Introduction
In China, breast cancer (BCa) is the most common
cancer among women (1), and the
incidence rate and mortality rate are rising (2,3) The
mortality of breast cancer reflects the poor prognosis of breast
cancer due to tumor metastasis (3,4).
However, the molecular mechanism underlying the process is still
unclear.
The long non-coding (lncRNAs) RNAs are a group of
sequences that have been recently used to study several cancer
types, such as liver cancer, breast cancer and colorectal carcinoma
(5-7).
lncRNAs are members of a family of >200-nucleotides long linear
transcripts (8) that lack
protein-encoding function; they were thus deemed genomic ‘noise’
and ‘junk’ (9). Several recent
reports reveal the implication of lncRNAs in a range of genetic
processes for gene expression modulation at epigenetic,
transcriptional and posttranscriptional levels (10,11)
and their dysfunction was closely associated with human diseases,
including cardiovascular diseases, neurodegenerative and endocrine
diseases (12). In BCa, the lncRNAs
are frequently abnormally expressed and are implicated in the BCa
occurrence and development (12-14).
Hence, a detailed study of the lncRNAs and their role in BCa may
lead to unraveling potential targets to enhance the patient
prognosis and therapy in this disease. A recently discovered
lncRNA, growth arrest-specific protein 6 (Gas6) antisense RNA 1
(GAS6-AS1) was found to play key role in several malignant tumors
(15). These studies suggest that
GAS6-AS1 can participate in cancer development and may be utilized
as biomarkers for cancer diagnosis and prognosis. Nonetheless, the
activity of GAS6-AS1 in BCa needs to be examined and established.
Thus, GAS6-AS1 was the focus of the present study, intending to
investigate the pathogenicity of BCa and offer insights for its
therapy.
Materials and methods
Patients and specimen of tissues
The research ethics committee of the Zhengxing
Hospital granted approval for this study (approval no. 2016-07-22),
which was carried out as per the Declaration of Helsinki. A total
of 36 patients (all female) with BCa (range, 35-76 years; mean, 56
years) were enrolled between January 2016 and December 2018; none
of these had undergone chemotherapy, preoperative radiotherapy, or
any other treatment for cancer. The sample tissues of BCa and
adjacent (>3 cm) normal tissues were surgically excised and
snap-frozen in liquid nitrogen and maintained at -80˚C. Written
informed consent was obtained from all participants before specimen
collection.
Culture and maintenance of cell
cultures
Two BCa cell lines (MDA-MB-231 and MCF-7) and normal
Hs578Bst cells were acquired from the American Type Culture
Collection and cultured in Dulbecco's Modified Eagle's medium
containing heat-inactivated fetal bovine serum (10%), 100 mg/ml
streptomycin and 100 U/ml penicillin. DMEM, FBS, and antibiotics
were purchased from Gibco (Thermo Fisher Scientific, Inc.). All the
mentioned cell lines were cultured at 37˚C in an atmosphere with
humidity and 5% CO2.
Cell transfection
To examine the loss-of-function effects, the small
interfering RNA (siRNA) si-GAS6-AS1 (against GAS6-AS1) and si-NC
(control; nontargeting siRNA) were procured from Guangzhou RiboBio
Co., Ltd. The design and synthesis of agomir-215-5p were by
Shanghai GenePharma Co., Ltd., and the agomir-NC was the negative
control. The 50 pmol/ml antagomir-215-5p was used for silencing
endogenous microRNA (miR)-215-5p, and the control for
antagomir-215-5p was 50 pmol/ml antagomir-NC. The synthesis of
vector 40 pmol/ml pcDNA3.1-SOX9 (pc-SOX9) to overexpress SRY-box
transcription factor 9 (SOX9) and empty 40 pmol/ml pcDNA3.1 vector
was done by Shanghai GenePharma Co., Ltd. One day prior to
transfection, 2x105 cells were seeded onto 6-well
plates. The transfection of the aforementioned molecular constructs
was done transiently into cells using Lipofectamine®
2000 reagent from Invitrogen (Thermo Fisher Scientific, Inc.) for
24-48 h at 37˚C in accordance with the manufacturer's protocol,
with downstream analyses conducted at 24-48 h post-transfection.
The primer sequences were: antagomir-215-5p, sense 5'-UGGAUUUC
AAUCACCA-3' and antisense 5'-GUGAUUCACAAAGAA AUCCAUU-3';
antagomir-NC, sense 5'-UUCUCCGUCACG UTT-3' and antisense
5'-ACGUGACACGUGAGAATT-3'
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from tissues and cells was isolated by
TRIzol® reagent from Invitrogen (Thermo Fisher
Scientific, Inc.), and a NanoDrop (NanoDrop Technologies; Thermo
Fisher Scientific, Inc.) was used to estimate its quality and
concentration. Reverse transcription was conducted using the Mir-X™
miRNA First-Strand Synthesis kit from Takara Biotechnology Co.,
Ltd., with 2 µg RNA, according to the manufacturer's instructions.
