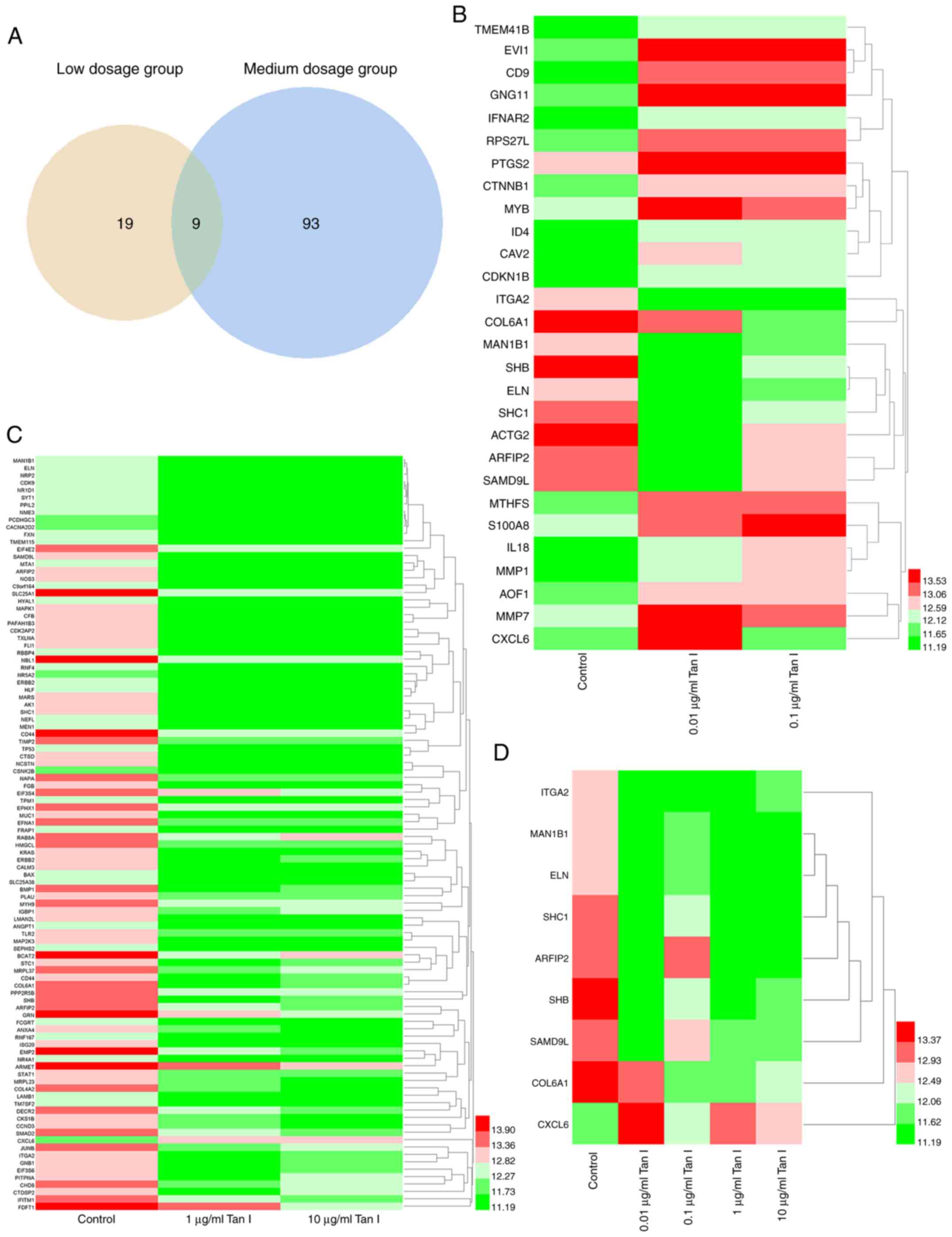
the medium dosage groups, respectively (Table I). Notably, there were nine
overlapping DEGs in the low and medium dosage groups (Fig. 1A), which were integrin subunit α
(ITGA2), endoplasmic reticulum mannosyl-oligosaccharide
1,2-α-mannosidase (MAN1B1), SH2 domain containing adaptor protein B
(SHB), elastin (ELN), Src homology 2 domain-containing)
transforming protein 1 (SHC1), collagen type VI α1 chain (COL6A1),
ADP ribosylation factor interacting protein 2 (ARFIP2), chemokine
(C-X-C motif) ligand 6 (CXCL6) and sterile α motif domain
containing 9 like (SAMD9L). The visualized heatmaps of the DEGs in
the individualized different dosage groups are presented in
Fig. 1B and C, which showed that some genes were
upregulated (such as CD9, IFNAR2 and PTGS2) and others were
downregulated (such as MAPK1, SLC25A1 and KRAS). In the heatmaps,
red and green represent upregulation and downregulation of gene
expression, respectively.
 | Table IDifferentially expressed genes
identified in the different dosage groups. |
Table I
Differentially expressed genes
identified in the different dosage groups.
