Introduction
Gastric cancer (GC), one of the most common fatal
types of cancer, was the 5th most regularly diagnosed tumor and the
3rd most common cause of tumor-related death worldwide in
2018(1). Advances have been made in
the diagnosis and treatment of GC; however, the prognosis of
patients with GC remains poor, with >70% of patients eventually
succumbing to the disease (2).
Therefore, early diagnosis and treatment is crucial for improving
prognosis (3). GC is the final
result of long-term biological processes, including the gradual
accumulation of genotypical and phenotypical changes (4). Precancerous lesions, pathologically
defined as dysplasia, have been associated with the development of
GC. Dysplasia can be further classified as low- or high-grade
intraepithelial neoplasia (HGIN) (5). HGIN is equivalent to severe allogeneic
hyperplasia and carcinoma in situ (4). A previous study reported that 59.1% of
patients with gastric high-grade intraepithelial neoplasia (GHGIN)
progress to gastric adenocarcinoma within one year after endoscopy
(6). Thus, early detection of
precancerous lesions, particularly GHGIN, is important for the
prevention of GC and reducing its associated mortality rate.
Currently, endoscopy followed by pathological examination is the
most reliable diagnostic tool for GHGIN; however, it is invasive
and uncomfortable. Therefore, non-invasive, cost-effective and
highly sensitive biomarkers are needed to diagnose GHGIN.
Some studies have explored the role of exosomes in
local and systemic intercellular communication during cancer
progression (7,8), and some studies suggest the potential
application of exosomes in cancer screening and diagnosis (9,10).
Exosomes are small extracellular vesicles (30-100 nm in diameter),
within a lipid bilayer membrane that encompasses cytoplasm without
organelles, and are highly heterogeneous, possibly reflecting the
phenotypic state of the cells that produced them (11). Exosomes can transfer bioactive
substances, such as proteins, coding RNAs, non-coding (nc)RNAs and
DNA from donor to recipient cells, resulting in the differentiation
of genetic and epigenetic factors and the reprogramming of target
cells (11,12). Some exosomes from cancer cells, such
as glioma, gastric cancer and breast cancer reportedly promote
angiogenesis, regulate the immune system and remodel the
microenvironment, all of which are factors that facilitate cancer
progression (13,14). In addition, exosomes are present in
almost all body fluids, which reinforces their potential
application as non-invasive biomarkers for the diagnosis of various
types of cancer (15).
Long non-coding (lnc)RNAs are transcripts >200
nucleotides in length, that have no or limited protein-coding
capacity; however, they play a crucial role in genetic regulation
at the epigenetic, transcriptional and post-transcriptional levels
(16). In addition to mutations or
abnormal expression levels of protein-coding genes, mutations and
mis-regulation of ncRNAs, particularly lncRNAs, appear to play
noteworthy roles in cancer, such as prostate cancer, stomach cancer
and lung cancer (17). Increasing
evidence indicated that lncRNAs were associated with the initiation
and development of tumors, such as non-small cell lung cancer and
gastric cancer, directly or indirectly via gene expression
(18,19). Furthermore, lncRNAs are selectively
distributed to exosomes and used as signal messengers for
intercellular communication (20).
Thus, exosomal lncRNAs can reshape the tumor microenvironment,
thereby promoting tumor development, metastasis, angiogenesis and
chemoresistance (15). In addition,
lncRNAs in exosomes are not affected by ribonuclease-mediated
degradation and are stable in body fluids (21). Abnormal lncRNAs have been found in
different types of tumors, suggesting their potential as molecular
markers (22-24).
The serum level of LINC00310 is increased in patients with breast
cancer compared with healthy controls, with a sensitivity and
specificity of 77.08 and 87.23%, respectively. These levels are
valuable for breast cancer diagnosis (25). Colorectal cancer-associated HOTAIR
also exhibited oncogenic properties. High expression levels of
HOTAIR in tumour tissues was closely associated with vascular
invasion and metastasis. Patients with high expression levels of
HOTAIR were more likely to have a poor prognosis (26). Therefore, lncRNAs have attracted
increasing attention in the field of tumor-derived exosome
research.
Some studies have revealed that plasma exosome
lncRNAs are potential biomarkers for GC (23,24).
However, the expression profile of lncRNAs in circulating exosomes
in patients with GHGIN remains to be elucidated. Therefore, the
current study aimed to discover new lncRNA biomarkers for GHGIN,
and investigate the carcinogenic mechanism of GHGIN and GC. The
expression profiles of circulating exosomal lncRNAs in patients
with GHGIN and healthy controls were compared using high-throughput
sequencing to identify differentially expressed (DE) lncRNAs. In
addition, the potential roles of lncRNAs and their predicted
targets were elucidated by constructing a lncRNA-micro(mi)RNA-mRNA
interaction network. The study findings suggested sensitive and
specific non-invasive diagnostic markers for GHGIN.
Materials and methods
Patients and sample collection
In the present study, 5 patients diagnosed with
GHGIN and free of any other type of cancer were recruited at The
First Hospital of Jilin University from February to June 2019. The
inclusion and exclusion criteria was as follows: i) All enrolled
patients were pathologically confirmed as GHGIN, and all
pathological diagnoses were reviewed by pathologists in our
hospital; ii) Patients exhibited no history of other tumors or
other serious underlying diseases; iii) Patients did not receive
any preoperative treatment; and iv) All samples were collected with
informed consent of patients and healthy volunteers, and approved
by the Ethics Committee of the First Hospital of Jilin University.
All the patients underwent endoscopic submucosal dissection for
treatment. All specimens were diagnosed as GHGIN by two independent
professional pathologists according to the World Health
Organization Classification of Tumors of the Digestive System
(27). A total of 5 healthy donors,
without a history of precancerous lesions or cancer, were included
in the control group, the healthy control (HC). Table I and Fig. S1 shows the clinical features and
the pathological examination images of the study participants,
respectively. Subsequently, ~10 ml of peripheral blood was
collected in an EDTA anticoagulation tube. The plasma was
immediately separated by centrifugation (1,900 x g; 10 min) at 4˚C,
then the supernatant (plasma) was collected and centrifuged again
(3,000 x g; 15 min) at 4˚C and stored as soon as possible at -80˚C
until use. The study protocol was approved by The Ethics Committee
of the First Hospital of Jilin University, and all the procedures
were performed in accordance with The Declaration of Helsinki. All
participants provided written informed consent.
 | Table IClinical features of the study
participants. |
Table I
Clinical features of the study
participants.
| Sample group | Sample ID | Age | Sex | Biopsy site |
|---|
| GHGIN | GHGIN 1 | 59 | Male | Antrum of
stomach |
| | GHGIN 2 | 68 | Female | Antrum of
stomach |
| | GHGIN 3 | 66 | Female | Angle of
stomach |
| | GHGIN 4 | 62 | Female | Antrum of
stomach |
| | GHGIN 5 | 69 | Male | Body of
stomach |
| HC | HC1 | 35 | Male | Antrum of
stomach |
| | HC2 | 27 | Male | Antrum of
stomach |
| | HC3 | 32 | Male | Antrum of
stomach |
| | HC4 | 28 | Male | Body of
stomach |
| | HC5 | 30 | Female | Antrum of
stomach |
Exosome isolation
The exosomes were isolated from the plasma samples
using an exoEasy Maxi kit (Qiagen GmbH) according to the
manufacturer's instructions and stored at -80˚C until use. Briefly,
buffer XBP was mixed with the plasma from the patients, then the
plasma/XBP mixture was added into the exoEasy spin column and
centrifuged at 500 x g for 1 min at 4˚C. Buffer XWP was added to
remove the residual buffer from the column. The flow-through was
discarded and transferred to the spin column in a fresh collection
tube, then 400 µl Buffer XE was added to the membrane and
centrifuged at 500 x g for 5 min at 4˚C to collect the exosome.
The exosome suspension was diluted to 0.5 mg/ml in
PBS (28). Then, it was loaded onto
a carbon-coated formvar grid and stained with 2% osmic acid. After
incubation for 10 min at room temperature, excess fluid was blotted
with filter paper and adsorbed exosomes were negatively stained
with 1% phosphotungstic acid for 5 min at room temperature.
Finally, the air-dried exosome-containing grids were observed using
a HT7700 transmission electron microscope (TEM; Hitachi, Ltd.).
The particle size distribution of exosomes was
detected using a nanoparticle tracking analyser (NanoFCM Profession
V1.0; Xiamen Fuliu Biotechnology Co., Ltd.).
