Introduction
Gastric cancer (GC) is one of the leading causes of
cancer-related mortality worldwide, demonstrating over 50% of
morbidity and mortality (1).
Despite significant advances, the overall survival rate of patients
with GC has not improved in the past 20 years (2). Although numerous molecular targets
have been reported to be involved in the progression of GC
(3,4), the mechanisms underlying GC
development remain to be further investigated.
Serine/arginine rich splicing factor (SRSF)1 is a
35-kDa serine/arginine-rich splicing factor. SRSF1 can regulate
pre-mRNA alternative splicing and maintain genome stability by
interacting with transcription factor E2F transcription factor 1 in
the cell cycle (5). Furthermore,
SRSF1 facilitates transcriptional elongation by recruiting positive
transcription elongation factor b kinase and promoting the
subsequent phosphorylation of serine 2 on the C-terminal domain of
RNA polymerase II at the post-transcriptional level (6).
Macrophage stimulating 1 receptor (MST1R) is a
membrane tyrosine kinase of the MET family, which has been reported
to be a novel potential target for cancer treatment (7). MST1R has been found to be positively
associated with the development and progression of multiple types
of epithelial cancer, including GC (8-10).
Moreover, several preclinical studies have revealed that MST1R
exhibited oncogenic properties, including the promotion of cellular
proliferation, migration, invasion and survival in several human
cancer cell lines (11,12). Despite exhibiting diverse functions
in numerous malignancies, the alternatively spliced products of
MST1R show considerable sequence homology with each other (11). In human cancer, the alterative
skipping of MST1R exon 5 and exon 6 promotes the loss of 106 amino
acids in the MST1R β-chain, which produces a 109 kDa MST1R splicing
variant known as recepteur d'origine nantais (RON) Δ160. This
isoform was identified to be involved in the activation of the
PI3K/AKT signaling pathway and cellular transformation in
vitro (13). Previous studies
found that SRSF1 played a key role in the regulation of MST1R
pre-mRNA splicing (8,13). Therefore, further investigation into
the association between SRSF1 and MST1R isoform splice variants in
GC may provide novel insights for targeted therapies.
Materials and methods
Cell lines and culture
The GC cell lines Kato III and MKN-45 and the
immortalized gastric epithelium cell line GES-1 were obtained from
the State Key Laboratory for Diagnosis and Treatment of Infectious
Diseases, The First Affiliated Hospital, Zhejiang University School
of Medicine (Hangzhou, China). Cells were cultured in RPMI-1640
medium (Thermo Fisher Scientific, Inc.), supplemented with 10% FBS
(Thermo Fisher Scientific, Inc.), 100 µg/ml streptomycin and
penicillin, and maintained at 37˚C in 5% CO2.
Vector construction and cell
transfection
Small interfering RNA (siRNA) targeting SRSF1
(SRSF1-siRNA; 10 nM), FENDRR-siRNA (10 nM), pcDNA3.1 vector for
MST1R overexpression (pcDNA3.1-MST1R, MST1R OE) and the
corresponding empty vector [(referred to as negative control (NC)]
were purchased from Sangon Biotech (Shanghai) Co., Ltd. SRSF1- and
FENDRR-siRNA were transfected into GC cells using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
transfection efficiency was analyzed using reverse
transcription-quantitative PCR (RT-qPCR). The sequences of the
siRNAs were as follows: NC siRNA (siRNA-ctrl),
5'-UUCUCCGAACGUGUCACGUTT-3'; SRSF1-siRNA,
5'-GGAAUGAAGCAACUGAGAUUU-3'; and FENDRR-siRNA,
5'-GGGTTACGATTGCCCAGAT-3'. For MST1R overexpression, GC cells were
plated into 60-mm plates at a density of 4x105
cells/well and incubated overnight. Upon cells reaching 50-60%
confluence, supernatants containing the pcDNA3.1 vector carrying
the MST1R gene were added directly to the cell culture and
incubated for 24 h. Following the incubation, GC cells were plated
in selection medium containing 2.5 µg/ml puromycin for a further 3
days.
Cell Counting Kit-8 (CKK-8) assay
A CCK-8 assay (Beyotime Institute of Biotechnology)
was used to determine cell viability. Briefly, GC cells were plated
into 96-well plates at a density of 5x103 cells/well and
transfected with NC, siRNA-FENDRR or siRNA-FENDRR + MST1R pcDNA3.1
vector for 72 h. Following the incubation, cells were incubated
with 10 µl CCK-8 reagent for another 2 h at 37˚C. The absorbance of
each well was measured at a wavelength of 450 nm using a microplate
reader (Thermo Fisher Scientific, Inc.).
Cell apoptosis
GC cells were seeded into six-well plates
(5x104 cells/well). Following centrifugation at 200 x g
for 5 min at 4˚C, the cell pellet was resuspended in 100 µl binding
buffer and incubated with 5 µl Annexin V-FITC (BD Biosciences) and
propidium iodide (BD Biosciences) at room temperature for 15 min.
The cell apoptotic rate was measured using a flow cytometer (BD
Biosciences) and the data were analyzed using WinMDI 2.9
software.
Agarose electrophoresis
Vectors at a concentration of 0.03 µg DNA/µl were
subjected to electrophoresis on an ethidium bromide-containing gel
(1% agarose). Subsequently, bands were photographed with a Vilber
E-BOX (Vilber Lourmat Sté). Meanwhile, 1 unit DNase and 1.2 µg DNA
(Sigma-Aldrich; Merck KGaA) were incubated at 37˚C with the vectors
and complexes for 30 min. Subsequently, 2% SDS solution was added
as a DNA release reagent. Samples were subjected to agarose gel
electrophoresis and compared with untreated DNA. The data were
quantified using Image Pro Plus (version 6.0; Media Cybernetics,
Inc.).
RT-qPCR
Total RNA was extracted from cell lines using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Total RNA was reverse transcribed into cDNA using a
PrimeScript RT Reagent kit (Takara Bio, Inc.) according to the
manufacturer's protocol. qPCR was subsequently analyzed using a
SYBR Premix Ex Taq II kit (Takara Bio, Inc.) on a 7900HT system
(Applied Biosystems; Thermo Fisher Scientific, Inc.) according to
the following conditions: 60˚C for 1 min, 90˚C for 15 min, followed
by 40 cycles of application at 90˚C for 15 sec and 55˚C for 60 sec.
The following primer sequences were used for the qPCR: FENDRR
forward, 5'-TTCATCGGCTGCGTATTCG-3' and reverse,
5'-TTGCCTTCTAGTCGCCTCC-3'; and β-actin forward,
5'-GTCCACCGCAAATGCTTCTA-3' and reverse, 5'-TGCTGTCACCTTCACCGTTC-3'.
