Introduction
Colorectal cancer (CRC) remains one of the most
common malignancies worldwide, with over 1 million cases diagnosed
in 2014. While patients with early stage CRC have a 5-year survival
rate of 90%, those with advanced disease have a survival rate of
just 13% (1). The mechanistic basis
of CRC development and progression remains to be fully elucidated
and numerous patients exhibit various associated risk factors
contributing to disease onset (2).
CRC also remains the fourth deadliest cancer type, although there
have been significant reductions in average mortality rates for
patients with CRC in recent decades owing to diagnostic and
therapeutic innovations (3). Poor
outcomes of CRC are most frequently a result of tumor metastasis or
the acquisition of a drug-resistant form of the disease, and such
phenotypes arise from cancer-associated gene dysregulation
(2,4-6).
In this light, a better understanding of the genes associated with
CRC development may be of value. In addition, it is essential that
novel diagnostic and prognostic biomarkers of CRC are identified in
order to better screen for this deadly disease and to predict its
progression in affected individuals.
Transmembrane p24 trafficking protein 3 [also known
as transmembrane Emp24 protein transport domain containing 3
(TMED3)] is an important protein associated with both innate immune
functionality and protein trafficking within the vesicles of cells
(7). There are 10 known TMED family
proteins (8), and most of them have
been studied in tumor-associated contexts (9-12),
whereas TMED3 has only been studied in select instances wherein its
role in the development and progression of prostate (13), colon (14) and liver (15) cancers was examined. TMED3 belongs to
a family of p24 proteins involved in selecting cargo in coat
protein complex vesicles in the secretory endoplasmic
reticulum-Golgi network (16).
Given the large diversity of cargo and the existence of only 10
TMED p24 proteins, it is likely that each is able to affect
multiple secretion events in direct and indirect context-dependent
manners. Furthermore, p24 proteins may exist as monomers or dynamic
complexes where one is able to affect the stability of others
(17-20).
They appear to be non-redundant (21) and affect multiple signaling pathways
in mammalian cells (10,22,23).
In flies and mammals, specific TMED proteins control WNT secretion
(14,24-26).
These studies all suggested that TMED3 is related to these
tumorigenic processes. Previous studies have reported that TMED3
act as metastatic suppressors in human colorectal cancer cells
through the WNT-TCF pathway (14,27).
The importance of this gene in CRC, however, has remained largely
elusive, with its association with patient prognosis being
completely undetermined. Thus, in the present study, TMED3
expression was examined at the protein level in CRC and normal
para-cancerous tissue samples, and furthermore, the relationship of
TMED3 with clinicopathological findings and survival outcomes in
these patients was assessed.
Patients and methods
Patients and samples
Between June 2006 and March 2009, a total of 176
formalin-fixed paraffin-embedded (FFPE) pairs of tumor and normal
para-cancerous tissue samples were collected from patients with
stage I-III CRC undergoing curative surgery at Changhai Hospital,
Second Military Medical University (Shanghai, China). These samples
were archived for immunohistochemistry (IHC) analyses and the CRC
diagnosis was confirmed by pathological examination of all samples.
Patients were not included in the present study cohort if they had
received prior anti-cancer therapy, suffered from abnormal cardiac,
lung, liver or renal function, had been previously diagnosed with
other cancers or died due to other causes. The clinicopathological
characteristics of this patient cohort are compiled in Table I.
 | Table IAssociation between TMED3 protein
expression and clinicopathologic characteristics of patients with
CRC in the first study cohort (n=176). |
Table I
Association between TMED3 protein
expression and clinicopathologic characteristics of patients with
CRC in the first study cohort (n=176).
| | TMED3 protein
level | |
|---|
| Characteristic | Patients (n) | High (n=88) | Low (n=88) | P-value |
|---|
| Sex | | | | 0.879 |
|
Female | 77 | 38 | 39 | |
|
Male | 99 | 50 | 49 | |
| Age (years) | | | | 0.868 |
|
<60 | 51 | 25 | 26 | |
|
≥60 | 125 | 63 | 62 | |
| Tumor location | | | | 0.649 |
|
Rectum | 79 | 38 | 41 | |
|
Colon | 97 | 50 | 47 | |
| Degree of
differentiation | | | | 0.724 |
|
Well +
moderate | 134 | 66 | 68 | |
|
Poor | 42 | 22 | 20 | |
| Tumor size
(cm) | | | | 0.006 |
|
<5 | 70 | 26 | 44 | |
|
≥5 | 106 | 62 | 44 | |
| Local invasion | | | | 0.013 |
|
pT1-T2 | 134 | 60 | 74 | |
|
pT3-T4 | 42 | 28 | 14 | |
| Lymph node
metastasis | | | | 0.003 |
|
N0+N1 | 109 | 45 | 64 | |
|
N2 | 67 | 43 | 24 | |
| TNM stage | | | | 0.282 |
|
I + Ⅱ | 105 | 49 | 56 | |
|
Ⅲ | 71 | 39 | 32 | |
| Adjuvant
chemotherapy | | | | 0.245 |
|
No | 51 | 29 | 22 | |
|
Yes | 125 | 59 | 66 | |
| CA19-9 (kU/l) | | | | 0.546 |
|
<40 | 82 | 39 | 43 | |
|
≥40 | 94 | 49 | 45 | |
| Serum CEA level
(ng/ml) | | | | 0.544 |
|
<10 | 78 | 41 | 37 | |
|
≥10 | 98 | 47 | 51 | |
An additional cohort of independent samples was
obtained between April and October 2013 from 63 patients with stage
I-III CRC undergoing curative surgery at this same institution.