Exosome identification
Specific exosomal markers [CD9 and tumor
susceptibility gene 101 (TSG101)] were detected using western blot
analysis. First, the exosomes were isolated from plasma samples
using an exoEasy Maxi kit (Qiagen GmbH). The exosomes were lysed in
standard RIPA buffer (Beyotime Institute of Biotechnology)
supplemented with protease and phosphatase inhibitors (Beyotime
Institute of Biotechnology) and oscillated several times. After the
exosomes were fully lysed, they were centrifuged at 13,800 x g for
5 min at 4˚C and the supernatant was collected. A BCA protein assay
kit (Beyotime Institute of Biotechnology) was used to measure the
protein concentration. A total of 30 µg protein per lane were
separated on 10% acrylamide/bisacrylamide gels and transferred to
PVDF membranes. Membranes were subsequently blocked with 5% skimmed
milk at 4˚C overnight and incubated with primary antibodies at 4˚C
overnight. Following primary incubation, membranes were incubated
with secondary antibodies at room temperature for 1 h. The
following antibodies were used: Rabbit anti-human CD9 (1:1,000;
cat. no. 98327S; Cell Signaling Technology, Inc.), rabbit
anti-human TSG101 (1:1,000; cat. no. ab125011; Abcam) and rabbit
anti-calnexin (1:1,000; cat. no. 2679S; Cell Signaling Technology,
Inc.). Calnexin, an endoplasmic reticulum marker, was used as the
negative control. Proteins were visualized using ChemiSignal™ ECL
Plus Chemical Luminescence agent (cat. no. 1810212Shanghai Qinxiang
Technology Co., Ltd.) using the Tanon 6100 chemiluminescent imaging
system (Tanon Science & Technology Co., Ltd.), according to the
manufacturer's instructions.
RNA isolation and quality control
Total RNA from the exosomes was extracted using
TRIzol® (Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. Briefly, 1 ml TRIzol®
was added to the exosomes collected from 1 ml plasma, then mixed
well and left to incubate for 5 min at room temperature. A total of
0.2 ml 99.8% chloroform (Sigma-Aldrich; Merck KGaA) was added, and
the samples were oscillated for 15 sec, incubated at 15-30˚C for
2-3 min and centrifuged at 12,000 x g for 15 min at 4˚C. The upper
aqueous phase was transferred to a new centrifuge tube and 0.5 ml
99.5% isopropanol (Sigma-Aldrich; Merck KGaA) was added. After
being oscillated, incubated at 15-30˚C for 10 min and centrifuged
at 12,000 x g at 4˚C for 10 min, the supernatant was removed.
Subsequently, 1 ml 75% ethanol was added to the centrifuge tube to
wash the RNA pellet; after shaking, the tube was centrifuged at
7,500 x g for 5 min at 4˚C. The ethanol solution was removed and
the RNA pellet dried in the air for 5-10 min, then RNase-free water
was added, mixed with the RNA pellet by pipetting several times and
incubated at 55-60˚C for 10 min. The RNA solution obtained was
stored at -80˚C. Qualitive and quantitative analyses of the RNA
samples were performed using a NanoDrop ND 1000 spectrophotometer
(Thermo Fisher Scientific, Inc.) at a wavelength of 260 nm.
Denaturing agarose gel (1%) electrophoresis was used to measure RNA
integrity and genomic DNA (gDNA) contamination. When the gDNA was
completely removed, the RNA integrity number was ≥7 and the
OD260/OD280 ratio was 1.8-2.1. RNA purity was
thus verified and the samples were used for further
experiments.
RNA library construction, and lncRNA
and mRNA sequencing
High-throughput RNA sequencing of all the samples
was conducted by Cloud-Seq Biotech, Co., Ltd. First, rRNAs were
eliminated from total RNA using the NEBNext® rRNA
depletion kit (cat. no. E6350S New England Biolabs, Inc.). Then,
the NEBNext® Ultra™ II Directional RNA Library Prep kit
(cat. no. E7760; New England Biolabs, Inc.) was used to construct
RNA libraries from the rRNA-depleted RNAs according to the
manufacturer's instructions. The BioAnalyzer 2100 system (Agilent
Technologies, Inc.) was used to evaluate the quality and quantity
of the libraries. 10 pM libraries were denatured as single-stranded
DNA molecules, captured on Illumina flow cells, amplified in
situ as clusters and finally sequenced for 150 cycles on
Illumina HiSeq Sequencer (Illumina, Inc.) according to the
manufacturer's instructions.
lncRNA sequencing analysis
Cutadapt software v1.9.3 (http://code.google.com/p/cutadapt/) was used to remove
connectors and low-quality reads to obtain high-quality clean
reads. The results were sequenced and compared with the reference
genome (UCSC hg19) using HISAT2 software v2.04 (http://www.ccb.jhu.edu/software/hisat/).
Differential expression profiles of the lncRNAs and mRNAs were
obtained using Cuffdiff software v2.2.1, which is part of the
Cufflinks package (http://cufflinks.cbcb.umd.edu/). Fold-change (FC) and
P-values were calculated based on fragments per kilobase million
mapped reads to screen DE lncRNAs and mRNAs. lncRNAs with logFC
≥2.0 and P≤0.05 in at least one sample were selected for target
gene prediction. Gene Ontology (GO; www.geneontology.org) functional enrichment analysis
(29) and Kyoto Encyclopedia of
Genes and Genomes (KEGG; www.genome.jp/kegg) pathway enrichment analyses
(30) of the candidate genes were
conducted using the Database for Annotation, Visualization and
Integrated Discovery (http://david.abcc.ncifcrf.gov). The top 10 enriched
pathways in the up- and downregulated lncRNAs and mRNAs, according
to the P-value cut-offs, were constructed using OmicShare tools
(www.omicsshare.com/toos/).
Reverse transcription-quantitative
RT-q(PCR)
Total RNA was isolated from exosomes using RNA
extraction buffer (Sigma-Aldrich; Merck KGaA), gDNA was removed and
cDNA was synthesized to form gDNA-free total RNA using the
Hifair® II 1st Strand cDNA Synthesis kit (Shanghai
Yeasen Biotechnology Co., Ltd.), following the manufacturer's
specifications. GAPDH was used as the internal control. The primers
for the 6 randomly selected lncRNAs for RT-qPCR are shown in
Table II. RT-qPCR was performed by
Cloud-Seq Biotech, Co., Ltd. using SYBR® Green Master
Mix (Thermo Fisher Scientific, Inc.) with the QuantStudio 5
Real-Time PCR System (Thermo Fisher Scientific, Inc.), following
the manufacturer's instructions. The program was setup as follows:
A total of 40 cycles at 95˚C for 10 sec and 60˚C for 1 min. lncRNA
expression levels were normalized to that of GAPDH and calculated
using the 2-ΔΔCq method (31). Samples were analyzed in triplicate
for each lncRNA.
 | Table IIRandomly selected lncRNAs for reverse
transcription-quantitative PCR and the primers used. |
Table II
Randomly selected lncRNAs for reverse
transcription-quantitative PCR and the primers used.
| lncRNA ID | Primer
direction | Primer sequence,
5'-3' |
|---|
|
ENST00000608199 | Forward |
CGTTCTGGACCAAGCCTAAA |
| | Reverse |
AGGAATTGGGAGGAGTGCTT |
|
ENST00000442783 | Forward |
TGGGGACAAAATGAGAGTCC |
| | Reverse |
TTACAGGTGTGAGCCACTGC |
|
ENST00000427153 | Forward |
GAGATGCAAAGCCAGGCTAC |
| | Reverse |
TCTCTGCAGCTGATTCTGGA |
|
ENST00000608625 | Forward |
CATCTCCCAGACCTGCTTGT |
| | Reverse |
TCCCCAGCTCCCTTCTTATT |
|
ENST00000518266 | Forward |
TTTGGTCATGTTTTCTTGTGGT |
| | Reverse |
TGGACACGCTGTCAAATTGT |
|
ENST00000523150 | Forward |
CAGCATATGGCCTTTGGACT |
| | Reverse |
TGAGGAGACCGCAGTAAACC |
| GAPDH | Forward |
GGCCTCCAAGGAGTAAGACC |
| | Reverse |
AGGGGAGATTCAGTGTGGTG |
Prediction of lncRNA-miRNA-mRNA
interactions
According to the competing endogenous (ce) RNA
theory (32), lncRNAs and mRNAs
that demonstrated significantly different expression were selected
(P<0.05). Subsequently, based on target lncRNA-miRNA and
mRNA-miRNA information from the MiRcode database v11 (www.mircode.org), in addition to common target miRNA
information (33), a
lncRNA-miRNA-mRNA interaction network was constructed. To further
increase the credibility of the network, only lncRNA-miRNA-mRNA
gene pairs with positive correlations between lncRNA and mRNA were
retained (34). The interaction
network was visualized using Cytoscape v3.8.0 software (35). Using PubMed, mRNA in the interactive
network was searched to determine any potential associations with
gastric cancer.
