Introduction
Spinal cord injuries (SCI) are defined as acute or
chronic damage to the spinal cord, respectively traumatic or caused
by comorbidities (1). SCI are
irreversible (2) and patients
suffer from several complications (3), including urinary tract infection and
neurogenic bladder (NGB) (4). It
is reported that 46% of NGB were caused by SCI (5). NGB refers to the dysfunction of
bladder secondary to any neurological disease, including SCI
(6). NGB results in elevated
detrusor pressure (7) and
continuous vesicoureteral reflux (VUR), which causes a series of
damage to the upper urinary tract, including ureter dilation,
hydronephrosis, kidney failure and even patients' death (8). Therefore, it is important to identify
VUR early and perform an active and effective intervention to
reduce or delay the occurrence of VUR and prevent upper urinary
tract damage, which may improve the prognosis of patients with NGB
and have a positive effect on improving patients' quality of
life.
Exosomes are small, cell-secreted vesicles ranging
in size from 30-100 nm (9).
Exosomes contain a variety of biomolecules, including proteins,
mRNAs, long non-coding RNAs and microRNAs (10), which not only reflect the
functional state of exosome-derived cells but also affect the
biological function of downstream target cells (11). As promising diagnostic candidates,
exosomes have been evaluated in several types of cancer, including
bladder (12) and renal cancer
(13). Urinary exosomes can
originate from the kidney, ureter, bladder or even prostate.
Therefore, urine exosomal proteins may contain important biological
information of disease pathophysiology (14) and can be used as non-invasive and
convenient potential markers (15). Some exosome-related studies have
been performed on non-cancerous urinary diseases, such as renal
injury (16), IgA nephropathy
(17) and urinary tract infection
(18). However, to the best of our
knowledge, no research has investigated urinary exosome proteins in
patients with NGB after SCI.
Accordingly, the present study was designed to
identify the different protein profiles of urine exosomes between
VUR and non-VUR patients, plus to explore potential biomarkers in
predicting diagnosis and guiding prognosis in patients with
SCI.
Materials and methods
Patients and experimental design
Multicenter patients with SCI were recruited into
the present study from September 2019 to November 2020. A total of
316 patients (age, 5-64, male, 182; female, 134) were initiatively
recruited. After screening, only 60 patients (male, 45; female, 15)
were included in the present study. Patient inclusion screening
criteria were as follows: i) Patients met the ASIA diagnostic
criteria (19) and subsequently
had NGB symptoms; ii) patients were aged over 18 years old
regardless of sex; iii) patients had no significant urinary tract
infections or symptoms of hematuria; and iv) patients had no
serious complications of other organs. The exclusion criteria
included the following: i) Patients with diabetes or hypertension
disease; ii) patients with severe chronic heart and lung disease or
with chronic liver, kidney and urinary system disease before being
investigated; and iii) patients with severe infectious diseases.
According to the urological ultrasound or urography results, the
included 60 patients were divided into either the VUR group or
non-VUR group. The guideline flowchart of the research protocol is
presented in Fig. 1. To illustrate
the differential exosome protein profile and to select promising
predictable biomarkers between VUR and non-VUR patients, a group of
15 VUR patients and 15 non-VUR patients were enrolled in the
preliminary screening study. Subsequently, five samples were
collected from each group for preliminary mass spectrometry
analysis, and then another 10 samples were used for target analysis
by parallel reaction monitoring (PRM). In the validation study, a
total of 25 patients with VUR and 35 non-VUR patients were enrolled
(including the patients in the first screening part).
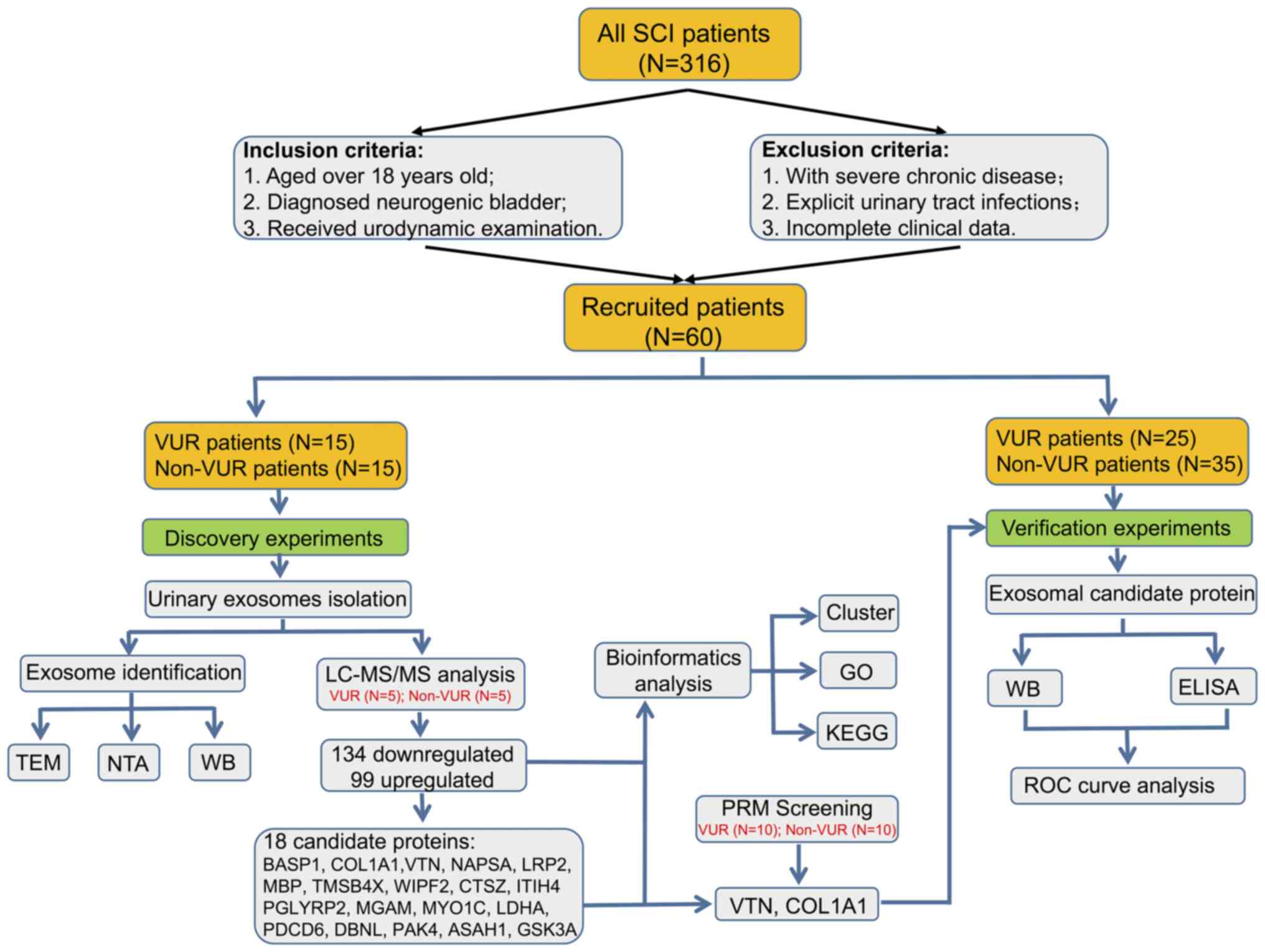 | Figure 1Workflow of the present study design
and brief experimental schedule. SCI, Spinal cord injury; VUR,
vesicoureteral reflux; non-VUR, non-vesicoureteral reflux;
LC-MS/MS, liquid chromatography-mass spectrometry; PRM, Parallel
Reaction Monitoring; TEM, Transmission Electron Microscope; NTA,
Nanoparticle Tracking Analysis; WB, Western blotting; GO, Gene
Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; ROC,
receiver operating characteristic. |
The present study was conducted by Shenzhen Hospital
of Southern Medical University where the study center is located.