Quantitative evaluation of the synthesized cDNA was done by qPCR
using the Mir-X™ miRNA RT-qPCR TB Green® kit from Takara
Biotechnology Co., Ltd. As an endogenous control, the small nuclear
RNA U6 normalized the expression of miR-215-5p. To measure
transcript levels of GAS6-AS1 and SOX9, the PrimeScript™ RT Reagent
kit from Takara Biotechnology Co., Ltd., was used to convert a
specified amount of total 2 µg RNA to cDNA. Then, the amplification
of cDNA products was done using the SYBR-Green PCR Master Mix from
Takara Biotechnology Co., Ltd. The level of SOX9 and GAS6-AS1
transcripts were normalized with that of GAPDH. The
2-ΔΔCq system was used to evaluate all genes expressions
(16). The qPCR reaction conditions
were as follows: Initial denaturation 10 min at 95˚C; denaturation
10 sec at 95˚C; annealing 20 sec at 60˚C; and extension 15 sec at
72˚C; for 40 cycles.
The primers used for qPCR were: SOX9: Forward,
5'-TCCGAGATTATGCCTCG-3', and reverse, 5'-TGACAG CGGAATGTTCTTCCC-3';
GAS6-AS1: Forward, 5'-TGACTTCAACAGCGACACCCA-3', and reverse,
5'-CACCCTGTTGCTGTAGCCAAA-3'; U6: Forward, 5'-TCC
GAATGTTGATTATGCCTCG-3', and reverse, 5'-TGACAG CGAATGTTCTTCCC-3';
GAPDH: Forward, 5'-TCCGAA TGTTGATTATGCCTCG-3', and reverse, 5'-TGA
CAGCGG AATGTTCTTCCC-3'; mir-215-5p: Forward, 5'-TCCGAA
TGTTGATTATGCCTCG-3', and reverse, 5'-TGACAGCAA CTTCCC-3';
Cell counting assay
Assessment of cell proliferation was done by the
Cell Counting Kit-8 (CCK-8) assay from Shanghai Haling
Biotechnology, Co., Ltd. After incubating the transfected cells for
one day, they were collected after trypsinization and seeded
(2x103 cells/well) onto 96-well plates. CCK-8 solution
(10 µl) was added per well and kept for 2 h at 37˚C. The absorbance
of the mixture was estimated in a microplate reader from Bio-Rad
Laboratories, Inc. at 450 nm.
Assay for colony formation
Each group of treated cells (1x103/well)
were seeded in a culture dish (10 cm) and cultured for 14 days.
Finally, colonies were stained for 15-20 min at room temperature by
crystal violet (1%) followed by the counting of colonies were
observed with an inverted light microscope (magnification, x200;
IX71; Olympus Corporation) and counted using ImageJ software
(version 4.0; National Institutes of Health).
Bioinformatics analysis
The putative GAS6-AS1 targets were predicted by
StarBase 3.0 (http://starbase.sysu.edu.cn/).
RNA immunoprecipitation (RIP)
assay
The RNA immunoprecipitation assay was performed
using the Magna RIP RNA-Binding Protein Immunoprecipitation kit
from EMD Millipore to assess the interaction of GAS6-AS1 and
miR-215-5p in BCa cells 2x107. BCa cell lysates were
incubated with magnetic beads conjugated with negative control
normal mouse IgG or human anti-Ago2 antibody (both from EMD
Millipore). The samples were digested with proteinase K to isolate
the immunoprecipitated RNA and RT-qPCR analysis was performed to
estimate the enrichment of GAS6-AS1 and miR-215-5p.
Assay for luciferase reporter
activity
The design and synthesis of GAS6-AS1 fragments
containing binding sites for WT (wild-type) and MUT (mutant) on
miR-215-5p was done by Shanghai GenePharma Co., Ltd. These were
cloned into the Target Expression Vector pmirGLO Dual-luciferase
from Promega Corporation to obtain the reporter plasmids
WT-GAS6-AS1 and MUT-GAS6-AS1. Likewise, the reporter plasmids
WT-SOX9 and MUT-SOX9 were prepared. One night prior to
transfection, seeding of cells (60-70% confluence) was done in
24-well plates. Transfection of these cells was performed with
reporter plasmids harboring WT or MUT in the presence of
agomir-215-5p or agomir-NC for 24 h at 37˚C using
Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) in accordance with the manufacturer's protocol.
After 48 h of transfection, luciferase activity of the cells was
estimated as per instructions using the Dual-Luciferase Reporter
Assay System from Promega Corporation. The data normalization was
done by the activity of Renilla luciferase.