Statistical analysis
Statistical analysis was conducted using GraphPad
Prism software v5.0 (GraphPad Software, Inc.). The data are
expressed as the mean ± standard deviation (unless otherwise
shown). Unpaired Student's t-test was used to assess differences
between two groups from three independent experiments. P<0.05
was considered to indicate a statistically significant
difference.
Results
Identification of isolated
exosomes
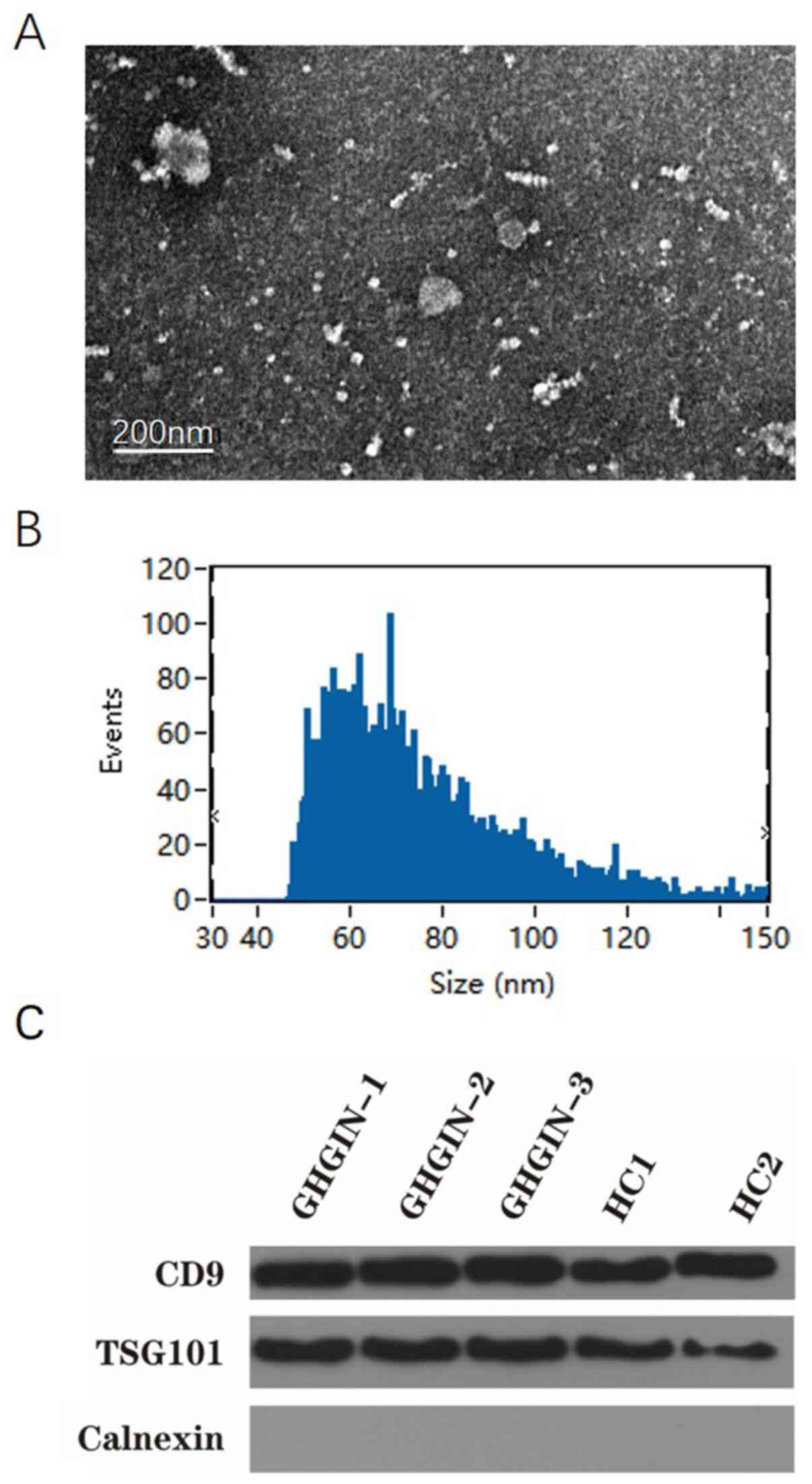
TEM revealed the morphology and size of the
extracted exosomes; they had a cup-shape appearance with a clearly
defined and relatively intact membrane, ranging between 30-100 nm
in size (Fig. 1A and B). The size of the exosomes was similar to
the results obtained using the grain diameter distribution map. The
exosome components, CD9 and TSG101, were detected in all samples
using western blot analysis, thus confirming the presence of
exosomes. However, the endoplasmic reticulum marker calnexin was
not detected, which verified the purity of the exosomes (Fig. 1C).
DE lncRNAs and mRNAs in plasma
exosomes from patients with GHGIN
To determine the expression profiles of lncRNAs in
GHGIN, high-throughput RNA sequencing was used to detect the
expression levels of exosomal lncRNAs and mRNAs in plasma obtained
from patients with GHGIN and HC. The expression levels of lncRNAs
and mRNAs in the plasma exosomes from patients with GHGIN differed
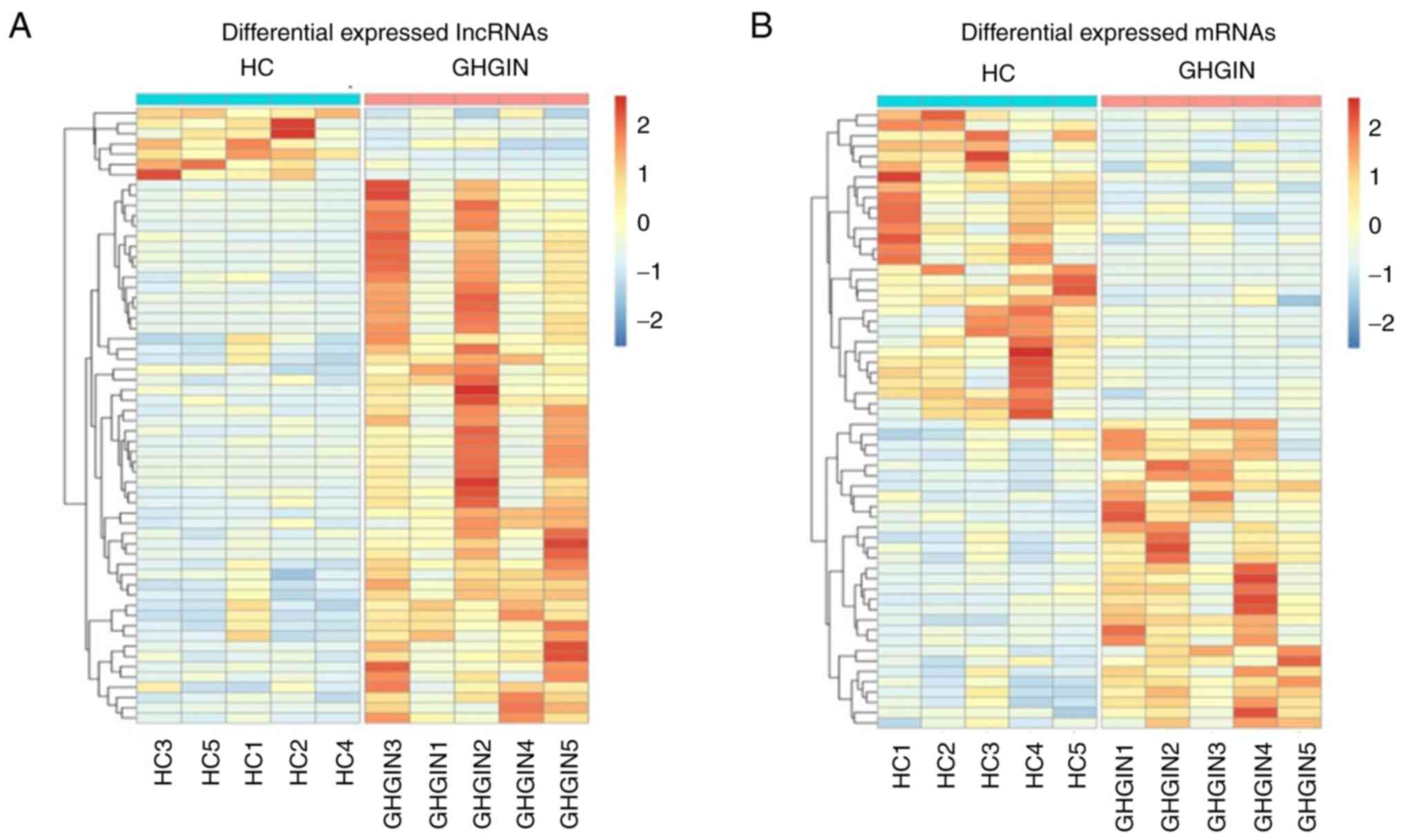
significantly from that in exosomes from HC. Fig. 2A and B show the heat map depicting the
expression levels of all DE lncRNAs and mRNAs, respectively. A
total of 25,145 lncRNAs and 20,254 mRNAs were identified in all the
samples; 83 DE lncRNAs and 233 DE mRNAs were identified (logFC
≥2.0; P≤0.05). Among these, 76 and 7 lncRNAs were up- and
downregulated, respectively. While 183 and 50 mRNAs were up- and
downregulated, respectively. Table
III lists the top 10 up- and downregulated lncRNAs and
mRNAs.
