Introduction
Tissue-specific mesenchymal stem cells (MSCs) play
an important role in tissue regeneration and homeostasis; they have
been isolated from various body tissues, such as bone marrow
(1), skeletal muscle (2), skin (3), adipose (4), umbilical cord blood (5), dental pulp (6) and periodontium (7). In particular, due to their less
invasive collection method compared with other tissue sources,
tooth-derived MSCs are expected to be a promising stem cell source
(7,8). However, tissue regeneration involves
not only the differentiation of MSCs into tissue-specific cells but
also the development of the microenvironment for the stem cell
niche, which is composed of various cytokines and immune cells
(9). The combination of optimal
MSC scaffolds with various cytokines that promote cell migration
and differentiation is an important strategy in tissue regenerative
medicine (10,11). In addition, tissue regeneration is
known to depend on the inflammatory response (12,13).
Interestingly, a number of studies have reported the accumulation
of MSCs in the early stages of inflammation in damaged tissues with
chronic inflammation (12,13), suggesting that the association
between MSCs and immune response is important during the
regeneration process.
Fibroblast growth factor 2 (FGF2) is a
heparin-binding protein that is expressed in a wide range of cells
and tissues, functioning as a pluripotent growth factor and serving
an important role in many physiological and pathological processes,
including limb development, neurogenesis, angiogenesis, wound
healing and tumor growth (14,15).
One of the properties of FGF2 is the regulation of proliferation in
various stem cells, such as embryonic stem cells and MSCs,
indicating that FGF2 plays crucial roles in embryogenesis and
tissue regeneration. Interestingly, FGF2-deficient mice were
viable, fertile and phenotypically indistinguishable from
homozygous FGF2 wild-type littermates upon gross examination
(16). However, FGF2-deficient
mice were characterized by a decrease in neuronal density in the
frontal motor cortex, modestly delayed skin wound healing and
decreased bone mass and new bone formation (16,17).
These results indicated that FGF2 played a key role in
neurogenesis, wound healing and bone formation. Therefore, FGF2 was
assumed to act as a tissue regeneration factor in bone defects
associated with periodontal disease. Furthermore, FGF2 has also
been studied as a therapeutic agent for diseases such as traumatic
skin wounds (18). Thus, FGF2 is
likely to play a central role in tissue regeneration.
Chemokine C-C motif ligand 11 (CCL11), also known as
eotaxin, is an eosinophil-specific chemoattractant cytokine that
plays an important role in inflammatory and allergic conditions
such as asthma and eosinophilia (19). Chemokine C-C motif receptor 3
(CCR3), which acts as a receptor for CCL11 and is involved in
allergic inflammation, is highly expressed in eosinophils,
basophils and type 2 helper lymphocytes (20). In addition, CCL11 has also been
reported to bind C-C chemokine receptor type 2 (CCR2) and CCR5;
however, it functions as an antagonist and agonist, respectively,
indicating that CCL11 plays an ambivalent role in the fine-tuning
of cellular responses at inflammation sites (21). Furthermore, CCL11 was also reported
to increase in animal plasma and cerebrospinal fluid samples with
age, correlating with a decrease in neurogenesis in heterochronic
parabiosis and aged mice (22).
Recently, increased levels of CCL11 were found to be correlated
with telomere length (23). Thus,
CCL11 was assumed to be an aging factor with a specific role in the
negative regulation of neurogenesis (24). In addition, circulating levels of
CCL11 are used in clinical studies as a biomarker of gastric cancer
and postmenopausal osteoporosis (25). A PABP-interacting motif 2-induced
inflammation mouse model indicated that osteoblasts expressed
CCL11, whereas osteoclasts expressed the CCL11-high affinity
receptor CCR3, with CCL11 promoting the migration of osteoclast
precursors (26). These
observations indicated that the CCL11/CCR3 axis is also involved in
pathological bone metabolism.
The present study aimed to characterize the changes
in cytokine and chemokine expression induced by FGF2 in human
dental pulp-derived MSCs.
Materials and methods
Reagents
FGF2 and AZD4547 (FGFR inhibitor) were purchased
from R&D Systems, Inc. and Santa Cruz Biotechnology, Inc.,
respectively. SB203580 (p38 MAPK inhibitor) and SP600125 (JNK
inhibitor) were purchased from FUJIFILM Wako Pure Chemical
Corporation. U0126 [MAPK kinase (MEK) inhibitor], anti-ERK1/2 (cat.
no. #9102), anti-phosphorylated (p)-ERK1/2 (cat. no. #9101),
anti-p38 MAPK (cat. no. #8690), anti-p-p38 MAPK (cat. no. #9211),
anti-stress-activated protein kinases (SAPK)/JNK (cat. no. #9252),
anti-p-SAPK/JNK (cat. no. #4668) and anti-β-actin antibodies (cat.
no. #4967) were purchased from Cell Signaling Technology, Inc.
Cells
SDP11 cells were used in the present study. We
previously established the cell line, SDP11, from the dental pulp
tissues of human deciduous teeth that were obtained from 3 healthy
children aged between 6-8 years old (27,28).
Informed consent was obtained from the donors' parents, following
the guidelines of Tokushima University Hospital (Tokushima, Tokyo,
Japan) prior to tooth extraction during orthodontic treatment. The
present study protocol was approved by the University of Tokushima
Hospital Ethical Review Committee (approval no. 1799). The tissues
sections were chopped into pulp using a surgical blade and digested
with collagenase (2 mg/ml) at 37˚C for 30 min. After being washed
with PBS (Sigma-Aldrich; Merck KGaA), the tissues were placed on a
cell culture dish and maintained in α-modified minimum essential
medium (α-MEM; Gibco; Thermo Fisher Scientific, Inc.) supplemented
with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.). Migrated
fibroblastic cells were used as dental pulp cells (DPCs). DPCs were
transfected with the pBABE-neo-hTERT plasmid (Addgene, Inc.) using
Lipofectamine™ LTX (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The transfected cells
were maintained 24 h after transfection using the selective
antibiotic G418 (Gibco; Thermo Fisher Scientific, Inc.) at a
concentration of 200 µg/ml for 12-15 days. Then, single-cell clones
were obtained by the limiting dilution cloning method. Clones
generated by single-cell clones were named single-cell clone
derived from human deciduous tooth pulp cells (SDP). SDP11 was
derived from clone number 11.
Cell culture
SDP11 cells were cultured in α-MEM supplemented with
10% FBS and 5% antibiotic-antimycotic (penicillin G, streptomycin
sulfate and amphotericin B) mixture (Gibco; Thermo Fisher
Scientific, Inc.). All cultures were maintained at 37˚C in a
humidified atmosphere containing 5% CO2.