| Groups | Gene symbol |
|---|
| Low dosage | TMEM41B, EVI1, CD9,
GNG11, IFNAR2, RPS27L, PTGS2, ITGA2, CTNNB1, MYB, ID4, CAV2, MTHFS,
CDKN1B, S100A8, IL18, MMP1, AOF1, MMP7, MAN1B1, SHB, ELN, SHC1,
COL6A1, ACTG2, ARFIP2, CXCL6, SAMD9L |
| Medium dosage | MAN1B1, ELN, NRP2,
CDK9, NR1D1, NME3, PPIL2, FXN, SYT1, PCDHGC3, CACNA2D2, TMEM115,
EIF4E2, SAMD9L, MTA1, ARFIP2, HYAL1, NOS3, MAPK1, C9orf164,
SLC25A1, CFB, PAFAH1B3, CDK2AP2, TXLNA, FLI1, RBBP4, NBL1, RNF4,
NR5A2, ERBB2, NCSTN, HLF, MARS, AK1, NAPA, SHC1, NEFL, MEN1, CD44,
FGB, EIF3S4, TPM1, EPHX1, TIMP2, CSNK2B, TP53, CTSD, MUC1, EFNA1,
FRAP1, RAB8A, ARFIP2, HMGCL, KRAS, GRN, ERBB2, CALM3, BAX,
SLC25A38, FCGRT, ANXA4, BMP1, PLAU, RNF167, MYH9, ISG20, IGBP1,
EMP2, NR4A1, ARMET, STAT1, LMAN2L, ANGPT1, MRPL23, TLR2, MAP2K3,
SEPHS2, BCAT2, STC1, MRPL37, CD44, COL6A1, COL4A2, LAMB1, CXCL6,
TM7SF2, DECR2, PPP2R5B, CKS1B, SHB, CCND3, SMAD2, JUNB, ITGA2,
GNB1, IFITM1, EIF3S6, PITPNA, CHD8, CTDSP2, FDFT1 |
Analysis of overlapping DEGs in the
low and medium dosage groups
There were nine overlapping DEGs in the low and
medium dosage groups, which were ITGA2, MAN1B1, SHB, ELN, SHC1,
COL6A1, ARFIP2, CXCL6 and SAMD9L. The visualization results of gene
expression were displayed using a heatmap (Fig. 1D). Among these nine genes, eight
were downregulated after four different dosages of Tanshinone I
were administrated (shown as green in the heatmap), compared with
the control group. Notably, the expression of these eight genes was
not dose-dependent. Only one of the nine overlapping genes (CXCL6)
was upregulated following Tanshinone I administration (shown as red
in the heatmap). The GO functional enrichment and KEGG pathway
analyses of each gene are presented in Table II, helping us understand the
functional enrichment of each gene. For example, SHC1 was enriched
in ‘epidermal growth factor receptor (ErbB) signaling pathway’ and
COL6A1 was enriched in ‘PI3K-Akt signaling pathway’, indicating
that Tanshinone I may treat lung adenocarcinoma by upregulating or
downregulating these genes and their enriched pathways.
 | Table IIGO functional enrichment and KEGG and
Genomes pathway analysis of overlapping genes in the low and medium
dosage groups. |
Table II
GO functional enrichment and KEGG and
Genomes pathway analysis of overlapping genes in the low and medium
dosage groups.
| Gene symbol | Category | ID | Description |
|---|
| ARFIP2 | BP | GO:0006928 | Movement of cell or
subcellular component |
| | BP | GO:0007264 | Small GTPase
mediated signal transduction |
| | CC | GO:0001726 | Ruffle |
| | CC | GO:0005737 | Cytoplasm |
| | MF | GO:0005515 | Protein
binding |
| | MF | GO:0005525 | GTP binding |
| | KEGG_PATHWAY | / | / |
| CXCL6 | BP | GO:0002446 | Neutrophil mediated
immunity |
| | BP | GO:0002690 | Positive regulation
of leukocyte chemotaxis |
| | CC | GO:0005576 | Extracellular
region |
| | CC | GO:0005615 | Extracellular
space |
| | MF | GO:0008009 | Chemokine
activity |
| | MF | GO:0008201 | Heparin
binding |
| | KEGG_PATHWAY | hsa04060 | Cytokine-cytokine
receptor interaction |
| SHB | BP | GO:0001525 | Angiogenesis |
| | BP | GO:0006469 | Negative regulation
of protein kinase activity |
| | CC | GO:0005829 | Cytosol |
| | CC | GO:0005886 | Plasma
membrane |
| | MF | GO:0001948 | Glycoprotein
binding |
| | MF | GO:0005070 | SH3/SH2 adaptor
activity |
| | KEGG_PATHWAY | / | / |
| SHC1 | BP | GO:0000165 | MAPK cascade |
| | BP | GO:0000187 | Activation of MAPK
activity |
| | CC | GO:0005622 | Intracellular |
| | CC | GO:0005759 | Mitochondrial
matrix |