 | Table IIITop 10 up- and downregulated lncRNAs
and mRNAs in patients with gastric high-grade intraepithelial
neoplasia compared with that in healthy controls. |
Table III
Top 10 up- and downregulated lncRNAs
and mRNAs in patients with gastric high-grade intraepithelial
neoplasia compared with that in healthy controls.
| A, Upregulated
lncRNAs |
|---|
| Name | logFC |
|---|
| RP11-290D2.6 | 1499.71 |
| XLOC_009961 | 282.17 |
| IGBP1-AS2 | 217.93 |
| XLOC_014415 | 153.39 |
| CTB-43P18.1 | 145.8 |
| RP11-223C24.1 | 105.82 |
| AC009133.15 | 95.31 |
| RP11-265N6.1 | 87.69 |
| XLOC_000702 | 84.28 |
| RP11-124N14.3 | 70.66 |
| B, Downregulated
lncRNAs |
| Name | logFC |
| JA760600 | -535.73 |
| RP5-1092A3.4 | -141.12 |
| AC007228.9 | -99.88 |
| RP3-470B24.5 | -69.09 |
| RP4-790G17.7 | -67.23 |
| MIR29A | -26.01 |
| RP11-728E14.3 | -22.34 |
| C, Upregulated
mRNAs |
| Name | logFC |
| BEST1 | 1041.40 |
| NRGN | 790.73 |
| FCGR2A | 244.57 |
| HLA-E | 236.44 |
| MTPN | 154.52 |
| RPL35A | 145.38 |
| BCR | 104.00 |
| WDR1 | 100.73 |
| HIST1H2BK | 95.73 |
| NUP88 | 72.43 |
| D, Downregulated
mRNAs |
| Name | logFC |
| MAP1LC3B | -200.72 |
| HINT3 | -104.05 |
| SCAND3 | -99.35 |
| SRRM2 | -32.84 |
| AGBL5 | -30.21 |
| PLEKHM1 | -19.42 |
| CLIP2 | -18.07 |
| CDS2 | -17.22 |
| TNNT1 | -14.028 |
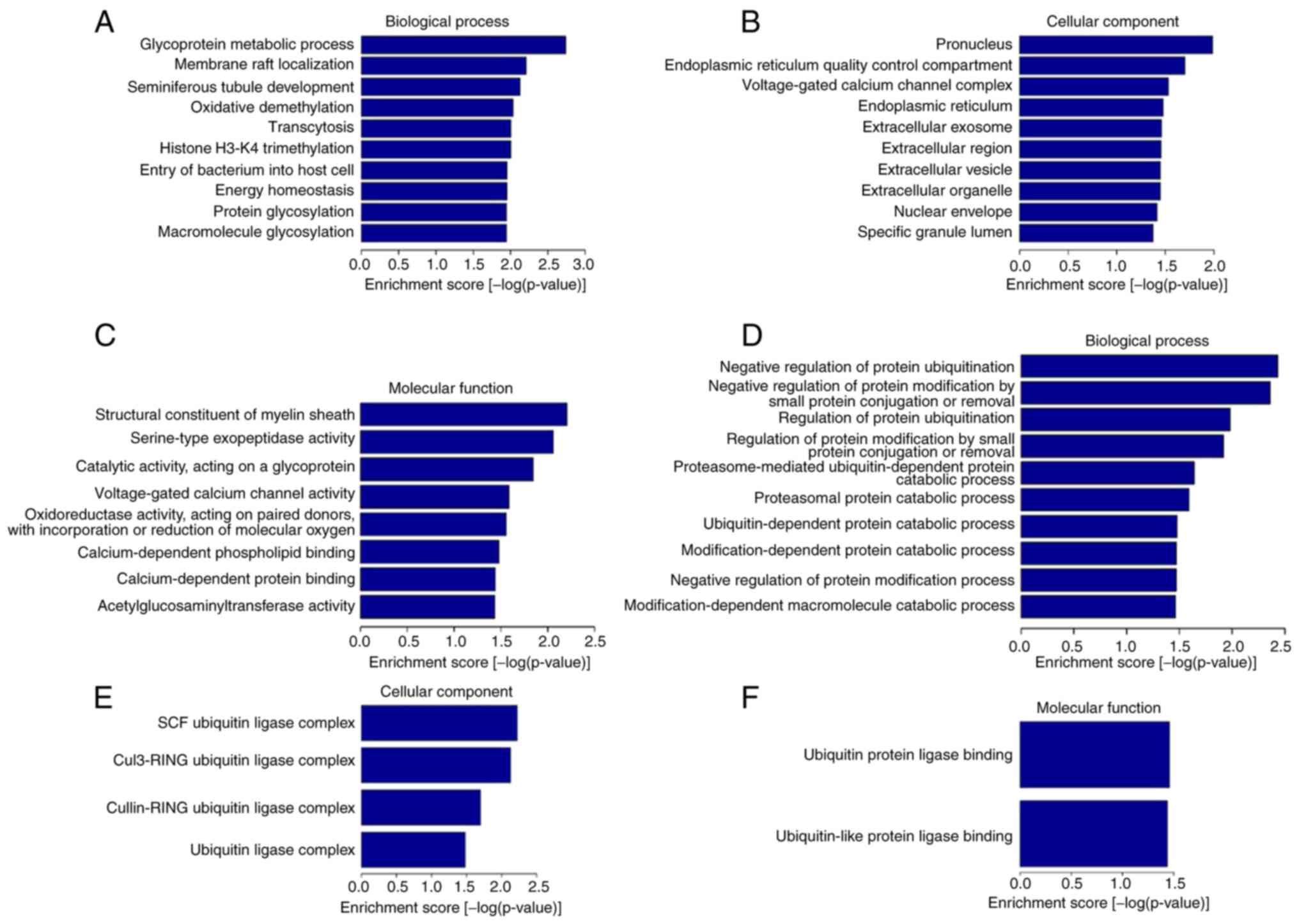
GO and KEGG enrichment of DE
lncRNAs
To identify the biological properties and functions
of the DE lncRNAs, GO and KEGG pathway enrichment analyses were
performed, as shown in Figs. 3A-F
and 4, respectively. GO molecular
function analysis indicated that upregulated DE lncRNAs were
associated with ‘structural constituent of myelin sheath’,
‘serine-type exopeptidase activity’, ‘catalytic activity, acting on
a glycoprotein’, ‘voltage-gated calcium channel activity’, etc.
Whereas GO molecular function analysis indicated downregulated DE
lncRNAs were enriched in ‘ubiquitin protein ligase binding’ and
‘ubiquitin-like protein ligase binding’.
For GO cellular component, the upregulated DE
lncRNAs were significantly enriched in ‘endoplasmic reticulum’,
‘endoplasmic reticulum quality control compartment’, ‘pronucleus’,
‘nuclear envelope’ and the extracellular space (‘extracellular
exosome’, ‘extracellular region’, ‘extracellular vesicle’ and
‘extracellular organelle’). While downregulated DE lncRNAs were
enriched in ‘ubiquitin ligase complex’ (‘SCF ubiquitin ligase
complex’, ‘cul3-RING ubiquitin ligase complex’ and ‘cullin-RING
ubiquitin ligase complex’).