The participating hospitals included Shenzhen Hospital of Southern
Medical University (Shenzhen, China), Shenzhen Xiao Chuanguo
Hospital (Shenzhen, China), Shenzhen Longcheng Hospital (Shenzhen,
China), Nanfang Hospital of Southern Medical University (Guangzhou,
China), People's Hospital of Mianzhu, Sichuan Province (Mianzhu,
China) and Bayi Rehabilitation Centre of Sichuan Province (Chengdu,
China). The study protocol was approved by the Scientific Ethics
Committee of Shenzhen Hospital of Southern Medical University
(approval no. NYSZYYEC20180002). All participants provided written
informed consent according to the principles of the Helsinki
Declaration. All data were kept confidential and processed
anonymously.
Exosome isolation and
identification
Urinary exosomes were isolated by serial
centrifugation as previously described (18). Briefly, urine samples were packed
with sterile centrifuge tubes and then transported to the
laboratory by icebox. Urine samples were passed through a 0.22-µm
polyvinylidene difluoride filter and subjected to
ultracentrifugation at 170,000 x g, 4˚C, for 60 min. After washing
in PBS, exosomes were resuspended in cell lysis buffer (containing
150 mM NaCl, 20 mM Tris-HCl pH 7.5 and 1% Triton X-100; cat. no.
P0013; Beyotime Institute of Biotechnology) for immediate further
use or stored at -80˚C. Before being used, the amount of protein
was measured using the Pierce™ Rapid Gold BCA assay kit
(cat. no. A53225; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions.
Transmission electron microscopy (TEM) and
Nanoparticle Tracking Analysis (NTA) were performed. After being
fixed in 4% formaldehyde for 10 min and deposited onto a copper
grid, the exosome samples were transferred into 1% glutaraldehyde
in PBS for 5 min and stained with 2.0% uranyl acetate in aqueous
suspension for 2 min. All the aforementioned procedures were
performed at room temperature unless clearly stated. Finally, the
JEM1400 (JEOL, Ltd.) electron microscope was used at 80 kV to
observing and capture images. For NTA, NanoSight NS3000 with 405 nm
blue laser (Malvern Panalytical, Ltd.) was used. Measurement data
were analyzed using NTA 3.0 analysis software (Malvern Panalytical,
Ltd.).
Tandem mass tag spectrometry analysis
and PRM
Tandem mass tag (TMT) labeling quantitative
proteomics analyses were carried out on an EASY-nLC 1000 UPLC
system (Thermo Fisher Scientific, Inc.). A total of 100 µg of
protein for each sample was digested with trypsin at 1:50
trypsin-to-protein mass ratio for the first digestion overnight
(37˚C) and 1:100 trypsin-to-protein mass ratio for a second 4
h-digestion (37˚C). After trypsin digestion, desalted peptide was
reconstituted in 0.5 M TEAB and processed according to the
manufacturer's protocol for the TMT10plex™ Isobaric
Label Reagent Set kit (cat. no. 90406; Thermo Fisher Scientific,
Inc.). The tryptic peptides were fractionated into fractions by
high-pH reverse-phase high-performance liquid chromatography using
BETASIL™ PREP C18 HPLC Columns (cat. no. 70105-259070A;
Thermo Fisher Scientific, Inc.). The gradient consisted of an
increase from 6 to 23% solvent B (0.1% formic acid in 98%
acetonitrile) over 26 min, 23 to 35% in 8 min and 35 to 80% in 3
min, then holding at 80% for the last 3 min, all at a constant flow
rate of 400 nl/min. The electrospray voltage applied was 2.0 kV.
The m/z scan range was 350 to 1800 for full scan and intact
peptides were detected in the Orbitrap at a resolution of 70,000. A
data-dependent procedure that alternated between one MS scan
followed by 20 MS/MS scans with 15.0s dynamic exclusion. Automatic
gain control was set at 5E4. Fixed first mass was set as 100
m/z.
The resulting peptides were subjected to a
nanoelectrospray ionization source (Nanospray Flex™ Ion
Source; cat. no. ES071; Thermo Fisher Scientific, Inc.) followed by
tandem mass spectrometry (MS/MS) in Q Exactive™ Plus
(Thermo Fisher Scientific, Inc.) coupled online to the UPLC.
Peptides were then selected for MS/MS using NCE setting at 28 and
the fragments were detected in the Orbitrap at a resolution of
17,500. The resulting MS/MS data were processed using Maxquant
search engine v.1.5.2.8 (https://maxquant.net/). The mass tolerance for
precursor ions was set as 20 ppm in the First search and 5 ppm in
the Main search, and the mass tolerance for fragment ions was set
as 0.02 Da. The false discovery rate of peptide identifications was
adjusted to <1% and the minimum score for modified peptides was
set to >40.
According to the TMT results, candidate proteins
containing ≥2 unique peptides were designed for PRM. The unique
peptides were used to identify the target proteins. Enzymatic
digestion is an important step to obtain peptides. However, some
proteins were quantified as containing only one peptide due to the
experimental error in enzyme digestion, which resulted in a lower
abundance of unique peptides. A total of 10 samples were included
in each group. The protein of each sample was enzymatically
hydrolyzed with an equal amount of standard protein, the volume was
adjusted to the same with lytic solution and then dithiothreitol
was added to reduce the final concentration to 5 mM at 56˚C for 30
min. The following procedure on Q Exactive™ Plus was the
same as the TMT as aforementioned. Fragment ion peak areas of the
selected peptides were used for quantitative analysis. The
quantitative data processing and proteomic analysis were processed
using Skyline v.3.6 (http://proteome.gs.washington.edu/software/skyline).
Bioinformatics analysis
In the bioinformatics analysis of TMT results, Gene
Ontology (GO) annotation proteome was derived from the UniProt-GOA
database (http://www.ebi.ac.uk/GOA/). Then
identified proteins were classified into three categories:
biological process, cellular component and molecular function.
Furthermore, Clusters of Orthologous Groups/euKaryotic Ortholog
Groups of proteins (COG/KOG) categories database (version ‘2003
COGs, 2014 update’; https://www.ncbi.nlm.nih.gov/research/cog-project/)
was used to identify the functional annotation of differential
proteins. Pathways were annotated using the Kyoto Encyclopedia of
Genes and Genomes database (Version, ‘Release 92.0, October 1,
2019’; http://www.genome.jp/kegg/). In the
bioinformatics analysis of PRM results, Gene function enrichment
(FunRich) analysis and Gene Ontology analysis of candidate proteins
were performed using the GenCLiP platform 3.0(20).
Western blotting
Typical loading controls such as cytoskeletal
elements (tubulin/actin) or metabolic enzymes (GAPDH) are lacking
in exosome samples (21).
Quantification of total protein is more effective and reliable
compared with typical housekeeping genes as loading controls
(22). Therefore, the present
study used total protein as a loading control by Coomassie G250
(cat. no. ST030; Beyotime Institute of Biotechnology). Exosome
samples were diluted in cell lysis buffer (cat. no. P0013; Beyotime
Institute of Biotechnology) to extract proteins. Protein
concentrations were measured using the BCA method. A total of 5 µg
protein was loaded in each lane, separated using 12% SDS-PAGE and
electrically transferred to PVDF membranes. After blocking for 1 h
at room temperature with 5% free-fat milk diluted with 0.2%
Tween-20 in PBS, the membranes were incubated at 4˚C overnight with
rabbit anti-vitronectin antibody (1:5,000; cat. no. ab45139;
Abcam), rabbit anti-α-1 type I collagen (COL1A1) antibody (1:1,000;
cat. no. 72026; Cell Signaling Technology, Inc.), rabbit anti-Alix
antibody (1:1,000; cat. no. ab88388; Abcam) or rabbit anti-CD63
antibody (1:5,000; cat. no. ab134045; Abcam). Goat anti-rabbit
horseradish peroxidase (HRP)-conjugated secondary antibodies
(1:5,000; cat. no. 7074S; Cell Signaling Technology, Inc.) were
co-incubated for 1 h at room temperature. Signals were detected and
captured in the Bio-Rad ChemiDoc Touch Imaging System (Bio-Rad
Laboratories, Inc.) using Pierce™ ECL Western Blotting
Substrate (Thermo Fisher Scientific, Inc.). Optical density values
were determined using ImageJ v1.53a software (National Institutes
of Health).
