Introduction
Acute myocardial infarction (AMI) is a disease that
has a 30% mortality rate, where ~50% mortality occur prior to
arrival at the hospital in the United States (1). AMI has been reported to be mainly
caused by myocardial ischemia-reperfusion (I/R) injury (1,2). The
incidence rate of AMI in the younger population (aged <45 years)
is rising annually in the United States (2,3).
Therefore, strategies to reduce the damage caused by myocardial I/R
injury have become a research focus. The pathogenesis of myocardial
I/R injury refers to a series of complex pathophysiological
processes, including the overproduction of reactive oxygen species
(4), an inflammatory response
(5) and mitochondrial dysfunction
(6). Furthermore, it has been
demonstrated that the inflammatory response is the main
pathological process leading to myocardial injury following
reperfusion (7,8).
The histone modifier lysine-specific demethylase 2B
(KDM2B) is a member of the JmjC domain-containing histone
demethylase family and has been reported to serve a role in
lymphomagenesis (9), adipogenesis
(10) and the self-renewal of
hematopoietic stem cells (10,11).
KDM2B is also associated with the occurrence and development of
tumors, such as colorectal cancer and T-cell acute lymphoblastic
leukemia (12-15).
Furthermore, KDM2B is also required for the regulation of choline
kinase-α during neuronal differentiation and in maintaining the
undifferentiated stage of neuroblasts (16). KDM2B also promotes IL-6 production
and the inflammatory response via gene-specific transcription
initiation (17). However, to the
best of our knowledge, the effects of KDM2B on the inflammatory
response in myocardial I/R injury and the corresponding mechanisms
are unknown.
Toll-like receptor 4 (TLR4) is a transmembrane
protein, the activation of which promotes NF-κB expression and the
release of inflammatory factors, such as IL-6, TNF-α and IL-1β
(18,19). Therefore, it can be hypothesized
that inhibition of the TLR4/NF-кB signaling pathway could reduce
myocardial I/R injury (20,21).
Thus, it is of particular significance to identify the role of
KDM2B in the inflammatory response in myocardial I/R injury.
Therefore, in the present study, a myocardial I/R injury rat model
was generated to further determine the potential function of KDM2B
following overexpression and silencing of KDM2B. The involvement of
the inflammatory mechanism was also investigated. The present study
provided information on an important therapeutic target for the
treatment of myocardial I/R injury.
Materials and methods
Ethics statement
The protocol for the use of animals in the present
study was approved by the Ethics Committee of The First Affiliated
Hospital of Gannan Medical University (Ganzhou, China).
Establishment of a myocardial I/R
injury model in rats and viral transduction
A total of 30 male Sprague Dawley rats (age, 3
months; weight, ~250 g) were purchased from Sibeifu (Beijing)
Biotechnology Co., Ltd. (license no. scxk 2019-0010; https://www.spf-tsinghua.com/) and housed under
consistent conditions at 20˚C with 30% humidity in a 12-h
light/dark cycle with free access to food and water. The rats were
anesthetized by intravenous injection of 1% pentobarbital sodium
(45 mg/kg). Following anesthesia, the skin was prepared at the neck
and cut to separate the subcutaneous fat, after which the trachea
was separated. A small hole was inserted into the trachea with a
needle 5-8 mm from the larynx. During surgery, the rats were
ventilated with room air using a rodent respirator (Columbus
Instruments International) set at 110-120 breaths/min. Left
anterior descending coronary artery was ligated with a catheter to
block the blood flow. After 50 min of ligation, the heart was
reperfused for 4 h by removing the catheter. The electrocardiogram
(ECG) was monitored. The catheter was removed after 30 min of
successful modeling as confirmed by ECG detection based on the
reduction of heart rate and elevation of ST segment. The injury of
heart tissue was also confirmed by the HE staining, which indicated
the heart infarction.
The rats were divided into the following six groups
(n=5 in each group): A sham group, a model group, an
adeno-associated virus (AAV)-small interfering RNA (si/siRNA)KDM2B
group, an AAV-siRNA scrambled negative control (NC,
pAdEasy-U6-CMV-EGFP) group, an AAV-KDM2B overexpression group and
an AAV-overexpression negative control (NC,
pAdEasy-EF1-MCS-3FLAG-CMV-EGFP) group. The AAV-encoding siKDM2B
(5'-UGGAAGAGGAAGAAGGCAATTUUGCCUUCUUCCUCUUCCATT-3'), overexpression
vectors and corresponding NC
(5'-UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT-3') were provided by
Jiangxi Zhonghong Boyuan Biotechnology Co., Ltd. During reperfusion
as previously described (22), 4
µl adenovirus overexpressing KDM2B (1.58x1010 PFU/ml) or
adenovirus silencing KDM2B (4.0x1010 PFU/ml) was locally
injected into the margin of the infarcted myocardium. In the sham
group, the animals received surgery without ligation and injection
of saline. A small hole was also inserted into the trachea as the
model group. After the injections, the thoracic cavity was sutured
and penicillin (160,000 units/kg) was injected intramuscularly. The
rats received 1% pentobarbital sodium (120 mg/kg) intraperitoneally
48 h after reperfusion and decapitated to collect the infarcted
heart tissues and 2 ml blood samples, which were stored at -80˚C
for further use.
H&E staining
Partial infarcted areas of the heart tissue from
three animals were fixed in 4% paraformaldehyde at 4˚C overnight.
Subsequently, the tissues were washed using water and dehydrated
with 70, 80 and 90% ethanol solutions and added into ethanol
absolute and xylene for 15 min. The tissue samples were added to a
mixture of xylenes and paraffin for 15 min and then embedded in
paraffin for 50-60 min at room temperature. The tissues were sliced
into 10-µm sections. The paraffin sections were dried, dewaxed,
hydrated and then stained in hematoxylin solution for 3 min at room
temperature, differentiated in ethanol differentiation solution for
15 sec, washed with water for 15 sec and stained with eosin for 3
min at room temperature. The sections were sealed and observed
under a light microscope (Olympus Corporation; magnification,
x200).