These samples were stored for reverse transcription-quantitative
(RT-q) PCR analyses. The clinicopathological characteristics of
this patient cohort are provided in Table II.
 | Table IIClinicopathological characteristics
of the second study cohort (n=63) for assessing TMED3 mRNA
level. |
Table II
Clinicopathological characteristics
of the second study cohort (n=63) for assessing TMED3 mRNA
level.
| Characteristic | N |
|---|
| Sex | |
|
Female | 30 |
|
Male | 33 |
| Age (years) | |
|
<60 | 20 |
|
≥60 | 43 |
| Tumor location | |
|
Rectum | 40 |
|
Colon | 23 |
| Degree of
differentiation | |
|
Well +
moderate | 50 |
|
Poor | 13 |
| Tumor size
(cm) | |
|
<5 | 38 |
|
≥5 | 25 |
| Local invasion | |
|
pT1-T2 | 22 |
|
pT3-T4 | 41 |
| Lymph node
metastasis | |
|
N0+N1 | 39 |
|
N2 | 24 |
| TNM stage | |
|
I + II | 36 |
|
III | 27 |
| Adjuvant
chemotherapy | |
|
No | 26 |
|
Yes | 37 |
| CA19-9 (kU/l) | |
|
<40 | 21 |
|
≥40 | 42 |
| Serum CEA level
(ng/ml) | |
|
<10 | 27 |
|
≥10 | 35 |
All patients provided written informed consent and
this study conformed to the Declaration of Helsinki and was
approved by the Institutional Ethics Committee of Changhai
Hospital, Second Military Medical University (Shanghai, China;
ethics approval no. 2017-148-01).
RT-qPCR
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.)
was used to extract total RNA from individual samples, and total
RNA (1 µg) was used for RT with PrimeScript™ RT reagent kit (Takara
Bio, Inc.) according to the manufacturer's protocol, using a
thermal cycler (i-Cycler; Bio-Rad Laboratories, Inc.). The RT
reaction was conducted in 40 µl reaction buffer at 37˚C for 15 min
and terminated by heating at 85˚C for 5 sec, followed by cooling at
4˚C. qPCR was performed with a 7500 Real-time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) and SYBR Premix Ex Taq™
(Takara Bio, Inc.) according to the manufacturer's protocol. PCR
was performed at 95˚C for 10 sec, followed by 40 cycles of 95˚C for
5 sec and 60˚C for 34 sec. Dissociation was initiated at 95˚C for
15 sec followed by 60˚C for 1 min and 95˚C for 15 sec. GAPDH served
as an internal standard. The comparative 2-ΔΔCq method
was used to determine the relative gene expression (28). Primers were as follows: TMED3,
forward 5'-GGGTTCTGTACCTGAGGAAA-3' and reverse
5'-CACCGAGGGTGAGCAGAT-3; GAPDH, forward 5'-TGTGGGCATCAATGGATTTGG-3'
and reverse 5'-ACACCATGTATTCCGGGTCAAT-3.
Immunohistochemistry (IHC)
IHC was performed as in previous studies (29). In brief, sections were heated and
then probed with anti-TMED3 antibody (1:50; cat. no. ab151056;
Abcam) for 60 min at 37˚C, followed by a 15 min incubation with
HRP-conjugated goat anti-rabbit polyclonal antibody solution (ready
to use; cat. no. SP-9001; OriGene Technologies, Inc.) at room
temperature for 15 min. Hematoxylin was then used to counterstain
samples and diaminobenzidine was used for sample development. A
total of three independent pathologists blinded to the patient
characteristics then examined and scored individual samples.
H-scores (30) were then assigned
at x200 magnification, with samples receiving scores of 0, 1, 2 or
3 corresponding to negative, weak, intermediate or strong staining,
respectively. Numbers of cells per field of view with a particular
staining intensity were then quantified, with H-scores being
assigned based on the following formula: H-score = (% of cells with
staining strength 1x1) + (% of cells with staining strength 2x2) +
(% of cells with staining strength 3x3). The final scores ranged
from 0-300, with 0 corresponding to 100% of cells being negative
for the antigen of interest and 300 corresponding to 100% strong
staining for that antigen. Median H-score values were used to
stratify patients into low- and high-expressing groups.