For GO biological process, upregulated DE lncRNAs
were particularly involved in ‘glycoprotein metabolic process’,
‘membrane raft localization’, ‘oxidative demethylation’, ‘histone
H3-K4 trimethylation’, ‘transcytosis’, ‘energy homeostasis’, ‘entry
of bacterium into host cell’ and ‘macromolecule and protein
glycosylation’. Whereas downregulated DE lncRNAs were enriched in
‘negative regulation of protein ubiquitination’, ‘negative
regulation of protein modification by small protein conjugation or
removal’ and ‘proteasome-mediated ubiquitin-dependent protein
catabolic process’, etc.
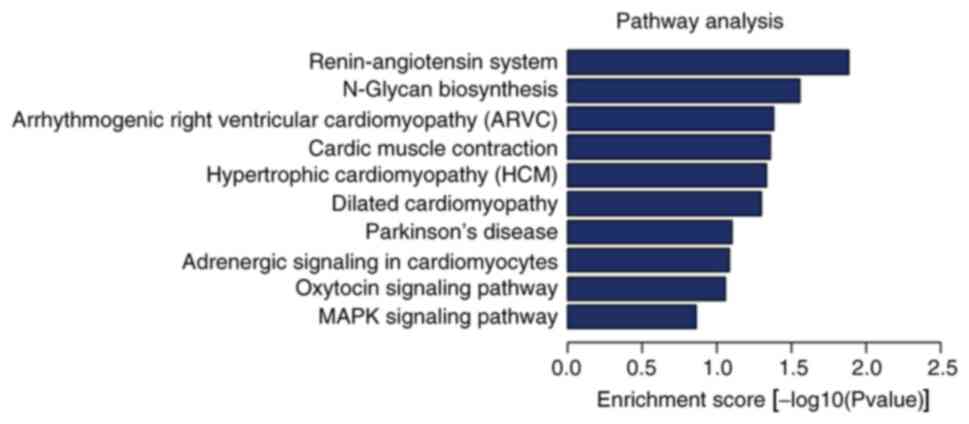
As seen in Fig. 4,
the top 10 enriched KEGG pathways were: ‘Renin-angiotensin system’
(RAS), ‘N-glycan biosynthesis’, ‘arrhythmogenic right ventricular
cardiomyopathy’, ‘cardiac muscle contraction’, ‘hypertrophic
cardiomyopathy’, ‘dilated cardiomyopathy’, ‘Parkinson's disease’,
‘adrenergic signaling in cardiomyocytes’, ‘oxytocin signaling
pathway’ and the ‘mitogen-activated protein kinase (MAPK) signaling
pathway’.
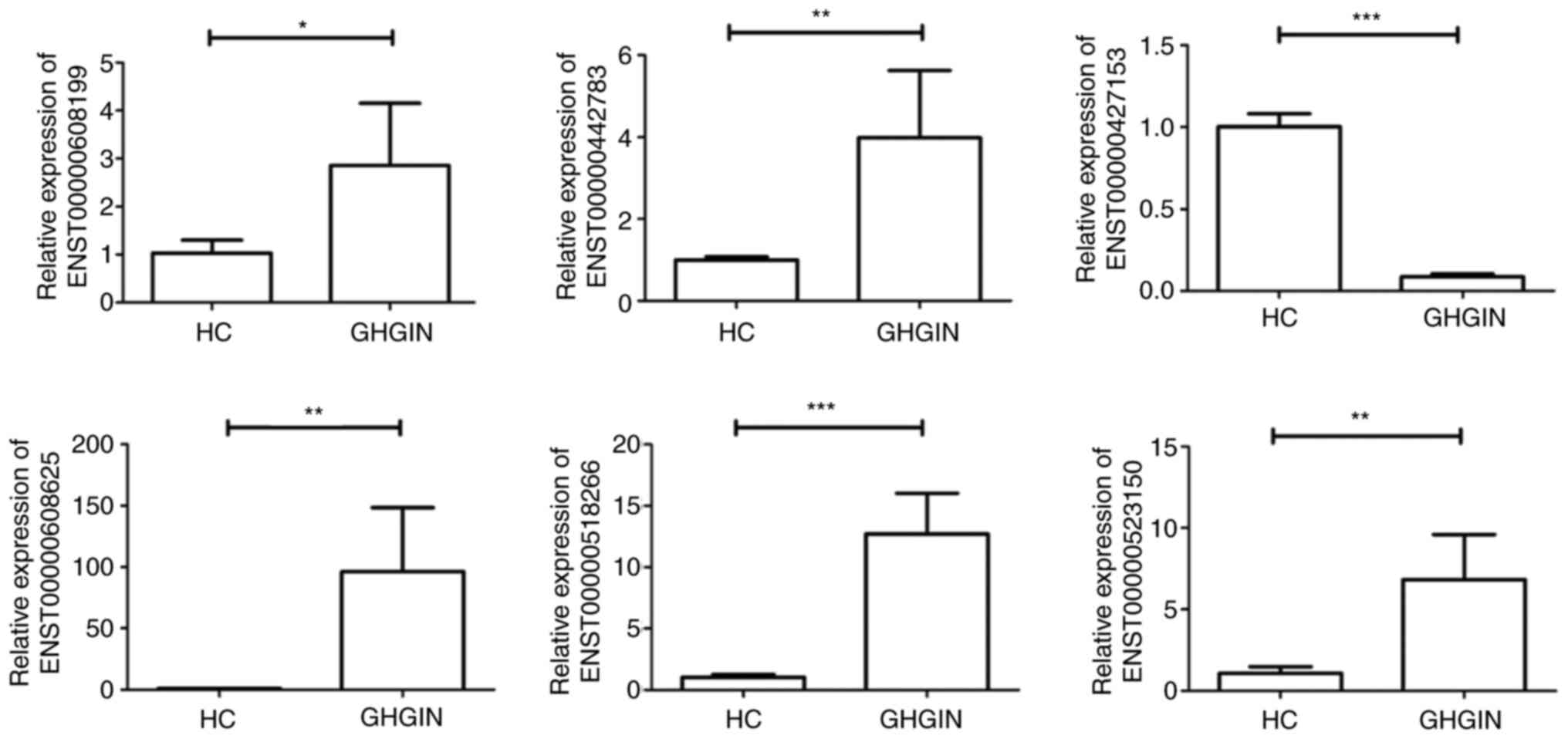
Validation of DE lncRNA expression
levels using RT-qPCR
RT-qPCR was performed to verify differential
expression of 6 lncRNAs. A total of 5 upregulated lncRNAs
(ENST00000608119, ENST00000442783, ENST00000608625, ENST00000518266
and ENST00000523150) and 1 downregulated lncRNA (ENST 00000427153)
were randomly selected for validation. Significant upregulation of
5 lncRNAs was observed in samples from patients with GHGIN compared
with that in the HC samples, while the expression level of
ESTN00000427153 was reduced in patients with GHGIN compared with
that in HC (Fig. 5), thus
confirming that the lncRNA expression profile was a reliable
indicator of GHGIN.