TUNEL assay
Partial infarcted areas of the heart tissue from
three animals were fixed in 4% paraformaldehyde at 4˚C overnight.
The tissues were embedded in paraffin and sectioned into 5-µm
thickness. The sectioned tissue was placed into a wet box and
incubated with 50 µg/ml proteinase K solution at 37˚C for 30 min.
Each slide was incubated with TUNEL detection solution (cat. no.
c1090; Beyotime Institute of Biotechnology) at 37˚C for 1 h in the
dark and double-stained with 5 µg/ml DAPI for 5 min at room
temperature. Following rinsing with PBS, the sections were
incubated with Antifade Mounting Medium solution (including
glycerol; cat. no. P0126; Beyotime Institute of Biotechnology) and
then observed under a fluorescence microscope (magnification,
x200). A researcher (LHL) who was blinded to the groups quantified
the number of TUNEL-positive cells in five fields in each section
and assessed any histological changes using the ImageJ 2x v2.0.0
(National Institutes of Health).
Reverse transcription-quantitative PCR
(RT-qPCR)
Myocardial tissue was ground into powder in liquid
nitrogen and was subsequently added to TRIzol reagent (Thermo
Fisher Scientific, Inc.). The concentration and purity of RNA
[optical density (OD) 260/OD280] were determined using a UV-Vis
spectrophotometer. cDNA was synthesized using the
HiScript® II Q RT SuperMix for qPCR (cat. no. r223-01;
Vazyme Biotech Co., Ltd.) for 50˚C 15 min, then 85˚C for 5 sec. The
products were used for qPCR, which was performed using the Applied
Biosystems StepOnePlus PCR System (Thermo Fisher Scientific, Inc.).
Each reaction contained the following: 9.5 µl RNase-free
dH2O, 1 µl cDNA, 1 µl forward primer, 1 µl reverse
primer and 12.5 µl 2X SYBR Green PCR Master Mix (cat. no. A4004M;
Xiamen Life Internet Technology Co., Ltd.; http://www.lifeint.cn/show_373.htm). The following
thermocycling conditions were used for qPCR: Initial denaturation
at 95˚C for 10 min; followed by 40 cycles of denaturation at 95˚C
for 10 sec, annealing at 58˚C for 30 sec and extension at 72˚C for
30 sec. The primer sequences were synthesized by General Biosystems
(Anhui) Co., Ltd. (Table I). The
relative mRNA expression levels of KDM2B, TLR4, NF-κB P65 and NOD-,
LRR- and pyrin domain-containing protein 3 (NLRP3) were quantified
and normalized to β-actin using the 2-ΔΔCq method
(23).
 | Table ISequences of primers used for reverse
transcription-quantitative PCR. |
Table I
Sequences of primers used for reverse
transcription-quantitative PCR.
| Gene | Sequence
(5'-3') | Primer length,
nt | Product length,
bp |
|---|
| KDM2B | F:
TTCAAACGTCCCCCGGTTC | 19 | 155 |
| | R:
CCAGGACCGCCGCTTT | 16 | |
| TLR4 | F:
CCAGAGCCGTTGGTGTATCT | 20 | 137 |
| | R:
GGCGATACAATTCGACCTGC | 20 | |
| NF-κB p65 | F:
GCAAAAGGACCTACGAGACC | 20 | 103 |
| | R:
CGGGAAGGCACAGCAATA | 18 | |
| NLRP3 | F:
GACCTCAACAGACGCTACACC | 21 | 102 |
| | R:
CCACATCTTAGTCCTGCCAAT | 21 | |
| β-actin | F:
GCCATGTACGTAGCCATCCA | 20 | 375 |
| | R:
GAACCGCTCATTGCCGATAG | 20 | |
Western blotting
Total protein was isolated from myocardial tissue
using a TriplePrep isolation kit (cat. no. 28-9425-44; Cytiva).
After 30 min lysis incubation on ice, the homogenates were
centrifuged at 11,058 x g for 15 min at 4˚C and the supernatant was
carefully extracted to obtain the total protein. Total protein
concentration was determined using a BCA kit. The protein was
denatured and total protein (25 µg) was separated using SDS-PAGE on
a 12% gel. The separated proteins were transferred onto
nitrocellulose membranes. Membranes were blocked in 5% non-fat milk
for 1 h at room temperature and incubated with primary antibodies
against KDM2B (cat. no. OM252731; OmnimAbs; 1:1,000; http://www.omnimabs.com/antibody_FBXL10_antibody_C_term-OM252731.html),
TLR4 (cat. no. 19811-1-ap; Proteintech Group, Inc.; 1:1,000), NF-κB
p65 (cat. no. 10745-1-ap; Proteintech Group, Inc.; 1:1,000),
phosphorylated (p)-p65 (cat. no. AF2006; Affinity Biosciences;
1:1,000), NLRP3 (cat. no. bs-10021R; BIOSS; 1:1,000) and tubulin
(cat. no. 10094-1-AP, Proteintech, 1:1,000) overnight at 4˚C.
Subsequently, membranes were incubated with the HRP-conjugated
anti-mouse IgG secondary antibody (dilution, 1:100; cat. no.
ab131368; Abcam) for 1-2 h at room temperature. Amersham ECL
Western Blotting Detection Reagent (cat. no. RPN2134; Cytiva) was
added to the membrane. The membrane was imaged using a Bio-Rad Gel
Imaging System (Bio-Rad Laboratories, Inc.). Gray density was
analyzed using Quantity One Analysis Software version 1.4.6
(Bio-Rad Laboratories, Inc.).
ELISA
ELISAs were used to detect the levels of TNF-α (cat.
no. PT516; Beyotime Institute of Biotechnology), IL-6 (cat. no. P
I328; Beyotime Institute of Biotechnology) and IL-1β (cat. no.
PI303; Beyotime Institute of Biotechnology) in the peripheral blood
according to the instructions of the kits.