Expression levels were quantified using the 2-ΔΔCq
method (14). β-actin was used as
the internal control for normalization.
Transwell assay
A Transwell assay was performed to analyze cell
migration and invasion. The upper chambers of the Transwell plates
were pretreated with 100 µl Matrigel (BD Biosciences) for 4 h at
37˚C, while Matrigel was not used in the migration assay GC cells
were seeded into the upper chamber of the plates in media
supplemented with 1% FBS at a density of 1x106
cells/chamber. RPMI-1640 medium supplemented with 10% FBS was added
into the lower chambers. Following 24 h of incubation at 37˚C, the
Transwell chamber was rinsed twice with PBS (5 min each time), then
the cells were fixed with 5% glutaraldehyde at 4˚C and stained at
37˚C with 0.1% crystal violet for 30 min. The Transwell chamber was
washed twice with PBS and observed under a light microscope
(magnification, x200). Three random fields were selected. The
number of cells invading the Matrigel was a reflection of the
invasive ability.
Western blotting
Total protein was extracted from cells using RIPA
lysis buffer (Beyotime Institute of Biotechnnology) and quantified
using a BCA protein assay kit (Beyotime Institute of
Biotechnology). Proteins (40 µg per lane) were separated via 10%
SDS-PAGE, then transferred onto PVDF membranes (Bio-Rad
Laboratories, Inc.). After blocking with 5% skimmed milk for 1 h at
room temperature, the membranes were incubated with the following
primary antibodies at 4˚C overnight: Anti-AKT (cat. no. ab18785;
1:1,000), anti-phosphorylated (p)-AKT (cat. no. ab38449; 1:1,000),
Anti-p-ERK (cat. no. ab201015; 1:1,000), anti-ERK (cat. no.
ab32081; 1:1,000), anti-MST1R (cat. no. ab52927; 1:1,000),
anti-cleaved caspase-3 (cat. no. ab32042; 1:1,000), anti-SRSF1
(Thermo Fisher Scientific, Inc.; cat. no. 32-4500, 1:1,000) and
anti-β-actin (cat. no. ab8226; 1:1,000; all Abcam except for
SRSF1). Following the primary antibody incubation, the membranes
were incubated with an anti-rabbit secondary antibody
(HRP-conjugated; Abcam; cat. no. ab7090, 1:5,000) at room
temperature for 1 h. Enhanced chemiluminescence reagent (Thermo
Fisher Scientific, Inc.) was used to visualize the protein bands.
ImageJ software (version 2.0; National Institutes of Health) was
used to quantify the intensity of the bands.
Bioinformatics prediction
The LncRNA2Targetv2.0 tool (http://123.59.132.21/lncrna2target/index.jsp) was
used to determine the interaction between long non-coding RNAs
(lncRNAs) and upstream of the SRSF1 coding region.
RNA pull-down
For the RNA pulldown assay, Biotin RNA Labeling Mix
(Roche Diagnostics) was used to transcribe and label probe-control
or probe-FENDRR in vitro. An RNA structure buffer (Thermo
Fisher Scientific, Inc.) was used to induce secondary structure
formation from the biotin-labeled RNAs. The biotinylated FENDRR
(Shanghai GenePharma Co., Ltd.) and negative control (bio-NC;
GenePharma) were coated with streptavidin-conjugated magnetic beads
(Roche Diagnostics). GC cells were lysed using lysis reagent (Roche
Diagnostics) and then incubated with the magnetic beads for 6 h.
The enrichment level of SRSF1 was detected by western blotting.
Immunofluorescence staining
GC cells were fixed with 4% paraformaldehyde for 10
min at room temperature and then fixed with pre-cooled methanol at
4˚C for a further 10 min. Subsequently, cells were incubated with
an anti-RON ∆160 primary antibody (Abcam; cat. no. ab124671,
1:1,000) overnight at 4˚C. Following the primary antibody
incubation, the cells were incubated with a goat anti-rabbit IgG
secondary antibody (Abcam; cat. no. ab6721, 1:5,000) for 1 h at
room temperature. DAPI (Beyotime Institute of Biotechnology) was
used to counterstain the nuclei. Samples were visualized using a
fluorescence microscope (model no. CX23; Olympus Corporation,
magnification, x200). Three random fields were selected.
Co-immunoprecipitation (co-IP)
assay
The co-IP assay was performed as previously
described with modifications (5).
Briefly, Kato III cell protein supernatants were pretreated with 50
µl A/G (Protein A/Protein G) beads (Selleck Chemicals) prior to
immunoprecipitation and then with 5 µg control IgG (Santa Cruz
Biotechnology, Inc.), anti-MST1R or anti-SRSF1 magnetic beads
(Shanghai ZE YE Biological Technology Co., Ltd.) overnight at 4˚C.
Following further incubation with 50 µl A/G beads at 4˚C for 6 h,
the immunoprecipitates were eluted with ice-cold PBS supplemented
with 0.2% NP-40 five times. Subsequently, the immunoprecipitated
proteins were separated via SDS-PAGE and visualized using western
blotting as aforementioned.