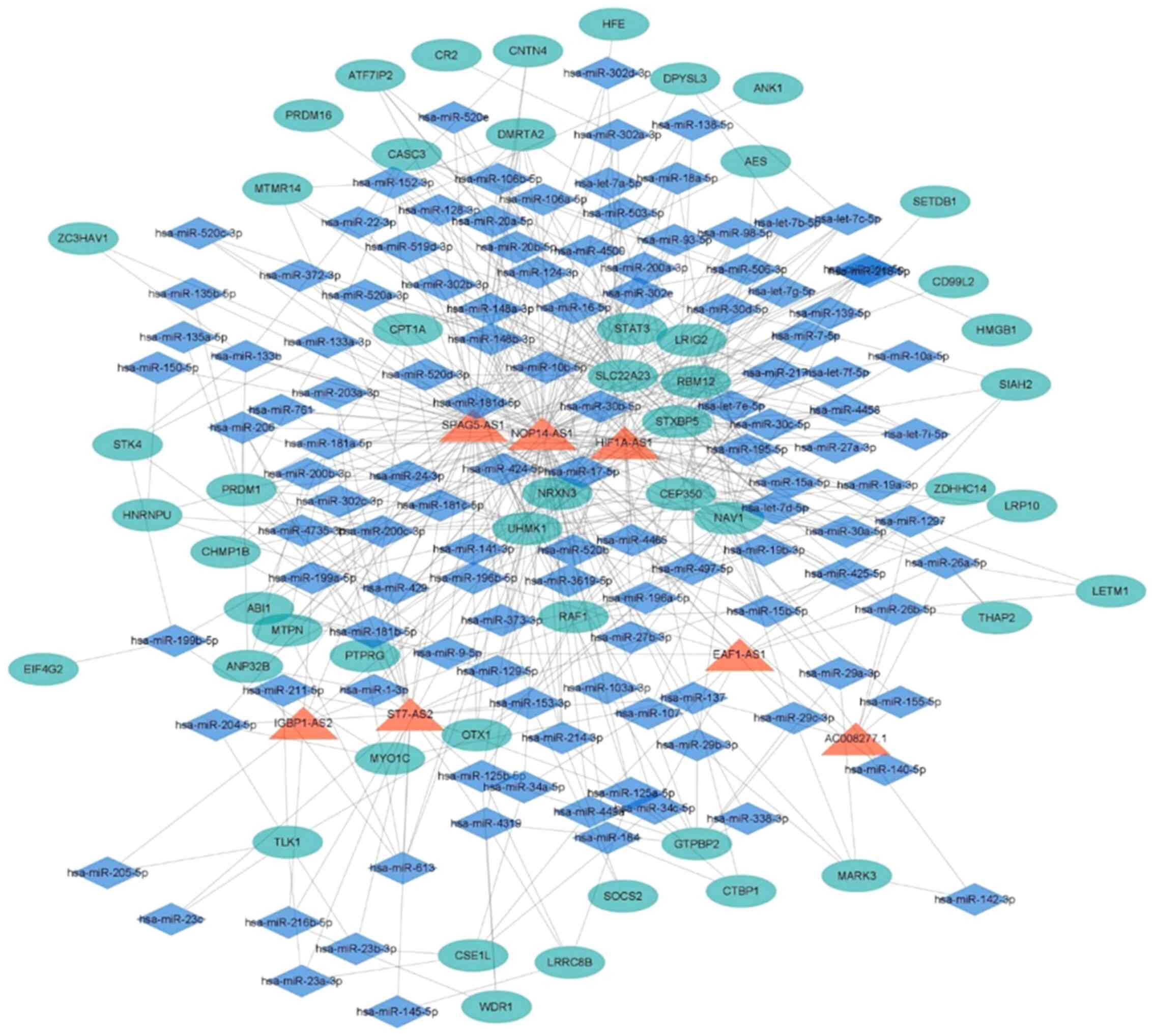
lncRNA-miRNA-mRNA interaction
network
ceRNAs mutually regulate transcripts at the
post-transcriptional level by competing for shared miRNAs (36,37).
The ceRNA network connects the function of protein-encoding mRNAs
to the function of ncRNAs, such as miRNAs, lncRNAs, pseudogenic
RNAs and circular RNAs (37). A
lncRNA-miRNA-mRNA interaction network was constructed in accordance
with the ceRNA theory, as shown in Fig.
6. A lncRNA-miRNA-mRNA interaction network with 585 edges and
178 nodes was finally obtained. A total of 7 lncRNAs, including
HIF1A-AS1, NOP14-AS1, SPAG5-AS1, EAF1-AS1, IGBP1-AS2, ST7-AS2 and
AC008277 were identified. In addition, 121 miRNAs and 50 mRNAs were
identified.
Discussion
The incidence rate of GC is declining; however, it
remains one of the most common and deadly cancer types worldwide,
accounting for over 700,000 deaths every year (1). Thus, early diagnosis and management of
preneoplastic conditions can reduce GC-related mortality (3). Generally, GHGIN is detected using
gastroscopy and biopsy, but procedure-induced discomfort causes
some individuals to avoid the examination (38). In addition, detection of GHGIN
requires technically skilled and experienced operators. These
problems lead to a low rate of detection for GHGIN, emphasizing the
requirement for non-invasive biomarkers. Elucidation of GHGIN
pathogenesis and development of non-invasive, sensitive and
specific biomarkers for GHGIN are required to improve the current
management of GC.
Exosomes are extracellular membranous vesicles
secreted by different types of cells, which are indispensable
promoters of information exchange between cells. More importantly,
exosomes play significant roles in multiple diseases, including
cancer (13). Previous studies have
demonstrated that exosomes are key molecules of cell-cell
communication between cancer and stromal cells in local and distant
microenvironments (39-41).
lncRNAs are endogenous ncRNAs that have been associated with the
occurrence, invasion and metastasis of cancer cells (15). Exosomal lncRNAs serve as messengers
for intercellular communication, reshape the tumor microenvironment
and have been associated with tumor proliferation, angiogenesis,
metastasis, drug resistance and other processes, such as epithelial
to mesenchymal transition and invasion (13). Gastric epithelial dysplasia is
hypothesized to progress through a series of histopathological
stages, from mild to severe dysplasia, to carcinoma in situ
and finally to invasive GC. These histopathological grading changes
are based on changes in the gross chromosome and changes in
expression of protein-coding genes and ncRNAs (42).
The present study identified 83 DE lncRNAs between
patients with GHGIN and HCs, including 76 upregulated and 7
downregulated lncRNAs. These DE lncRNAs, screened using RNA
sequencing analysis, were further verified using RT-qPCR. GO and
KEGG enrichment analyses were performed to predict the potential
functions of these lncRNAs, with respect to GHGIN occurrence, and a
corresponding lncRNA-miRNA-mRNA association network was
constructed. According to GO analysis, the DE lncRNAs were located
in the endoplasmic reticulum and extracellular space, and were
associated with biological processes involved in tumorigenesis,
such as protein and macromolecule glycosylation and regulation of
protein ubiquitination.
Glycosylation is the most common and complex
post-translational modification of cell surface and secreted
proteins. Considering its critical functions and biological
effects, changes in protein glycosylation are fundamental to
tumorigenic transformation (43).
This can decisively stimulate the progress of more malignant
features, such as impaired cell-cell adhesion, enhanced migration
and promotion of lymphatic metastasis (44). Aberrant glycosylation in tumor
biology indicated more aggressive phenotypes in gastric cancer cell
lines (45). The significant role
of glycosylation in cancer is highlighted by the fact that changes
in glycosylation regulate the development and progression of
cancer; therefore, genes and pathways associated with glycosylation
may serve as significant biomarkers, as well as specific
therapeutic targets (46).
The ubiquitin-proteasome system is one of the
noteworthy regulatory mechanisms that participate in protein
stability (47) and cell signal
transduction (48). Proteins are
ubiquitinated by the synergistic effect of ubiquitin-activating
enzymes. The ubiquitin-conjugating enzyme, E2O, is regularly
amplified or mutated in numerous tumors, such as breast, gastric,
kidney and ovarian cancer, and its high expression level was
associated with low survival rates in patients with gastric, lung,
breast and prostate carcinoma (49). Furthermore, the expression of
ubiquitin ligase tripartite motif 59 (TRIM59) was upregulated in
human gastric tumor tissues compared with that in non-tumor
tissues, and its levels have been associated with cancer
development and patient survival period (50). Further mechanism studies showed that
the interaction between TRIM59 and P53 promoted ubiquitination and
degradation, and promoted the progress of GC.
Some signaling pathways enriched in the KEGG
analysis in the current study were also associated with tumor
progression in GC, such as the RAS and MAPK signaling pathway.
Angiotensin-converting enzyme (ACE) is the main regulatory factor
of RAS and genetic polymorphisms of ACE have been associated with
the risk of developing different types of human cancer, such as
esophageal, gallbladder, colorectal and gastric cancer (51). RAS plays a notable role not only in
physiological homeostasis but also in carcinogenesis; it is
involved in numerous aspects of GC progression associated with
Helicobacter pylori infection. Furthermore, RAS inhibitors
reduce GC tumor development, progression and metastasis (52). Another study found that ACE and
angiotensin II receptor type 2 (AT2R) were significantly
upregulated in GC and metastatic tissues, while angiotensin II
receptor type 1 (AT1R) expression was higher in metastatic cancer
tissues compared with that found in previous investigations
(53). This study found evidence of
the local angiotensin II system expression in lymph node
metastasis, and that ACE, AT1R and AT2R activity promoted GC cell
invasion.