Follow-up of cases
The patients enrolled in the present study were
followed up from the date of surgery until death or most recent
follow-up, with those remaining alive as of the last follow-up
being censored. None was lost to follow-up. Overall survival (OS)
was determined based on the period of time between surgery and
death, while recurrence-free survival (RFS) was determined as the
period of time between surgery and CRC recurrence, with patients
not exhibiting recurrence being censored on the date of death or
most recent follow-up. The 7th edition of the tumor-node-metastasis
(TNM) system was used to stage patients' tumors based on the
criteria defined by the American Joint Committee on Cancer Staging
(31). All patients with stage III
CRC as well as those with stage II disease and either poorly
differentiated or pT4 tumors were administered 5-fluorouracil-based
adjuvant chemotherapy. In all patients, RFS and OS were calculated
monthly through to December 2017.
Online database
In the present study, the gene expression database
for normal and tumor tissues (GENT) (http://medicalgenome.kribb.re.kr/GENT/; http://genome.kobic.re.kr/GENT/) was used.
Statistical analysis
SPSS 24.0 (IBM Corp.) was used for all statistical
testing. DifferencesinTMED3 mRNA expression and H-scores between
the CRC tissues and their corresponding normal para-cancerous
tissues were analyzed for statistical significance using the paired
t-test. The relationship between TMED3 expression and
clinicopathological characteristics of patients was assessed using
the χ2 test. Differences in patient overall survival
(OS) and recurrence-free survival (RFS) as a function of TMED3
expression were compared via the Kaplan-Meier method, with the
significance determined with the log-rank test. A Cox proportional
hazards model was used to determine the independent factors of OS
and RFS based on variables selected after the univariate analysis.
P<0.05 (two-tailed) was considered to indicate statistical
significance.
Results
Assessment of TMED3 expression in CRC
tissue samples
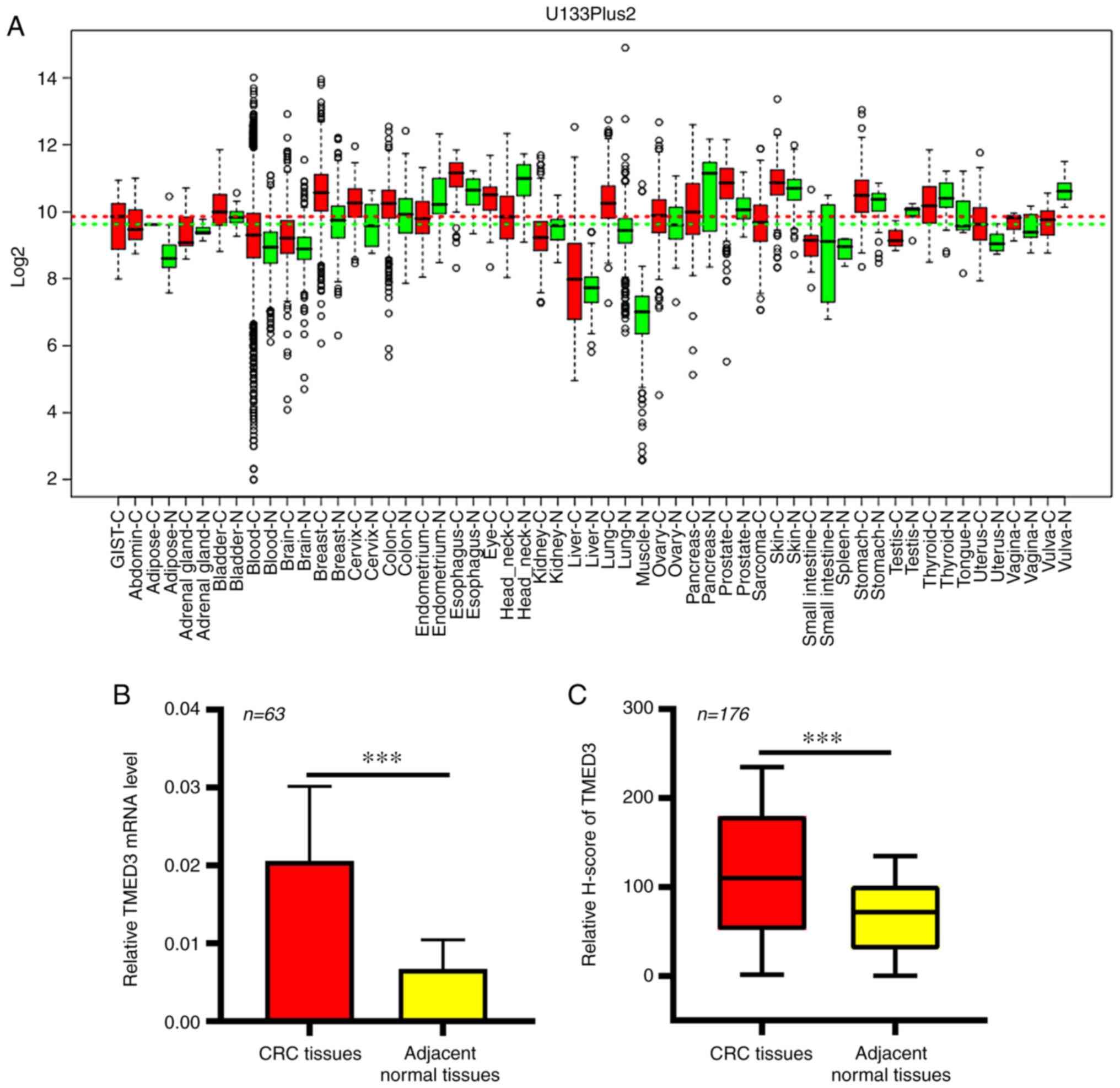
In the GENT database, significant increases in the
expression of TMED3 at the mRNA level were observed in numerous
cancer types, including CRC (Fig.