Statistical analysis
Data are presented as the mean ± SD. Statistical
differences between two groups were determined using unpaired
Student's t-test. Comparisons among multiple groups were made using
an ANOVA followed by a Tukey's post hoc test. Statistical analysis
was performed using GraphPad Prism 7 software (GraphPad Software,
Inc.). P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression levels of MST1R and RON
∆160 are upregulated in Kato III cells
To determine the biological function of MST1R and
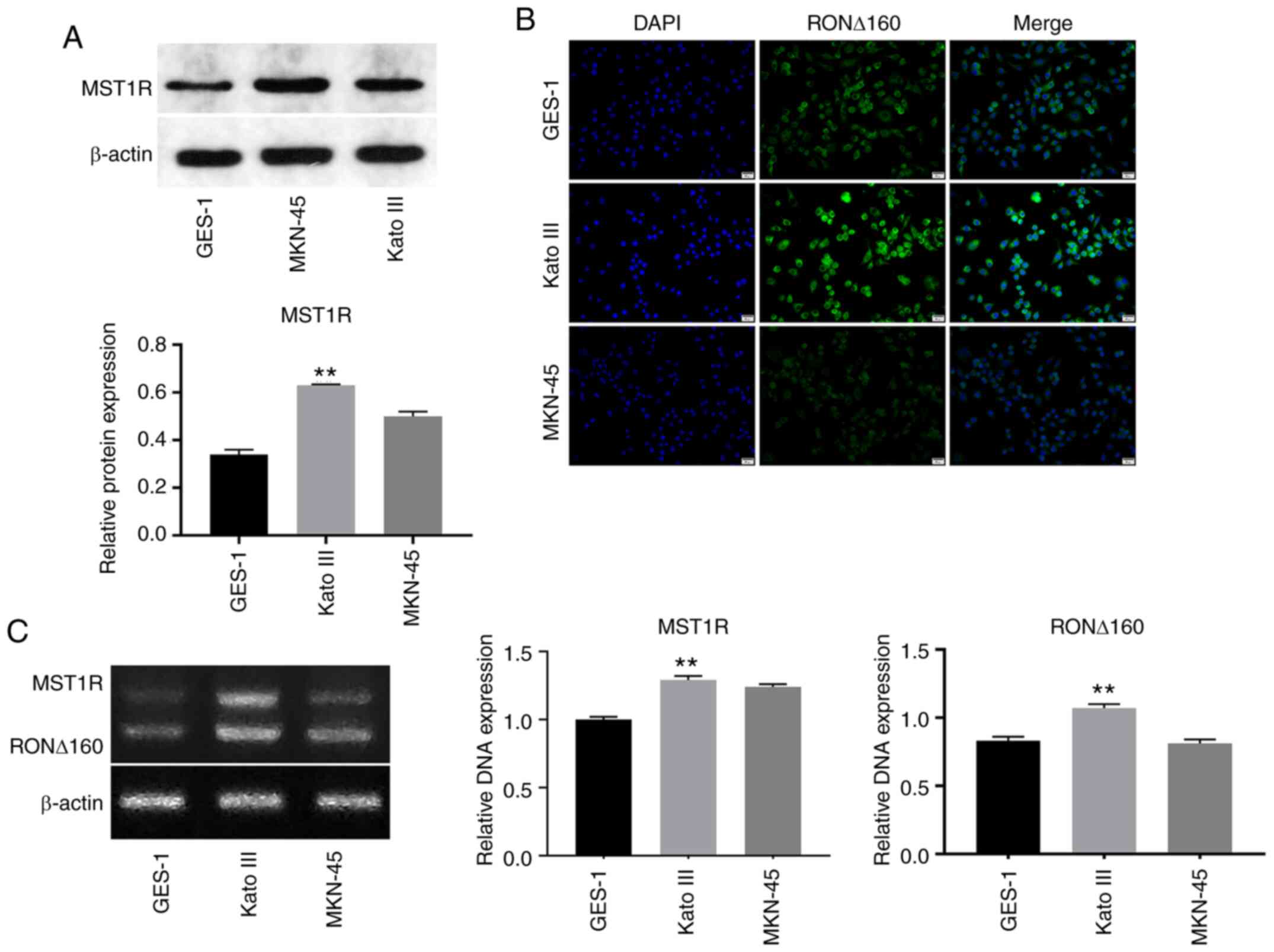
RON ∆160 in GC, western blotting was performed. As indicated in
Fig. 1A, the expression levels of
MST1R were significantly upregulated in Kato III cells compared
with the other cell lines. The expression levels of RON ∆160 in
Kato III cells were higher compared with GES-1 cells (Fig. 1B). Moreover, the mRNA expression
levels of MST1R and RON ∆160 were also notably upregulated in Kato
III cells compared with GES-1 cells (Fig. 1C). Therefore, Kato III cells were
selected for use in subsequent experiments. These results revealed
that the expression levels of MST1R and RON ∆160 may be upregulated
in Kato III cells.
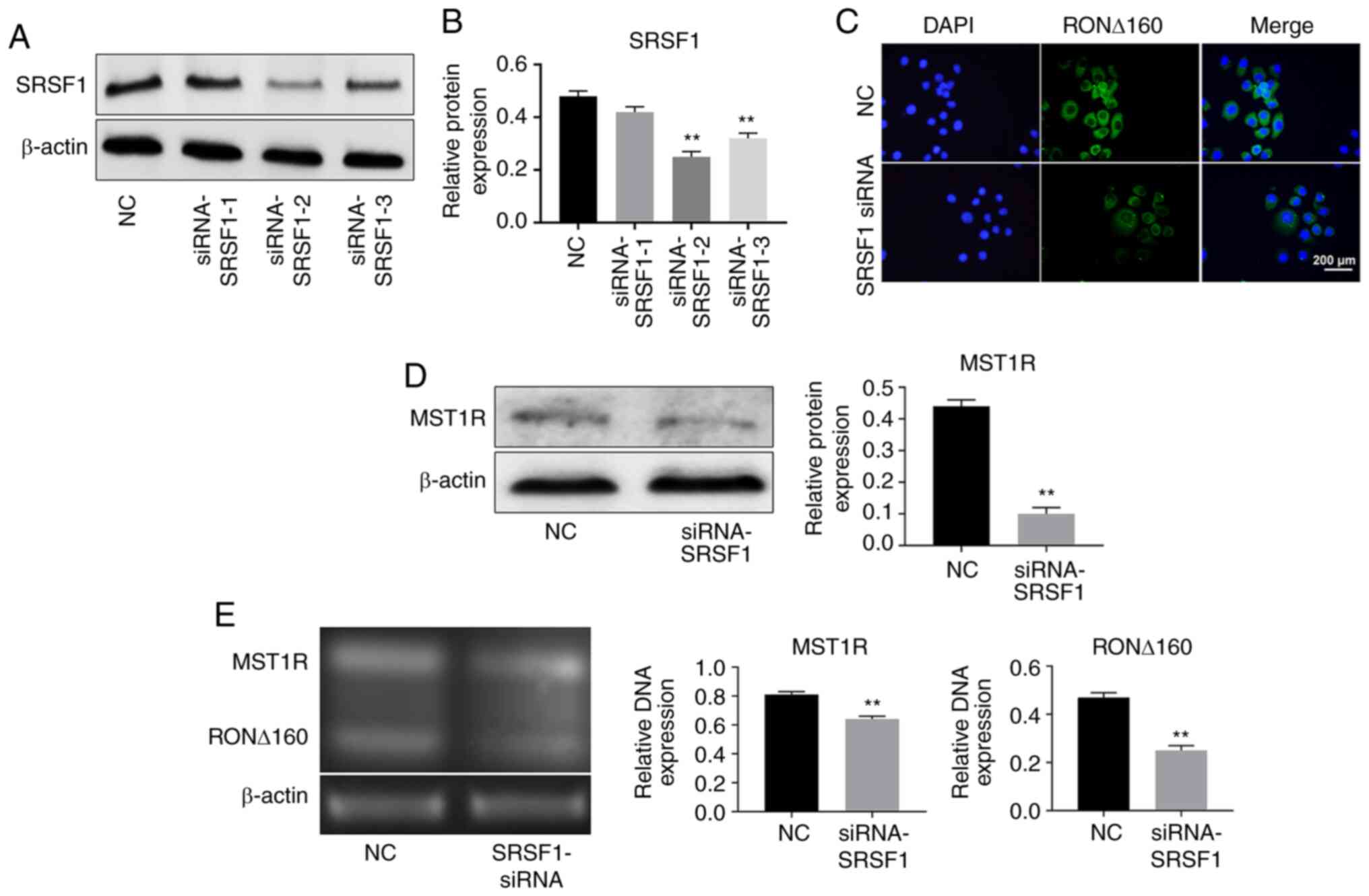
Knockdown of SRSF1 significantly
downregulates MST1R and RON ∆160 expression levels in Kato III
cells
To determine the transfection efficiency of
SRSF1-siRNA, RT-qPCR was performed. As shown in Fig. 2A and B, SRSF1 expression levels were
significantly downregulated in Kato III cells following SRSF1
knockdown. Moreover, Kato III cells were more sensitive to
SRSF1-2-siRNA. Thus, SRSF1-2- siRNA was selected for use in
subsequent experiments. Similarly, the expression levels of RON
∆160 were significantly downregulated following the transfection
with SRSF1-2- siRNA in Kato III cells (Fig. 2C). The expression levels of MST1R
were also downregulated in GC cells following the knockdown of
SRSF1 (Fig. 2D). As expected, the
mRNA expression levels of MST1R and RON ∆160 in GC cells were also
notably downregulated following transfection with SRSF1-2-siRNA
(Fig. 2E). Taken together, these
findings indicated that the knockdown of SRSF1 may significantly
downregulate MST1R and RON ∆160 in Kato III cells.