Statistical analysis
All results of independent experiments (conducted
thrice) are presented as the mean ± SD. Two groups were compared
using the Student's t-test; variations among multiple groups were
compared through one-way ANOVA and then by Tukey's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
GAS6-AS1 is increased in BCa
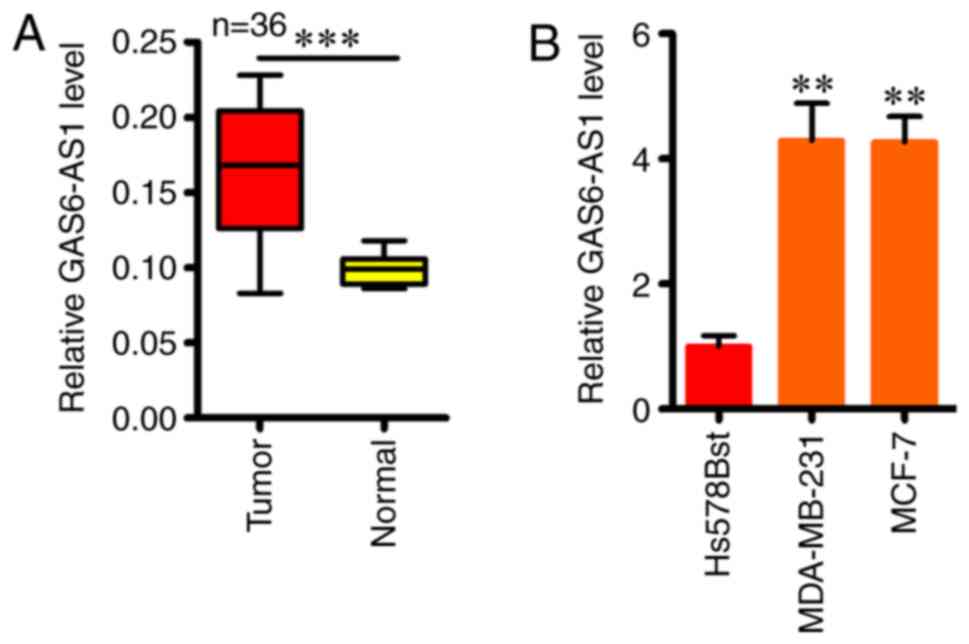
The function of GAS6-AS1 in BCa was first assessed
by estimating the expression of GAS6-AS1 by RT-qPCR in 36 BCa
tissues and the adjacent healthy tissues. An enhanced level of
GAS6-AS1 was observed in BCa tissues compared with the adjacent
healthy tissues (Fig. 1A).
Furthermore, RT-qPCR was done in two BCa cell lines (MDA-MB-231 and
MCF-7 cells) and normal Hs578Bst cells, which detected higher
GAS6-AS1 expression in BCa cell lines compared with normal Hs578Bst
cells (Fig. 1B). Thus, GAS6-AS1 may
play an important part in BCa malignancy.
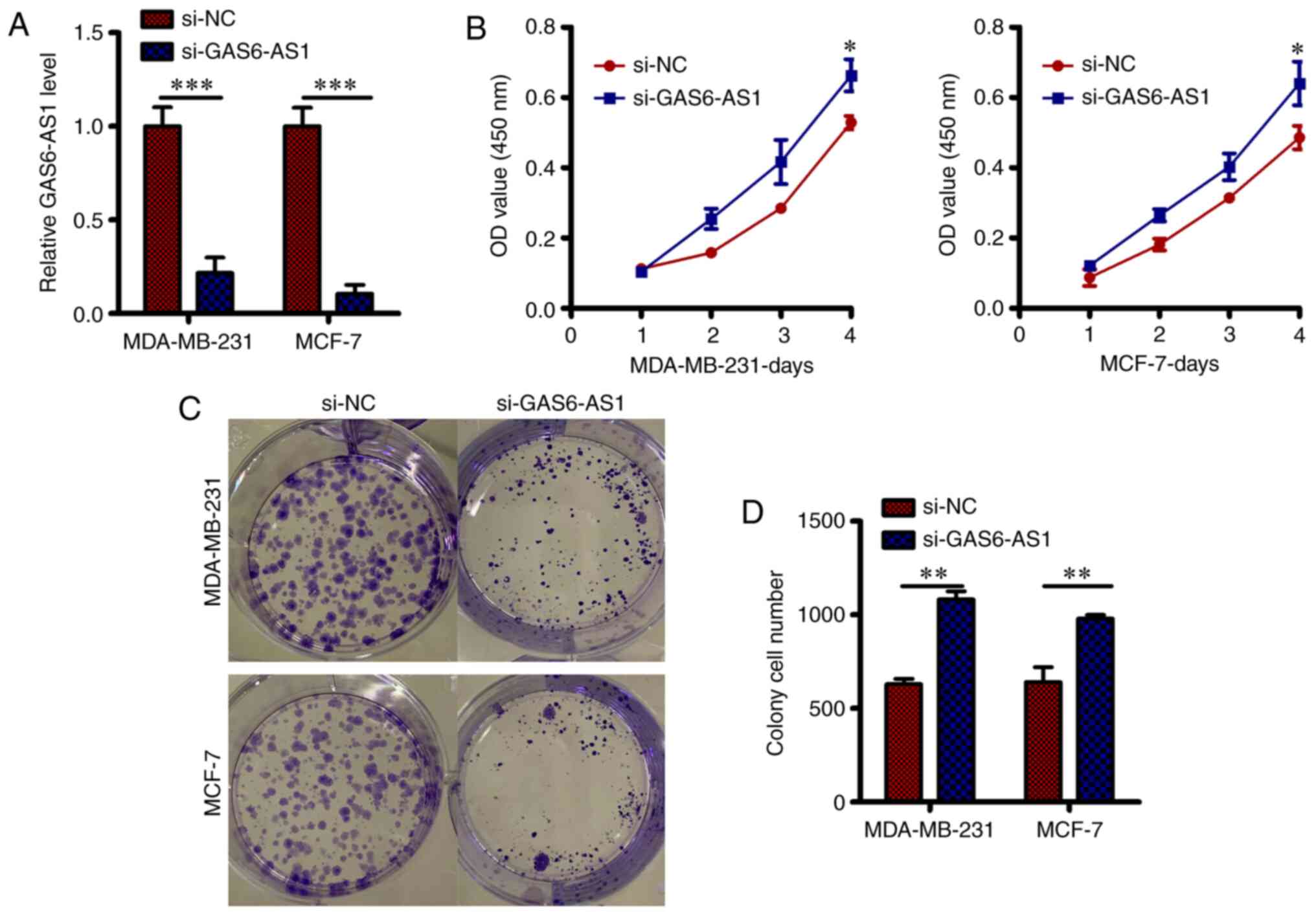
In vitro GAS6-AS1 silencing led to
inhibition of the BCa cell malignant phenotype
For examining the function of GAS6-AS1 in the
progression of BCa, si-GAS6-AS1 was transfected in MDA-MB-231 and