The MAPK signaling pathway is widely expressed in
multicellular organisms and plays a key role in various biological
processes, such as cell proliferation, apoptosis, differentiation,
migration and invasion (54). Yang
and Huang (55) demonstated that
MAPK signaling was associated with GC invasion and metastasis
(55). CD97 promoted the
proliferation and invasion of GC cells via the exosome-mediated
MAPK signaling pathway in vitro, and exosomal miRNAs may be
involved in the activation of CD97-related pathways (56). LINC00483 is an oncogenic lncRNA in
GC which was found to activate the MAPK signaling pathway in GC
cells and promoted GC cell proliferation, invasiveness and
metastasis in vitro and in vivo (57). By regulating the key components of
the aforementioned signaling pathways, lncRNAs could control GC
progression.
Previous studies have demonstrated that RNAs
cross-regulate each other with miRNA response elements, known as
the ceRNA hypothesis (32,37). lncRNAs are suggested to play notable
roles in cancer. Several studies have reported that lncRNAs
interact with miRNAs and regulate their expression levels as ceRNAs
(58-60).
Bioinformatics analysis was used to predict whether these lncRNAs
may affect the expression of differential mRNAs by high-throughput
sequencing through the miRNA sponge pathway. The lncRNA-miRNA-mRNA
interaction network constructed in the current study identified 7
lncRNAs, including HIF1A-AS1, NOP14-AS1, SPAG5-AS1, EAF1-AS1,
IGBP1-AS2, ST7-AS2 and AC008277.1. HIF1A-AS1, an oncogene,
reportedly participates in the occurrence and development of
multiple types of cancer, including lung cancer, colorectal cancer
and hepatocellular carcinoma (61-64).
HIF1A-AS1 is elevated in a variety of cancers. In the present
study, results obtained using high-throughput RNA sequencing
demonstrated that HIF1A-AS1 was significantly increased in plasma
exosomes of patients with GHIGN, prompting further study of the
expression and role of this gene in GC. In addition, NOP14-AS1 was
found to be upregulated in lung and liver cancer cells following
exposure to DNA damage (65). Among
the 50 target mRNAs shown in the interaction network, 20 were
associated with the occurrence and progression of GC in PubMed.
Taken together, these findings suggested that the lncRNAs and mRNAs
found in GHGIN plasma exosomes were associated with the occurrence
and progression of GC.
In summary, a series of exosomal DE lncRNAs were
identified from the plasma of patients diagnosed with GHGIN using
high-throughput RNA sequencing and their potential functions were
investigated through bioinformatics analysis. The current study
provides insight into the mechanism of GHGIN and aids in the
development of potential biomarkers for its diagnosis, which will
provide improved diagnosis and treatment of GC in the future.
Supplementary Material
Pathological examination images of 5
patients with GHGIN. GHGIN, gastric high-grade intraepithelial
neoplasia.
Acknowledgements
The authors would like to thank Dr Huanfa Yi for
invaluable advice and guidance on this manuscript for English
language editing.
Funding
The present study was funded by The National Natural Science
Foundation of China (grant nos. 30972610, 81273240, 91742107 and
81570002), National Key Research and Development Program (grant
nos. 2017YFC0910000 and 2017YFD0501300) and Jilin Province Science
and Technology Agency (grant nos. 20200403084SF, JLSWSRCZX2020-009,
20200901025SF, 20190101022JH, 2019J026, 20170622009JC, 2017C021,
2017J039, SXGJXX2017-8, JJKH20180197KJ, DBXM154-2018 and
2018SCZWSZX-015).
Availability of data and materials
The datasets generated in the present study have
been uploaded to NCBI GEO (accession no. GSE153413; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE153413).
Authors' contributions
FH and YJ designed the study and collected samples.
MW, XZ and LD analyzed the data. FH, MR, MZ and QM collected data
and conducted the literature review. FH and MZ drafted the
manuscript. FH, MR and YJ confirm the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The study protocol was approved by The Ethics
Committee of the First Hospital of Jilin University, and all the
procedures were performed in accordance with The Declaration of
Helsinki. All participants provided written informed consent prior
to the procedures.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Allemani C, Weir HK, Carreira H, Harewood
R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A,
et al: Global surveillance of cancer survival 1995-2009: Analysis
of individual data for 25,676,887 patients from 279
population-based registries in 67 countries (CONCORD-2). Lancet.
385:977–1010. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Eusebi LH, Telese A, Marasco G, Bazzoli F
and Zagari RM: Gastric cancer prevention strategies: A global
perspective. J Gastroenterol Hepatol. 35:1495–1502. 2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Fassan M, Baffa R and Kiss A: Advanced
precancerous lesions within the GI tract: The molecular background.
Best Pract Res Clin Gastroenterol. 27:159–169. 2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Sung JK: Diagnosis and management of
gastric dysplasia. Korean J Intern Med. 31:201–209. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Li D, Bautista MC, Jiang SF, Daryani P,
Brackett M, Armstrong MA, Hung YY, Postlethwaite D and Ladabaum U:
Risks and predictors of gastric adenocarcinoma in patients with
gastric intestinal metaplasia and dysplasia: A population-based
study. Am J Gastroenterol. 111:1104–1113. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wortzel I, Dror S, Kenific CM and Lyden D:
Exosome-mediated metastasis: Communication from a distance. Dev
Cell. 49:347–360. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Meldolesi J: Exosomes and ectosomes in
intercellular communication. Curr Biol. 28:R435–R444.
2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Wang J, Liu Y, Sun W, Zhang Q, Gu T and Li
G: Plasma exosomes as novel biomarker for the early diagnosis of
gastric cancer. Cancer Biomark. 21:805–812. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kahroba H, Hejazi MS and Samadi N:
Exosomes: From carcinogenesis and metastasis to diagnosis and
treatment of gastric cancer. Cell Mol Life Sci. 76:1747–1758.
2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kalluri R: The biology and function of
exosomes in cancer. J Clin Invest. 126:1208–1215. 2016.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Abak A, Abhari A and Rahimzadeh S:
Exosomes in cancer: Small vesicular transporters for cancer
progression and metastasis, biomarkers in cancer therapeutics.
PeerJ. 6(e4763)2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Becker A, Thakur BK, Weiss JM, Kim HS,
Peinado H and Lyden D: Extracellular vesicles in cancer:
Cell-to-cell mediators of metastasis. Cancer Cell. 30:836–848.
2016.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Maia J, Caja S, Strano Moraes MC, Couto N
and Costa-Silva B: Exosome-Based Cell-Cell communication in the
tumor microenvironment. Front Cell Dev Biol. 6(18)2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang M, Zhou L, Yu F, Zhang Y, Li P and
Wang K: The functional roles of exosomal long non-coding RNAs in
cancer. Cell Mol Life Sci. 76:2059–2076. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long Noncoding RNAs.
Cell. 172:393–407. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Bhan A, Soleimani M and Mandal SS: Long
Noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Jiang C, Yang Y, Yang Y, Guo L, Huang J,
Liu X, Wu C and Zou J: Long Noncoding RNA (lncRNA) HOTAIR affects
tumorigenesis and metastasis of non-small cell lung cancer by
upregulating miR-613. Oncol Res. 26:725–734. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Gu L, Lu LS, Zhou DL and Liu ZC: UCA1
promotes cell proliferation and invasion of gastric cancer by
targeting CREB1 sponging to miR-590-3p. Cancer Med. 7:1253–1263.
2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Sun Z, Yang S, Zhou Q, Wang G, Song J, Li
Z, Zhang Z, Xu J, Xia K, Chang Y, et al: Emerging role of
exosome-derived long non-coding RNAs in tumor microenvironment. Mol
Cancer. 17(82)2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhou R, Chen KK, Zhang J, Xiao B, Huang Z,
Ju C, Sun J, Zhang F, Lv XB and Huang G: The decade of exosomal
long RNA species: An emerging cancer antagonist. Mol Cancer.
17(75)2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Sarfi M, Abbastabar M and Khalili E: Long
noncoding RNAs biomarker-based cancer assessment. J Cell Physiol.
234:16971–16986. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhao R and Zhang Y, Zhang X, Yang Y, Zheng
X, Li X, Liu Y and Zhang Y: Exosomal long noncoding RNA HOTTIP as
potential novel diagnostic and prognostic biomarker test for
gastric cancer. Mol Cancer. 17(68)2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Dong L, Lin W, Qi P, Xu MD, Wu X, Ni S,
Huang D, Weng WW, Tan C, Sheng W, et al: Circulating long RNAs in
serum extracellular vesicles: Their characterization and potential
application as biomarkers for diagnosis of colorectal cancer.