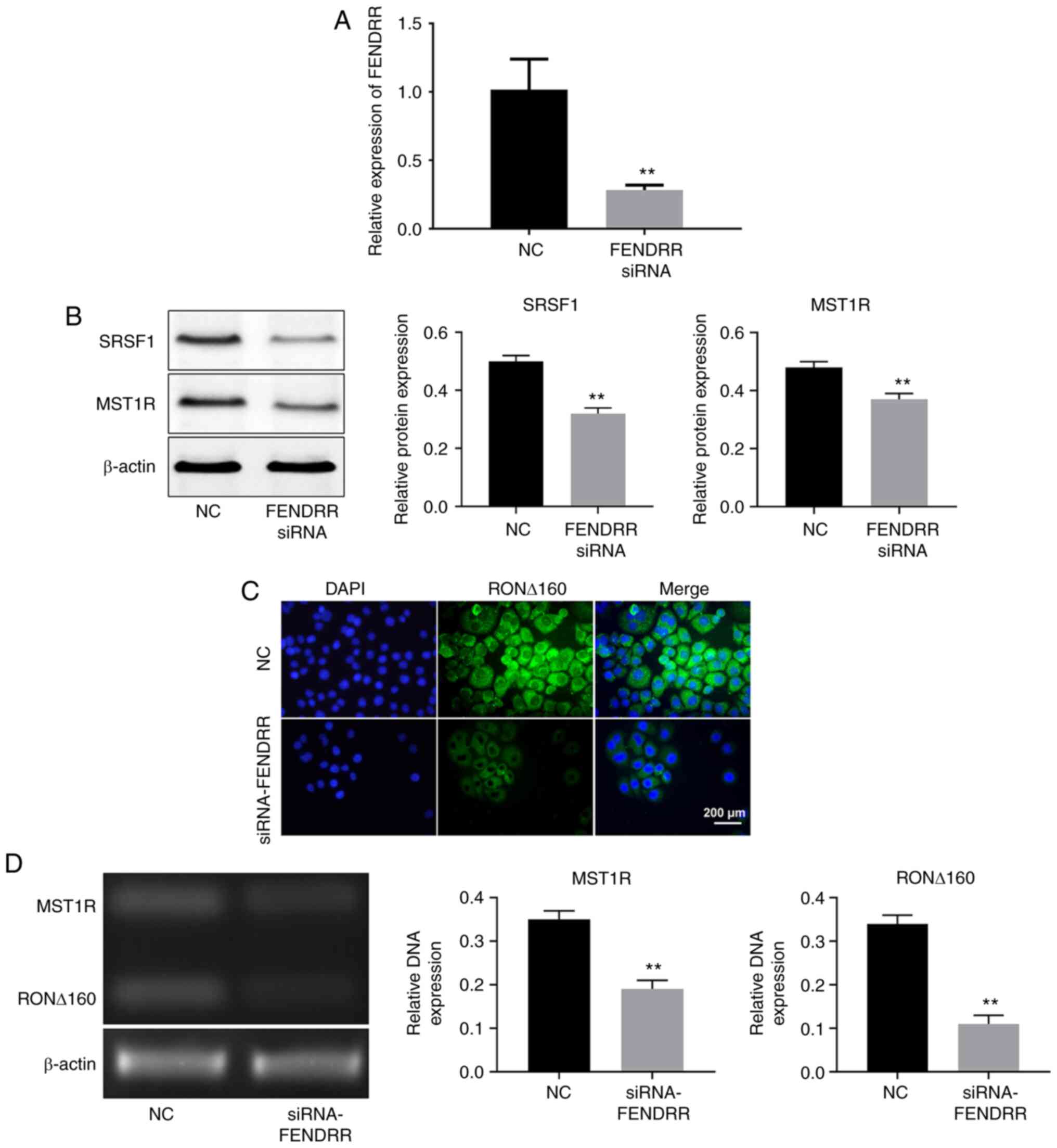
Knockdown of FENDRR suppresses the
expression levels of MST1R and alters the distribution of RON ∆160
in Kato III cells
FENDRR was identified to directly interact with
SRSF1. RT-qPCR was used to analyze the transfection efficiency of
FENDRR-siRNA transfection. As shown in Fig. 3A, the expression levels of FENDRR
were downregulated in Kato III cells in the presence of
FENDRR-siRNA. In addition, the knockdown of FENDRR significantly
downregulated the expression levels of SRSF1 and MST1R in Kato III
cells (Fig. 3B). Immunofluorescence
staining revealed that RON ∆160 expression was significantly
upregulated in Kato III cells in the presence of FENDRR-siRNA
(Fig. 3C). mRNA expression levels
of MST1R and RON ∆160 in Kato III cells were also markedly
upregulated following the transfection with FENDRR-siRNA (Fig. 3D). Altogether, these findings
suggested that the silencing of FENDRR may regulate the expression
levels of MST1R and alter the distribution of RON ∆160 in Kato III
cells by suppressing SRSF1 expression.