Comprehensive expression profiling and
quantitative reverse transcription-quantitative PCR (RT-qPCR)
SDP11 cells were plated at 6.0x104
cells/cm2 in a 35-mm culture dish and reached 100%
confluence after 24 h. Then, cells were treated with 20 ng/ml FGF2
for 48 h for comprehensive expression profiling and for 6, 12, 24,
48 and 72 h for CCL11 expression. To test the effect of FGF2
concentration on the expression of CCL11, SDP11 cells were cultured
with 0.1, 1, 10, 20 and 40 ng/ml FGF2 for 24 h. To examine the
effect of AZD4547, SB203580, U0126 or SP600125, SDP11 cells were
cultured in the presence or absence of 20 ng/ml FGF2 and with or
without 10 µM inhibitor for 24 h. Total RNA was extracted from
pretreated SDP11 cells using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. A total of 2 µg extracted total RNA
was used to produce first-strand cDNA using the PrimeScript™ RT
Master Mix (Perfect Real Time; Takara Bio, Inc.) in accordance with
the manufacturer's instructions. qPCRwas performed using TB Green™
Premix Ex Taq™ II (Takara Bio, Inc.), according to the
manufacturer's instructions, and the Thermal Cycler Dice Real Time
system (Takara Bio, Inc.) under the following conditions: 95˚C for
5 sec, followed by 45 cycles of 95˚C for 5 sec, 62˚C for 30 sec and
72˚C for 20 sec, with a final cycle at 95˚C for 15 sec, 62˚C for 30
sec and 95˚C for 15 sec. Regarding primer information,
comprehensive gene expression profiling was performed using the
PrimerArray® (human cytokine-cytokine receptor
interactions, cat. no. PH001; Takara Bio, Inc.) that was packaged
into premixed 96 primer pairs (Table
I) in a single-use 96-well plate. The primer sequences used for
CCL11 and GAPDH were as follows: CCL11 forward,
5'-CTTCAGCCTCCAACATGAAGGTC-3' and reverse,
5'-CTATTGGCCAGGTTAAAGCAGCA-3'; GAPDH forward,
5'-GCACCGTCAAGGCTGAGAAC-3' and reverse, 5'-TGGTGAAGACGCCAGTGG-3'.
The predicted size of each fragment was 130 and 267 base pairs,
respectively. CCL11 expression was corrected using the housekeeping
gene GAPDH. Relative gene expression was analyzed using the
2-ΔΔcq method (29).
 | Table IGene list in the
PrimerArray® of human cytokine-cytokine receptor
interactions. |
Table I
Gene list in the
PrimerArray® of human cytokine-cytokine receptor
interactions.
| Category | Gene name |
|---|
| Cytokine genes | IL2RG, KIT, MET,
IFNGR1, IL1A, IL8, IL6, CXCL12, IL10RB, IL8RA, TGFB1, CSF1, CSF3R,
IL2RB, KITLG, PRLR, LEPR, HGF, TGFBR2, VEGFA, TNFRSF1A, ACVR1,
ACVR2B, BMPR1B, BMPR2, CCR1, CX3CR1, IL13RA1, IL18, CXCL10, CCR7,
CNTFR, EGF, FLT4, CXCL2, IL1RAP, IL7R, IL13, INHBA, INHBB, KDR,
LTBR, CXCL9, NGFR, TNFRSF11B, PDGFB, CCL2, CCL5, CCL11, CCL18,
CCL21, CCL22, CCL24, CXCL5, CX3CL1, TGFB2, TGFB3, VEGFB, TNFSF12,
TNFSF10, TNFRSF10D, TNFRSF10B, TNFRSF10A, OSMR, FLT3, ACVR1B,
BMPR1A, CCR6, CCL20, TGFBR1, CXCL14, CSF1R, CCL13, PDGFRA, CXCR6,
IL17RA, IL20RA, PDGFC, TNFRSF12A, EDA2R, CXCL16, PLEKHQ1, TSLP,
CCL28, CD40, IL28RA, IL6ST, EGFR |
| Housekeeping
genes | GUSB, HPRT1, PGK1,
ACTB, GAPDH, TBP, B2M, PPIA |
Western blotting
SDP11 cells were plated at a density of
3.0x104 cells/cm2 in a 60-mm culture dish, in
which cells reached a confluence of 80% after 24 h. Prior to FGF2
treatment, the culture dish was washed thrice with PBS, replaced
with serum-free α-MEM and incubated at 37˚C for 1 h. Cells were
then incubated with or without 20 ng/ml FGF2 and in the presence or
absence of 10 µM SB203580, U0126 or SP600125 for 5 min. After these
treatments, SDP11 cells were washed thrice with PBS containing 1 mM
sodium vanadate Na3VO4 (Sigma-Aldrich; Merck
KGaA) and lysed in 100 µl Cellytic M reagent solution buffer
(Sigma-Aldrich; Merck KGaA) supplemented with complete mini
protease inhibitor cocktail tablets (Roche Diagnostics) and
PhosSTOP™ (Roche Diagnostics). Lysed cells were collected and
centrifuged at 13,205 x g for 5 min at 4˚C. Protein concentration
was measured using the micro-BCA protein assay reagent (Pierce;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
instructions. Subsequently, 10 µg of lysate protein denatured in
LDS sample buffer (Invitrogen; Thermo Fisher Scientific, Inc.)
containing a NuPAGE™ Sample Reducing Agent (Invitrogen; Thermo
Fisher Scientific, Inc.) was loaded onto each well of NuPAGE™ 4-12%
Bis-Tris Protein gels (Invitrogen; Thermo Fisher Scientific, Inc.).
After loading onto the NuPAGE 4-12% Bis-Tris Protein gels, proteins
were transferred to PVDF membranes (Invitrogen; Thermo Fisher
Scientific, Inc.) and blocked with 5% skimmed milk in Tris-buffered
saline containing 0.05% Tween-20 (Bio-Rad Laboratories, Inc.) for
40 min at room temperature. Membranes were then immunoblotted for
40 min at room temperature with primary antibodies (anti-ERK1/2,
anti-p-ERK1/2, anti-p38 MAPK, anti-p-p38 MAPK, anti-SAPK/JNK,
anti-p-SAPK/JNK and anti-β-actin; all, 1:1,000). Membranes were
subsequently incubated for 40 min at room temperature with the
HRP-conjugated anti-rabbit IgG secondary antibody (Cell Signaling
Technology, Inc.; #7074; 1:1,000). Membranes were visualized using
an ECL kit (Cytiva), according to the manufacturer's instructions.
Blot images were acquired using an Amersham™ Imager 600
(Cytiva).
Statistical analysis
Each experiment was run in triplicate. For the
analyses presented in Figs. 2,
3, 4 and 5D,
data were pooled from three independent experiments, and the
results were presented as the mean ± standard error of the mean. In
Fig. 5A-C, the data from three
independent experiments with similar results are shown as
representatives. Error bars indicate standard deviations.
Statistical analysis was performed with one-way ANOVA and Tukey
post hoc test using MS Excel (version 16.52; Microsoft
Corporation). P<0.05 was considered to indicate a statistically
significant difference.