ELISA
A modified lysis method was performed as described
previously (23). Notably, the
amount of protein in exosomes resuspended in PBS was measured using
a BCA assay kit (cat. no. ZJ101; Epizyme, Inc.). Subsequently,
similar protein amounts of urinary exosomes were dissolved in a
lysis buffer (cat. no. P0013; Beyotime Institute of Biotechnology).
To detect exosomal vitronectin (VTN) and COL1A1, 10 µg total
protein was used. Human VTN ELISA (cat. no. CSB-E08983h; CUSABIO
Technology LLC) and human COL1A1 ELISA (cat. no. RK01149; ABclonal
Biotech Co., Ltd.) kits were used following the manufacturer's
protocol.
Statistical analysis
Statistical analysis was conducted using SPSS
version 18.0 (SPSS, Inc.) and GraphPad Prism 9 (GraphPad Software,
Inc.). Bioinformatics figures in the LC-MS/MS results were exported
by R studio (v1.3.1093; RStudio, Inc.) (24). The homogeneity tests between the
VUR and non-VUR groups were validated using chi-square, Fisher's
exact or t-tests. Comparison between two groups was made using
paired t-test or Mann-Whitney U test. Receiver operating
characteristic (ROC) curves were used to calculate the overall
diagnostic performance of candidate biomarkers. The data are
presented as means ± SD. All statistical tests were two-tailed, and
P<0.05 was considered to indicate a statistically significant
difference. All the validation experiments were performed in
triplicates.
Results
Socio-demographic and clinical
characteristics
Clinical data from the VUR and non-VUR groups were
summarized in Table I. In total,
25 VUR patients and 35 non-VUR patients were included in the
present study. No significant difference appeared in age, sex
ratio, injury region, AIS level and urination methods between the
two groups (Table I), suggesting
statistical comparability between the two groups. As expected, the
course of diseases, residual bladder urine and intravesical
pressure in the VUR group were significantly higher compared with
the non-VUR group (Table I).
Notably, the bladder safe-capacity (295.20±75.17 ml vs.
342.29±167.12 ml) revealed no significance between the two groups.
In addition, a bladder safe capacity significantly <300 ml in
the VUR group suggests that the effective bladder volume was
shrinking, which is a sign of decreased bladder compliance and
contracture.
 | Table IClinical characteristics between the
VUR and non-VUR groups. |
Table I
Clinical characteristics between the
VUR and non-VUR groups.
| Parameter | VUR (n=25) | Non-VUR (n=35) | X2 or
t-test | P-value |
|---|
| Sex (n, %) | | | 1.120 | 0.290a |
|
Male | 17 | 28 | | |
|
Female | 8 | 7 | | |
| Age, years | 34.64±12.98 | 40.63±12.42 | -1.807 | 0.076 |
| Course of diseases,
days | 540.44±533.04 | 167.09±297.28 | 3.464 | 0.001 |
| Damage level | | | | 0.156b |
|
Cervical
spine | 6 | 12 | | |
|
Thoracic
vertebrae | 8 | 15 | | |
|
Lumbar
spine | 8 | 3 | | |
|
Cauda
equina | 3 | 5 | | |
| AISA level | | | | 0.128b |
|
A | 9 | 20 | | |
|
B | 4 | 3 | | |
|
C | 10 | 6 | | |
|
D | 2 | 6 | | |
| Urination
methods | | | 1.714 | 0.190a |
|
Automatic/Leakage | 7 | 5 | | |
|
Catheterization/Cystostomy | 18 | 30 | | |
| Bladder
compliance | | | |
1.44x10-4b |
|
Normal | 0 | 14 | | |
|
Abnormal | 25 | 21 | | |
| Residual bladder
urine, ml | 139.20±82.86 | 78.54±41.93 | 3.723 |
4.48x10-4 |
| Safe bladder
capacity, ml | 295.20±75.17 | 342.29±167.12 | -1.315 | 0.879 |
| Intravesical
pressure, cm H2O | 95.56±23.53 | 40.46±21.60 | 9.386 |
3.08x10-13 |
The clinical data suggested that the bladder
functional status was significantly different between the VUR and
non-VUR groups, and worse lower urinary tract damage was observed
in patients with VUR.
Urinary exosome characterization and
identification
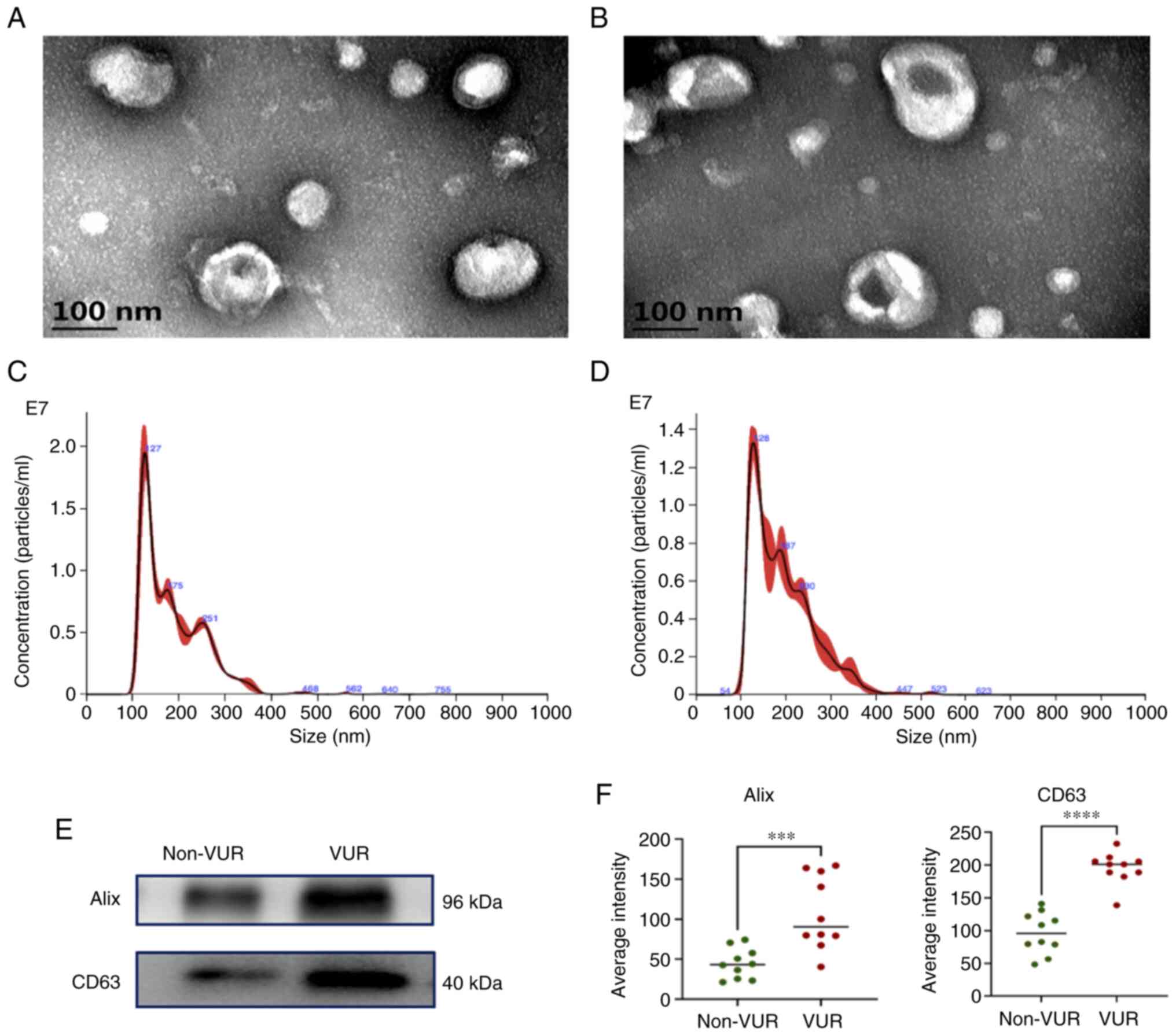
Exosomes were characterized using TEM, NTA and
western blotting. Morphologically, exosomes appeared as
spherical-like structureS surrounded by a layer of membrane-like
material under TEM (Fig. 2A and
B). NTA illustrated that the mean
diameters of exosomes were 106.7±60.0 nm and 186.4±67.4 in the
non-VUR and VUR groups, respectively (Fig. 2C and D). Exosome markers Alix and CD63 were
detected using western blotting, which confirmed that the
effectiveness of exosome sample isolation. Notably, Alix and CD63
excretion were significantly enhanced in VUR patients compared with
non-VUR patients (Fig. 2E and
F).