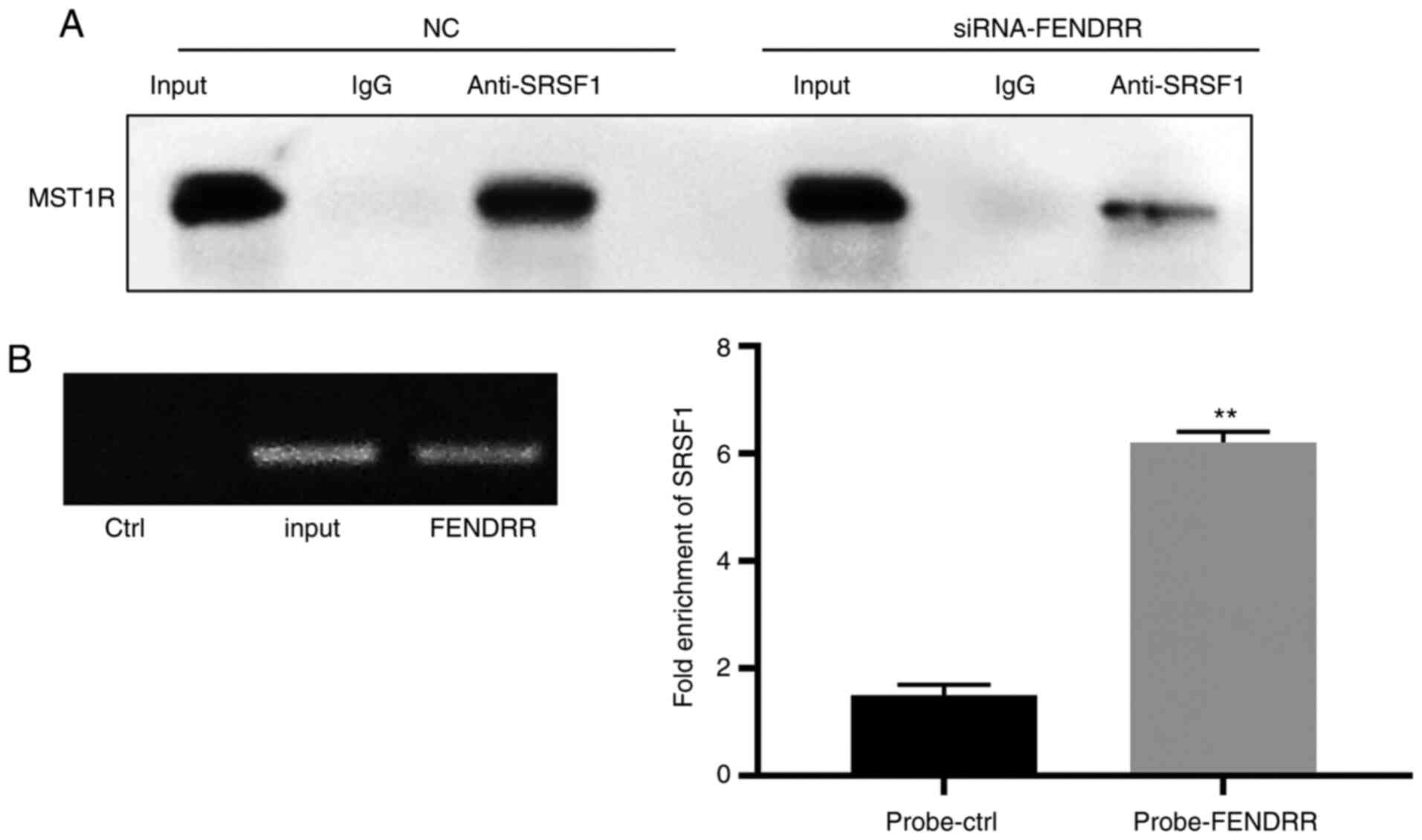
Silencing of FENDRR suppresses the
proliferation of GC cells by regulating MST1R
To determine the effect of FENDRR siRNA on
interaction between SRSF1 and MST1R, co-IP assays were performed.
As demonstrated in Fig. 4A, SRSF1
was found to directly bind with MST1R, while FENDRR siRNA reversed
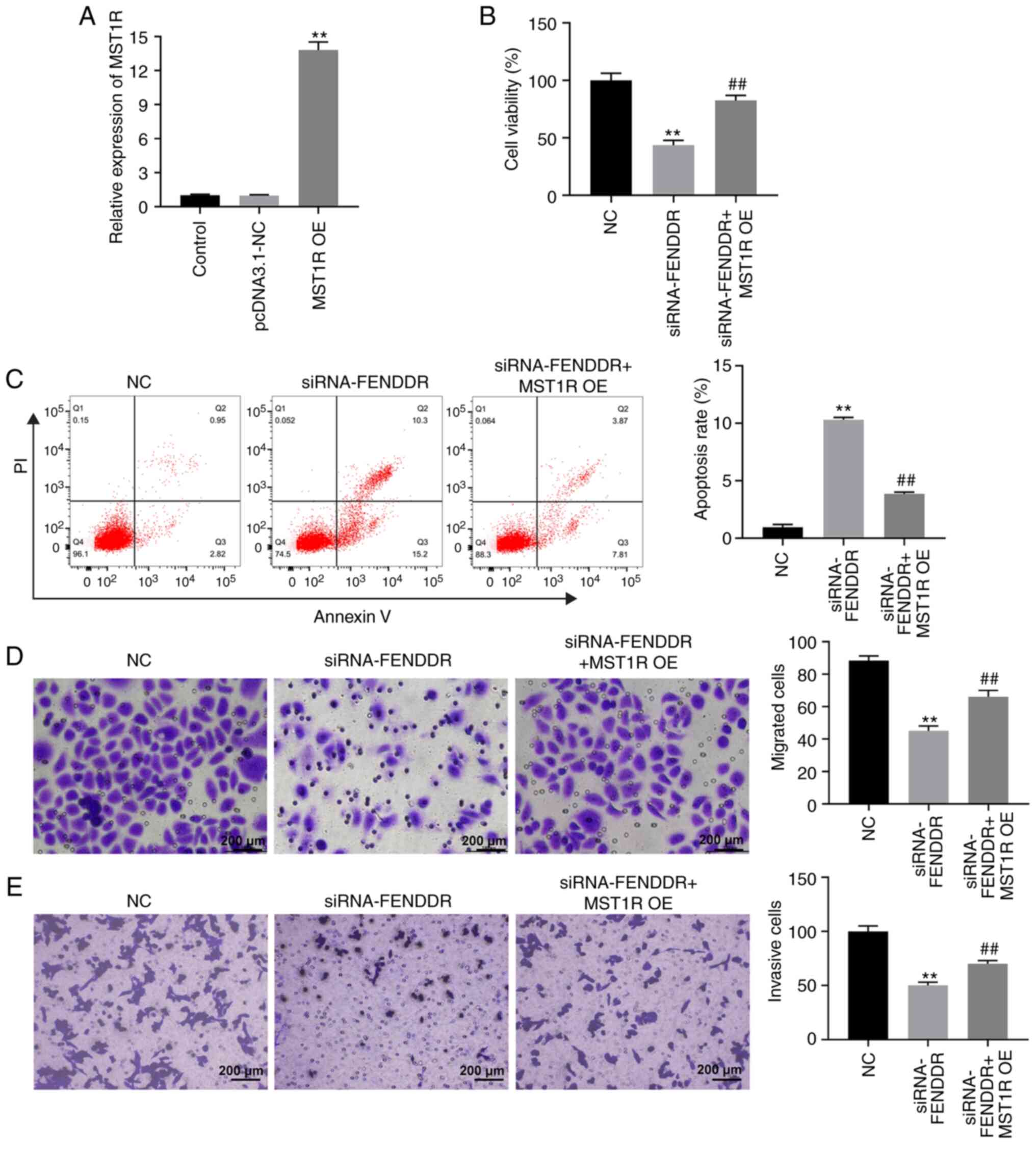
this phenomenon. Meanwhile, FENDRR could bind with SRSF1 (Fig. 4B). A CCK-8 assay was used to analyze
cell viability. The results revealed that the knockdown of FENDRR
notably inhibited the viability of Kato III cells, while the
reduction in viability induced by FENDRR-siRNA was partially
rescued by the overexpression of MST1R (Fig. 5A). Similarly, the apoptosis of Kato
III cells was markedly increased following the genetic silencing of
FENDRR, which was significantly reversed by the overexpression of
MST1R (Fig. 5B). In addition, Kato
III cell migration and invasion levels were suppressed in the
presence of FENDRR-siRNA. However, the inhibitory effect of
FENDRR-siRNA on cell invasion was significantly abrogated by MST1R
overexpression (Fig. 5C and
D). Altogether, these findings
suggested that the knockdown of FENDRR may suppress the
proliferation of GC cells by regulating MST1R expression.