| | MF | GO:0005068 | Transmembrane
receptor protein tyrosine kinase adaptor activity |
| | MF | GO:0005088 | Ras
guanyl-nucleotide exchange factor activity |
| | KEGG_PATHWAY | hsa04012 | Erbb signaling
pathway |
| COL6A1 | BP | GO:0001649 | Osteoblast
differentiation |
| | BP | GO:0007155 | Cell adhesion |
| | CC | GO:0005576 | Extracellular
region |
| | CC | GO:0005581 | Collagen
trimer |
| | MF | GO:0048407 | Platelet-derived
growth factor binding |
| | KEGG_PATHWAY | hsa04151 | PI3K-Akt signaling
pathway |
| ELN | BP | GO:0007519 | Skeletal muscle
tissue development |
| | BP | GO:0007585 | Respiratory gaseous
exchange |
| | CC | GO:0005576 | Extracellular
region |
| | CC | GO:0005578 | Proteinaceous
extracellular matrix |
| | MF | GO:0005201 | Extracellular
matrix structural constituent |
| | MF | GO:0005515 | Protein
binding |
| | KEGG_PATHWAY | hsa04974 | Protein digestion
and absorption |
| ITGA2 | BP | GO:0001666 | Response to
hypoxia |
| | BP | GO:0002687 | Positive regulation
of leukocyte migration |
| | CC | GO:0005634 | Nucleus |
| | CC | GO:0005886 | Plasma
membrane |
| | MF | GO:0001618 | Virus receptor
activity |
| | MF | GO:0005178 | Integrin
binding |
| | KEGG_PATHWAY | hsa04145 | Phagosome |
| MAN1B1 | BP | GO:0006491 | N-glycan
processing |
| | BP | GO:0008152 | Metabolic
process |
| | CC | GO:0005783 | Endoplasmic
reticulum |
| | CC | GO:0005789 | Endoplasmic
reticulum membrane |
| | MF | GO:0004559 | Alpha-mannosidase
activity |
| | MF | GO:0004571 |
Mannosyl-oligosaccharide
1,2-alpha-mannosidase activity |
| | KEGG_PATHWAY | hsa00510 | N-Glycan
biosynthesis |
| SAMD9L | BP | GO:0034058 | Endosomal vesicle
fusion |
| | CC | GO:0005769 | Early endosome |
| | MF | / | / |
| | KEGG_PATHWAY | / | / |
GO functional enrichment analysis of
DEGs
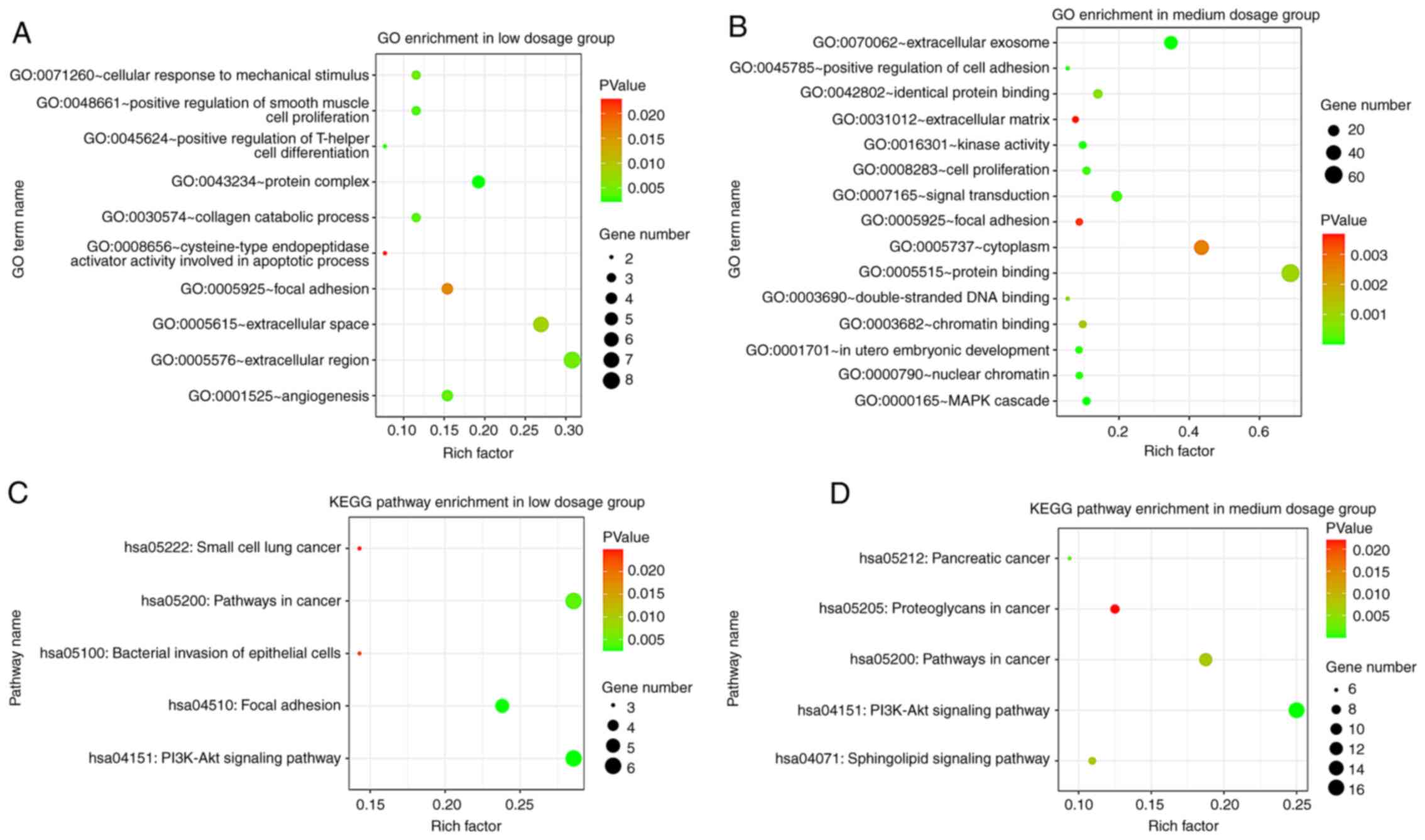
Significant terms in the GO enrichment analysis are
presented in Fig. 2A and B. The DEGs in the low dosage group
(Fig. 2A) were particularly
enriched in the biological processes (BP) including, ‘positive
regulation of T-helper cell differentiation’, ‘positive regulation
of smooth muscle cell proliferation’, ‘collagen catabolic process’,
‘angiogenesis’ and ‘cellular response to mechanical stimulus’. As
for the cellular components (CC), the genes were enriched in
‘protein complex’, ‘extracellular region’, ‘extracellular space’
and ‘focal adhesion’. The molecular function (MF) results showed
that DEGs were most enriched in ‘cysteine-type endopeptidase
activator activity involved in apoptotic process’.
In the medium dosage group (Fig. 2B), BP genes were primarily enriched
in the ‘MAPK cascade’, ‘in utero embryonic development’,
‘positive regulation of cell adhesion’, ‘signal transduction’ and
‘cell proliferation’. For CC, the DEGs were mainly enriched in
‘extracellular exosome’, ‘nuclear chromatin’, ‘cytoplasm’, ‘focal
adhesion’ and ‘extracellular matrix’. MF genes were predominantly
enriched in ‘kinase activity’, ‘identical protein binding’,
‘protein binding’, ‘double-stranded DNA binding’ and 'chromatin
binding'.
KEGG pathway analysis of DEGs
The KEGG pathway enrichment analysis suggested that
the DEGs in the low dosage group were mainly enriched in ‘PI3K-Akt
signaling pathway’, ‘Focal adhesion’, ‘Pathways in cancer’,
‘Bacterial invasion of epithelial cells’ and ‘Small cell lung
cancer’ (Fig. 2C). As for the
medium dosage group (Fig. 2D), the
DEGs were mainly enriched in ‘PI3K-Akt signaling pathway’,
‘Pancreatic cancer’, ‘Sphingolipid signaling pathway’, ‘Pathways in
cancer’ and ‘Proteoglycans in cancer’.