Briefly, patients with VUR excreted exosomes with
bigger diameters containing more exosome markers, such as Alix and
CD63.
Proteomic profile and bioinformatics
analysis
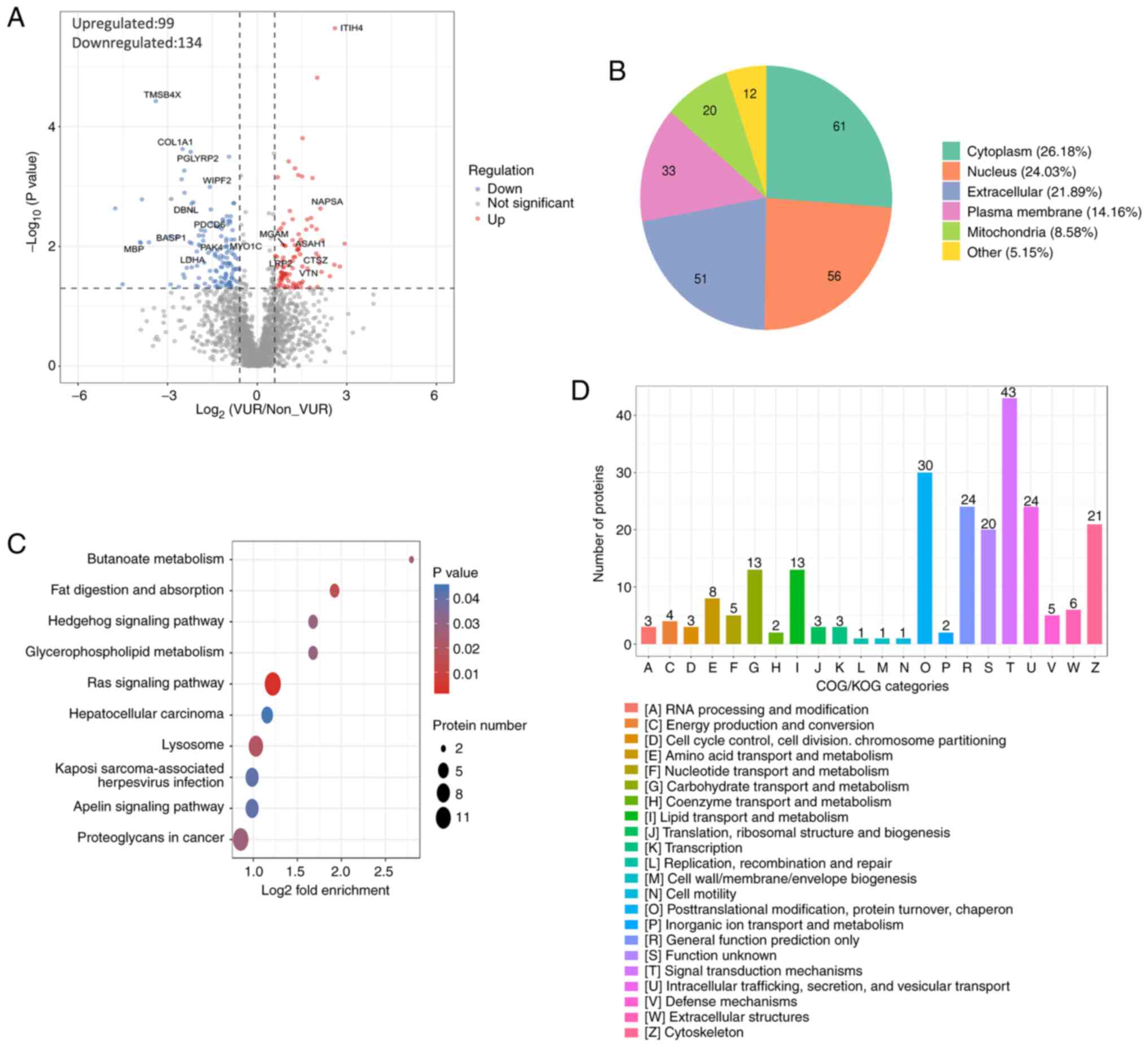
According to the TMT results, a total of 15,208.0
peptide segments were identified, of which the specific peptide
segment was 14,648.0. The present study identified 2,622.0
proteins, of which 2,024.0 were quantifiable (Table SI). EV-specific proteins such as
CD63, CD9 and Alix were expressed in all samples (Table SI). Compared with the non-VUR
group, 134 protein expressions were upregulated and 99 protein
expressions were downregulated in the VUR group with 1.5-fold as
the threshold of differential expression change and P<0.05 as
the significant threshold (Fig.
3A; Table SII). Subcellular
structural localization indicated that 21.89% of differentially
expressed proteins were localized extracellularly and 26.18% were
located in the cytoplasm (Fig.
3B). Kyoto Encyclopedia of Genes and Genomes analysis revealed
these differentially expressed proteins were mainly associated with
the ‘RAS signaling pathway’ and ‘lysosome’ system, which are the
most active pathways (Fig. 3C).
For a deep study, COG/KOG category analysis reported that 43
proteins were associated with the signal transduction mechanisms
(Fig. 3D). In addition, 24
proteins demonstrated a close relationship with intracellular
trafficking, secretion and vesicular transport (Fig. 3D).
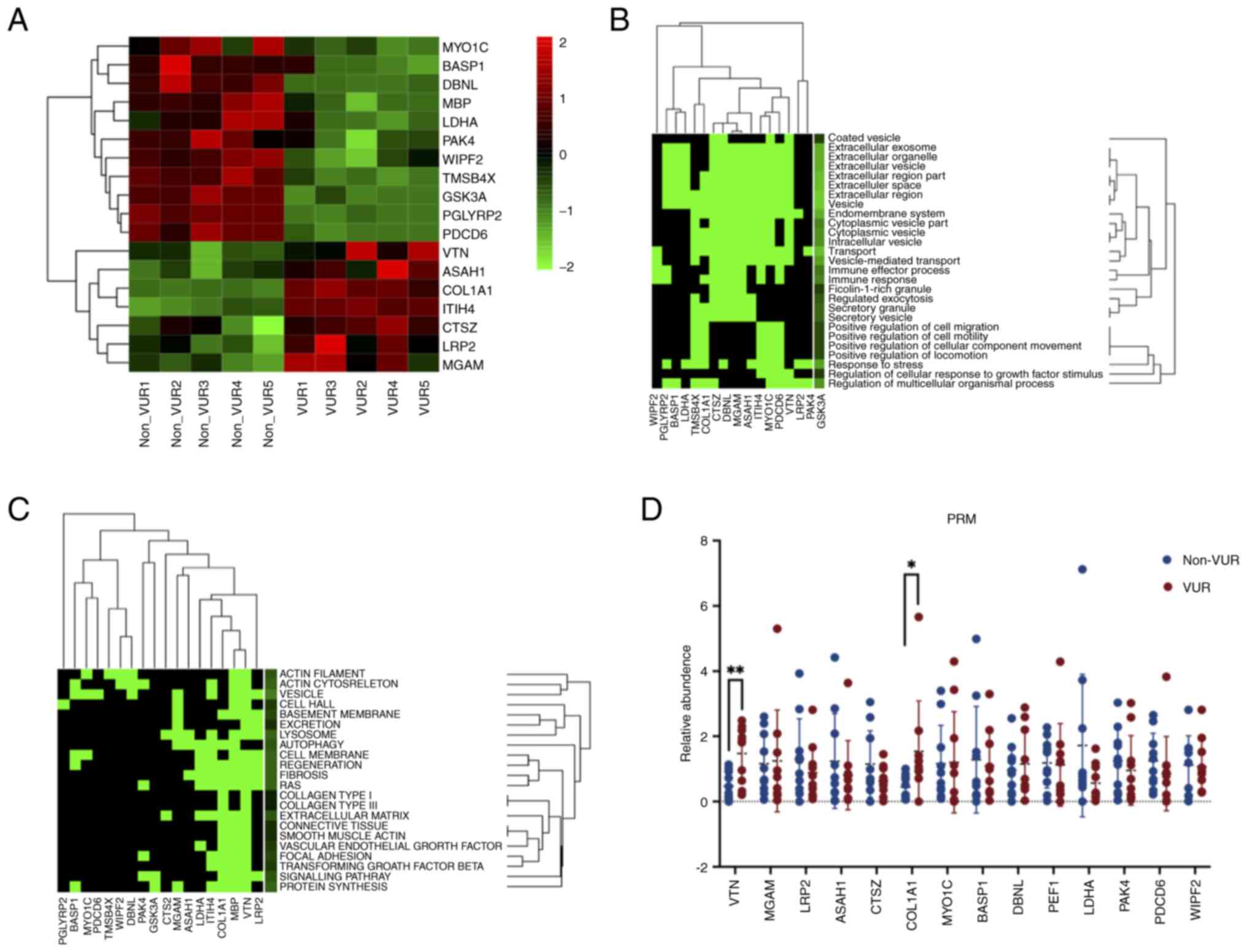
Based on the peak of peptide segments and
bioinformatics analyses aforementioned, 18 proteins were selected
for PRM verification. The molecular location and functions of the
18 candidate proteins were listed in Table II according to the UniProt
database. For example, thymosin β-4 (TMSB4X), WAS/WASL interacting
protein family member 2, unconventional myosin-Ic, drebrin-like
protein and p21-activated kinase 4 (PAK4) were associated with the
cytoskeleton. Other proteins were associated with apoptosis,
regeneration or migration, such as brain acid soluble protein 1
(BASP1), VTN, cathepsin Z, L-lactate dehydrogenase A chain,
programmed cell death protein 6, PAK4, N-acylsphingosine
amidohydrolase 1 and inter-α-trypsin inhibitor heavy chain H4
(ITIH4). Moreover, the present study also revealed that BASP1 and
low density lipoprotein-related protein 2 were directly associated
with renal function, indicating possible abnormalities in renal
function. The expression differences of these 18 candidate proteins
are presented in the heatmap (Fig.