Knockdown of FENDRR significantly
inhibits the progression of GC in vitro via inactivation of
PI3K/AKT signaling
To further investigate the mechanism through which
FENDRR mediated the progression of GC, western blotting was
performed. As shown in Fig. 6A-E,
the protein expression levels of MST1R and the p-AKT/AKT and
p-ERK/ERK ratios were notably downregulated in Kato III cells
following the knockdown of FENDRR, and were partially reversed
following MST1R overexpression. In contrast, the genetic silencing
of FENDRR markedly upregulated the ratio of cleaved
caspase-3/procaspase-3 in GC cells, while the promoting effect of
FENDRR-siRNA on the cleaved caspase-3/procaspase-3 ratio was
notably abrogated following MST1R overexpression. Altogether, these
results suggested that the knockdown of FENDRR may inhibit the
progression of GC in vitro by inhibiting PI3K/AKT
signaling.
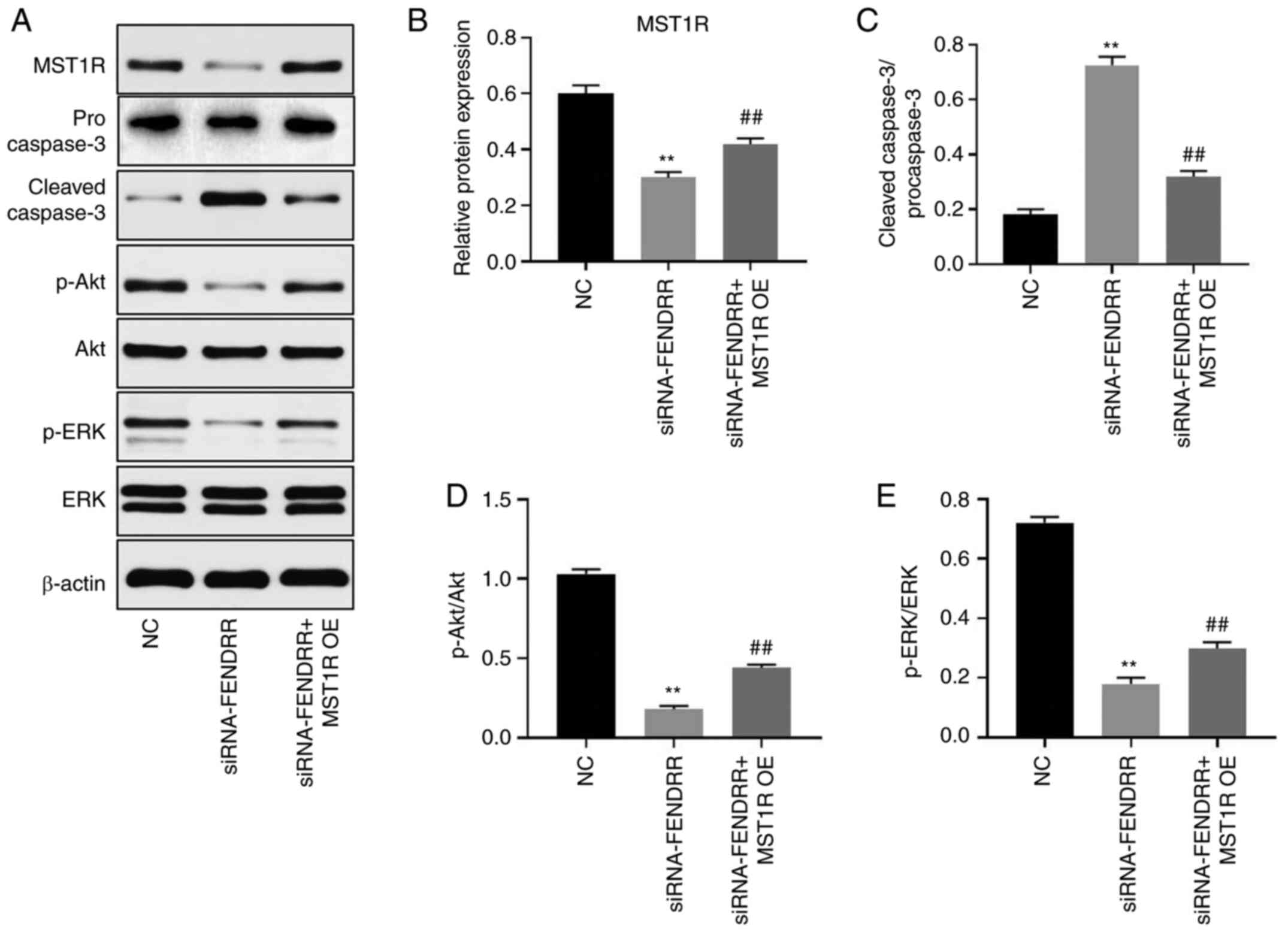 | Figure 6Silencing of FENDRR inhibits the
tumorigenesis of gastric cancer cells in vitro via
inactivation of PI3K/Akt signaling. (A) The protein expression
levels of MST1R, caspase-3, p-Akt, Akt, p-ERK and ERK in Kato III
cells were determined by western blot. (B) The relative protein
expression of MST1R was quantified by normalizing to β-actin. (C)
The ratio of cleaved caspase-3/procaspase-3 was calculated. (D) The
ratio of p-Akt/Akt was calculated. (E) The ratio of p-ERK/ERK was
calculated. FENDRR, FOXF1 adjacent non-coding developmental
regulatory RNA; MST1R, macrophage stimulating 1 receptor; OE,
overexpression vector; NC, non-coding control; siRNA, short
interfering RNA; ERK, extracellular signal related kinase; p,
phosphorylated **P<0.01 vs. NC;
##P<0.01 vs. siRNA-FENDRR. |
Discussion
MST1R has been reported to play a key role in the
pathogenesis of GC (9). The
alternative splicing variant of precursor MST1R mRNA, RON ∆160, has
been identified as an oncogenic transcription factor that regulates
numerous signaling pathways associated with tumorigenesis,
including cellular transformation activities such as focus
formation and anchorage-independent growth (8). A previous study demonstrated that the
upregulation of variant RON Δ160 in GC altered the phenotype and
enhanced the invasive ability of GC cells. Furthermore, RON Δ160
was found to be closely associated with tumorigenesis, and both
regional lymph node and widespread metastasis (13). The findings of the present study
revealed that the expression levels of MST1R were regulated by
FENDRR knockdown. These findings further validated the results of
previous studies (8,13), indicating that FENDRR and SRSF1 may
promote the tumorigenesis of GC by regulating RON ∆160
expression.