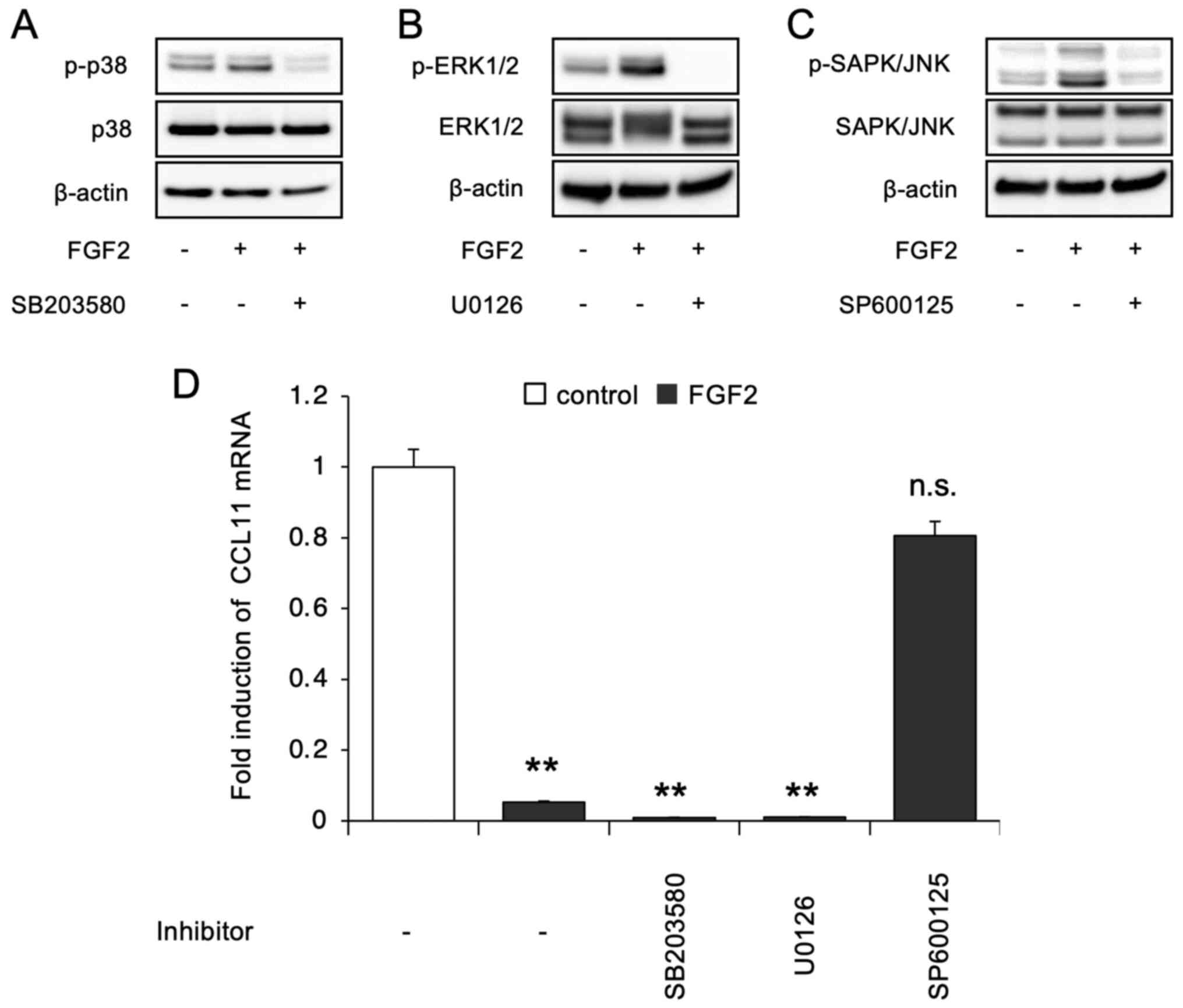 | Figure 5Phosphorylation levels of p38 MAPK,
ERK1/2 and SAPK/JNK after treatment of SDP11 cells with FGF2. SDP11
cells were treated with FGF2 for 5 min, after which they were
immunoblotted with (A) anti-p-p38 MAPK and anti-p38 MAPK, (B)
anti-p-ERK1/2 and anti-ERK1/2, and (C) anti-p-SAPK/JNK and
anti-SAPK/JNK. Anti-β-actin antibodies were used for normalization.
The effect of each specific kinase inhibitor, namely, p38 MAPK
inhibitor (SB203580), MEK inhibitor (U0126) and JNK inhibitor
(SP600125) was evaluated. FGF2 induced the phosphorylation of p38
MAPK, ERK1/2 and SAPK/JNK, which was blocked by each respective
inhibitor when co-administered. (D) SDP11 cells were cultured with
or without 20 ng/ml FGF2 in the presence of 10 µM SB203580, 10 µM
U0126 or SP600125 for 24 h. The expression of CCL11 was then
examined by reverse transcription-quantitative PCR. Only the JNK
inhibitor blocked the FGF2-mediated suppression in the expression
of CCL11. Data are presented as the mean ± SEM.
**P<0.01. SAPK, stress-activated protein kinases;
FGF2, fibroblast growth factor 2; CCL11, C-C motif ligand 11; n.s.,
not significant |
Results
Altered expression of cytokines in
FGF2-treated cells
To clarify the interactions between FGF2 and the
immune system in dental pulp-derived MSCs, the expression of
cytokines in FGF2-treated SDP11 cells was examined using a
PrimerArray® consisting of human cytokine-cytokine
receptor interactions (Table I).
The cells were treated with 20 ng/ml FGF2 for 48 h, after which
gene expression was assessed by RT-qPCR using the
PrimerArray® as aforementioned. A total of 48 molecules
involved in the immune response were found to be expressed in SDP11
cells and, interestingly, FGF2 suppressed the expression of
chemokine C-C motif ligand 11 (CCL11) among expressed cytokines
(Fig. 1). No satisfactory
amplification products were found in 40 cytokine genes.