4A). GO enrichment analysis of 18 candidate markers suggested
that the majority of functions were associated with the biological
mechanism of exosomes, such as ‘extracellular exosome’ and
‘vesicle-mediated transport’ (Fig.
4B). The results also revealed that these proteins were
significantly associated with, for example, ‘vesicle’, ‘RAS’ and
‘lysosome’ systems, ‘extracellular matrix’ and ‘signaling pathway’
(Fig. 4C). The present results
were highly consistent with the bioinformatics results of 233
differential proteins in Fig. 3C.
Furthermore, functions including ‘fibrosis’, ‘transforming growth
factor beta’, ‘focal adhesion’ and ‘smooth muscle actin’ also
showed strong associations with the present 18 candidate proteins
(Fig. 4C). Among these proteins,
it was revealed that the aforementioned functional enrichment
results were mainly contributed to by VTN, myelin basic protein
(MBP), COL1A1 and ITIH4 (Fig. 4C).
The present results seemed to indicate that the four candidate
proteins were more likely to predict disease progression. Hence,
the PRM validation experiments were expected to corroborate with
these results.
 | Table IICell localization and biological
function of the 18 candidate biomarkers. |
Table II
Cell localization and biological
function of the 18 candidate biomarkers.
| Name | Location | Molecular
function |
|---|
| BASP1 | Nucleus | Mesenchymal to
epithelial transition, glomerular visceral epithelial cell
differentiation |
| COL1A1 | Extracellular | A member of the
group I collagen |
| VTN | Extracellular | Cell adhesion and
spreading factor |
| LRP2 |
Plasma/membranee | Important for the
functional integrity of the kidney |
| MBP | Nucleus | The most abundant
protein components of the myelin membrane in the CNS |
| TMSB4X | Nucleus | Plays an important
role in the organization of the cytoskeleton and inhibits actin
polymerization |
| WIPF2 | Nucleus | Plays an active
role in the formation of cell surface protrusions and
reorganization of the actin filament system |
| CTSZ | Extracellular | Positive regulation
of neuron apoptotic process, regulation of neuron death |
| PGLYRP2 | Extracellular | Regulation of
inflammatory response, innate immune response |
| MGAM | Golgi
apparatus | An alternate
pathway for starch digestion |
| MYO1C | Cytoplasm | Regulating movement
of intracellular vesicles to the plasma membrane. Links the actin
cytoskeleton to cellular membranes |
| LDHA | Cytoplasm | Positive regulation
of the apoptotic process |
| PDCD6 | Cytoplasm
nucleusus | Plays a key role in
endoplasmic reticulum-Golgi vesicular transport, endosomal
biogenesis or membrane repair. |
| DBNL | Nucleus | Plays a role in the
reorganization of the actin cytoskeleton, formation of cell
projections and in neuron morphogenesis |
| PAK4 | Nucleus | Cytoskeleton
regulation, cell migration, growth, proliferation or cell survival
and stabilization of actin filaments. |
| ASAH1 | Extracellular | Mediate cellular
signaling pathways including cell proliferation, apoptosis and
differentiation |
| GSK3A | Nucleus | Regulation of
transcription factors and microtubules anti-apoptotic function |
| ITIH4 | Extracellular | Involved in
inflammatory responses to trauma and plays a role in
regeneration |
In the PRM validation, only 14 of the 18 proteins
were quantitatively analyzed, while the remainder consisting of
MBP, TMSB4X, peptidoglycan recognition protein 2 and glycogen
synthase kinase-3α were excluded because of failure to be
quantified. PRM analysis revealed that only VTN (P=0.005), and
COL1A1 (P=0.043) had significant differences between the two groups
(Fig. 4D; Table SIII). Therefore, VTN and COL1A1
were selected for further validation.
Collectively, significantly different protein
profiles of urinary exosomes were uncovered between the VUR and
non-VUR groups. Signal transduction-related and vesicle-related
proteins represented the most significant proteins, especially VTN
and COL1A1.
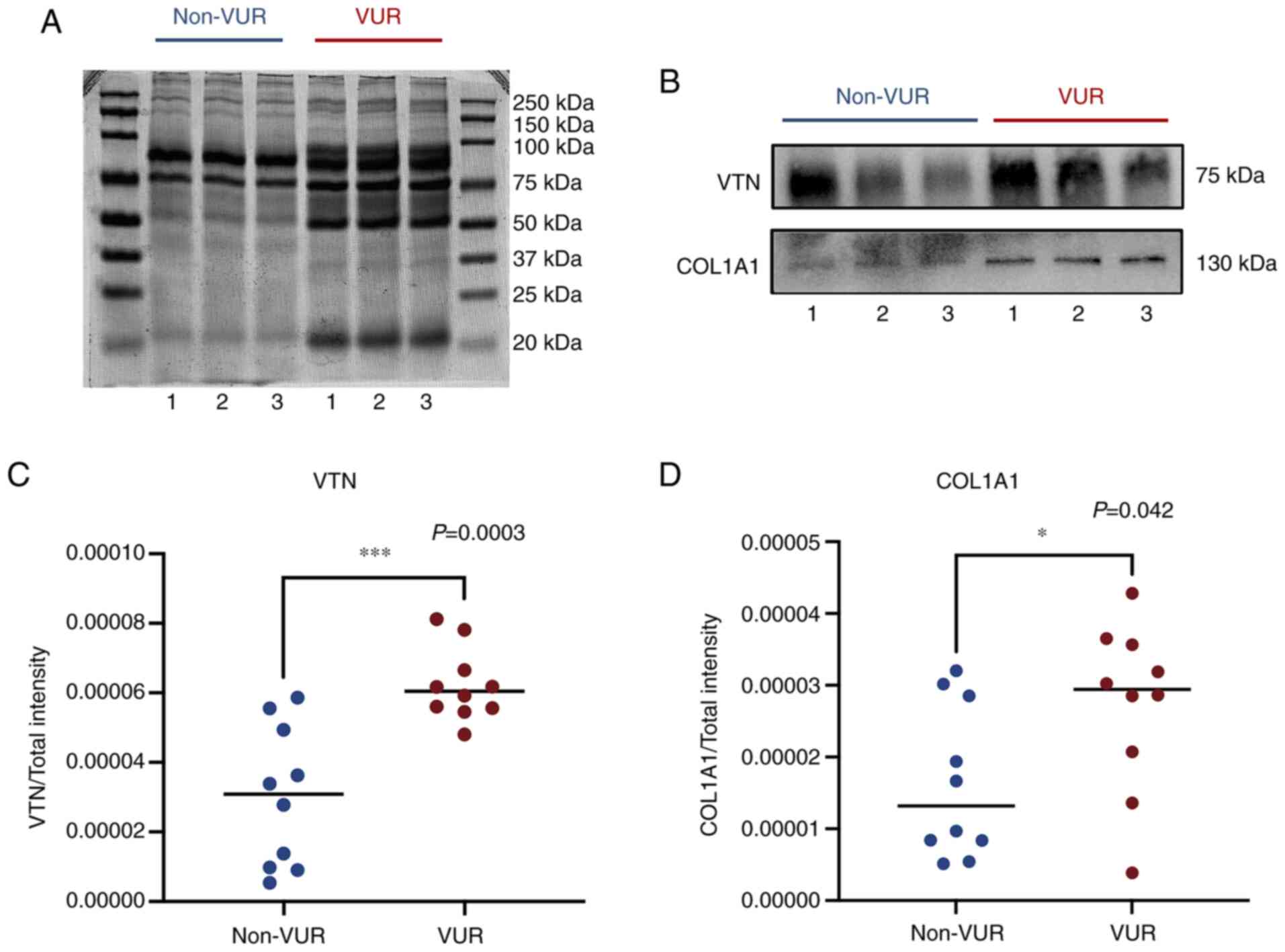
Validation by western blotting
As presented in Fig.