1A). This thus suggested a potential role for TMED3 as a
regulator in the development and/or progression of CRC. This
finding was then further confirmed in an independent cohort of
patients with CRC (Table II), in
which elevated TMED3 expression in CRC tissue samples relative to
the levels in normal para-cancerous tissues from the same patients
was observed (P<0.01; Fig. 1B).
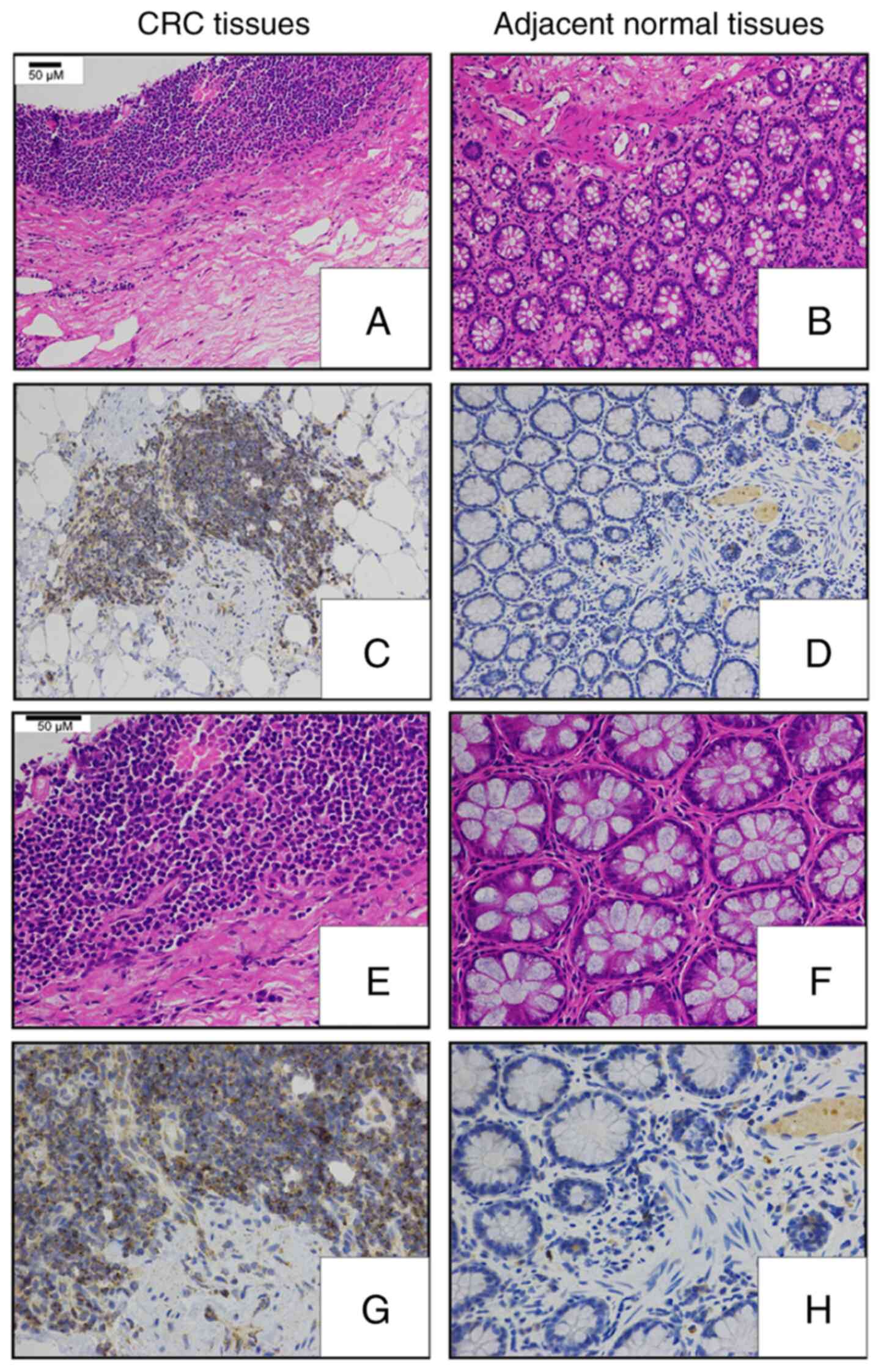
Next, an IHC-based approach was used in order to assess TMED3
protein levels in 176 pairs of FFPE CRC and normal para-cancerous
tissue sections (Table I). These
analyses revealed that TMED3 was localized to both the nucleus and
the cytoplasm of tumor cells and confirmed the elevated expression
of this protein specifically in tumor cells relative to normal
para-cancerous cells (Figs. 1C and
2).