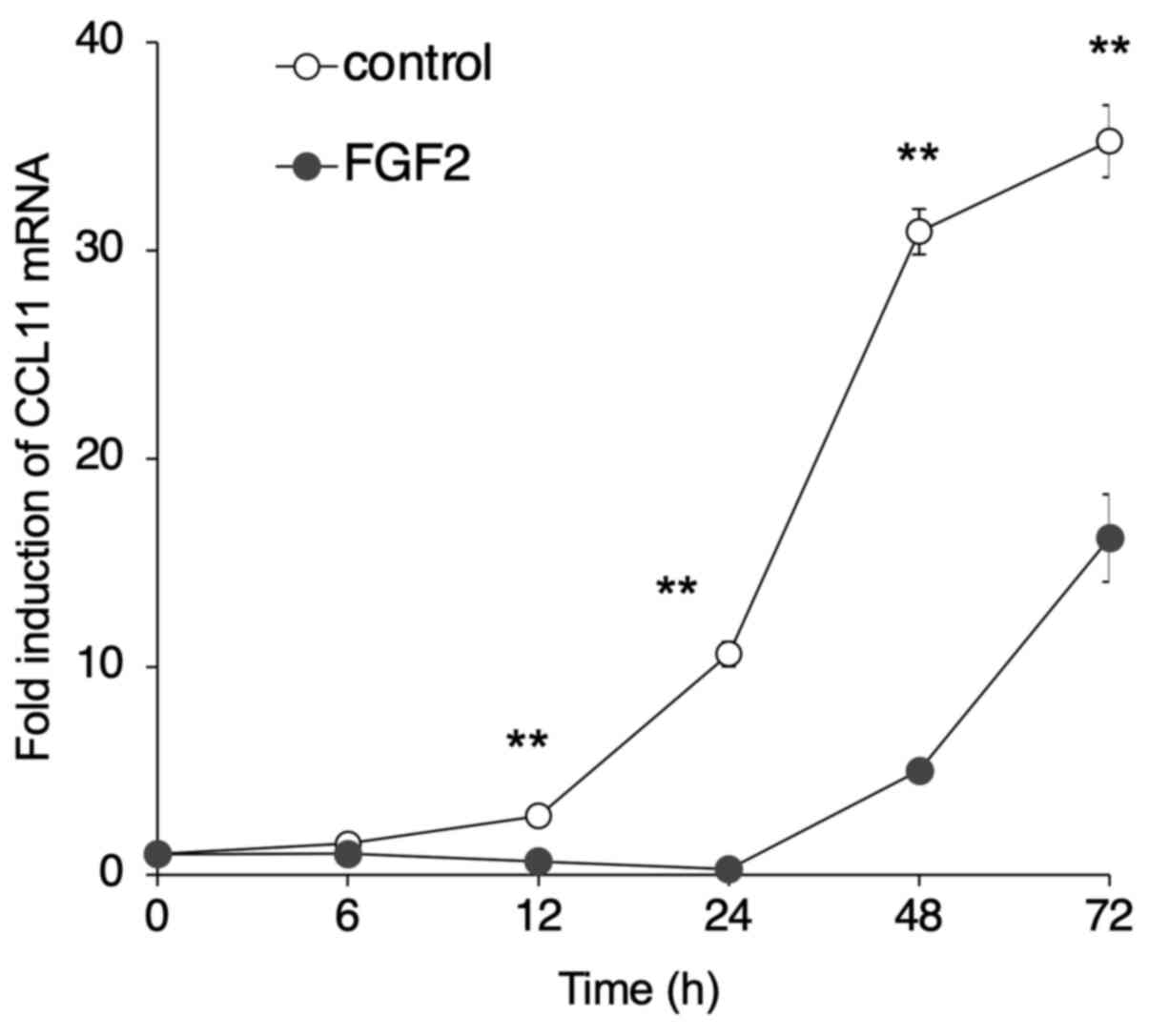
FGF2-induced suppression of CCL11
To confirm that FGF2 suppressed CCL11 expression in
SDP11 cells, cells were cultured in the presence of FGF2 for 72 h.
The expression of CCL11 was then examined using RT-qPCR. CCL11
expression level decreased in cells following treatment with FGF2
for 12, 24, 48 and 72 h compared with untreated control cells
(Fig. 2). Subsequently, the effect
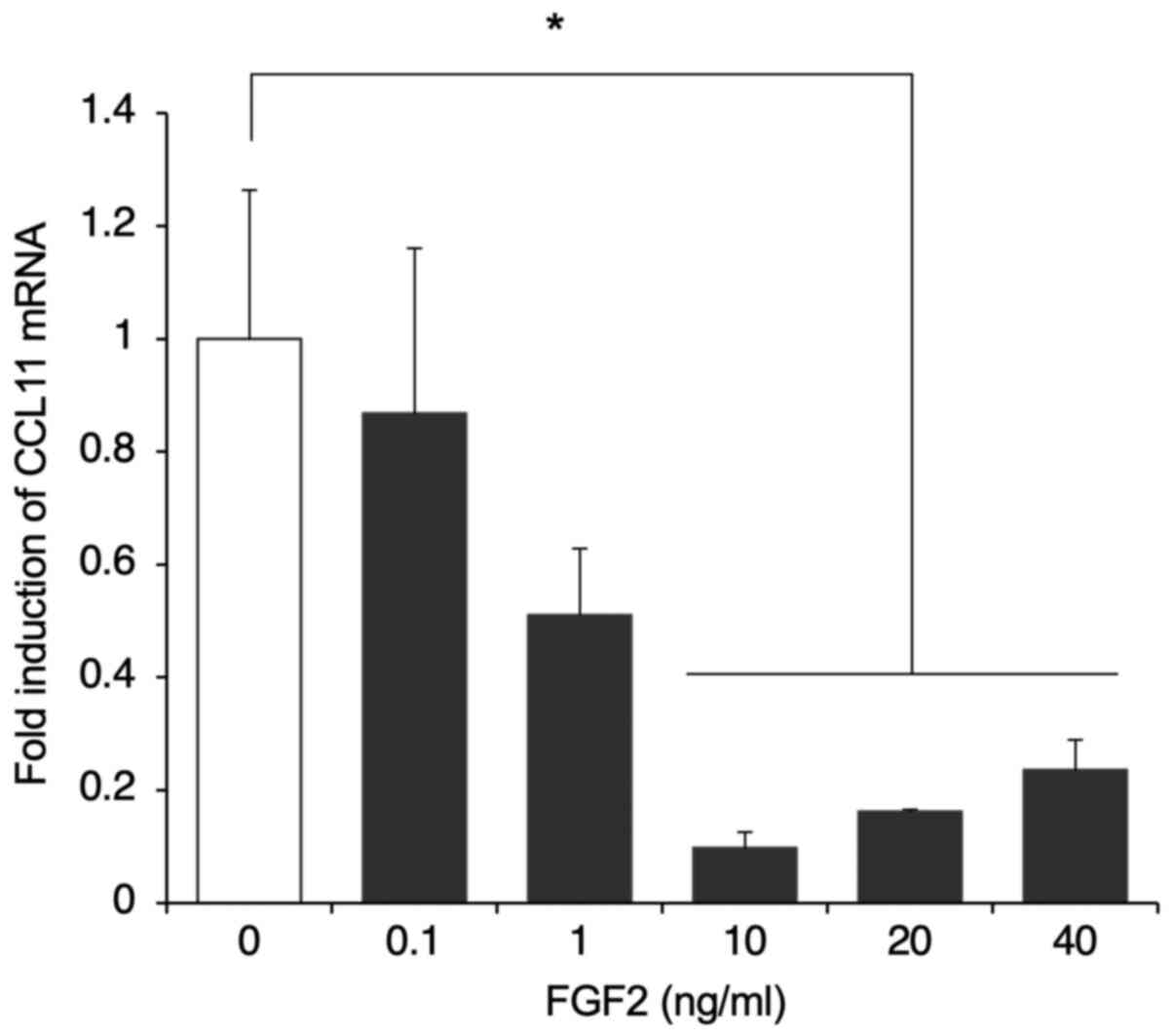
of FGF2 concentration on the expression of CCL11 was examined. To
this end, SDP11 cells were cultured with 0.1, 1, 10, 20 and 40
ng/ml FGF2 for 24 h. The administration of ≥10 ng/ml FGF2
suppressed the expression of CCL11 compared with that in the
control (Fig. 3). There were no
significant differences at 0.1 and 1 ng/ml FGF2 (Fig. 3). These results suggested that FGF2
negatively regulated the expression of CCL11 in dental pulp-derived
MSCs in a time- and dose-dependent manner.