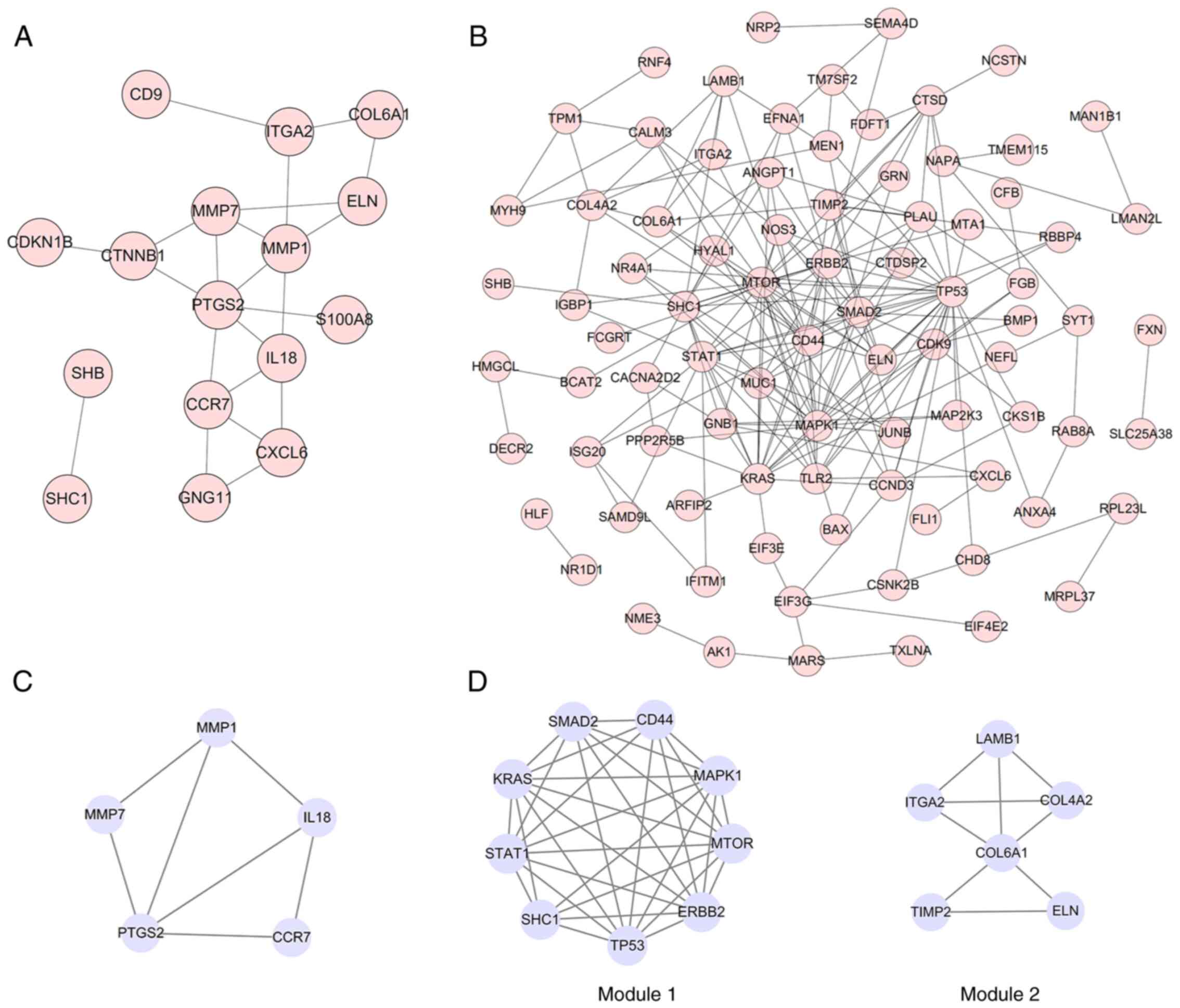
PPI network construction, module
analysis and hub genes identification
As presented in Fig.
3A and B, the final PPI network
of DEGs in the low dosage group was composed of 16 nodes and 22
edges after excluding the isolated nodes, whilst the PPI in the
medium group contained 85 nodes and 201 edges. MCODE was used to
analyze the key module (composed of genes with a relatively high
degree of association) that consisted of five nodes and seven edges
in the low dosage group (Fig. 3C)
and two key modules in the medium dosage group (Fig. 3D). Module 1 consisted of nine nodes
and 34 edges, where the genes were enriched in the KEGG pathway,
including ‘Pancreatic cancer’, ‘Central carbon metabolism in
cancer’ and ‘Glioma’ (data not shown). Module 2 consisted of six
nodes and nine edges, where the genes were enriched in the KEGG
pathways, including ‘ECM-receptor interaction’, ‘Focal adhesion’
and ‘PI3K-Akt signaling pathway’ (data not shown).