Association between TMED3 levels and
clinicopathological characteristics of patients with CRC
Next, the relationship between TMED3 expression
levels and clinicopathological characteristics of patients with CRC
was examined. The 176 patients were divided into a TMED3-high
(n=88) and a TMED3-low (n=88) group according to the median TMED3
expression levels in this cohort. Elevated TMED3 expression was
indicated to be significantly associated with larger tumor size
(P=0.006), depth of local invasion (P=0.013) and lymph node
metastasis (P=0.003; Table I). By
contrast, TMED3 expression levels were not significantly associated
with patient sex, age, tumor location, tumor differentiation grade,
TNM stage, adjuvant chemotherapy status, carbohydrate antigen 19-9
(CA19-9) levels or serum carcinoembryonic antigen levels. These
results thus suggested that TMED3 expression may be significantly
linked to CRC metastasis.
Association between TMED3 expression
and postoperative survival of patients with CRC
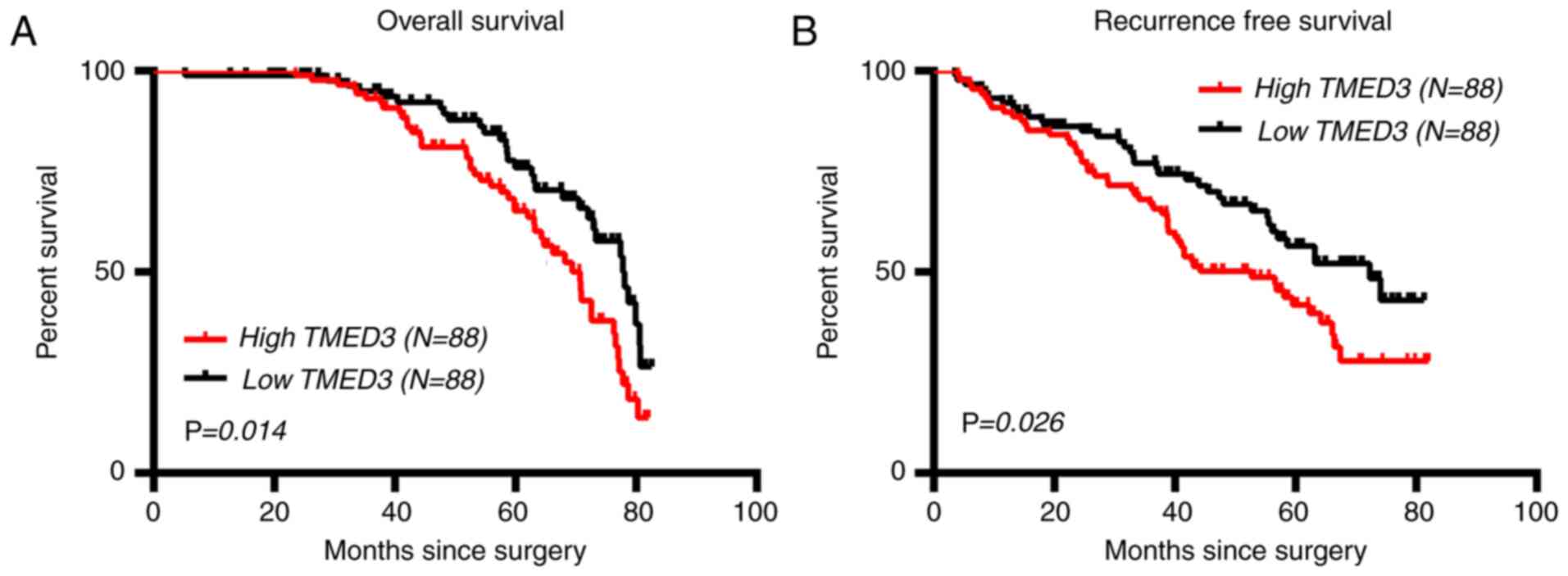
The association between the expression of TMED3 and
postoperative survival outcomes in the 176 patients with CRC was
then explored. The results indicated that the TMED3-low group had a
significantly lower median OS time than theTMED3-high group (78.10
vs. 70.70 months; 95% CI, 1.115-2.715, P=0.014; Fig. 3A). Similarly, TMED3 expression was
associated with RFS of the patients with CRC, with the TMED3-low
group having a significantly longer median RFS time than
theTMED3-high group (72.30 vs. 52.70 months, 95% CI, 1.059-2.433,
P=0.026; Fig. 3B).