Statistical analysis
Data are presented as the mean ± SD. There were five
animals in each group. SPSS 21.0 (IBM Corp.) was used to analyze
the differences among ≥ three groups using one-way ANOVA. In cases
where differences were found among groups, Bonferroni's post hoc
test was further applied to analyze the difference between two
groups. P<0.05 was considered to indicate a statistically
significant difference.
Results
Effects of KDM2B on pathological
changes in myocardial I/R injury in rats
The cells in the control group were regularly and
orderly arranged, with complete fibers. Cell edema or necrosis was
also not observed in the control group. In the model group, siRNA
NC group and overexpression NC group, the cell arrangement was
disordered, with a large area of necrosis accompanied by neutrophil
infiltration. In the si-KDM2B group, the pathological changes were
aggravated, whereas the pathological features were improved with no
evidence of necrosis or neutrophil infiltration in the KDM2B
overexpression group (Fig. 1).
These results suggested that KDM2B overexpression may prevent
pathological changes in the heart tissue upon I/R injury.
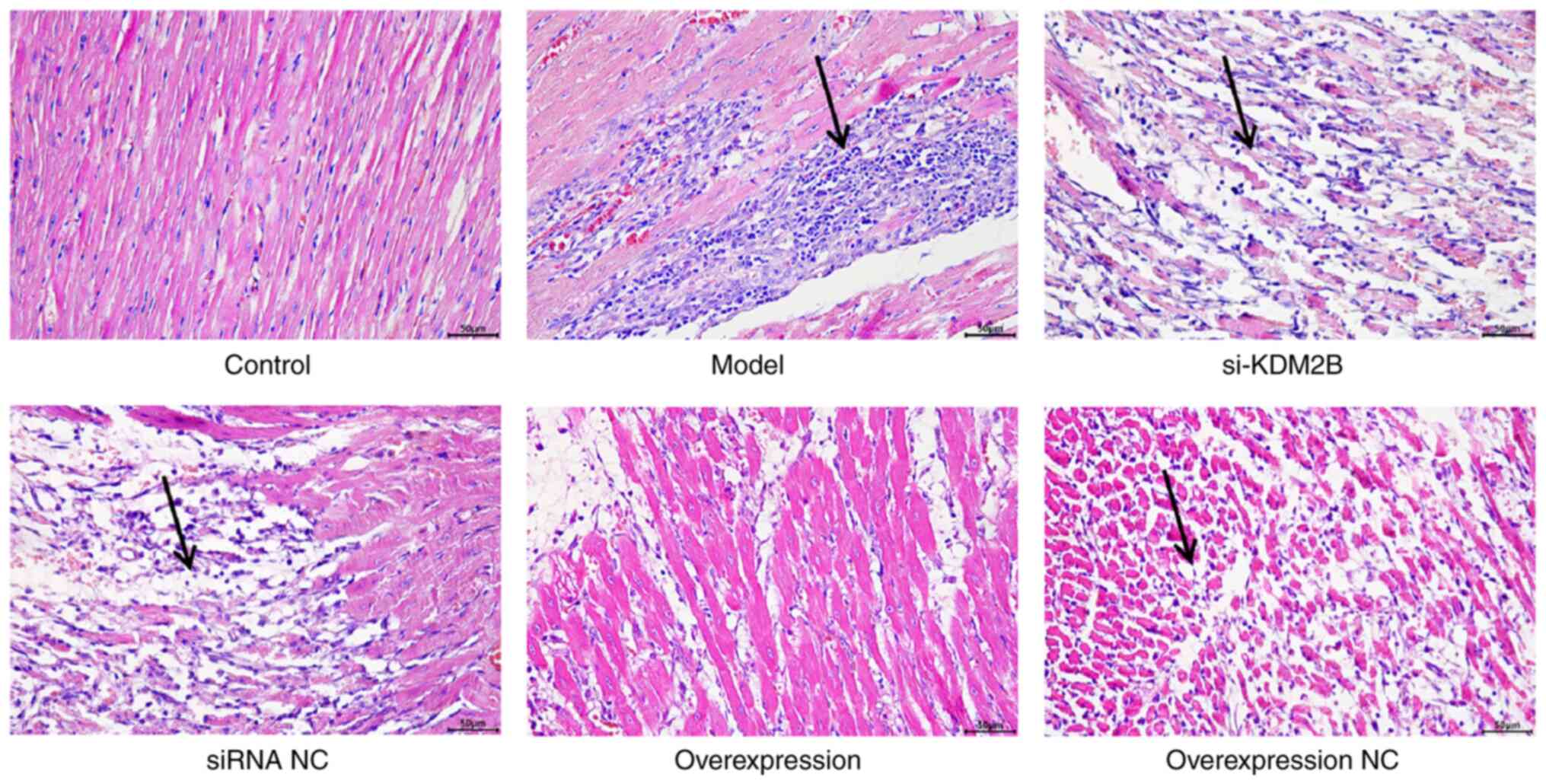 | Figure 1Effects of KDM2B on pathological
changes in myocardial ischemia-reperfusion injury in rats. In
total, six groups, including the sham control group, model group,
si-KDM2B group, siRNA NC group, KDM2B overexpression group and
overexpression NC group, were included in the present study.
H&E staining was applied to determine the pathological changes.
Arrows indicate sites of necrosis and neutrophil infliltration.