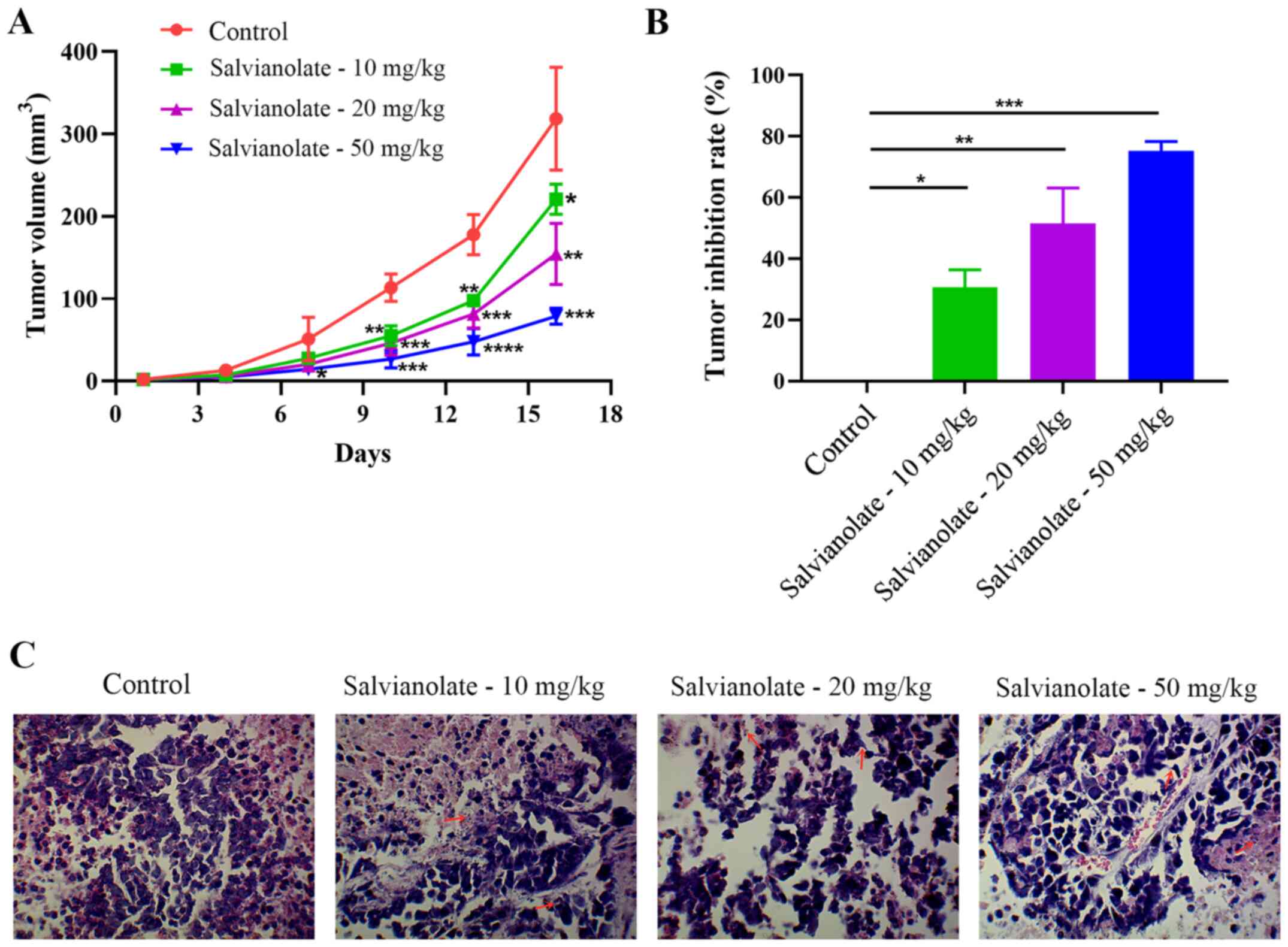
Salvianolate attenuates xenograft
tumors in nude mice
Animal models were used to verify the effect of
salvianolate on lung adenocarcinoma in vivo. In the present
study, the maximum tumor diameter exhibited by any nude mouse was
10.8 mm and the specific data on the tumor volume and diameter over
the entire experimental time period were shown in Table SI. The data in the present study
revealed that salvianolate significantly reduced the tumor volume
at 10, 13 and 16 days, compared with that in the control group
(P<0.05; Fig. 4A). In addition,
the inhibition rate of salvianolate on tumors was significantly
increased in a dose-dependent manner (Fig. 4B). H&E staining showed that the
salvianolate administration groups exhibited nuclear pyknosis and
fragmentation, suggesting apoptosis, compared with that in the
control group (Fig. 4C).
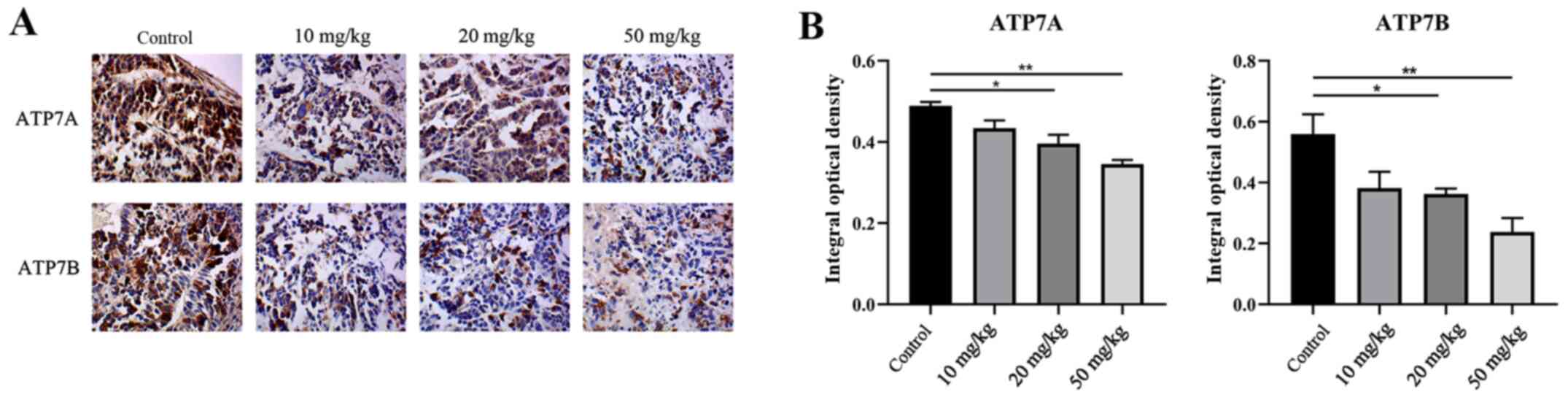
Salvianolate treatment downregulates
the expression of ATP7A/7B
IHC staining was performed to detect the expression
levels of ATP7A and ATP7B. IHC showed that salvianolate
significantly decreased the expression levels of ATP7A and ATP7B
(Fig. 5A and B).