Identification of factors associated
with prognosis of patients with CRC
Next, logistic regression analysis using the Cox
proportional hazards model was used to identify factors associated
with OS and RFS outcomes for patients with CRC. According to the
univariate analysis, tumor size, degree of differentiation, local
invasion status, lymph node metastasis, TNM staging and TMED3
expression levels were all significantly associated with OS
(P<0.05; Table III). These
same factors were also associated with post-operative RFS in the
patients with CRC (P<0.05). However, patient sex, age and CA19-9
levels had no significant effect on the RFS and OS of the patients
with CRC (P>0.05). A multivariate analysis was next performed,
with all factors identified in the univariate analyses being
incorporated into this multivariate model. This analysis revealed
that elevated TMED3 protein levels were associated with reduced OS
[hazard ratio (HR), 1.875; 95% CI, 1.305-2.641; P=0.036)] and RFS
(HR, 1.776; 95% CI, 1.374-2.661; P=0.048) of patients with CRC.
Taken together, these results indicated that TMED3 may be an
independent predictor of survival outcomes for patients with
CRC.
 | Table IIIUnivariate and multivariate analyses
of factors influencing patient survival in the study cohort
(Table I). |
Table III
Univariate and multivariate analyses
of factors influencing patient survival in the study cohort
(Table I).
| A, Overall
survival |
|---|
| | Univariate
analysis | Multivariate
analysis |
|---|
| Variable | Comparison | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Sex | Male/female | 1.236 | 0.298 | 2.131 | 0.901 | - | - | - | - |
| Age (years) | ≥60/<60 | 0.522 | 0.679 | 1.655 | 0.537 | - | - | - | - |
| Tumor location | Colon/rectum | 1.412 | 0.264 | 1.974 | 0.141 | - | - | - | - |
| Tumor size
(cm) | ≥5/<5 | 1.139 | 1.241 | 1.987 | 0.044 | 1.109 | 0.887 | 1.397 | NS |
| Degree of
differentiation | Poor/well +
moderate | 1.593 | 1.295 | 3.692 | 0.015 | 1.761 | 1.286 | 2.962 | 0.039 |
| Local invasion | pT3-4/pT1-2 | 1.588 | 1.887 | 4.153 | 0.011 | 1.471 | 1.388 | 1.655 | 0.043 |
| Lymph node
metastasis | N2/N0+N1 | 1.954 | 1.216 | 2.366 | 0.039 | 1.204 | 1.229 | 1.912 | 0.041 |
| TNM stage | Ⅲ/I + Ⅱ | 2.181 | 1.451 | 2.957 | 0.021 | 1.769 | 1.545 | 2.697 | 0.019 |
| CA19-9 (kU/l) | ≥37/<37 | 1.041 | 0.361 | 1.483 | 0.172 | - | - | - | - |
| CEA (ng/ml) | ≥5/<5 | 1.368 | 0.849 | 1.933 | 0.051 | - | - | - | - |
| TMED3 protein
level | High/low | 2.128 | 1.821 | 3.928 | 0.021 | 1.875 | 1.305 | 2.641 | 0.036 |
| B, Recurrence-free
survival |
| | Univariate
analysis | Multivariate
analysis |
| Variable | Comparison | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Sex | Male/female | 1.334 | 0.513 | 1.726 | 0.417 | - | - | - | - |
| Age (years) | ≥60/<60 | 1.231 | 0.321 | 1.631 | 0.322 | - | - | - | - |
| Tumor location | Colon/rectum | 1.279 | 0.550 | 1.675 | 0.926 | - | - | - | - |
| Tumor size
(cm) | ≥5/<5 | 1.515 | 1.317 | 2.975 | 0.035 | 0.812 | 0.663 | 1.781 | NS |
| Degree of
differentiation | Poor/well +
moderate | 1.912 | 1.527 | 2.701 | 0.024 | 1.207 | 1.162 | 2.718 | 0.046 |
| Local invasion | pT3-4/pT1-2 | 1.564 | 1.115 | 2.565 | 0.017 | 0.991 | 0.869 | 1.718 | NS |
| Lymph node
metastasis | N2/N0+N1 | 1.805 | 1.247 | 2.992 | 0.043 | 1.238 | 1.186 | 2.118 | 0.039 |
| TNM stage | Ⅲ/I + Ⅱ | 2.155 | 1.443 | 2.787 | 0.032 | 1.032 | 0.789 | 1.878 | NS |
| CA19-9 (kU/l) | ≥37/<37 | 1.291 | 0.847 | 2.386 | 0.735 | - | - | - | - |
| CEA (ng/ml) | ≥5/<5 | 1.317 | 0.907 | 2.483 | 0.493 | - | - | - | - |
| TMED3 protein
level | High/low | 1.438 | 1.302 | 2.643 | 0.028 | 1.776 | 1.374 | 2.661 | 0.048 |
Discussion
TMED3 is a protein that is well-known to have key
roles in vesicular trafficking, particularly in the context of
early secretory pathways (12). To
date, only three studies have performed a comprehensive examination
of the significance of TMED3 in human cancer, proving its relevance
in prostate (13), colon (14) and liver cancer (15). These studies, however, have provided
contradictory results that suggest that the role of TMED3 is
strongly dependent on the tumor type. Vainio et al (13) first observed high TMED3 mRNA
expression levels in prostate cancer and found this expression to
correlate with that of the oncogenes ETS transcription factor ERG
and androgen receptor, leading to the conclusion that TMED3 may
represent a potential therapeutic target in this cancer type.