Scale bars, 50 µm. AAV, adeno-associated virus; KDM2B,
lysine-specific demethylase 2B; NC, negative control; si/siRNA,
small interfering RNA; si-KDM2B, AAV-siKDM2B; siRNA NC, AAV-siRNA
scrambled negative control. |
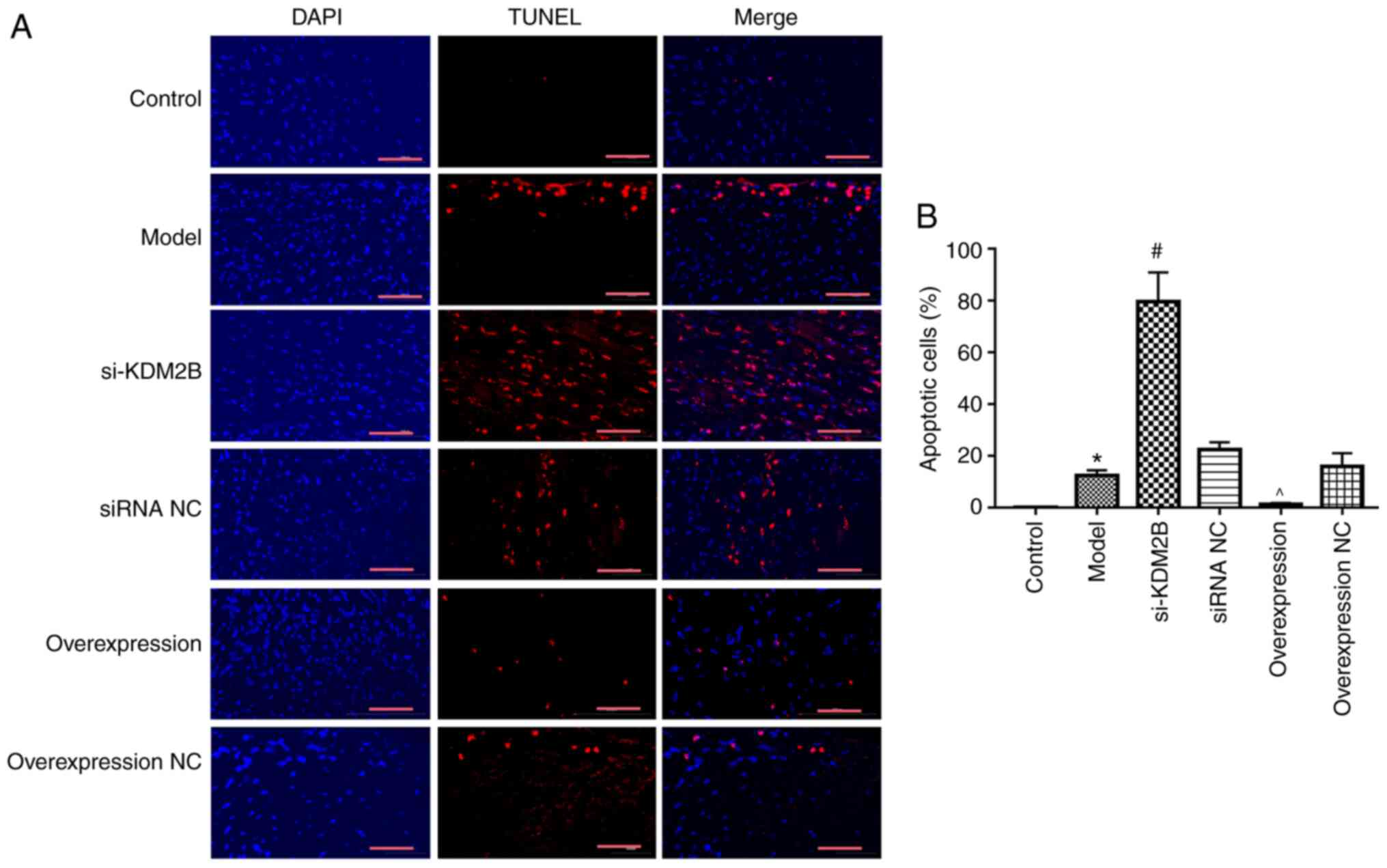
Effects of KDM2B on myocardial
apoptosis in myocardial I/R injury in rats
The effect of KDM2B on myocardial apoptosis in rats
with myocardial I/R injury was investigated. Compared with the
control group, the percentage of apoptotic cells in the heart
tissue in the model group was significantly increased (P<0.05).
si-KDM2B further promoted this significant increase compared with
that in the siRNA NC group, whereas KDM2B overexpression
significantly reduced the extent of apoptosis compared with that in
the overexpression NC group (P<0.05; Fig. 2). These results indicated that
KDM2B may serve a protective role in the apoptosis of myocardial
tissues upon myocardial I/R injury.
Effects of KDM2B on the inflammatory
response in myocardial I/R injury in rats
Compared with those of the control group, the levels
of IL-1β, IL-6 and TNF-α in the peripheral blood of the model group
were significantly increased (P<0.05; Fig. 3). Compared with the siRNA NC group,
the si-KDM2B group exhibited a further significant increase in the
levels of IL-1β, IL-6 and TNF-α (P<0.05), whereas overexpression
of KDM2B significantly reduced the levels of IL-1β, IL-6 and TNF-α
compared with those in the overexpression NC group (P<0.05).
These results suggested that overexpression of KDM2B may inhibit
the inflammatory response in myocardial I/R injury in rats.