Discussion
Salvia miltiorrhiza has been applied as a TCM
since it has been reported to possess anticancer properties
(11). Components of Salvia
miltiorrhiza can be divided into fat-soluble compounds that are
comprised of diterpenoid quinones and water-soluble compounds, such
as hydrophilic phenolic acids (13). The present study mainly investigated
the effects of Tanshinone I (a diterpenoid quinones compound) and
salvianolate (a water-soluble compound) on lung adenocarcinoma.
Tanshinone I is a notable component of the
fat-soluble compounds that can be isolated from Salvia
miltiorrhiza (28). In the
present study, hub genes and key pathways involved in lung
adenocarcinoma following administration of different dosages of
Tanshinone I were identified. In the low dosage group (0.01 and
0.10 µg/m Tanshinone I vs. control group) and the medium dosage
group (1 and 10 µg/ml Tanshinone I vs. control group), 28 DEGs and
58 DEGs were identified, respectively. Notably, the KEGG pathway
analysis indicated that DEGs in the two groups were both enriched
in the PI3K/Akt signaling pathway. Previous studies showed that
Tanshinone I induced apoptosis by inhibiting the PI3K/Akt/mTOR
pathway in ovarian cancer cells (29) and human breast cancer cell lines
(30). Although there is some
evidence to suggest that Tanshinone I can inhibit the proliferation
of lung adenocarcinoma cell lines A549, CL1-0 and CL1-5 (14,22),
further in vitro experiments are required to investigate the
underlying mechanism and molecular target of Tanshinone I in lung
adenocarcinoma.
Notably, nine overlapping DEGs were identified in
the low and medium dosage groups, which warrant further study.
ITGA2 is the α subunit of a transmembrane receptor that has been
reported to regulate cell migration and differentiation (31). Previous studies have shown that
ITGA2 expression was upregulated in several different types of
cancer (32), including gastric
(33) and breast cancer (34). The present study indicated that
ITGA2 expression was downregulated by Tanshinone I in lung
adenocarcinoma, which may be a potential target. MAN1B1 encodes
α-1,2-mannosidase, which mediates protein glycosylation
modification and glycoprotein polysaccharide hydrolysis (35). It has been reported that high
expression levels of MAN1B1 were associated with poor prognosis in
bladder cancer (36). However, the
effects of MAN1B1 downregulation induced by Tanshinone I on
responses in lung adenocarcinoma require further investigation.
Other studies have shown that SHB knockdown increased the
susceptibility of the SVR angiosarcoma cell line to cisplatin and
staurosporine (37) whilst
impairing the growth of tumors in mice injected with Lewis lung
carcinoma cells or T241 fibrosarcoma cells (38). The present study demonstrated the
association between Tanshinone I treatment and the downregulation
of SHB in lung adenocarcinoma. ELN is a signature extracellular
matrix protein in the lungs, where ELN-derived fragments have been
shown to be pro-tumorigenic (39).
The SHC1 protein exists in three isoforms, p46SHC, p52SHC and
p66SHC (40). Xu et al
(41) demonstrated that salvianolic
acid A pretreatment increased the expression of sirtuin 1, which
was associated with downregulation of p66SHC, a growth factor
adapter SHC, in a drug-induced liver injury mouse model. COL6A1,
which contributes to maintaining the integrity of tissues, was
found to be upregulated in cervical cancer (42) and pancreatic cancer tissues
(43) compared with that in
adjacent non-tumor tissues. It has been reported that
overexpression of ARFIP2 inhibited tumor necrosis
factor-α-stimulated NF-κB signaling by interacting with inhibitor
of NF-κB kinase β/NF-κB essential modulator (44). CXCL6 is a chemokine that
participates in cancer angiogenesis, metastasis and the immune
response (45), where it has been
reported to promote NSCLC cell survival and metastasis (46). CXCL6 expression was demonstrated to
be upregulated by Tanshinone I treatment in a lung adenocarcinoma
cell line in the present study. SAMD9L had been demonstrated to be
a tumor suppressor in breast, hepatocellular and squamous cell
carcinoma (47). However, its
functional role remains poorly understood. In the present study,
bioinformatics analysis revealed that these nine genes were DEGs
following the administration of different doses of Tanshinone I,
the mechanistic details of which warrants further
investigation.
Salvianolate is comprised of salvianolic acids A and
B and is a major water-soluble component in the extract of
Salvia miltiorrhiza. The present study showed that
salvianolate significantly decreased the tumor volumes of xenograft
nude mice, compared with those in the control group. It would
therefore be useful to investigate if Tanshinone I and salvianolate
exert synergistic effects on lung adenocarcinoma in future studies.
Although the IHC expression of ATP7A in 20 mg/kg salvianolate
appeared to be higher compared with that in 10 mg/kg, it can still
be concluded that, overall, salvianolate (10, 20 and 50 mg/kg)
decreased the expression of ATP7A and ATP7B in a
concentration-dependent manner. ATP7A/7B are copper efflux
transporters that maintain cellular copper homeostasis (15). Whether copper homeostasis or other
pathways are part of the underlying mechanism mediated by the
treatment of salvianolate remains to be investigated.
To conclude, the present study revealed the DEGs and
underlying pathways in lung adenocarcinoma following treatment with
different doses of Tanshinone I using bioinformatics tools. In
addition, the present study also verified the antitumor effects of
salvianolate, another active ingredient of Salvia
miltiorrhiza, in animals and lung adenocarcinoma cells. These
findings suggest the potential value of applying the Salvia
miltiorrhiza extract for lung adenocarcinoma treatment.
Supplementary Material
The specific data of length and width
of tumors in each nude mouse
Acknowledgements
Not applicable.
Funding
Funding: The present study was financially supported by the
Hunan Provincial Natural Science Foundation of China (grant no.
2019JJ40519) and Hunan Province Traditional Chinese Medicine
Research Project (grant no. 201179).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HT, YL, JC and XL conceived and designed the study.
HT and JM performed animal experiments and collected data. HT, YL
and LC, JC and XL analyzed and interpreted the data. JC and XL
revised the manuscript. JY and ZL interpreted the data and checked
the revised version of the manuscript. All authors read and
approved the manuscript.