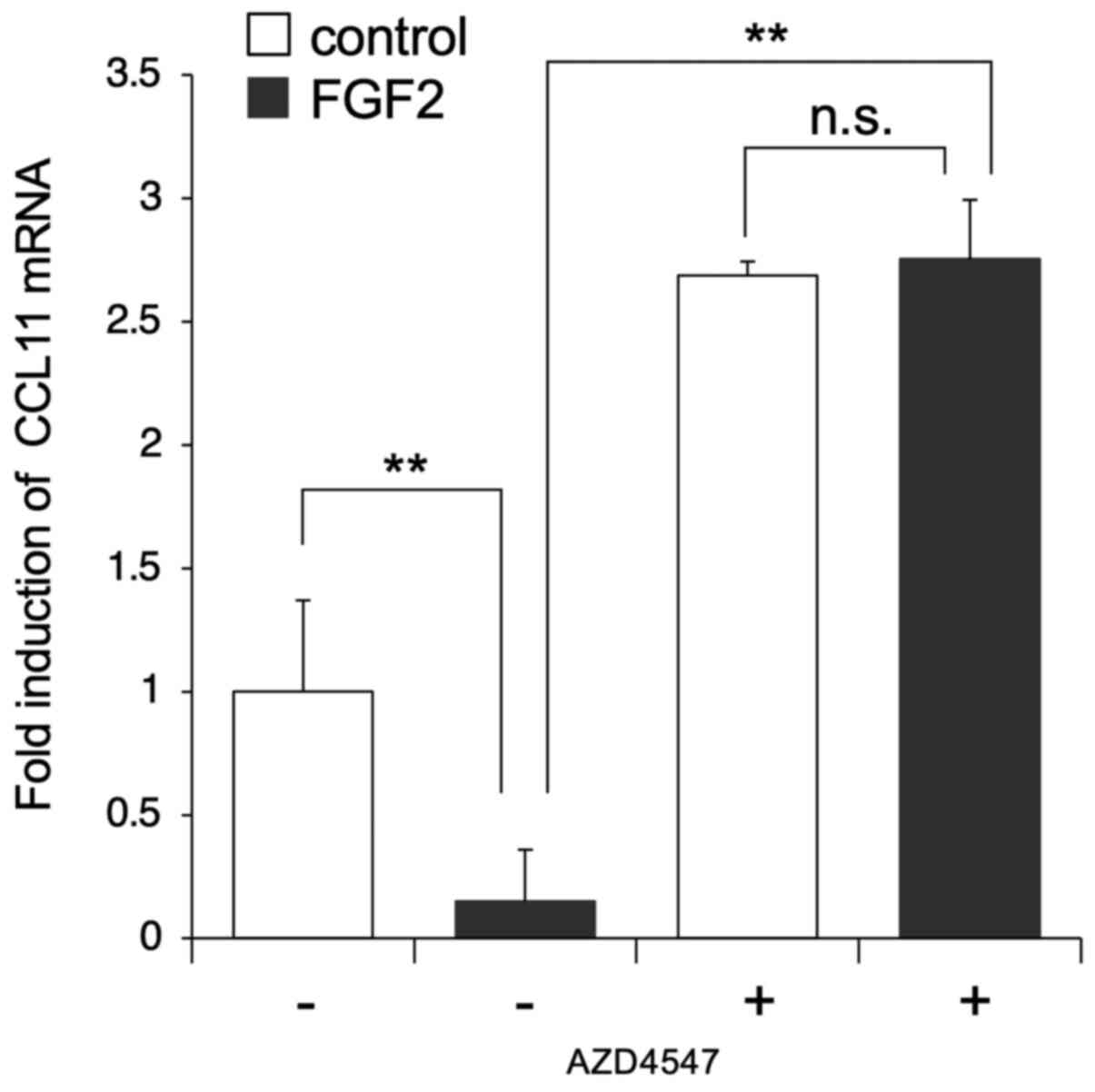
Regulation of expression of CCL11 by
FGFR signaling
The subsequent aim of the present study was to
characterize the effect of FGF2 on the suppression of CCL11
expression. To assess whether the FGF2-induced suppression of CCL11
was mediated by FGF receptor signaling, AZD4547, a selective
FGFR1/2/3 inhibitor, was used in cell culture. SDP11 cells were
cultured in the presence or absence of 20 ng/ml FGF2 and with or
without 10 µM AZD4547 for 24 h. The expression of CCL11 was then
examined by RT-qPCR. The results indicated that AZD4547 abolished
the FGF2-induced suppression of CCL11 (Fig. 4). Furthermore, the results
demonstrated that AZD4547 increased the expression of CCL11 in the
absence of exogenous administration of FGF2, which may have been
due to the suppression of endogenous FGF signaling (Fig. 4). These results suggested that
FGFR-mediated signaling played an important role in the regulation
of the expression of CCL11 in dental pulp-derived MSCs.
FGF2 mediates the suppression of CCL11
via JNK signaling
To identify the signaling pathways involved in the
FGF2-mediated suppression of CCL11 expression in SDP11 cells,
western blotting was performed to examine the phosphorylation of
p38 MAPK, ERK1/2 and JNK, which are different members of the MAP
kinase family activated downstream of FGFR. The results indicated
that FGF2 stimulated the phosphorylation of p38 MAPK, ERK1/2 and
SAPK/JNK in SDP11 cells, which was suppressed following treatment
with each specific kinase inhibitor, namely, the p38 MAPK
(SB203580), MEK (U0126) and JNK (SP600125) inhibitors (Fig. 5A-C), respectively. To further
clarify the pathway mediating the suppression of CCL11 expression
downstream of FGF2, the levels of CCL11 were examined in cells
cultured with FGF2 in the presence of each specific kinase
inhibitor. The results indicated that SP600125, but not SB203580 or
U0126, inhibited the FGF2-induced suppression in the expression of
CCL11 (Fig. 5D). The present
results suggested that the JNK pathway was responsible for the
inhibitory effect on the expression of CCL11 in human dental
pulp-derived MSCs.
Discussion
The present study used an RT-qPCR-based primer array
technique to analyze the FGF2-induced altered expression of
cytokines in the SDP11 human dental pulp-derived MSC line. The
results indicated that FGF2 regulated the expression of
inflammatory cytokines in SDP11 cells, suggesting that FGF2 was
involved in the regulation of inflammatory cytokines in
pulp-derived MSCs. Furthermore, the expression level of CCL11 was
significantly decreased by FGF2 in a time- and dose-dependent
manner, and its inhibition was blocked by the AZD4547 FGFR
inhibitor. The present study also reported that SP600125, an
inhibitor of JNK, abolished the FGF2-induced suppression of CCL11.