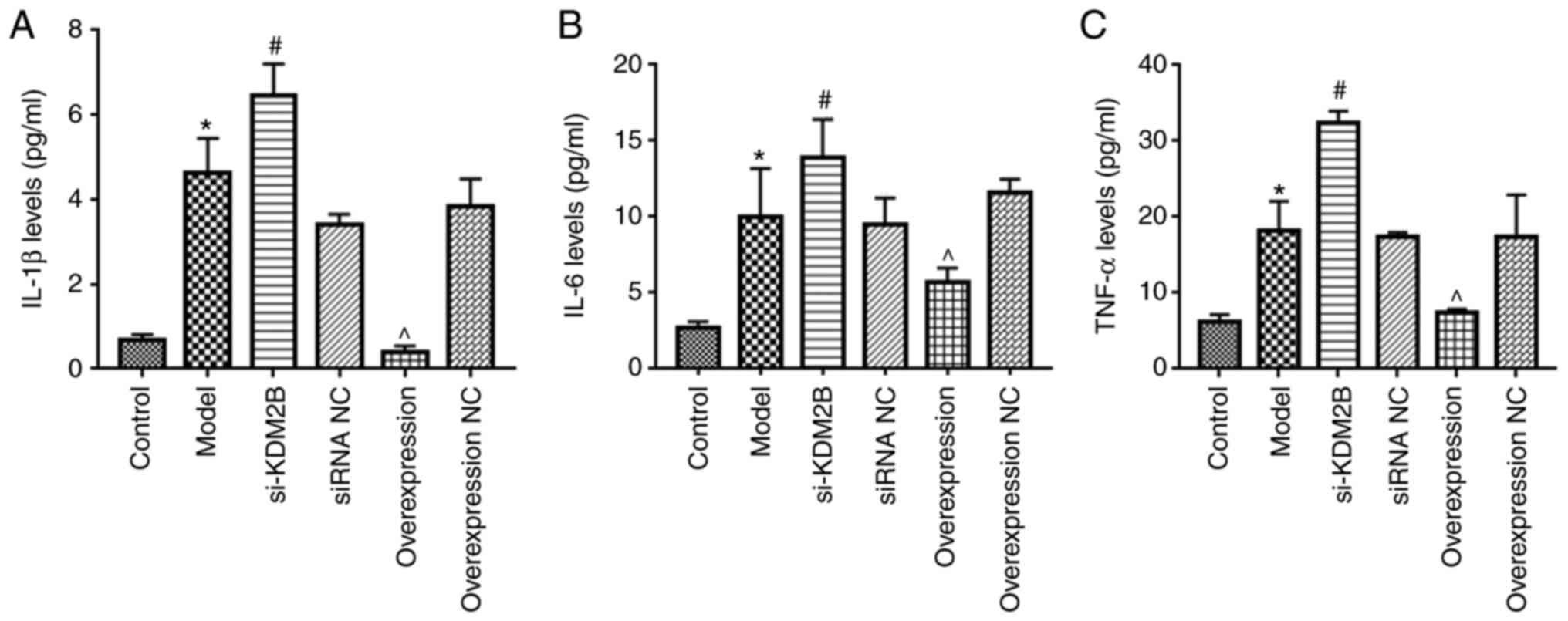 | Figure 3Effects of KDM2B on the inflammatory
response in myocardial ischemia-reperfusion injury in rats. Five
groups, including the sham control group, model group, si-KDM2B
group, siRNA NC group, AAV-KDM2B overexpression group and
AAV-overexpression NC group, were included in the present study.
The levels of (A) IL-1β, (B) IL-6 and (C) TNF-α in peripheral blood
were detected via ELISA. *P<0.05 vs. control;
#P<0.05 vs. siRNA NC; ^P<0.05 vs.
overexpression NC. AAV, adeno-associated virus; KDM2B,
lysine-specific demethylase 2B; NC, negative control; si/siRNA,
small interfering RNA; si-KDM2B, AAV-siKDM2B; siRNA NC, AAV-siRNA
scrambled negative control. |
Effects of KDM2B on TLR4, NF-κB p65
and NLRP3 expression in myocardial I/R injury in rats
Western blotting demonstrated that KDM2B protein
expression was significantly decreased in the model group compared
with the control group (P<0.05). Compared with the corresponding
siRNA NC group, si-KDM2B significantly reduced KDM2B protein
expression levels further (P<0.05), whereas KDM2B overexpression
significantly promoted KDM2B protein expression compared with that
in the overexpression NC group (P<0.05).
The protein expression levels of NLRP3, NF-κB
p65/p-p65 ratio and TLR4 in the model group were significantly
higher compared with those of the control group (P<0.05).
Compared with the corresponding siRNA NC group, the protein
expression levels of NLRP3, NF-κB p65/p-p65 ratio and TLR4 in the
si-KDM2B group were significantly increased further (P<0.05),
whereas the protein expression levels in the overexpression group
were significantly reduced compared with those in the
overexpression NC group (P<0.05; Fig. 4A and B).
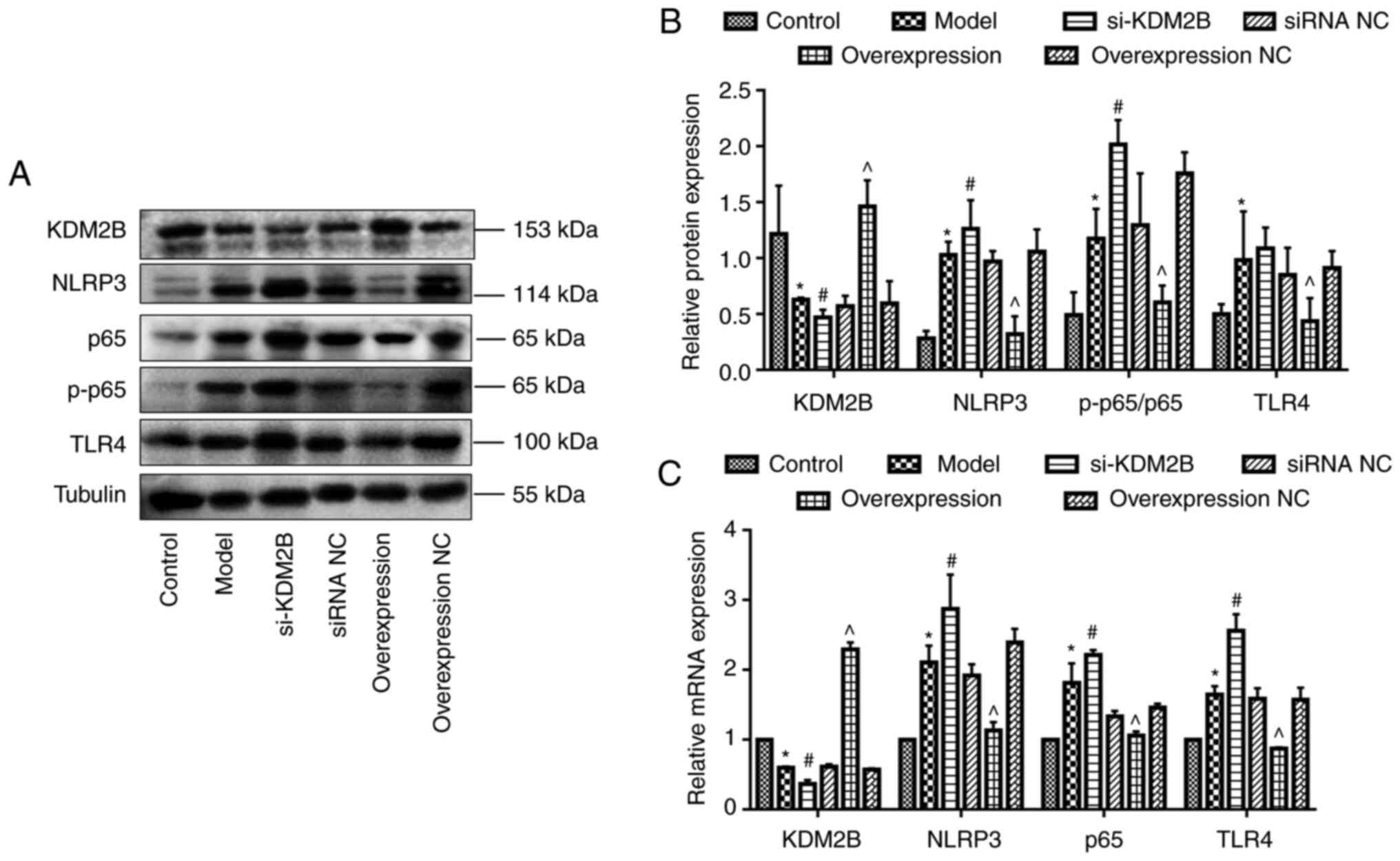 | Figure 4Effects of KDM2B on TLR4, NF-κB p65
and NLRP3 expression levels in myocardial ischemia-reperfusion
injury in rats. Five groups, including the sham control group,
model group, si-KDM2B group, siRNA NC group, AAV-KDM2B
overexpression group and AAV-overexpression NC group, were included
in the present study. (A) Western blotting and (B)
semi-quantification of protein expression levels. (C)
Quantification of mRNA expression levels. *P<0.05 vs.