5, exosomal proteins VTN and COL1A1 were both detected using
western blotting despite VTN expression being significantly
stronger compared with COL1A1. Total protein was set as the loading
control to ensure comparability between the two groups (Fig. 5A). The intensity of the bands for
each protein was then compared with the total protein, and the
ratios are presented in Fig. 5C.
Both exosomal VTN (P=0.0003) and COL1A1 (P=0.042) were
significantly higher in VUR patients compared with those in non-VUR
patients (Fig. 5B-D). The
variation tendency of exosomal VTN and COL1A1 between VUR and
non-VUR patients was very similar to that observed by TMT.
The present results suggested that patients with VUR
excreted more VTN and COL1A1 compared with non-VUR patients, and
that VTN and COL1A1 in urinary exosomes may be potential diagnostic
markers for predicting VUR.
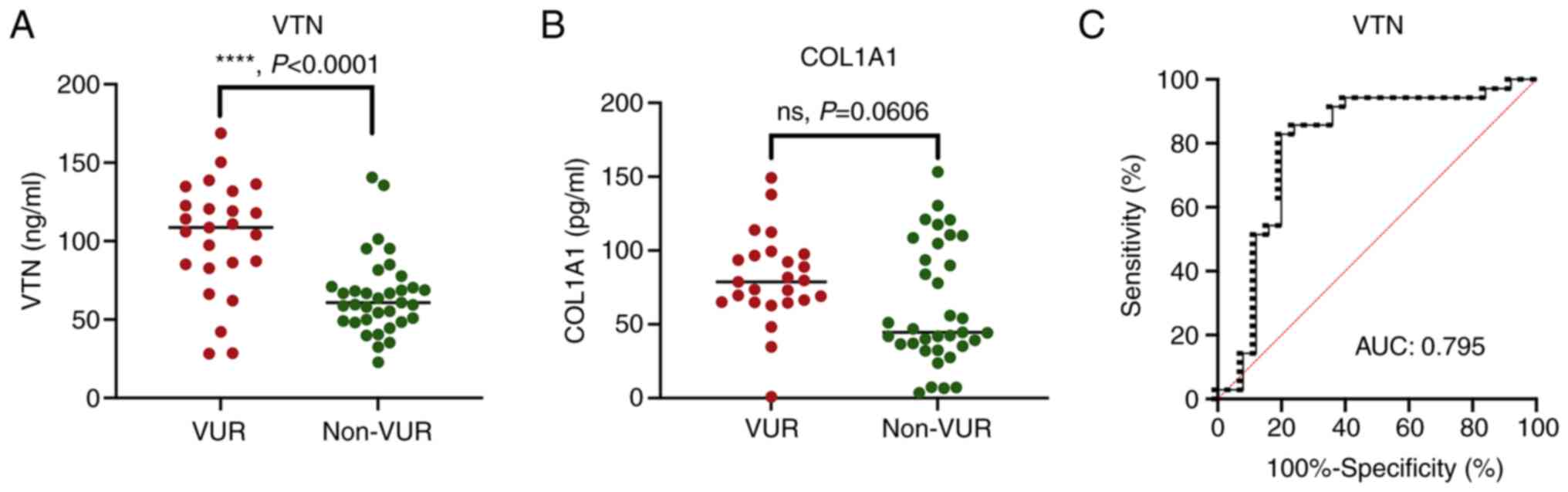
ELISA validation and ROC
ELISA results demonstrated that the VTN
concentrations in patients with VUR were significantly higher
compared with those in the non-VUR patients (P<0.0001; Fig. 6A). However, no significance was
revealed in exosomal COL1A1 between the two groups by ELISA
(P=0.1499; Fig. 6B). It was
suggested that COL1A1 may not be stably expressed in exosomes.
Accordingly, only the ELISA results of the VTN were used to
generate the ROC curve analysis. The ROC curve had an AUC of 0.795
(95% CI, 0.667-0.923) for VTN, with 80% sensitivity at 82.9%
specificity (Fig. 6C).
Overall, the above data demonstrated that urinary
exosomal VTN could distinguish and predict VUR in patients with
NGB.
Discussion
The present study identified the differences in
urine exosomal protein profiles between VUR and non-VUR patients
and revealed that exosomal VTN could be used as a potential marker
in predicting disease progression in patients with NGB. At present,
there are no recognized non-invasive, effective biochemical
indicators to predict urinary reflux (25). The present results demonstrated
that urine exosomal VTN may be an early biomarker for monitoring
VUR.
Decreases in bladder compliance and bladder
remodeling are the main causes of urine reflux (26). Studies have indicated that bladder
overfilling and abnormally high pressure can lead to bladder
structure injury and an increase in bladder wall thickness
(27,28). NGB causes a loss of normal
urination function, resulting in long-term overfilling and
abnormally high pressure of bladder tissue, damaged bladder tissue
structure, compensatory thickening of smooth muscle, increased
deposition of collagen fibers in bladder tissue and eventually
leads to bladder tissue fibrosis (29-32).
Based on these findings, the present study hypothesized that the
increase of urine exosomal VTN may indicate a poor prognosis of
bladder fibrosis and remodeling.
VTN, also known as S-protein, is one of the main
components of the extracellular matrix that participates in cell
spreading (33), cell adhesion
(34) and cell migration (35). VTN is highly associated with
various types of tissue fibrosis (36,37),
including the fibrosis of kidney disease (38). More recently, a study focused on
urine exosomal VTN revealed a positive relationship with renal
fibrosis (39). Urine exosomal VTN
may be secreted from the kidney or the bladder. However, to the
best of our knowledge, no studies have demonstrated that VTN is
associated with bladder fibrosis at present. The present study
firstly proposed that exosomal VTN is highly associated with NGB
fibrosis, instead of renal fibrosis. The main reasons are as
follows: i) Bladder remodeling and fibrosis caused by bladder
dysfunction are the main pathological mechanisms of bladder
compliance decline; ii) the decrease in bladder compliance causes
urine reflux and kidney damage and, as a result, bladder fibrosis
and remodeling should be an earlier warning event of urine reflux.
Consequently, it was hypothesized that elevated VTN may be more
associated with bladder fibrosis compared with renal fibrosis.
Furthermore, VTN mediates tissue fibrosis progression by
upregulating TGF-β1(40).