MCF-7 cells to deplete GAS6-AS1 (Fig.
2A; P<0.05) and the controls were si-NC transfected cells.
The proliferative ability, measured by CCK-8 assay, of both
MDA-MB-231 and MCF-7 cells harboring si-GAS6-AS1 showed inhibition
compared with that of the control cells (Fig. 2B; P<0.05). Furthermore, the
colony-forming capacity of MDA-MB-231 and MCF-7 cells deficient in
GAS6-AS1 (following transfection with si-GAS6-AS1) was greatly
inhibited (Fig. 2C and D; P<0.05), thus, strongly suggesting
the cancer-enhancing activity of lncRNA GAS6-AS1 in BCa.
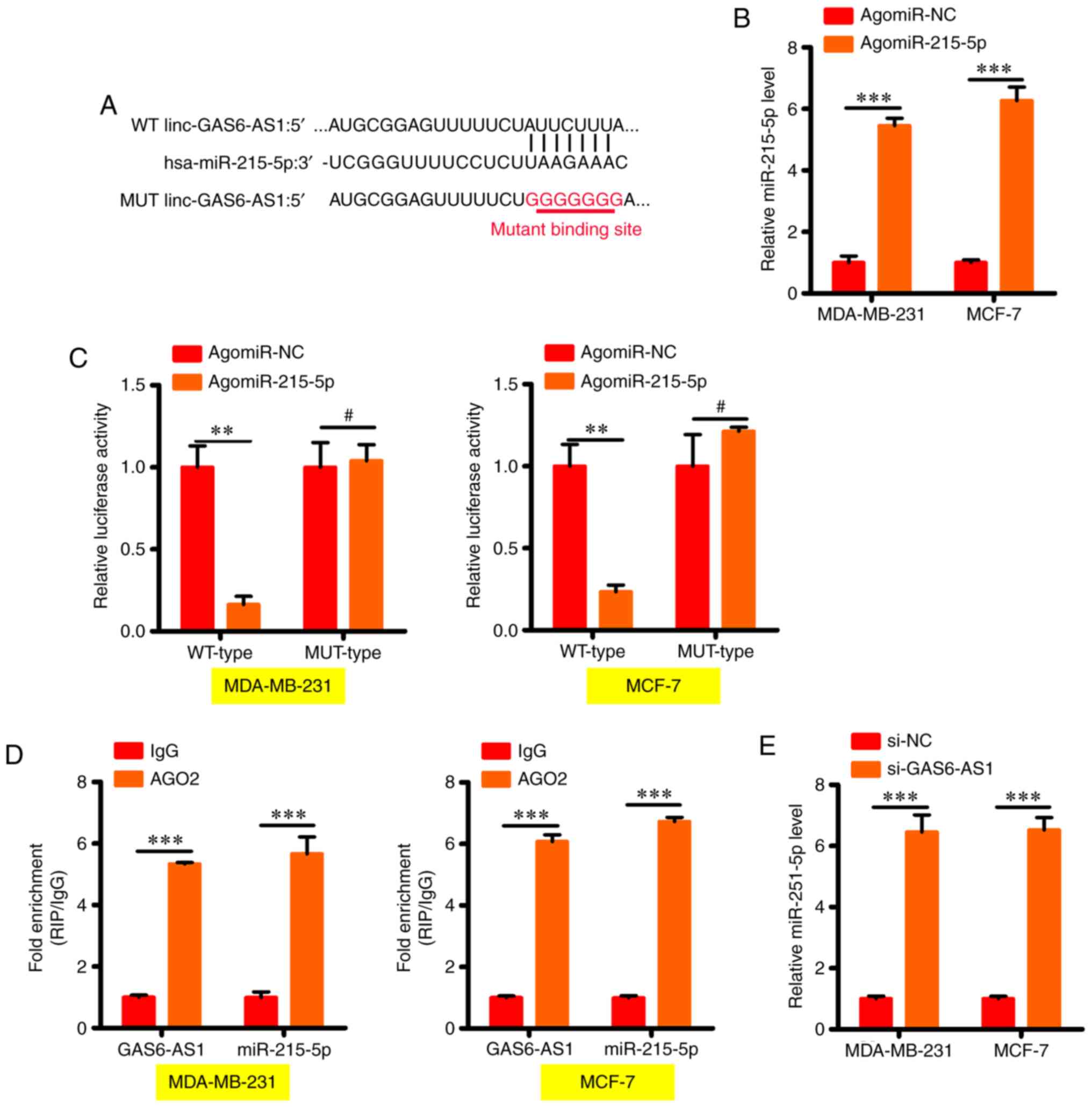
GAS6-AS1 interacts directly with
miR-215-5p in BCa cells as a quencher of miRNA
lncRNAs participate in several biological processes
by functioning as competing endogenous RNA (ceRNA) against miRNAs.
Next, StarBase 3.0, a publicly available algorithm, was used to
predict the miRNAs that interact directly with GAS6-AS1. The
binding sites on miR-215-5p were found to be complementary
(Fig. 3A) with those on GAS6-AS1,
and was thus used for further verification; more so, because the
progression of BCa was impeded by this miRNA (17). The prediction of GAS6-AS1 and
miR-215-5p binding in BCa cells was further confirmed by luciferase
reporter activity. MDA-MB-231 and MCF-7 cells were transfected with
WT- or MUT-GAS6-AS1 when either agomie-215-5p or agomir-NC was
present in the test cells. A notable increase in miR-215-5p
expression was detected in MDA-MB-231 and MCF-7 cells following
agomir-215-5p transfection (Fig.
3B). In addition, the luciferase activity of cells harboring
WT-GAS6-AS1 declined drastically after agomir-215-5p transfection;
however, there was no change in MUT-GAS6-AS1 activity when
miR-215-5p was upregulated (Fig.
3C). Next, RIP assay was conducted to assess the miR-215-5p and
GAS6-AS1 interactions, and miR-215-5p and GAS6-AS1 were found to be
enriched in immunoprecipitates containing Ago2 in comparison with
the IgG control (Fig. 3D).
Subsequently, the expression of miR-215-5p in response to GAS6-AS1
modulation was tested in BCa cells, which was indeed fount to be
enhanced in MDA-MB-231 and MCF-7 cells in response to the silencing
of GAS6-AS1 (Fig. 3E). These
outcomes thus indicate the miR-215-5p quenching function of
GAS6-AS1 in BCa cells.