control; #P<0.05 vs. siRNA NC; ^P<0.05
vs. overexpression NC. AAV, adeno-associated virus; KDM2B,
lysine-specific demethylase 2B; NC, negative control; NLRP3, NOD-,
LRR- and pyrin domain-containing protein 3; p-, phosphorylated;
si/siRNA, small interfering RNA; si-KDM2B, AAV-siKDM2B; siRNA NC,
AAV-siRNA scrambled negative control; TLR4, toll-like receptor
4. |
Compared with the control group, the mRNA expression
levels of KDM2B were significantly decreased, whereas the mRNA
expression levels of NLRP3, NF-κB p65 and TLR4 in the model group
were significantly increased (P<0.05). Compared with the
corresponding siRNA NC group, mRNA expression levels of KDM2B in
the si-KDM2B group were significantly reduced, and this was
accompanied by a significant increase in the expression levels of
NLRP3, NF-κB p65 and TLR4. Furthermore, the mRNA expression levels
of KDM2B in the KDM2B overexpression group were significantly
increased, whereas the mRNA expression levels of NLRP3, NF-κB p65
and TLR4 were significantly decreased compared with compared with
those in the overexpression NC group (P<0.05; Fig. 4C).
Discussion
In the present study, KDM2B was overexpressed or
silenced in the I/R injury rat model. The results demonstrated that
reducing KDM2B expression upregulated the protein and mRNA
expression levels of TLR4, NLRP3 and NF-κB p65 and promoted the
apoptosis of myocardial cells. However, KDM2B overexpression
significantly downregulated the mRNA and protein expression levels
of TLR4, NLRP3 and NF-κB p65 and inhibited the apoptosis of
myocardial tissues. Therefore, the present study suggest that KDM2B
may be a treatment target for myocardial I/R injury.
Histone modification could affect the innate immune
response by changing the transcription level of genes (24). KDM2B is a member of the histone
lysine demethylase KDM family, which serves important regulatory
roles in cell differentiation, development and tumorigenesis
(25,26), as well as the inflammatory response
(27,28). KDM2B promotes the production of
IL-6 and the inflammatory response via brahma-related gene
1-mediated chromatin remodeling (17). Additionally, KDM2B-deficient mice
exhibit stronger resistance to endotoxic shock and colitis, a
lighter inflammatory phenotype and reduced serum IL-6 production
(17). Furthermore, KDM2B
expression is reduced in the nasal mucosa of patients with chronic
atrophic rhinitis, where reduced KDM2B facilitates the development
of nasal mucosa (29). Therefore,
KDM2B serves a dual role in the regulation of inflammation in
epithelial cells (29). The
differential function of KDM2B may be explained by distinctive
mechanisms of KDM2B in different cells or diseases. Therefore, the
role of KDM2B in inflammation will be a focus of future research.
In the present study, KDM2B was silenced or overexpressed in
myocardial I/R injury rat models. The results demonstrated that
si-KDM2B significantly increased the levels of inflammation in the
model rats, which was indicated by the increased levels of IL-1β,
IL-6 and TNF-α. However, overexpression of KDM2B inhibited the
expression of these inflammatory genes. Pathological results also
demonstrated that the overexpression of KDM2B improved myocardial
tissue injury. The inflammatory response after initial ischemic
injury is a key mechanism of secondary degeneration (30,31).
The key role of the NLRP3 inflammasome in regulating
the inflammatory response has been demonstrated in numerous studies
(32,33). The NLRP3 inflammasome can also
increase brain injury and neuroinflammation in ischemic stroke
(34,35). In the present study, NLRP3
expression levels were detected and the results demonstrated that
myocardial I/R injury promoted the mRNA and protein expression of
the NLRP3 inflammasome. si-KDM2B further promoted NLRP3 expression.
By contrast, overexpression of KDM2B reduced NLRP3 expression.
Activation of the NLRP3 inflammasome requires the TLR4/NF-κB
signaling pathway to promote the transcription of NLRP3(36). TLR4 is highly expressed in
myocardial cells and vascular endothelial cells, which is closely
related to the occurrence and development of cardiovascular disease
(37). TLR4 activates the immune
and inflammatory response by regulating NF-κB (32) and aggravates myocardial injury
(38,39). The results of the present study
demonstrated that silencing and overexpression of KDM2B promoted or
inhibited the protein and mRNA expression levels of TLR4, NLRP3 and
NF-κB p65, respectively, which is opposite with the trend of KDM2B.