SRSF1 is known to serve roles in RNA splicing and
genome stability (6,15,16).
The RNA recognition motif of SRSF1 for RNA binding can promote
spliceosome assembly at adjacent splice sites to facilitate
appropriate exon inclusion (17,18).
The aberrant spliceosome function of SRSF1 was previously
associated with the mis-splicing of multiple genes, including MST1R
and enhancer of zeste 2 polycomb repressive complex 2 subunit,
which have been implicated in the pathogenesis of myeloid neoplasms
(19,20). The results of the current study
demonstrated that the knockdown of SRSF1 could lead to the
inactivation of MST1R and RON Δ160 in GC cells. In addition, the
results of the co-IP assay found that SRSF1 directly bound to
MST1R. These data provided novel insights into the biological
function of SRSF1, suggesting that SRSF1 may regulate MST1R and RON
Δ160 expression. According to Bonomi et al (21), the involvement of SRSF1 in
epithelial-to-mesenchymal transition derives from its ability to
affect the splicing program of the proto-oncogene MST1R. Previous
research has shown that SRSF1 could promote the production of MST1R
through skipping of exon 11. More specifically, SRSF1 acts by
directly binding to an exonic splicing enhancer (ESE) located in
the constitutive exon 12(22).
Therefore, SRSF1 may interact with MST1R in GC.
An increasing number of studies have reported the
important role of non-coding RNAs in cellular biological functions
(23,24). Previous research indicated that
FENDRR was associated with the development of osteosarcoma
(25). The present study found that
the knockdown of FENDRR downregulated MST1R and RON ∆160 expression
levels in GC cells. Based on these findings, it was hypothesized
that the knockdown of FENDRR expression may result in the
downregulation of RON ∆160 in GC.
Furthermore, the current research revealed that the
genetic silencing of FENDRR inactivated the PI3K/AKT signaling
pathway in GC cells. PI3K/AKT signaling is known to be involved in
the tumorigenesis of cancer (26,27). A
previous study reported that PI3K/AKT signaling played a key role
in cancer progression, drug resistance and treatment (28). Xu et al (29) found that PI3K/AKT signaling led to
reduced apoptosis and increased proliferation in GC cells. The
findings of the present study were consistent with the
aforementioned studies. Moreover, the present study demonstrated
that MST1R overexpression partially rescued the inhibitory effect
of FENDRR-siRNA on the PI3K/AKT signaling pathway in vitro.
Ling et al (30)
demonstrated that MST1R promoted PI3K/AKT signaling during the
development of colorectal cancer. These findings were consistent
with the present data, suggesting that FENDRR may mediate the
expression levels of MST1R and RON ∆160 by inhibiting PI3K/AKT
signaling.
In conclusion, the results of the present study
suggested that lncRNA FENDRR may function as an oncogene during the
progression of GC by mediating the alternative splicing of MST1R
and SRSF1 expression. Therefore, lncRNA FENDRR may serve as a
potential target for the diagnosis and treatment of GC.
Acknowledgements
Not applicable.
Funding
Funding: This study was supported by a grant from the National
Natural Science Foundation of China (grant no. 81272680) and
Zhejiang Provincial Department of Science and Technology (grant no.
2020C03112).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DZ and XW conceived and supervised the study. DZ, XZ
and XW designed the study. LT, JZ and YZ performed the experiments
and analyzed the data. DZ, XZ and XW were responsible for
confirming the authenticity of all the raw data. All authors
reviewed the results and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Not applicable
Patient consent for publication
Not applicable.
Competing interests
These authors declare that they have no competing
interests.
References
|
1
|
Rawicz-Pruszyński K, van Sandick JW,
Mielko J, Ciseł B and Polkowski WP: Current challenges in gastric
cancer surgery: European perspective. Surg Oncol. 27:650–656.
2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Miao L, Qi J, Zhao Q, Wu QN, Wei DL, Wei
XL, Liu J, Chen J, Zeng ZL, Ju HQ, et al: Targeting the STING
pathway in tumor-associated macrophages regulates innate immune
sensing of gastric cancer cells. Theranostics. 10:498–515.
2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Barok M, Le Joncour V, Martins A, Isola J,
Salmikangas M, Laakkonen P and Joensuu H: ARX788, a novel anti-HER2
antibody-drug conjugate, shows anti-tumor effects in preclinical
models of trastuzumab emtansine-resistant HER2-positive breast
cancer and gastric cancer. Cancer Lett. 473:156–163.
2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wang Y, Lu JH, Wang F, Wang YN, He MM, Wu
QN, Lu YX, Yu HE, Chen ZH, Zhao Q, et al: Inhibition of fatty acid
catabolism augments the efficacy of oxaliplatin-based chemotherapy
in gastrointestinal cancers. Cancer Lett. 473:74–89.
2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Anczuków O, Akerman M, Cléry A, Wu J, Shen
C, Shirole NH, Raimer A, Sun S, Jensen MA, Hua Y, et al:
SRSF1-regulated alternative splicing in breast cancer. Mol Cell.