Ethics approval and consent to
participate
All animal research protocols were following with
the guidelines of the Ethics Committee of Institutional Animal Care
and Use in Central South University and approved by Medical Ethics
Committee of Xiangya Hospital, Central South University (approval
no. 2017121170; Changsha, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Barta JA, Powell CA and Wisnivesky JP:
Global epidemiology of lung cancer. Ann Glob Health.
85(85)2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
World Health Organization: Cancer Today.
2021. Accessed from https://gco.iarc.fr/today/home.
|
|
3
|
Blandin Knight S, Crosbie PA, Balata H,
Chudziak J, Hussell T and Dive C: Progress and prospects of early
detection in lung cancer. Open Biol. 7(7)2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhou K, Zhao S, Guo W and Ding L: Efficacy
and safety of erlotinib combined with bevacizumab in the treatment
of non-small cell lung cancer: A systematic review and
meta-analysis. Medicine (Baltimore). 99(e18771)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Atal S, Asokan P and Jhaj R: Recent
advances in targeted small-molecule inhibitor therapy for
non-small-cell lung cancer-An update. J Clin Pharm Ther.
45:580–584. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Zugazagoitia J, Guedes C, Ponce S, Ferrer
I, Molina-Pinelo S and Paz-Ares L: Current challenges in cancer
treatment. Clin Ther. 38:1551–1566. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Liao YH, Li CI, Lin CC, Lin JG, Chiang JH
and Li TC: Traditional Chinese medicine as adjunctive therapy
improves the long-term survival of lung cancer patients. J Cancer
Res Clin Oncol. 143:2425–2435. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wang Y, Liu Y, Du X, Ma H and Yao J: The
anti-cancer mechanisms of berberine: A Review. Cancer Manag Res.
12:695–702. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Guo Q, Li J and Lin H: Effect and
molecular mechanisms of traditional Chinese medicine on regulating
tumor immunosuppressive microenvironment. BioMed Res Int.
2015(261620)2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Lin YY, Lee IY, Huang WS, Lin YS, Kuan FC,
Shu LH, Cheng YC, Yang YH and Wu CY: Danshen improves survival of
patients with colon cancer and dihydroisotanshinone I inhibit the
proliferation of colon cancer cells via apoptosis and skp2
signaling pathway. J Ethnopharmacol. 209:305–316. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Lin YS, Shen YC, Wu CY, Tsai YY, Yang YH,
Lin YY, Kuan FC, Lu CN, Chang GH, Tsai MS, et al: Danshen Improves
Survival of Patients With Breast Cancer and Dihydroisotanshinone I
Induces Ferroptosis and Apoptosis of Breast Cancer Cells. Front
Pharmacol. 10(1226)2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Su CY, Ming QL, Rahman K, Han T and Qin
LP: Salvia miltiorrhiza: Traditional medicinal uses,
chemistry, and pharmacology. Chin J Nat Med. 13:163–182.
2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Tung YT, Chen HL, Lee CY, Chou YC, Lee PY,
Tsai HC, Lin YL and Chen CM: Active component of Danshen (Salvia
miltiorrhiza Bunge), tanshinone I, attenuates lung
tumorigenesis via inhibitions of VEGF, Cyclin A, and Cyclin B
expressions. Evid Based Complement Alternat Med.
2013(319247)2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
La Fontaine S and Mercer JF: Trafficking
of the copper-ATPases, ATP7A and ATP7B: Role in copper homeostasis.
Arch Biochem Biophys. 463:149–167. 2007.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Shanbhag V, Jasmer-McDonald K, Zhu S,
Martin AL, Gudekar N, Khan A, Ladomersky E, Singh K, Weisman GA and
Petris MJ: ATP7A delivers copper to the lysyl oxidase family of
enzymes and promotes tumorigenesis and metastasis. Proc Natl Acad
Sci USA. 116:6836–6841. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Yang T, Chen M, Chen T and Thakur A:
Expression of the copper transporters hCtr1, ATP7A and ATP7B is
associated with the response to chemotherapy and survival time in
patients with resected non-small cell lung cancer. Oncol Lett.
10:2584–2590. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Li YQ, Chen J, Yin JY, Liu ZQ and Li XP:
Gene expression and single nucleotide polymorphism of ATP7B are
associated with platinum-based chemotherapy response in non-small
cell lung cancer patients. J Cancer. 9:3532–3539. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Samimi G, Safaei R, Katano K, Holzer AK,
Rochdi M, Tomioka M, Goodman M and Howell SB: Increased expression
of the copper efflux transporter ATP7A mediates resistance to
cisplatin, carboplatin, and oxaliplatin in ovarian cancer cells.
Clin Cancer Res. 10:4661–4669. 2004.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Li YQ, Zhang XY, Chen J, Yin JY and Li XP:
ATP7B rs9535826 is associated with gastrointestinal toxicity of
platinum-based chemotherapy in nonsmall cell lung cancer patients.
J Cancer Res Ther. 14:881–886. 2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Li YQ, Yin JY, Liu ZQ and Li XP: Copper
efflux transporters ATP7A and ATP7B: Novel biomarkers for platinum
drug resistance and targets for therapy. IUBMB Life. 70:183–191.
2018.PubMed/NCBI View
Article : Google Scholar
|
|
22
|
Lee CY, Sher HF, Chen HW, Liu CC, Chen CH,
Lin CS, Yang PC, Tsay HS and Chen JJ: Anticancer effects of
tanshinone I in human non-small cell lung cancer. Mol Cancer Ther.