Activation of TGF-β/Smad signaling pathways appear to be an
important mechanism of tissue fibrosis (41), including in the kidney (42) and bladder (43). Consistent with this theory, the
present study also revealed that fibrosis and TGF-β signaling
pathways were highly activated. However, whether urine exosomal VTN
also promotes fibrosis through the TGF-β/Smad signaling pathway in
NGB requires further investigation.
According to the TMT results, the expression of
COL1A1 was significantly downregulated. However, its expression was
significantly elevated in the VUR group as demonstrated by PRM and
western blotting validation. Moreover, the significant difference
disappeared when ELISA was performed. It was hypothesized that the
possible explanations for these contradictory results mainly
include the three following reasons. First, the different sample
sizes may contribute to those inconsistencies, as only five cases
in each group were used in the mass spectrometry analysis
experiment while 10 samples were used in the PRM validation
experiments. When ELISA was performed among the 60 included
samples, the difference of COL1A1 between groups disappeared.
Therefore, COL1A1 was not selected to be a candidate biomarker.
Accordingly, it was hypothesized that this was directly associated
with sample size and individual differences between samples.
Second, these may be associated with different progression statuses
of diseases. COL1A1 is a part of type I collagen, which is an
important component of the extracellular matrix (ECM). Abnormal ECM
deposition is a major pathological change in fibrosis (44). That is to say that COL1A1 would be
significantly elevated when significant fibrosis exists in the
bladder wall. In the present study, patients were grouped by urine
reflux. The patients with significant fibrosis were included in the
VUR group, which resulted in great individual variability of COL1A1
in the VUR group. Consistent with this interference, the ELISA
results of COL1A1 in the VUR group revealed high variability.
Thirdly, the results of the ELISA validation may be limited by the
sensitivity of the ELISA kits. Accordingly, future studies will
need to increase the sample sizes and develop more cost-efficient
and sensitive kits.
Although COL1A1 was not selected to be an effective
biomarker, it may still be associated with disease progression.
Functional enrichment analysis in the present study revealed that
extracellular matrix-related functions were also significantly
activated. Collagen I and III are the main factors of bladder
compliance (45). Once collagen is
heavily deposited in the bladder wall, elastin becomes relatively
reduced, leading to thicker bladder walls, decreased smooth muscle
function, reduced bladder compliance and even bladder fibrosis
(46). Moreover, the present study
revealed that the RAS pathway was significantly activated. The
RAS-MAPK pathway is an important mechanism for causing
epithelial-mesenchymal transition and has a significant role in the
development of fibrosis (47).
Although the final ELISA results were not satisfactory, COL1A1
still may play a notable role in the decline of bladder compliance
and bladder fibrosis, and may be used as a biological marker to
predict the occurrence of VUR. Following studies should focus on
the relevance of COL1A1 to disease progression.
Several limitations should be noted in the present
study. First, the sample size was limited, and bigger cohort
studies are required to confirm these observations. Secondly,
exosomal COL1A1 revealed no significance in the ELISA validation.
These may be limited by the sensitivity of the ELISA kits.
Therefore, developing a more cost-efficient and sensitive method
for the detection of proteins is important and worthwhile. Third,
exosomal VTN was identified to be an effective biomarker for the
diagnosis of VUR. Whether VTN may be associated with the
progression of bladder remodeling and fibrosis is still an
important factor and this is another limitation of the present
study. According to the aforementioned evidence of VTN and COL1A1
being involved in bladder remodeling, subsequent studies should
further explore the impact of different degrees of damage on the
profile of exosome secretion. Additionally, future studies will
investigate whether urine exosomal VTN is involved in bladder
remodeling and fibrosis via the TGF-β/Smad signaling pathway in
NGB.
Overall, to the best of our knowledge, the present
study proposed for the first time that the increase of urinary
exosomal VTN could be a potential marker in predicting VUR in
patients with NGB. Alterations in these exosomal proteins may
suggest early urinary tract remodeling. The mechanism of VTN
(perhaps including COL1A1) involved in urinary tract remodeling
needs to be verified, and a large-scale prospective study is
needed.
Supplementary Material
Raw data of all the quantifiable
proteins identified by mass spectrometry analysis.
Raw data of the differential proteins
identified by mass spectrometry analysis between the VUR group and
the non-VUR groups.
Raw data of the quantifiable proteins
identified by parallel reaction monitoring validation.
Acknowledgements
The authors would like to thank Professor Xin Li,
chief director of Central Laboratory of Shenzhen Hospital, Southern
Medical University (Shenzhen, China), for providing the rabbit
anti-CD63 antibody.
Funding
Funding: This work was supported by Sanming Project of Medicine
in Shenzhen, China (grant no. SZSM201612018).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WC conceived the initial idea, designed the study
and revised the manuscript. JL wrote the manuscript and performed
the majority of the experiments. SC and CZ collected the urinary
samples and performed some of the experiments. LC, CZ and YH were
responsible for the data analysis and conception of this article.
JL and SC confirmed the authenticity of all the raw data. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Scientific
Ethics Committee of Shenzhen Hospital, Southern Medical University
(approval no. NYSZYYEC20180002; Shenzhen, China). All participants
provided written informed consent according to the principles of
the Helsinki Declaration.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ahuja CS, Wilson JR, Nori S, Kotter MRN,
Druschel C, Curt A and Fehlings MG: Traumatic spinal cord injury.
Nat Rev Dis Primers. 3(17018)2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Cristante AF, Barros Filho TE, Marcon RM,
Letaif OB and Rocha ID: Therapeutic approaches for spinal cord
injury. Clinics (Sao Paulo). 67:1219–1224. 2012.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Furlan JC, Noonan V, Singh A and Fehlings
MG: Assessment of impairment in patients with acute traumatic
spinal cord injury: A systematic review of the literature. J
Neurotrauma. 28:1445–1477. 2011.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Noreau L, Noonan VK, Cobb J, Leblond J and
Dumont FS: Spinal cord injury community survey: A national,
comprehensive study to portray the lives of canadians with spinal
cord injury. Top Spinal Cord Inj Rehabil. 20:249–264.
2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Dave CN, Khalaf A, Patel HD, Kohn TP and
Burnett AL: Neurogenic bladder is an independent risk factor for
complications associated with inflatable penile prosthesis
implantation. Int J Impot Res. 32:520–524. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Li LF, Ka-Kit Leung G and Lui WM: Sacral
nerve stimulation for neurogenic bladder. World Neurosurg.
90:236–243. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Nseyo U and Santiago-Lastra Y: Long-term
complications of the neurogenic bladder. The Urol Clin North Am.
44:355–366. 2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhang T, Liu H, Liu Z and Wang L:
Acupuncture for neurogenic bladder due to spinal cord injury: A
systematic review protocol. BMJ Open. 4(e006249)2014.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Théry C, Zitvogel L and Amigorena S:
Exosomes: Composition, biogenesis and function. Nat Rev Immunol.
2:569–579. 2002.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Dai J, Su Y, Zhong S, Cong L, Liu B, Yang
J, Tao Y, He Z, Chen C and Jiang Y: Exosomes: Key players in cancer
and potential therapeutic strategy. Signal Transduct Target Ther.
5(145)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
He C, Zheng S, Luo Y and Wang B: Exosome
theranostics: Biology and translational medicine. Theranostics.
8:237–255. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Chen C, Luo Y, He W, Zhao Y, Kong Y, Liu
H, Zhong G, Li Y, Li J, Huang J, et al: Exosomal long noncoding RNA
LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin
Invest. 130:404–421. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Cimadamore A, Gasparrini S, Santoni M,
Cheng L, Lopez-Beltran A, Battelli N, Massari F, Giunchi F,
Fiorentino M, Scarpelli M and Montironi R: Biomarkers of
aggressiveness in genitourinary tumors with emphasis on kidney,
bladder, and prostate cancer. Expert Rev Mol Diagn. 18:645–655.
2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Quaglia M, Merlotti G, Guglielmetti G,
Castellano G and Cantaluppi V: Recent advances on biomarkers of
early and late kidney graft dysfunction. Int J Mol Sci.
21(5404)2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Dear JW, Street JM and Bailey MA: Urinary
exosomes: A reservoir for biomarker discovery and potential
mediators of intrarenal signalling. Proteomics. 13:1572–1580.