60:105–117. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Comiskey DF Jr, Montes M, Khurshid S,
Singh RK and Chandler DS: SRSF2 regulation of MDM2 reveals splicing
as a therapeutic vulnerability of the p53 pathway. Mol Cancer Res.
18:194–203. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yoon JY, Wang JY and Roehrl MH: A combined
FAK, c-MET, and MST1R three-protein panel risk-stratifies
colorectal cancer patients. Transl Oncol. 13(100836)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Krishnaswamy S, Mohammed AK, Tripathi G,
Alokail MS and Al-Daghri NM: Splice variants of the extracellular
region of RON receptor tyrosine kinase in lung cancer cell lines
identified by PCR and sequencing. BMC Cancer.
17(738)2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chakedis J, French R, Babicky M, Jaquish
D, Howard H, Mose E, Lam R, Holman P, Miyamoto J, Walterscheid Z,
et al: A novel protein isoform of the RON tyrosine kinase receptor
transforms human pancreatic duct epithelial cells. Oncogene.
35:3249–3259. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Milan M, Benvenuti S, Balderacchi AM,
Virzì AR, Gentile A, Senetta R, Cassoni P, Comoglio PM and Stella
GM: RON tyrosine kinase mutations in brain metastases from lung
cancer. ERJ Open Res. 4(4)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhao S, Cao L and Freeman JW: Knockdown of
RON receptor kinase delays but does not prevent tumor progression
while enhancing HGF/MET signaling in pancreatic cancer cell lines.
Oncogenesis. 2(e76)2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Paronetto MP, Passacantilli I and Sette C:
Alternative splicing and cell survival: From tissue homeostasis to
disease. Cell Death Differ. 23:1919–1929. 2016.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhou DH, Li C and Yang LN: Variant RONΔ160
of the RON receptor tyrosine kinase promotes the growth and
invasion in vitro and in vivo in gastric cancer cell lines. Cancer
Cell Int. 15(9)2015.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-ΔΔC(T)) method. Methods. 25:402–408. 2001.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yoon M and Rikkerink EHA: Rpa1 mediates an
immune response to avrRpm1Psa and confers resistance against
Pseudomonas syringae pv actinidiae. Plant J. 102:688–702.
2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhang D, Liu K, Hu W, Lu X, Li L, Zhang Q,
Huang H and Wang H: Prenatal dexamethasone exposure caused fetal
rats liver dysplasia by inhibiting autophagy-mediated cell
proliferation. Toxicology. 449(152664)2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
van Bergeijk P, Seneviratne U,
Aparicio-Prat E, Stanton R and Hasson SA: SRSF1 and PTBP1 are
trans-acting factors that suppress the formation of a CD33 splicing
isoform linked to Alzheimer's disease Risk. Mol Cell Biol.
39(39)2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Coomer AO, Black F, Greystoke A, Munkley J
and Elliott DJ: Alternative splicing in lung cancer. Biochim
Biophys Acta Gene Regul Mech. 1862(194388)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Barbagallo D, Caponnetto A, Brex D,
Mirabella F, Barbagallo C, Lauretta G, Morrone A, Certo F, Broggi
G, Caltabiano R, et al: CircSMARCA5 regulates VEGFA mRNA splicing
and angiogenesis in glioblastoma multiforme through the binding of
SRSF1. Cancers (Basel). 11(11)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhou X, Wang R, Li X, Yu L, Hua D, Sun C,
Shi C, Luo W, Rao C, Jiang Z, et al: Splicing factor SRSF1 promotes
gliomagenesis via oncogenic splice-switching of MYO1B. J Clin
Invest. 129:676–693. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Bonomi S, di Matteo A, Buratti E, Cabianca
DS, Baralle FE, Ghigna C and Biamonti G: HnRNP A1 controls a
splicing regulatory circuit promoting mesenchymal-to-epithelial
transition. Nucleic Acids Res. 41:8665–8679. 2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Ghigna C, Giordano S, Shen H, Benvenuto F,
Castiglioni F, Comoglio PM, Green MR, Riva S and Biamonti G: Cell
motility is controlled by SF2/ASF through alternative splicing of
the Ron protooncogene. Mol Cell. 20:881–890. 2005.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Lu H, Han N, Xu W, Zhu Y, Liu L, Liu S and
Yang W: Screening and bioinformatics analysis of mRNA, long
non-coding RNA and circular RNA expression profiles in
mucoepidermoid carcinoma of salivary gland. Biochem Biophys Res
Commun. 508:66–71. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Yang J, Sun G, Hu Y, Yang J, Shi Y, Liu H,
Li C, Wang Y, Lv Z, Niu J, et al: Extracellular vesicle lncRNA
metastasis-associated lung adenocarcinoma transcript 1 released
from glioma stem cells modulates the inflammatory response of
microglia after lipopolysaccharide stimulation through regulating
miR-129-5p/high mobility group box-1 protein axis. Front Immunol.
10(3161)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Kun-Peng Z, Chun-Lin Z and Xiao-Long M:
Antisense lncRNA FOXF1-AS1 promotes migration and invasion of
osteosarcoma cells through the FOXF1/MMP-2/-9 pathway. Int J Biol
Sci. 13:1180–1191. 2017.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhao SJ, Kong FQ, Jie J, Li Q, Liu H, Xu
AD, Yang YQ, Jiang B, Wang DD, Zhou ZQ, et al: Macrophage MSR1
promotes BMSC osteogenic differentiation and M2-like polarization
by activating PI3K/AKT/GSK3β/β-catenin pathway. Theranostics.
10:17–35. 2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wu CY, Zheng C, Xia EJ, Quan RD, Hu J,
Zhang XH and Hao RT: Lysophosphatidic acid receptor 5 (LPAR5) plays
a significance role in papillary thyroid cancer via
phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin
(mTOR) pathway. Med Sci Monit. 26(e919820)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Jiang L, Liu XQ, Ma Q, Yang Q, Gao L, Li
HD, Wang JN, Wei B, Wen J, Li J, et al: hsa-miR-500a-3P alleviates
kidney injury by targeting MLKL-mediated necroptosis in renal
epithelial cells. FASEB J. 33:3523–3535. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Xu R, Wu J, Zhang X, Zou X, Li C, Wang H,
Yuan M, Chen M, Sun Q and Liu S: Modified Bu-zhong-yi-qi decoction
synergies with 5 fluorouracile to inhibits gastric cancer progress
via PD-1/PD-L1-dependent T cell immunization. Pharmacol Res.
152(104623)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ling Y, Kuang Y, Chen LL, Lao WF, Zhu YR,
Wang LQ and Wang D: A novel RON splice variant lacking exon 2
activates the PI3K/AKT pathway via PTEN phosphorylation in
colorectal carcinoma cells. Oncotarget. 8:39101–39116.
2017.PubMed/NCBI View Article : Google Scholar
|