The current results suggested that FGF2 modulated the expression of
CCL11 via the FGFR-JNK cascade in human dental pulp-derived
MSCs.
Of note, FGF2 is known to play an important role in
tooth development and dentin repair. The local application of
gelatin hydrogel-delivered FGF2 to the exposed pulps of teeth with
dentin defects was reported to induce the formation of dentin-like
particles in vivo (30).
Exogenous FGF2 was also shown to promote the activity of alkaline
phosphatase, the formation of calcified nodules and the expression
of odontogenic marker genes in human DPCs in vitro,
suggesting that FGF2 positively regulated odontoblast
differentiation (31). In
contrast, it has also been reported that FGF2 inhibited the
terminal differentiation of odontoblasts in pulp cells (32). Our preliminary study suggested that
FGF2 is not involved in the formation of calcified nodules in SDP11
cells (data not shown). A previous study reported that continuous
stimulation with FGF2 inhibited odontoblast differentiation,
whereas early and limited stimulation with FGF2 markedly increased
odontoblast differentiation in pulp cells (33). Similarly, FGF2-deficient mice were
characterized by decreased osteoblast differentiation (17), while non-specific overexpression of
FGF2 in transgenic mice under a phosphoglycerate kinase promoter
lead to skeletal abnormalities, including shortening and flattening
of long bones and moderate macrocephaly (34), indicating that FGF2 could act both
as a positive and negative regulator of osteogenesis. Thus, the
effect of FGF2 on odontoblast differentiation appears to be
specific to the cell differentiation stage.
Another important property of FGF2 is its role in
inflammation. In particular, FGF2 was found to prevent the
neuroinflammation-induced decrease in the phosphorylation of ERK1/2
and alleviated the neuroinflammation-induced impairment in
hippocampal neurogenesis (35). In
another study, FGF2 improved the survival rate of septic mice by
inhibiting the inflammatory response, alleviating lung injury
(36). Moreover, FGF2 was reported
to stimulate the migration and proliferation of endothelial cells
in vivo and to relieve inflammation by inducing the
expression of inflammation-related genes, such as proinflammatory
cytokines and chemokines in endothelial cells (36-38).
In human DPCs, FGF2 was demonstrated to induce the expression of
chemokines, such as IL-6, IL-8, monocyte chemoattractant protein-1α
(MIP-1α) and MIP-3α (31). These
observations suggested that the pluripotent growth factor FGF2
efficiently directed cell populations toward the maintenance of
tissue homeostasis and regeneration, depending on the inflammatory
state. The present study also reported that FGF2 altered the
cytokine expression profile of the SDP11 human dental pulp-derived
MSC cell line. Interestingly, there have been numerous studies
indicating that the expression of cytokines was increased by FGF2
(31,39,40),
whereas only a few studies have reported a decrease (31). Therefore, considering the
multifunctionality of FGF2, the present study subsequently focused
on the cytokines that were decreased following FGF2 treatment. The
expression of CCL11 was found to be suppressed by exogenous FGF2 in
SDP11 cells. The present results suggested that FGF2 negatively
regulated the expression of CCL11 in human dental pulp-derived
MSCs.
CCL11, a chemoattractant cytokine, is known to
affect the cellular response to inflammatory conditions (19). CCL11 has also been indicated to
correlate with aging and negative regulation of neurogenesis
(22,24). More specifically, CCL11 has been
associated with chronic inflammation and the attenuation of nerve
regeneration in old age (41).
These results suggested that CCL11 and CCL11 receptors, such as
CCR3, could serve as targets for anti-inflammatory therapies. In
fact, ASM8, an antisense nucleotide targeting CCR3, was reported to
enable a consistent protective effect against allergen-induced
asthmatic responses (42). R321,
another peptide-based CCR3 antagonist, which targets the second
transmembrane helix and first extracellular loop of both human and
mouse CCR3, has been suggested to be effective in the treatment of
eosinophilic inflammation (43).
Moreover, the administration of a CCL11-neutralizing antibody was
indicated to induce tooth pulp regeneration by reducing the number
of M1 macrophages and improving the M1/M2 ratio (44). M1 macrophages are typically
activated during inflammation, whereas M2 macrophages are activated
during anti-inflammatory responses and tissue regeneration
(45). Accordingly, the present
in vitro results suggested that the FGF2-mediated
downregulation of CCL11 expression in dental MSCs had
anti-inflammatory and tissue regeneration effects. Consistently,
FGF was reported to be increased in reversible pulpitis but
decreased in irreversible pulpitis, which significantly increased
the expression of CCL11(46).
FGF2 was reported to interact with specific cell
surface receptors, including FGFR1, FGFR2, FGFR3 and FGFR4(47). Accordingly, FGF2 signaling plays
important roles in a multitude of physiological and pathological
processes by regulating cell proliferation, migration,
differentiation and survival (47). In addition, FGFR is a family of
tyrosine kinases that can activate signaling cascades, such as
RAS-MAPK, PI3K-AKT, phospholipase C γ and STAT (48). Among the downstream mediators of
FGF2 signaling pathways, the MAPK pathway has been demonstrated to
play a pivotal role in the function of stem cells, including
mesenchymal stem cells (49-51).
Thus, the present study focused on the MAPK pathway and found that
FGF2 induced the phosphorylation of p38 MAPK, ERK1/2 and JNK in
SDP11 cells. Likewise, in a previous study, treatment with FGF2,
limited to the early stages of undifferentiated DPCs, promoted the
formation of functional odontoblasts via the FGFR/MEK/ERK and bone
morphogenetic protein signaling pathways (33). Moreover, ERK and p38 MAPK were
found to induce the expression of osteogenic marker genes and
promote osteoblast differentiation (52). The present study indicated that
AZD4547 and SP600125, but not SB203580 or U0126, inhibited the
FGF2-induced suppression of CCL11 in SDP11 cells. The present
results suggested that the FGF2/FGFR/JNK pathway is involved in the
FGF2-mediated suppression of the expression of CCL11 in human
dental pulp-derived MSCs.
The present study demonstrated that FGF2 regulated
the expression of cytokines in SDP11 cells and to the best of our
knowledge, indicated for the first time that FGF2 may play a role
in the suppression of the expression of CCL11 through the
FGF2/FGFR/JNK signaling pathway. There are several limitations in
the present study, mainly due to the use of a cell line as a mimic
of human dental pulp-derived MSCs. However, analysis using a simple
model is essential for clarifying the function of FGF2, as FGF2
contributes to tissue homeostasis and regeneration by recognizing
its surrounding spatiotemporal environment and cross-talking with
various cells, cytokines and signaling molecules (31). Further studies using primary cell
cultures or in vivo models are required to confirm the
results obtained in the present study. Finally, the present
findings on the FGF2-mediated suppression of the expression of
CCL11 could facilitate the delineation of the molecular mechanisms
of tissue regeneration in teeth.