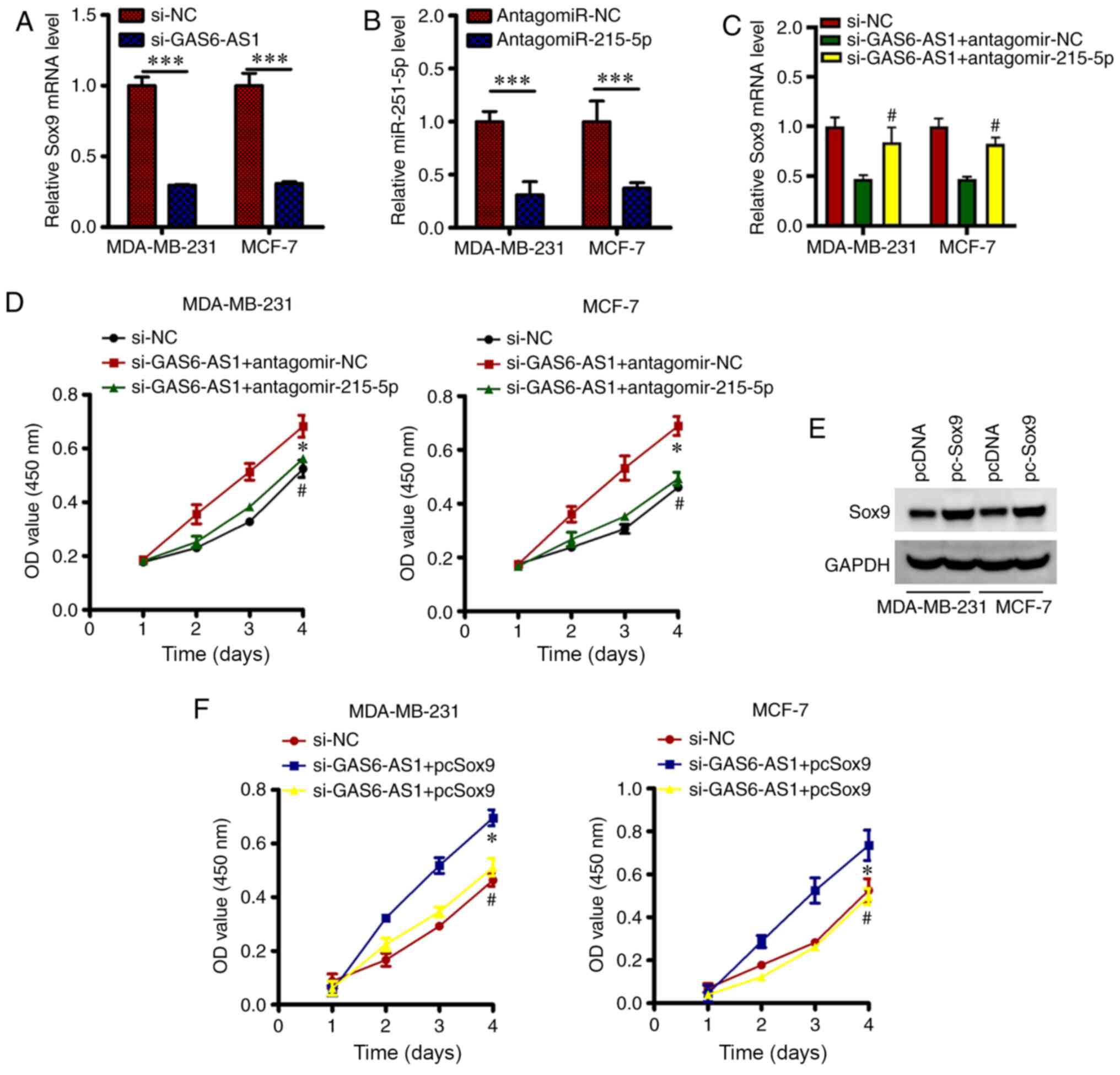
GAS6-AS1 sequesters miR-215-5p and
positively modulates the expression of SOX9 in BCa cells
miR-215-5p directly targets the SOX9 gene in BCa
cells (17). On observing that
GAS6-AS1 sequesters miR-215-5p, the function of GAS6-AS1 was
assessed in SOX9 regulation in BCa cells. For this, MDA-MB-231 and
MCF-7 cells were transfected with si-GAS6-AS1 or si-NC, and the
level of SOX9 expression was estimated. GAS6-AS1 silencing notably
decreased the expression of SOX9 at transcript level (Fig. 4A) in these two transfected cells. To
explore whether GAS6-AS1 sequesters miR-215-5p and regulates SOX9
expression, MDA-MB-231-si GAS6-AS1 and MCF-7- GAS6-AS1 cells were
transfected with antagomir-215-5p or antagomir-NC. On evaluating
the extent of antagomir-215-5p transfection by RT-qPCR, a
significant decline in the level of miR-215-5p was observed in
MDA-MB-231 and MCF-7 cells transfected with antagomiR-215-5p
(Fig. 4B). Moreover, the decline in
the SOX9 transcript (Fig. 4C;
P<0.05) due to GAS6-AS1 was reversed by re-introduction of
antagomiR-215-5p in MDA-MB-231 and MCF-7 cells. Therefore, GAS6-AS1
positively regulates the levels of SOX9 in BCa cells by
sequestering miR-215-5p. Furthermore, rescue experiments elucidated
whether the miR-215-5p/SOX9 axis mediated GAS6-AS1 activity in BCa
cells. For this, the co-transfection of si-GAS6-AS1 along with
antagomiR-215-5p or antagomir-NC was done in MDA-MB-231 and MCF-7
cells, and proliferative capacity of cells was assessed. GAS6-AS1
silencing impeded the proliferation of MDA-MB-231 and MCF-7 cells
and inhibition of miR-215-5p in these cells partly neutralized the
outcomes of GAS6-AS1 silencing (Fig.