Cancer Epidemiol Biomarkers Prev. 25:1158–1166. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Li J, Peng W, Du L, Yang Q, Wang C and Mo
YY: The oncogenic potentials and diagnostic significance of long
non-coding RNA LINC00310 in breast cancer. J Cell Mol Med.
22:4486–4495. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Svoboda M, Slyskova J, Schneiderova M,
Makovicky P, Bielik L, Levy M, Lipska L, Hemmelova B, Kala Z,
Protivankova M, et al: HOTAIR long non-coding RNA is a negative
prognostic factor not only in primary tumors, but also in the blood
of colorectal cancer patients. Carcinogenesis. 35:1510–1515.
2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Bosman FT, Carneiro F, Hruban RH and
Theise ND (eds): WHO Classification of Tumours of the Digestive
System. 4th ed. IARC Press, Lyon, 2010.
|
|
28
|
Piao HY, Guo S, Wang Y and Zhang J:
Exosomal long non-coding RNA CEBPA-AS1 inhibits tumor apoptosis and
functions as a non-invasive biomarker for diagnosis of gastric
cancer. Onco Targets Ther. 13:1365–1374. 2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
The Gene Ontology Consortium. Expansion of
the Gene Ontology knowledgebase and resources. Nucleic Acids Res.
45:D331–D338. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361.
2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Jeggari A, Marks DS and Larsson E:
MiRcode: A map of putative microRNA target sites in the long
non-coding transcriptome. Bioinformatics. 28:2062–2063.
2012.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Paci P, Colombo T and Farina L:
Computational analysis identifies a sponge interaction network
between long non-coding RNAs and messenger RNAs in human breast
cancer. BMC Syst Biol. 8(83)2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
36
|
The research progress of ceRNA in the head
and neck carcinoma. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi.
32:634–638. 2018.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
37
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
He CZ and Zhang KH: Serum protein and
genetic tumor markers of gastric carcinoma. Asian Pac J Cancer
Prev. 14:3437–3442. 2013.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Wang S, Su X, Xu M, Xiao X, Li X, Li H,
Keating A and Zhao RC: Exosomes secreted by mesenchymal
stromal/stem cell-derived adipocytes promote breast cancer cell
growth via activation of Hippo signaling pathway. Stem Cell Res
Ther. 10(117)2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Kahlert C, Melo SA, Protopopov A, Tang J,
Seth S, Koch M, Zhang J, Weitz J, Chin L, Futreal A and Kalluri R:
Identification of double-stranded genomic DNA spanning all
chromosomes with mutated KRAS and p53 DNA in the serum exosomes of
patients with pancreatic cancer. J Biol Chem. 289:3869–3875.
2014.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Wang S, Xu M, Li X, Su X, Xiao X, Keating
A and Zhao RC: Exosomes released by hepatocarcinoma cells endow
adipocytes with tumor-promoting properties. J Hematol Oncol.
11(82)2018.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Gibb EA, Enfield KS, Stewart GL, Lonergan
KM, Chari R, Ng RT, Zhang L, MacAulay CE, Rosin MP and Lam WL: Long
non-coding RNAs are expressed in oral mucosa and altered in oral
premalignant lesions. Oral Oncol. 47:1055–1061. 2011.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Stowell SR, Ju T and Cummings RD: Protein
glycosylation in cancer. Annu Rev Pathol. 10:473–510.
2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Rasheduzzaman M, Kulasinghe A, Dolcetti R,
Kenny L, Johnson NW, Kolarich D and Punyadeera C: Protein
glycosylation in head and neck cancers: From diagnosis to
treatment. Biochim Biophys Acta Rev Cancer.
1874(188422)2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Ferreira JA, Magalhães A, Gomes J, Peixoto
A, Gaiteiro C, Fernandes E, Santos LL and Reis CA: Protein
glycosylation in gastric and colorectal cancers: Toward cancer
detection and targeted therapeutics. Cancer Lett. 387:32–45.
2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Pinho SS and Reis CA: Glycosylation in
cancer: Mechanisms and clinical implications. Nat Rev Cancer.
15:540–555. 2015.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Cheng J, North BJ, Zhang T, Dai X, Tao K,
Guo J and Wei W: The emerging roles of protein
homeostasis-governing pathways in Alzheimer's disease. Aging Cell.
17(e12801)2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Gutierrez GJ and Ronai Z: Ubiquitin and
SUMO systems in the regulation of mitotic checkpoints. Trends
Biochem Sci. 31:324–332. 2006.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Ullah K, Zubia E, Narayan M, Yang J and Xu
G: Diverse roles of the E2/E3 hybrid enzyme UBE2O in the regulation
of protein ubiquitination, cellular functions, and disease onset.
FEBS J. 286:2018–2034. 2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Zhou Z, Ji Z, Wang Y, Li J, Cao H, Zhu HH
and Gao WQ: TRIM59 is up-regulated in gastric tumors, promoting
ubiquitination and degradation of p53. Gastroenterology.
147:1043–1054. 2014.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Abdeahad H, Avan A, Khazaei M,
Soleimanpour S, Ferns GA, Fiuji H, Ryzhikov M, Bahrami A and
Hassanian SM: Angiotensin-converting enzyme gene polymorphism and
digestive system cancer risk: A meta-analysis based on 9656
subjects. J Cell Biochem. 120:19388–19395. 2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Sugimoto M, Yamaoka Y, Shirai N and Furuta
T: Role of renin-angiotensin system in gastric oncogenesis. J
Gastroenterol Hepatol. 27:442–451. 2012.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Carl-McGrath S, Ebert MP, Lendeckel U and
Röcken C: Expression of the local angiotensin II system in gastric
cancer may facilitate lymphatic invasion and nodal spread. Cancer
Biol Ther. 6:1218–1226. 2007.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Yang SH, Sharrocks AD and Whitmarsh AJ:
MAP kinase signalling cascades and transcriptional regulation.
Gene. 513:1–13. 2013.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Yang M and Huang CZ: Mitogen-activated
protein kinase signaling pathway and invasion and metastasis of
gastric cancer. World J Gastroenterol. 21:11673–11679.
2015.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Li C, Liu DR, Li GG, Wang HH, Li XW, Zhang
W, Wu YL and Chen L: CD97 promotes gastric cancer cell
proliferation and invasion through exosome-mediated MAPK signaling
pathway. World J Gastroenterol. 21:6215–6228. 2015.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Li D, Yang M, Liao A, Zeng B, Liu D, Yao
Y, Hu G, Chen X, Feng Z, Du Y, et al: Linc00483 as ceRNA regulates
proliferation and apoptosis through activating MAPKs in gastric
cancer. J Cell Mol Med. 22:3875–3886. 2018.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Guo LL, Song CH, Wang P, Dai LP, Zhang JY
and Wang KJ: Competing endogenous RNA networks and gastric cancer.
World J Gastroenterol. 21:11680–11687. 2015.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Tay Y, Kats L, Salmena L, Weiss D, Tan SM,
Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al:
Coding-independent regulation of the tumor suppressor PTEN by
competing endogenous mRNAs. Cell. 147:344–357. 2011.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Wu Y, Liu H, Shi X, Yao Y, Yang W and Song
Y: The long non-coding RNA HNF1A-AS1 regulates proliferation and
metastasis in lung adenocarcinoma. Oncotarget. 6:9160–9172.
2015.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Zhu W, Zhuang P, Song W, Duan S, Xu Q,
Peng M and Zhou J: Knockdown of lncRNA HNF1A-AS1 inhibits oncogenic
phenotypes in colorectal carcinoma. Mol Med Rep. 16:4694–4700.
2017.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Gao J, Cao R and Mu H: Long non-coding RNA
UCA1 may be a novel diagnostic and predictive biomarker in plasma
for early gastric cancer. Int J Clin Exp Pathol. 8:12936–12942.
2015.PubMed/NCBI
|
|
64
|
Hong F, Gao Y, Li Y, Zheng L, Xu F and Li
X: Inhibition of HIF1A-AS1 promoted starvation-induced
hepatocellular carcinoma cell apoptosis by reducing
HIF-1α/mTOR-mediated autophagy. World J Surg Oncol.
18(113)2020.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Goyal A, Fiškin E, Gutschner T,
Polycarpou-Schwarz M, Groß M, Neugebauer J, Gandhi M,
Caudron-Herger M, Benes V and Diederichs S: A cautionary tale of
sense-antisense gene pairs: Independent regulation despite inverse
correlation of expression. Nucleic Acids Res. 45:12496–12508.
2017.PubMed/NCBI View Article : Google Scholar
|