7:3527–3538. 2008.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Walter W, Sánchez-Cabo F and Ricote M:
GOplot: An R package for visually combining expression data with
functional analysis. Bioinformatics. 31:2912–2914. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Doncheva NT, Morris JH, Gorodkin J and
Jensen LJ: Cytoscape StringApp: Network analysis and visualization
of proteomics data. J Proteome Res. 18:623–632. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ministry of Science and Technology of the
People's Republic of China: 2021. Accessed from: http://www.most.gov.cn/xxgk/xinxifenlei/fdzdgknr/fgzc/gfxwj/gfxwj2010before/201712/t20171222_137025.html.
|
|
28
|
Fu L, Han B, Zhou Y, Ren J, Cao W, Patel
G, Kai G and Zhang J: The anticancer properties of tanshinones and
the pharmacological effects of their active ingredients. Front
Pharmacol. 11(193)2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zhou J, Jiang YY, Chen H, Wu YC and Zhang
L: Tanshinone I attenuates the malignant biological properties of
ovarian cancer by inducing apoptosis and autophagy via the
inactivation of PI3K/AKT/mTOR pathway. Cell Prolif.
53(e12739)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang L, Wu J, Lu J, Ma R, Sun D and Tang
J: Regulation of the cell cycle and PI3K/Akt/mTOR signaling pathway
by tanshinone I in human breast cancer cell lines. Mol Med Rep.
11:931–939. 2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Adorno-Cruz V and Liu H: Regulation and
functions of integrin α2 in cell adhesion and disease. Genes Dis.
6:16–24. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ren D, Zhao J, Sun Y, Li D, Meng Z, Wang
B, Fan P, Liu Z, Jin X and Wu H: Overexpressed ITGA2 promotes
malignant tumor aggression by up-regulating PD-L1 expression
through the activation of the STAT3 signaling pathway. J Exp Clin
Cancer Res. 38(485)2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Chuang YC, Wu HY, Lin YL, Tzou SC, Chuang
CH, Jian TY, Chen PR, Chang YC, Lin CH, Huang TH, et al: Blockade
of ITGA2 Induces Apoptosis and Inhibits Cell Migration in Gastric
Cancer. Biol Proced Online. 20(10)2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ding W, Fan XL, Xu X, Huang JZ, Xu SH,
Geng Q, Li R, Chen D and Yan GR: Epigenetic silencing of ITGA2 by
MiR-373 promotes cell migration in breast cancer. PLoS One.
10(e0135128)2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Van Scherpenzeel M, Timal S, Rymen D,
Hoischen A, Wuhrer M, Hipgrave-Ederveen A, Grunewald S, Peanne R,
Saada A, Edvardson S, et al: Diagnostic serum glycosylation profile
in patients with intellectual disability as a result of MAN1B1
deficiency. Brain. 137:1030–1038. 2014.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Wang HF, Wu JH, Gai JW, Yang SQ, Ma QT, Ma
HS and Feng Q: MAN1B1 is associated with poor prognosis and
modulates proliferation and apoptosis in bladder cancer. Gene.
679:314–319. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Funa NS, Reddy K, Bhandarkar S, Kurenova
EV, Yang L, Cance WG, Welsh M and Arbiser JL: Shb gene knockdown
increases the susceptibility of SVR endothelial tumor cells to
apoptotic stimuli in vitro and in vivo. J Invest Dermatol.
128:710–716. 2008.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Funa NS, Kriz V, Zang G, Calounova G,
Akerblom B, Mares J, Larsson E, Sun Y, Betsholtz C and Welsh M:
Dysfunctional microvasculature as a consequence of shb gene
inactivation causes impaired tumor growth. Cancer Res.
69:2141–2148. 2009.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Thorlacius-Ussing J, Kehlet SN, Rønnow SR,
Karsdal MA and Willumsen N: Non-invasive profiling of
protease-specific elastin turnover in lung cancer: Biomarker
potential. J Cancer Res Clin Oncol. 145:383–392. 2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wright KD, Miller BS, El-Meanawy S, Tsaih
SW, Banerjee A, Geurts AM, Sheinin Y, Sun Y, Kalyanaraman B, Rui H,
et al: The p52 isoform of SHC1 is a key driver of breast cancer
initiation. Breast Cancer Res. 21(74)2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Xu X, Hu Y, Zhai X, Lin M, Chen Z, Tian X,
Zhang F, Gao D, Ma X, Lv L, et al: Salvianolic acid A
preconditioning confers protection against concanavalin A-induced
liver injury through SIRT1-mediated repression of p66shc in mice.
Toxicol Appl Pharmacol. 273:68–76. 2013.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Hou T, Tong C, Kazobinka G, Zhang W, Huang
X, Huang Y and Zhang Y: Expression of COL6A1 predicts prognosis in
cervical cancer patients. Am J Transl Res. 8:2838–2844.
2016.PubMed/NCBI
|
|
43
|
Owusu-Ansah KG, Song G, Chen R, Edoo MIA,
Li J, Chen B, Wu J, Zhou L, Xie H, Jiang D, et al: COL6A1 promotes
metastasis and predicts poor prognosis in patients with pancreatic
cancer. Int J Oncol. 55:391–404. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
You DJ, Park CR, Furlong M, Koo O, Lee C,
Ahn C, Seong JY and Hwang JI: Dimer of arfaptin 2 regulates NF-κB
signaling by interacting with IKKβ/NEMO and inhibiting IKKβ kinase
activity. Cell Signal. 27:2173–2181. 2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Verbeke H, Struyf S, Berghmans N, Van
Coillie E, Opdenakker G, Uyttenhove C, Van Snick J and Van Damme J:
Isotypic neutralizing antibodies against mouse GCP-2/CXCL6 inhibit
melanoma growth and metastasis. Cancer Lett. 302:54–62.
2011.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Li J, Tang Z, Wang H, Wu W, Zhou F, Ke H,
Lu W, Zhang S, Zhang Y, Yang S, et al: CXCL6 promotes non-small
cell lung cancer cell survival and metastasis via down-regulation
of miR-515-5p. Biomed Pharmacother. 97:1182–1188. 2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Davidsson J, Puschmann A, Tedgård U,
Bryder D, Nilsson L and Cammenga J: SAMD9 and SAMD9L in inherited
predisposition to ataxia, pancytopenia, and myeloid malignancies.
Leukemia. 32:1106–1115. 2018.PubMed/NCBI View Article : Google Scholar
|