Duquet et al (14) performed
a study on TMED3 in a xenograft model of colon cancer, indicating
that TMED3 suppressed tumor metastasis via regulating WNT/TCF
pathway signaling, thus suggesting that in this context, TMED3
suppresses tumor progression. In contrast to these results,
however, Zheng et al (15)
determined that elevated TMED3 expression in hepatocellular
carcinoma (HCC) was associated with more aggressive disease and
unfavorable patient prognosis, with this protein promoting enhanced
tumor cell invasion at least in part via modulating IL-11/STAT3
signaling. Duquet et al (14) performed an extensive functional
examination of the role of TMED3 in murine models and colon cancer
cell lines, while the role of this protein in clinical tissue
samples was not directly assessed.
In the present study, TMED3 levels were
significantly higher in CRC tissue samples relative to those in
normal para-cancerous tissues. This was the case at both the mRNA
level, as confirmed via RT-qPCR analysis of 63 patient sample
pairs, and at the protein level, as confirmed via IHC assessment of
176 patient sample pairs. In all cases, significantly elevated
TMED3 levels were observed in tumor tissue samples relative to
those in the matched controls (P<0.001). These results confirmed
that TMED3 expression was significantly elevated in CRC tissues at
the mRNA and protein level.
Next, the association between the expression of
TMED3 and the clinicopathological characteristics of patients with
CRC were explored in the IHC cohort (n=176). When patients were
stratified according to TMED3 expression levels, it was indicated
that high TMED3 expression was significantly associated with larger
tumor size (P=0.006), depth of local invasion (P=0.013) and lymph
node metastasis (P=0.003), suggesting that elevated TMED3
expression is associated with more aggressive and malignant
behaviors of CRC. Postoperative survival outcomes were then
assessed in these 176 patients, revealing that TMED3-low patients
had significantly longer OS and RFS relative to TMED3-high CRC
patients. Furthermore, univariate and multivariate models were used
to explore the relationship between specific variables and
postoperative patient survival. The analysis revealed that elevated
TMED3 expression was an independent predictor of reduced CRC
patient survival. This suggests that TMED3 may have a role in the
development of CRC or in its progression, thus making it a
potential prognostic biomarker for CRC. Further research regarding
the relevance of TMED3 in CRC and whether it is a viable
therapeutic target is thus warranted and such analyses may have the
potential to further improve the OS and RFS of patients with CRC.
Previous studies have confirmed that TMED3 expression levels are
linked to the prognosis of patients with HCC (15), but the present study was the first,
to the best of our knowledge, to examine its prognostic value in
CRC. The present results are consistent with those of Zheng et
al (15), but not with those of
Duquet et al (14), possibly
due to small sample sizes. The present results are, however, the
first to highlight the prognostic relevance of TMED3 expression
levels in CRC.
In conclusion, the present results revealed that
TMED3 expression levels are predictive of survival outcomes for
patients with CRC, with elevated expression of this protein being
independently predictive of poor OS. As such, TMED3 represents a
valuable novel prognostic biomarker that may be used for monitoring
and/or the treatment of patients with CRC. However, as the present
study had a relatively small sample size, it has the potential to
be susceptible to bias. Therefore, future studies with a larger
independent sample of patients with CRC are required in order to
validate these results.
Acknowledgements
Not applicable.
Funding
Funding: This study was supported by a grant from the Natural
Science Foundation of the Shanghai Science and Technology
Commission (grant no. 16ZR1400800).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
RFW and HTY contributed to the study design. YGH
performed data analysis. LQH contributed to the collection of
tissue samples and patient data. YGH and HTY wrote the manuscript.
RFW, HTY, LQH and YGH confirmed the authenticity of the raw data.
All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The Ethics Committee of Changhai Hospital, Second
Military Medical University (Shanghai, China) approved the study.
Written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Welch HG and Robertson DJ: Colorectal
cancer on the cecline - Why screening can't explain it all. N Engl
J Med. 374:1605–1607. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Glunde K, Bhujwalla ZM and Ronen SM:
Choline metabolism in malignant transformation. Nat Rev Cancer.
11:835–848. 2011.PubMed/NCBI View
Article : Google Scholar
|
|
5
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Guo P, Huang ZL, Yu P and Li K: Trends in
cancer mortality in China: An update. Ann Oncol. 23:2755–2762.
2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Connolly DJ, O'Neill LA and McGettrick AF:
The GOLD domain-containing protein TMED1 is involved in
interleukin-33 signaling. J Biol Chem. 288:5616–5623.
2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Strating JR, Hafmans TG and Martens GJ:
Functional diversity among p24 subfamily members. Biol Cell.
101:207–219. 2009.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zakariyah A, Hou W, Slim R and
Jerome-Majewska L: TMED2/p24β1 is expressed in all gestational
stages of human placentas and in choriocarcinoma cell lines.