4D; P<0.05). Moreover, the co-transfection in MDA-MB-231 and
MCF-7 cells was done with si-GAS6-AS1 and the empty pcDNA3.1 vector
(control) or pc-SOX9 and the efficiency of transformation was
analyzed by western blotting. There was a significant increase in
SOX9 level (Fig. 4E) in MDA-MB-231
and MCF-7 cells. Furthermore, CCK-8 assay demonstrated that pc-SOX9
co-transfection was found to ameliorate the decline in the
proliferation (Fig. 4F; P<0.05)
of GAS6-AS1-silenced MDA-MB-231 and MCF-7 cells. These
observations, hence, suggest the oncogenic effect of GAS6-AS1 in
BCa advancement by acting as a ceRNA for miR-215-5p and
consequently enhancing SOX9 expression.
Discussion
lncRNAs have attracted a large interest because of
their vital function in the progression of cancer (18-20).
In BCa, there has been a report that aberrant expression of lncRNAs
also contribute to the onset and advancement of BCa (18). Therefore, potential treatment
targets may be revealed by studying the roles of lncRNAs in BCa
tumorigenesis. Until recently, the function of GAS6-AS1 in the
malignancy of BCa has not been studied in detail. Hence, the
present study evaluated the GAS6-AS1 expression level and its
specific role in the malignancy of BCa cells. The tumor-enhancing
function of GAS6-AS1 in BCa was also elucidated. A high expression
of GAS6-AS1 was demonstrated in BCa tissue samples as well as in
the tested BCa cell lines. The functional experiments revealed
inhibition of BCa cell proliferative and colony-forming capacities
as a result of GAS6-AS1 silencing.
lncRNAs participate in sophisticated mechanisms to
perform vital roles in cancer occurrence and progression.
Currently, the predominant mechanism is regulated by ceRNA by which
lncRNAs and miRNAs interact competitively and upregulate specific
miRNA target genes. After revealing the tumor-enhancing role of
GAS6-AS1 in BCa, the mechanisms of this aggressive behavior lncRNA
were investigated. Firstly, it was predicted that miR-215-5p
possesses a binding site complementary to that on GAS6-AS1, and
this was further established by RIP and luciferase reporter assays.
Secondly, the downregulation of GAS6-AS1 enhanced miR-215-5p level
and a consequent decline in the expression of SOX9. Thirdly, the
outcomes of GAS6-AS1 silencing on the malignant phenotypes of BCa
could be abrogated by miR-215-5p inhibition or SOX9 restoration. A
previous study reported that miR-215-5p was decreased in BCa
tissue, and an in vitro and in vivo model indicated
that miR-215-5p inhibits breast cancer cell growth and metastatic
through targeting Sox9(17). Taken
together, the present study proposes a ceRNA model that includes
GAS6-AS1, SOX9 and miR-215-5p in BCa cells.
Among multiple potential target genes, SOX9 was
selected for further validation, since SOX9 has been demonstrated
to be involved in BCa carcinogenesis and progression (21-24).
For example, SOX9 could enhance BCa cell endocrine resistance
(21). Another study indicated that
miRNA-511 might suppress BCa progression via the Sox9/ PI3K/Akt
pathway (23). Meanwhile, SLUG and
SOX9 cooperate to promote the progression of breast cancer
(24). The present study identified
for the first time an upstream mechanism modulating the axis of
GAS6-AS1/miR-215-5p/SOX9 in BCa cells, in vitro. GAS6-AS1
possesses a miR-215-5p binding site, and acts as a ceRNA and
sequesters miR-215-5p in BCa cells, leading to an enhanced level of
SOX9. Identification of the regulatory network of
GAS6-AS1/miR-215-5p /SOX9 will possibly aid in fully determining
the stage of BCa and provide possible targets for the therapy of
patients suffering from BCa.
The present study identified the mode of action of
GAS6-AS1 in enhancing BCa progression, for which there are no
earlier reports. GAS6-AS1 silencing enhanced the malignancy of BCa
cells. GAS6-AS1 positively regulated SOX9 expression by
sequestering miR-215-5p in BCa cells.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YX and XW wrote the manuscript and contributed to
the conception of the study. ZX, WX and YLL performed the data
analysis. KH and YNL contributed to data acquisition and analysis,
and revised the manuscript. YJ and ZX worked on the part of the
study related to patients. YX and XW confirm the authenticity of
all the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by the research ethics
committee of the Zhengxing Hospital and performed in accordance
with the Declaration of Helsinki. Written informed consent was
obtained from all participants before specimen collection.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zhao J, Mao Z, Fedewa SA, Nogueira L,
Yabroff KR, Jemal A and Han X: The affordable care act and access
to care across the cancer control continuum: A review at 10 years.