Acknowledgements
Not applicable.
Funding
The present work was supported by JSPS KAKENHI (grant nos.
17H04414, 20H03898, 17K17332, 20K10205, 20K18760 and 16K11804).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TH and TIwam conceived and designed the study. RK,
YA, TK and TH participated in patient recruitment and preparation
of primary cells. RK, TH, KI, AS, KYU, AM, AN, KK and HN carried
out the molecular analyses and interpreted the data. RK, TH, TIwas
and TIwam carried out the data analyses. TH, YA, RK, and TIwam
confirm the authenticity of all the raw data. TIwam and RK wrote
the manuscript. All authors read and approved the manuscript.
Ethics approval and consent to
participate
Ethical approval was obtained from the Ethics
Committee of Tokushima University Hospital (approval no. 1799).
Written informed consent was obtained from all patients or their
guardians, based on the guidelines set by the Ethics Committee of
Tokushima University Hospital (Tokushima, Japan).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Friedenstein AJ, Chailakhjan RK and
Lalykina KS: The development of fibroblast colonies in monolayer
cultures of guinea-pig bone marrow and spleen cells. Cell Tissue
Kinet. 3:393–403. 1970.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Williams JT, Southerland SS, Souza J,
Calcutt AF and Cartledge RG: Cells isolated from adult human
skeletal muscle capable of differentiating into multiple mesodermal
phenotypes. Am Surg. 65:22–26. 1999.PubMed/NCBI
|
|
3
|
Toma JG, Akhavan M, Fernandes KJ,
Barnabé-Heider F, Sadikot A, Kaplan DR and Miller FD: Isolation of
multipotent adult stem cells from the dermis of mammalian skin. Nat
Cell Biol. 3:778–784. 2001.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Zuk PA, Zhu M, Mizuno H, Huang J, Futrell
JW, Katz AJ, Benhaim P, Lorenz HP and Hedrick MH: Multilineage
cells from human adipose tissue: Implications for cell-based
therapies. Tissue Eng. 7:211–228. 2001.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Erices A, Conget P and Minguell JJ:
Mesenchymal progenitor cells in human umbilical cord blood. Br J
Haematol. 109:235–242. 2000.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Gronthos S, Mankani M, Brahim J, Robey PG
and Shi S: Postnatal human dental pulp stem cells (DPSCs) in vitro
and in vivo. Proc Natl Acad Sci USA. 97:13625–13630.
2000.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Seo BM, Miura M, Gronthos S, Bartold PM,
Batouli S, Brahim J, Young M, Robey PG, Wang CY and Shi S:
Investigation of multipotent postnatal stem cells from human
periodontal ligament. Lancet. 364:149–155. 2004.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Miura M, Gronthos S, Zhao M, Lu B, Fisher
LW, Robey PG and Shi S: SHED: Stem cells from human exfoliated
deciduous teeth. Proc Natl Acad Sci USA. 100:5807–5812.
2003.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ferraro F, Celso CL and Scadden D: Adult
stem cels and their niches. Adv Exp Med Biol. 695:155–168.
2010.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Cheung AS, Zhang DK, Koshy ST and Mooney
DJ: Scaffolds that mimic antigen-presenting cells enable ex vivo
expansion of primary T cells. Nat Biotechnol. 36:160–169.
2018.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Padma AM, Carrière L, Krokström Karlsson
F, Sehic E, Bandstein S, Tiemann TT, Oltean M, Song MJ, Brännström
M and Hellström M: Towards a bioengineered uterus: Bioactive sheep
uterus scaffolds are effectively recellularized by enzymatic
preconditioning. NPJ Regen Med. 6(26)2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Martin I, Galipeau J, Kessler C, Le Blanc
K and Dazzi F: Challenges for mesenchymal stromal cell therapies.
Sci Transl Med. 11(11)2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Galipeau J and Sensébé L: Mesenchymal
stromal cells: Clinical challenges and therapeutic opportunities.
Cell Stem Cell. 22:824–833. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Guillonneau X, Tassin J, Berrou E,
Bryckaert M, Courtois Y and Mascarelli F: In vitro changes in
plasma membrane heparan sulfate proteoglycans and in perlecan
expression participate in the regulation of fibroblast growth
factor 2 mitogenic activity. J Cell Physiol. 166:170–187.
1996.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Brewer JR, Mazot P and Soriano P: Genetic
insights into the mechanisms of Fgf signaling. Genes Dev.
30:751–771. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ortega S, Ittmann M, Tsang SH, Ehrlich M
and Basilico C: Neuronal defects and delayed wound healing in mice
lacking fibroblast growth factor 2. Proc Natl Acad Sci USA.
95:5672–5677. 1998.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Montero A, Okada Y, Tomita M, Ito M,
Tsurukami H, Nakamura T, Doetschman T, Coffin JD and Hurley MM:
Disruption of the fibroblast growth factor-2 gene results in
decreased bone mass and bone formation. J Clin Invest.
105:1085–1093. 2000.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Lou ZC and Wang YB: Healing outcomes of
large (>50%) traumatic membrane perforations with inverted edges
following no intervention, edge approximation and fibroblast growth
factor application; a sequential allocation, three-armed trial.
Clin Otolaryngol. 38:289–296. 2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Mattoli S, Stacey MA, Sun G, Bellini A and
Marini M: Eotaxin expression and eosinophilic inflammation in
asthma. Biochem Biophys Res Commun. 236:299–301. 1997.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Kitaura M, Nakajima T, Imai T, Harada S,
Combadiere C, Tiffany HL, Murphy PM and Yoshie O: Molecular cloning
of human eotaxin, an eosinophil-selective CC chemokine, and
identification of a specific eosinophil eotaxin receptor, CC
chemokine receptor 3. J Biol Chem. 271:7725–7730. 1996.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ogilvie P, Bardi G, Clark-Lewis I,
Baggiolini M and Uguccioni M: Eotaxin is a natural antagonist for
CCR2 and an agonist for CCR5. Blood. 97:1920–1924. 2001.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Villeda SA, Luo J, Mosher KI, Zou B,
Britschgi M, Bieri G, Stan TM, Fainberg N, Ding Z, Eggel A, et al:
The ageing systemic milieu negatively regulates neurogenesis and
cognitive function. Nature. 477:90–94. 2011.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Czepielewski LS, Massuda R, Panizzutti B,
Grun LK, Barbé-Tuana FM, Teixeira AL, Barch DM and Gama CS:
Telomere length and CCL11 levels are associated with gray matter
volume and episodic memory performance in schizophrenia: Evidence
of pathological accelerated aging. Schizophr Bull. 44:158–167.