These results suggested that KDM2B may regulate the expression
levels of proteins associated with the TLR4/NF-κB signaling
pathway.
The present study demonstrated that KDM2B regulated
the TLR4/NF-κB p65 axis by potentially reducing the inflammatory
response, inhibiting cardiomyocyte apoptosis and improving
myocardial injury in rats with myocardial I/R injury. However, the
regulatory mechanism of KDM2B on the TLR4/NF-κB signaling pathway,
whether it depends on the activity of demethylase or directly
regulates non-histone proteins, is still unclear. A limitation of
the present study was that there are numerous types of cells in
myocardial tissues, and thus, the function of KDM2B in specific
cell types should be further investigated. Furthermore, the
potential of KDM2B as a therapeutic target for myocardial I/R
injury will need to be further clarified. In conclusion,
overexpression of KDM2B may prevent myocardial I/R injury in rats
by reducing the inflammatory response via regulation of the
KDM2B/TLR4/NF-κB p65 axis.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81960326) and Key R & D
Plan of Jiangxi Provincial Department of Science and Technology
(grant no. 20192BBG70039).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZW, LL, SH and RT performed the experiments and
analyzed the data. ZW and ZL confirmed the authenticity of all the
raw data. ZW and ZL designed the study and wrote the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Gannan Medical
University (approval no. LLSC-2021101202; Ganzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Anderson JL and Morrow DA: Acute
myocardial infarction. N Engl J Med. 376:2053–2064. 2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Yussman MG, Toyokawa T, Odley A, Lynch RA,
Wu G, Colbert MC, Aronow BJ, Lorenz JN and Dorn GW II:
Mitochondrial death protein Nix is induced in cardiac hypertrophy
and triggers apoptotic cardiomyopathy. Nat Med. 8:725–730.
2002.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Lam CK, Zhao W, Cai W, Vafiadaki E, Florea
SM, Ren X, Liu Y, Robbins N, Zhang Z, Zhou X, et al: Novel role of
HAX-1 in ischemic injury protection involvement of heat shock
protein 90. Circ Res. 112:79–89. 2013.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Chouchani ET, Pell VR, Gaude E,
Aksentijević D, Sundier SY, Robb EL, Logan A, Nadtochiy SM, Ord
ENJ, Smith AC, et al: Ischaemic accumulation of succinate controls
reperfusion injury through mitochondrial ROS. Nature. 515:431–435.
2014.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zhang Z, Qin P, Deng Y, Ma Z, Guo H, Guo
H, Hou Y, Wang S, Zou W, Sun Y, et al: The novel estrogenic
receptor GPR30 alleviates ischemic injury by inhibiting
TLR4-mediated microglial inflammation. J Neuroinflammation.
15(206)2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Shanmugam K, Ravindran S, Kurian GA and
Rajesh M: Fisetin confers cardioprotection against myocardial
ischemia reperfusion injury by suppressing mitochondrial oxidative
stress and mitochondrial dysfunction and inhibiting glycogen
synthase kinase 3β activity. Oxid Med Cell Longev.
2018(9173436)2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Huang Z, Zhao D, Wang Y, Li X, Li J, Han
J, Jiang L, Ai F and Zhou Z: C1q/TNF-related protein 9 decreases
cardiomyocyte hypoxia/reoxygenation-induced inflammation by
inhibiting the TLR4/MyD88/NF-κB signaling pathway. Exp Ther Med.
22(1139)2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Xu XN, Jiang Y, Yan LY, Yin SY, Wang YH,
Wang SB, Fang LH and Du GH: Aesculin suppresses the NLRP3
inflammasome-mediated pyroptosis via the Akt/GSK3β/NF-κB pathway to
mitigate myocardial ischemia/reperfusion injury. Phytomedicine.
92(153687)2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Vargas-Ayala RC, Jay A, Manara F, Maroui
MA, Hernandez-Vargas H, Diederichs A, Robitaille A, Sirand C,
Ceraolo MG, Romero-Medina MC, et al: Interplay between the
epigenetic enzyme lysine (K)-specific demethylase 2B and
epstein-barr virus infection. J Virol. 93:e00273–19.
2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Inagaki T, Iwasaki S, Matsumura Y,
Kawamura T, Tanaka T, Abe Y, Yamasaki A, Tsurutani Y, Yoshida A,
Chikaoka Y, et al: The FBXL10/KDM2B scaffolding protein associates
with novel polycomb repressive complex-1 to regulate adipogenesis.
J Biol Chem. 290:4163–4177. 2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Liang G, He J and Zhang Y: Kdm2b promotes
induced pluripotent stem cell generation by facilitating gene
activation early in reprogramming. Nat Cell Biol. 14:457–466.
2012.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Yan M, Yang X, Wang H and Shao Q: The
critical role of histone lysine demethylase KDM2B in cancer. Am J
Transl Res. 10:2222–2233. 2018.PubMed/NCBI
|
|
13
|
Zacharopoulou N, Tsapara A, Kallergi G,
Schmid E, Alkahtani S, Alarifi S, Tsichlis PN, Kampranis SC and
Stournaras C: The epigenetic factor KDM2B regulates EMT and small
GTPases in colon tumor cells. Cell Physiol Biochem. 47:368–377.
2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Chen L, Fu L, Kong X, Xu J, Wang Z, Ma X,
Akiyama Y, Chen Y and Fang J: Jumonji domain-containing protein 2B
silencing induces DNA damage response via STAT3 pathway in
colorectal cancer. Br J Cancer. 110:1014–1026. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Isshiki Y, Nakajima-Takagi Y, Oshima M,
Aoyama K, Rizk M, Kurosawa S, Saraya A, Kondo T, Sakaida E,
Nakaseko C, et al: KDM2B in polycomb repressive complex 1.1
functions as a tumor suppressor in the initiation of T-cell
leukemogenesis. Blood Adv. 3:2537–2549. 2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Domizi P, Malizia F, Chazarreta-Cifre L,
Diacovich L and Banchio C: KDM2B regulates choline kinase
expression and neuronal differentiation of neuroblastoma cells.