2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhou H, Pisitkun T, Aponte A, Yuen PS,
Hoffert JD, Yasuda H, Hu X, Chawla L, Shen RF, Knepper MA and Star
RA: Exosomal Fetuin-A identified by proteomics: A novel urinary
biomarker for detecting acute kidney injury. Kidney Int.
70:1847–1857. 2006.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Feng Y, Lv LL, Wu WJ, Li ZL, Chen J, Ni
HF, Zhou LT, Tang TT, Wang FM, Wang B, et al: Urinary exosomes and
exosomal CCL2 mRNA as biomarkers of active histologic injury in IgA
nephropathy. Am J Pathol. 188:2542–2552. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Mizutani K, Kawakami K, Horie K, Fujita Y,
Kameyama K, Kato T, Nakane K, Tsuchiya T, Yasuda M, Masunaga K, et
al: Urinary exosome as a potential biomarker for urinary tract
infection. Cell Microbiol. 21(e13020)2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Roberts TT, Leonard GR and Cepela DJ:
Classifications in brief: American spinal injury association (ASIA)
impairment scale. Clin Orthop Relat Res. 475:1499–1504.
2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Wang JH, Zhao LF, Wang HF, Wen YT, Jiang
KK, Mao XM, Zhou ZY, Yao KT, Geng QS, Guo D and Huang ZX: GenCLiP
3: Mining human genes' functions and regulatory networks from
PubMed based on co-occurrences and natural language processing.
Bioinformatics: Nov 4, 2019 (Epub ahead of print).
|
|
21
|
Jeppesen DK, Fenix AM, Franklin JL,
Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q,
Evans R, et al: Reassessment of exosome composition. Cell.
177:428–445.e18. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Eaton SL, Roche SL, Llavero Hurtado M,
Oldknow KJ, Farquharson C, Gillingwater TH and Wishart TM: Total
protein analysis as a reliable loading control for quantitative
fluorescent western blotting. PLoS One. 8(e72457)2013.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wang L, Skotland T, Berge V, Sandvig K and
Llorente A: Exosomal proteins as prostate cancer biomarkers in
urine: From mass spectrometry discovery to immunoassay-based
validation. Eur J Pharm Sci. 98:80–85. 2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
RStudio Team: RStudio: Integrated
development for R. RStudio, Inc., Boston MA, 2015.
|
|
25
|
Valério FC, Lemos RD, de C Reis AL,
Pimenta LP, Vieira ÉL and Silva ACE: Biomarkers in vesicoureteral
reflux: An overview. Biomark Med. 14:683–696. 2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wu CQ and Franco I: Management of
vesicoureteral reflux in neurogenic bladder. Investig Clin Urol. 58
(Suppl 1):S54–S58. 2017.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ekiz A, Özdemir-Kumral ZN, Erşahin M,
Tuğtepe H, Öğünç AV, Akakın D, Kıran D, Özsavcı D, Biber N, Hakan
T, et al: Functional and structural changes of the urinary bladder
following spinal cord injury; treatment with alpha lipoic acid.
Neurourol Urodyn. 36:1061–1068. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ozsoy O, Ozsoy U, Stein G, Semler O,
Skouras E, Schempf G, Wellmann K, Wirth F, Angelova S, Ankerne J,
et al: Functional deficits and morphological changes in the
neurogenic bladder match the severity of spinal cord compression.
Restor Neurol Neurosci. 30:363–381. 2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Hamid R, Averbeck MA, Chiang H, Garcia A,
Al Mousa RT, Oh SJ, Patel A, Plata M and Del Popolo G: Epidemiology
and pathophysiology of neurogenic bladder after spinal cord injury.
World J Urol. 36:1517–1527. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ginsberg D: The epidemiology and
pathophysiology of neurogenic bladder. Am J Manag Care. 19 (Suppl
10):s191–s196. 2013.PubMed/NCBI
|
|
31
|
Hu HZ, Granger N and Jeffery ND:
Pathophysiology, clinical importance, and management of neurogenic
lower urinary tract dysfunction caused by suprasacral spinal cord
injury. J Vet Intern Med. 30:1575–1588. 2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Compérat E, Reitz A, Delcourt A, Capron F,
Denys P and Chartier-Kastler E: Histologic features in the urinary
bladder wall affected from neurogenic overactivity-a comparison of
inflammation, oedema and fibrosis with and without injection of
botulinum toxin type A. Eur Urol. 50:1058–1064. 2006.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Preissner KT and Reuning U: Vitronectin in
vascular context: Facets of a multitalented matricellular protein.
Semin Thromb Hemost. 37:408–424. 2011.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Madsen CD and Sidenius N: The interaction
between urokinase receptor and vitronectin in cell adhesion and
signalling. Eur J Cell Biol. 87:617–629. 2008.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Naik MU and Naik UP: Junctional adhesion
molecule-A-induced endothelial cell migration on vitronectin is
integrin alpha v beta 3 specific. J Cell Sci. 119:490–499.
2006.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Shen TL, Liu MN, Zhang Q, Feng W, Yu W, Fu
XL and Cai XW: The positive role of vitronectin in radiation
induced lung toxicity: The in vitro and in vivo mechanism study. J
Transl Med. 16(100)2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Hayashida M, Hashimoto K, Ishikawa T and
Miyamoto Y: Vitronectin deficiency attenuates hepatic fibrosis in a
non-alcoholic steatohepatitis-induced mouse model. Int J Exp
Pathol. 100:72–82. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
López-Guisa JM, Rassa AC, Cai X, Collins
SJ and Eddy AA: Vitronectin accumulates in the interstitium but
minimally impacts fibrogenesis in experimental chronic kidney
disease. Am J Physiol Renal Physiol. 300:F1244–F1254.
2011.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Carreras-Planella L, Cucchiari D, Cañas L,
Juega J, Franquesa M, Bonet J, Revuelta I, Diekmann F, Taco O,
Lauzurica R and Borràs FE: Urinary vitronectin identifies patients
with high levels of fibrosis in kidney grafts. J Nephrol.
34:861–874. 2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Ciregia F, Deroyer C, Cobraiville G,
Plener Z, Malaise O, Gillet P, Fillet M, Malaise MG and de Seny D:
Modulation of αVβ6 integrin in
osteoarthritis-related synovitis and the interaction with
VTN(381-397 a.a.) competing for TGF-β1 activation. Exp
Mol Med. 53:210–222. 2021.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Su J, Morgani SM, David CJ, Wang Q, Er EE,
Huang YH, Basnet H, Zou Y, Shu W, Soni RK, et al: TGF-β
orchestrates fibrogenic and developmental EMTs via the RAS effector
RREB1. Nature. 577:566–571. 2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Ma TT and Meng XM: TGF-β/Smad and renal
fibrosis. Adv Exp Med Biol. 1165:347–364. 2019.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Chen Y, Ma Y, He Y, Xing D, Liu E, Yang X,
Zhu W, Wang Q and Wen JG: The TGF-β1 pathway is early involved in
neurogenic bladder fibrosis of juvenile rats. Pediatr Res.
90:759–767. 2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Alyaseer AAA, de Lima MHS and Braga TT:
The role of NLRP3 inflammasome activation in the epithelial to
mesenchymal transition process during the fibrosis. Front Immunol.
11(883)2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Liu C, Wan X, Gu M, Chen Y, Cai Z, Zhou J,
Chen Q and Wang Z: Effect of sulforaphane on bladder compliance in
a rat model of partial bladder outlet obstruction. Oxid Med Cell
Longev. 2019(6026719)2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Kraft M, Oussoren Y, Stewart FA, Dörr W
and Schultz-Hector S: Radiation-induced changes in transforming
growth factor beta and collagen expression in the murine bladder
wall and its correlation with bladder function. Radiat Res.
146:619–627. 1996.PubMed/NCBI
|
|
47
|
Feng YL, Chen DQ, Vaziri ND, Guo Y and
Zhao YY: Small molecule inhibitors of epithelial-mesenchymal
transition for the treatment of cancer and fibrosis. Med Res Rev.
40:54–78. 2020.PubMed/NCBI View Article : Google Scholar
|