Placenta. 33:214–219. 2012.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Liaunardy-Jopeace A, Bryant CE and Gay NJ:
The COP II adaptor protein TMED7 is required to initiate and
mediate the delivery of TLR4 to the plasma membrane. Sci Signal.
7(ra70)2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Hou W and Jerome-Majewska LA: TMED2/emp24
is required in both the chorion and the allantois for placental
labyrinth layer development. Dev Biol. 444:20–32. 2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hasegawa H, Liu L and Nishimura M:
Dilysine retrieval signal-containing p24 proteins collaborate in
inhibiting γ-cleavage of amyloid precursor protein. J Neurochem.
115:771–781. 2010.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Vainio P, Mpindi JP, Kohonen P, Fey V,
Mirtti T, Alanen KA, Perälä M, Kallioniemi O and Iljin K:
High-throughput transcriptomic and RNAi analysis identifies AIM1,
ERGIC1, TMED3 and TPX2 as potential drug targets in prostate
cancer. PLoS One. 7(e39801)2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Duquet A, Melotti A, Mishra S, Malerba M,
Seth C, Conod A and Ruiz i Altaba A: A novel genome-wide in vivo
screen for metastatic suppressors in human colon cancer identifies
the positive WNT-TCF pathway modulators TMED3 and SOX12. EMBO Mol
Med. 6:882–901. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zheng H, Yang Y, Han J, Jiang WH, Chen C,
Wang MC, Gao R, Li S, Tian T, Wang J, et al: TMED3 promotes
hepatocellular carcinoma progression via IL-11/STAT3 signaling. Sci
Rep. 6(37070)2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Strating JR and Martens GJ: The p24 family
and selective transport processes at the ER-Golgi interface. Biol
Cell. 101:495–509. 2009.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Füllekrug J, Suganuma T, Tang BL, Hong W,
Storrie B and Nilsson T: Localization and recycling of gp27
(hp24gamma3): Complex formation with other p24 family members. Mol
Biol Cell. 10:1939–1955. 1999.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Jenne N, Frey K, Brugger B and Wieland FT:
Oligomeric state and stoichiometry of p24 proteins in the early
secretory pathway. J Biol Chem. 277:46504–46511. 2002.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Jerome-Majewska LA, Achkar T, Luo L, Lupu
F and Lacy E: The trafficking protein Tmed2/p24beta(1) is required
for morphogenesis of the mouse embryo and placenta. Dev Biol.
341:154–166. 2010.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Montesinos JC, Sturm S, Langhans M,
Hillmer S, Marcote MJ, Robinson DG and Aniento F: Coupled transport
of Arabidopsis p24 proteins at the ER-Golgi interface. J Exp Bot.
63:4243–4261. 2012.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Denzel A, Otto F, Girod A, Pepperkok R,
Watson R, Rosewell I, Bergeron JJ, Solari RC and Owen MJ: The p24
family member p23 is required for early embryonic development. Curr
Biol. 10:55–58. 2000.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Doyle SL, Husebye H, Connolly DJ, Espevik
T, O'Neill LA and McGettrick AF: The GOLD domain-containing protein
TMED7 inhibits TLR4 signalling from the endosome upon LPS
stimulation. Nat Commun. 3(707)2012.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wang X, Yang R, Jadhao SB, Yu D, Hu H,
Glynn-Cunningham N, Sztalryd C, Silver KD and Gong DW:
Transmembrane emp24 protein transport domain 6 is selectively
expressed in pancreatic islets and implicated in insulin secretion
and diabetes. Pancreas. 41:10–14. 2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Buechling T, Chaudhary V, Spirohn K, Weiss
M and Boutros M: p24 proteins are required for secretion of Wnt
ligands. EMBO Rep. 12:1265–1272. 2011.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Port F, Hausmann G and Basler K: A
genome-wide RNA interference screen uncovers two p24 proteins as
regulators of Wingless secretion. EMBO Rep. 12:1144–1152.
2011.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Li X, Wu Y, Shen C, Belenkaya TY, Ray L
and Lin X: Drosophila p24 and Sec22 regulate Wingless
trafficking in the early secretory pathway. Biochem Biophys Res
Commun. 463:483–489. 2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Mishra S, Bernal C, Silvano M, Anand S,
Ruiz I and Altaba A and Altaba A: The protein secretion modulator
TMED9 drives CNIH4/TGFα/GLI signaling opposing TMED3-WNT-TCF to
promote colon cancer metastases. Oncogene. 38:5817–5837.
2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Tao QF, Yuan SX, Yang F, Yang S, Yang Y,
Yuan JH, Wang ZG, Xu QG, Lin KY, Cai J, et al: Aldolase B inhibits
metastasis through Ten-Eleven Translocation 1 and serves as a
prognostic biomarker in hepatocellular carcinoma. Mol Cancer.
14(170)2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Petroski MD and Deshaies RJ: Function and
regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol.
6:9–20. 2005.PubMed/NCBI View
Article : Google Scholar
|
|
31
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474.
2010.PubMed/NCBI View Article : Google Scholar
|