PLoS One. 14(e0210207)2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhou Q, Zhang Y, Wang B, Zhou W, Bi Y,
Huai W, Chen X, Chen Y, Liu Z, Liu X and Zhan Z: KDM2B promotes
IL-6 production and inflammatory responses through Brg1-mediated
chromatin remodeling. Cell Mol Immunol. 17:834–842. 2020.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhou XY, Liu J, Xu ZP, Fu Q, Wang PQ and
Zhang H: Dexmedetomidine inhibits the lipopolysaccharide-stimulated
inflammatory response in microglia through the pathway involving
TLR4 and NF-κB. Kaohsiung J Med Sci. 35:750–756. 2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Song Z, Shen F, Zhang Z, Wu S and Zhu G:
Calpain inhibition ameliorates depression-like behaviors by
reducing inflammation and promoting synaptic protein expression in
the hippocampus. Neuropharmacology. 174(108175)2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhao H, Chen Z, Xie LJ and Liu GF:
Suppression of TLR4/NF-κB signaling pathway improves cerebral
ischemia-reperfusion injury in rats. Mol Neurobiol. 55:4311–4319.
2018.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Arumugam TV, Okun E, Tang SC, Thundyil J,
Taylor SM and Woodruff TM: Toll-like receptors in
ischemia-reperfusion injury. Shock. 32:4–16. 2009.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhang Z, Song Z, Shen F, Xie P, Wang J,
Zhu AS and Zhu G: Ginsenoside Rg1 prevents PTSD-like behaviors in
mice through promoting synaptic proteins, reducing Kir4.1 and TNF-α
in the hippocampus. Mol Neurobiol. 58:1550–1563. 2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Mehta S and Jeffrey KL: Beyond receptors
and signaling: Epigenetic factors in the regulation of innate
immunity. Immunol Cell Biol. 93:233–244. 2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Chiu WT, Huang YF, Tsai HY, Chen CC, Chang
CH, Huang SC, Hsu KF and Chou CY: FOXM1 confers to
epithelial-mesenchymal transition, stemness and chemoresistance in
epithelial ovarian carcinoma cells. Oncotarget. 6:2349–2365.
2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wang JJ, Dong R, Wang LP, Wang JS, Du J,
Wang SL, Shan ZC and Fan ZP: Histone demethylase KDM2B inhibits the
chondrogenic differentiation potentials of stem cells from apical
papilla. Int J Clin Exp Med. 8:2165–2173. 2015.PubMed/NCBI
|
|
27
|
Souto JA, Sarno F, Nebbioso A, Papulino C,
Álvarez R, Lombino J, Perricone U, Padova A, Altucci L and de Lera
ÁR: A new family of jumonji C domain-containing KDM inhibitors
inspired by natural product purpurogallin. Front Chem.
8(312)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Kang MK, Mehrazarin S, Park NH and Wang
CY: Epigenetic gene regulation by histone demethylases: Emerging
role in oncogenesis and inflammation. Oral Dis. 23:709–720.
2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Liu CC, Sun C, Zheng X, Zhao MQ, Kong F,
Xu FL, Chen XJ, Wang XX, Zhang M and Xia M: Regulation of KDM2B and
Brg1 on inflammatory response of nasal mucosa in CRSwNP.
Inflammation. 42:1389–1400. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Jin R, Yang G and Li G: Inflammatory
mechanisms in ischemic stroke: Role of inflammatory cells. J Leukoc
Biol. 87:779–789. 2010.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Hamzei Taj S, Kho W, Aswendt M, Collmann
FM, Green C, Adamczak J, Tennstaedt A and Hoehn M: Dynamic
modulation of microglia/macrophage polarization by miR-124 after
focal cerebral ischemia. J Neuroimmune Pharmacol. 11:733–748.
2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Shen F, Song Z, Xie P, Li L, Wang B, Peng
D and Zhu G: Polygonatum sibiricum polysaccharide prevents
depression-like behaviors by reducing oxidative stress,
inflammation, and cellular and synaptic damage. J Ethnopharmacol.
275(114164)2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Song Z, Bian Z, Zhang Z, Wang X, Zhu A and
Zhu G: Astrocytic Kir4.1 regulates NMDAR/calpain signaling axis in
lipopolysaccharide-induced depression-like behaviors in mice.
Toxicol Appl Pharmacol. 429(115711)2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Ma C, Liu S, Zhang S, Xu T, Yu X, Gao Y,
Zhai C, Li C, Lei C, Fan S, et al: Evidence and perspective for the
role of the NLRP3 inflammasome signaling pathway in ischemic stroke
and its therapeutic potential (Review). Int J Mol Med.
42:2979–2990. 2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Sun J, Chi L, He Z, Gao Y, Gao Y, Huang Y
and Nan G: NLRP3 inflammasome contributes to neurovascular unit
damage in stroke. J Drug Target. 27:866–875. 2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Kopitar-Jerala N: Innate immune response
in brain, NF-kappa B signaling and cystatins. Front Mol Neurosci.
8(73)2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Goulopoulou S, McCarthy CG and Webb RC:
Toll-like receptors in the vascular system: Sensing the dangers
within. Pharmacol Rev. 68:142–167. 2016.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zhang Y, Lu Y, Ma L, Cao X, Xiao J, Chen
J, Jiao S, Gao Y, Liu C, Duan Z, et al: Activation of vascular
endothelial growth factor receptor-3 in macrophages restrains
TLR4-NF-κB signaling and protects against endotoxin shock.
Immunity. 40:501–514. 2014.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Li J, Xie C, Zhuang J, Li H, Yao Y, Shao C
and Wang H: Resveratrol attenuates inflammation in the rat heart
subjected to ischemia-reperfusion: Role of the TLR4/NF-κB signaling
pathway. Mol Med Rep. 11:1120–1126. 2015.PubMed/NCBI View Article : Google Scholar
|