2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Huber AK, Giles DA, Segal BM and Irani DN:
An emerging role for eotaxins in neurodegenerative disease. Clin
Immunol. 189:29–33. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wang W, Huang CY, Wang ZP, Xu SS, Qian TY,
Chen YD and Wu WG: Serum C-C motif ligand 11/eotaxin-1 may serve as
a candidate biomarker for postmenopausal osteoporosis. J Med
Biochem. 38:353–360. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Kindstedt E, Holm CK, Sulniute R,
Martinez-Carrasco I, Lundmark R and Lundberg P: CCL11, a novel
mediator of inflammatory bone resorption. Sci Rep.
7(5334)2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Akazawa Y, Hasegawa T, Yoshimura Y, Chosa
N, Asakawa T, Ueda K, Sugimoto A, Kitamura T, Nakagawa H, Ishisaki
A, et al: Recruitment of mesenchymal stem cells by stromal
cell-derived factor 1α in pulp cells from deciduous teeth. Int J
Mol Med. 36:442–448. 2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Iwamoto T, Nakamura T, Ishikawa M,
Yoshizaki K, Sugimoto A, Ida-Yonemochi H, Ohshima H, Saito M,
Yamada Y and Fukumoto S: Pannexin 3 regulates proliferation and
differentiation of odontoblasts via its hemichannel activities.
PLoS One. 12(e0177557)2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kikuchi N, Kitamura C, Morotomi T, Inuyama
Y, Ishimatsu H, Tabata Y, Nishihara T and Terashita M: Formation of
dentin-like particles in dentin defects above exposed pulp by
controlled release of fibroblast growth factor 2 from gelatin
hydrogels. J Endod. 33:1198–1202. 2007.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Kim YS, Min KS, Jeong DH, Jang JH, Kim HW
and Kim EC: Effects of fibroblast growth factor-2 on the expression
and regulation of chemokines in human dental pulp cells. J Endod.
36:1824–1830. 2010.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Sagomonyants K, Kalajzic I, Maye P and
Mina M: FGF signaling prevents the terminal differentiation of
odontoblasts. J Dent Res. 96:663–670. 2017.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Sagomonyants K, Kalajzic I, Maye P and
Mina M: Enhanced dentinogenesis of pulp progenitors by early
exposure to FGF2. J Dent Res. 94:1582–1590. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Coffin JD, Florkiewicz RZ, Neumann J,
Mort-Hopkins T, Dorn GW II, Lightfoot P, German R, Howles PN, Kier
A and O'Toole BA: Abnormal bone growth and selective translational
regulation in basic fibroblast growth factor (FGF-2) transgenic
mice. Mol Biol Cell. 6:1861–1873. 1995.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Tang MM, Lin WJ, Zhang JT, Zhao YW and Li
YC: Exogenous FGF2 reverses depressive-like behaviors and restores
the suppressed FGF2-ERK1/2 signaling and the impaired hippocampal
neurogenesis induced by neuroinflammation. Brain Behav Immun.
66:322–331. 2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Pan X, Xu S, Zhou Z, Wang F, Mao L, Li H,
Wu C, Wang J, Huang Y, Li D, et al: Fibroblast growth factor-2
alleviates the capillary leakage and inflammation in sepsis. Mol
Med. 26(108)2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Garmy-Susini B, Delmas E, Gourdy P, Zhou
M, Bossard C, Bugler B, Bayard F, Krust A, Prats AC, Doetschman T,
et al: Role of fibroblast growth factor-2 isoforms in the effect of
estradiol on endothelial cell migration and proliferation. Circ
Res. 94:1301–1309. 2004.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Presta M, Andrés G, Leali D, Dell'Era P
and Ronca R: Inflammatory cells and chemokines sustain FGF2-induced
angiogenesis. Eur Cytokine Netw. 20:39–50. 2009.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Gronthos S, Brahim J, Li W, Fisher LW,
Cherman N, Boyde A, DenBesten P, Robey PG and Shi S: Stem cell
properties of human dental pulp stem cells. J Dent Res. 81:531–535.
2002.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Vaseenon S, Chattipakorn N and
Chattipakorn SC: The possible role of basic fibroblast growth
factor in dental pulp. Arch Oral Biol. 109(104574)2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Büttner R, Schulz A, Reuter M, Akula AK,
Mindos T, Carlstedt A, Riecken LB, Baader SL, Bauer R and Morrison
H: Inflammaging impairs peripheral nerve maintenance and
regeneration. Aging Cell. 17(e12833)2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Gauvreau GM, Boulet LP, Cockcroft DW,
Baatjes A, Cote J, Deschesnes F, Davis B, Strinich T, Howie K,
Duong M, et al: Antisense therapy against CCR3 and the common beta
chain attenuates allergen-induced eosinophilic responses. Am J
Respir Crit Care Med. 177:952–958. 2008.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Pease JE and Williams TJ: Tipping the
balance: A biased nanobody antagonist of CCR3 with potential for
the treatment of eosinophilic inflammation. J Allergy Clin Immunol.
143:552–553. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Hayashi Y, Kawamura R, Nishimatsu SI,
Fukuta O and Nakashima M: Stem cell-induced pulp regeneration can
be enhanced by administration of CCL11-neutralizing antibody in the
ectopic tooth transplantation model in the aged mice. Rejuvenation
Res. 22:51–59. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Mills CD, Kincaid K, Alt JM, Heilman MJ
and Hill AM: M-1/M-2 macrophages and the Th1/Th2 paradigm. J
Immunol. 164:6166–6173. 2000.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Abd-Elmeguid A, Abdeldayem M, Kline LW,
Moqbel R, Vliagoftis H and Yu DC: Osteocalcin expression in pulp
inflammation. J Endod. 39:865–872. 2013.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Bikfalvi A, Klein S, Pintucci G and Rifkin
DB: Biological roles of fibroblast growth factor-2. Endocr Rev.
18:26–45. 1997.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Powers CJ, McLeskey SW and Wellstein A:
Fibroblast growth factors, their receptors and signaling. Endocr
Relat Cancer. 7:165–197. 2000.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Kunath T, Saba-El-Leil MK, Almousailleakh
M, Wray J, Meloche S and Smith A: FGF stimulation of the Erk1/2
signalling cascade triggers transition of pluripotent embryonic
stem cells from self-renewal to lineage commitment. Development.
134:2895–2902. 2007.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Haghighi F, Dahlmann J, Nakhaei-Rad S,
Lang A, Kutschka I, Zenker M, Kensah G, Piekorz RP and Ahmadian MR:
bFGF-mediated pluripotency maintenance in human induced pluripotent
stem cells is associated with NRAS-MAPK signaling. Cell Commun
Signal. 16(96)2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Ma Y, Kakudo N, Morimoto N, Lai F,
Taketani S and Kusumoto K: Fibroblast growth factor-2 stimulates
proliferation of human adipose-derived stem cells via Src
activation. Stem Cell Res Ther. 10(350)2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Franceschi RT and Ge C: Control of the
Osteoblast Lineage by Mitogen-Activated Protein Kinase Signaling.
Curr Mol Biol Rep. 3:122–132. 2017.PubMed/NCBI View Article : Google Scholar
|