CA Cancer J Clin. 70:165–181. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
DeSantis CE, Ma J, Gaudet MM, Newman LA,
Miller KD, Goding Sauer A, Jemal A and Siegel RL: Breast cancer
statistics, 2019. CA Cancer J Clin. 69:438–451. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Schnitt SJ: The diagnosis and management
of pre-invasive breast disease: Flat epithelial atypia -
classification, pathologic features and clinical significance.
Breast Cancer Res. 5:263–268. 2003.PubMed/NCBI View
Article : Google Scholar
|
|
5
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Hauptman N and Glavač D: Long non-coding
RNA in cancer. Int J Mol Sci. 14:4655–4669. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Bao Z, Yang Z, Huang Z, Zhou Y, Cui Q and
Dong D: LncRNADisease 2.0: An updated database of long non-coding
RNA-associated diseases. Nucleic Acids Res. 47 (D1):D1034–D1037.
2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Han S, Cao D, Sha J, Zhu X and Chen D:
LncRNA ZFPM2-AS1 promotes lung adenocarcinoma progression by
interacting with UPF1 to destabilize ZFPM2. Mol Oncol.
14:1074–1088. 2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhao M, Xin XF, Zhang JY, Dai W, Lv TF and
Song Y: LncRNA GMDS-AS1 inhibits lung adenocarcinoma development by
regulating miR-96-5p/CYLD signaling. Cancer Med. 9:1196–1208.
2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Han L, Kong R, Yin DD, Zhang EB, Xu TP, De
W and Shu YQ: Low expression of long noncoding RNA GAS6-AS1
predicts a poor prognosis in patients with NSCLC. Med Oncol.
30(694)2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yang S, Liu T, Sun Y and Liang X: The long
noncoding RNA LINC00483 promotes lung adenocarcinoma progression by
sponging miR-204-3p. Cell Mol Biol Lett. 24(70)2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Luo Z, Han Z, Shou F, Li Y and Chen Y:
LINC00958 accelerates cell proliferation and migration in non-small
cell lung cancer through JNK/c-JUN signaling. Hum Gene Ther
Methods. 30:226–234. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li W, Zhang B, Jia Y, Shi H, Wang H, Guo Q
and Li H: LncRNA LOXL1-AS1 regulates the tumorigenesis and
development of lung adenocarcinoma through sponging miR-423-5p and
targeting MYBL2. Cancer Med. 9:689–699. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ai J, Sun J, Zhou G, Zhu T and Jing L:
Long non-coding RNA GAS6-AS1 acts as a ceRNA for microRNA-585,
thereby increasing EIF5A2 expression and facilitating
hepatocellular carcinoma oncogenicity. Cell Cycle. 19:742–757.
2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Gao JB, Zhu MN and Zhu XL: miRNA-215-5p
suppresses the aggressiveness of breast cancer cells by targeting
Sox9. FEBS Open Bio. 9:1957–1967. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Liu X, Song J, Kang Y, Wang Y and Chen A:
Long noncoding RNA SOX21-AS1 regulates the progression of
triple-negative breast cancer through regulation of
miR-520a-5p/ORMDL3 axis. J Cell Biochem. 121:4601–4611.
2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chu F, Xue L and Miao H: Long noncoding
RNA TP73-AS1 in human cancers. Clin Chim Acta. 500:104–108.
2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Gourvest M, Brousset P and Bousquet M:
Long noncoding RNAs in acute myeloid leukemia: Functional
characterization and clinical relevance. Cancers (Basel).
11(E1638)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Jeselsohn R, Cornwell M, Pun M, Buchwalter
G, Nguyen M, Bango C, Huang Y, Kuang Y, Paweletz C, Fu X, et al:
Embryonic transcription factor SOX9 drives breast cancer endocrine
resistance. Proc Natl Acad Sci USA. 114:E4482–E4491.
2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Jana S, Madhu Krishna B, Singhal J, Horne
D, Awasthi S, Salgia R and Singhal SS: SOX9: The master regulator
of cell fate in breast cancer. Biochem Pharmacol.
174(113789)2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhao Y, Pang W, Yang N, Hao L and Wang L:
MicroRNA-511 inhibits malignant behaviors of breast cancer by
directly targeting SOX9 and regulating the PI3K/Akt pathway. Int J
Oncol. 53:2715–2726. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Fazilaty H, Gardaneh M, Akbari P, Zekri A
and Behnam B: SLUG and SOX9 cooperatively regulate tumor initiating
niche factors in breast cancer. Cancer Microenviron. 9:71–74.
2016.PubMed/NCBI View Article : Google Scholar
|