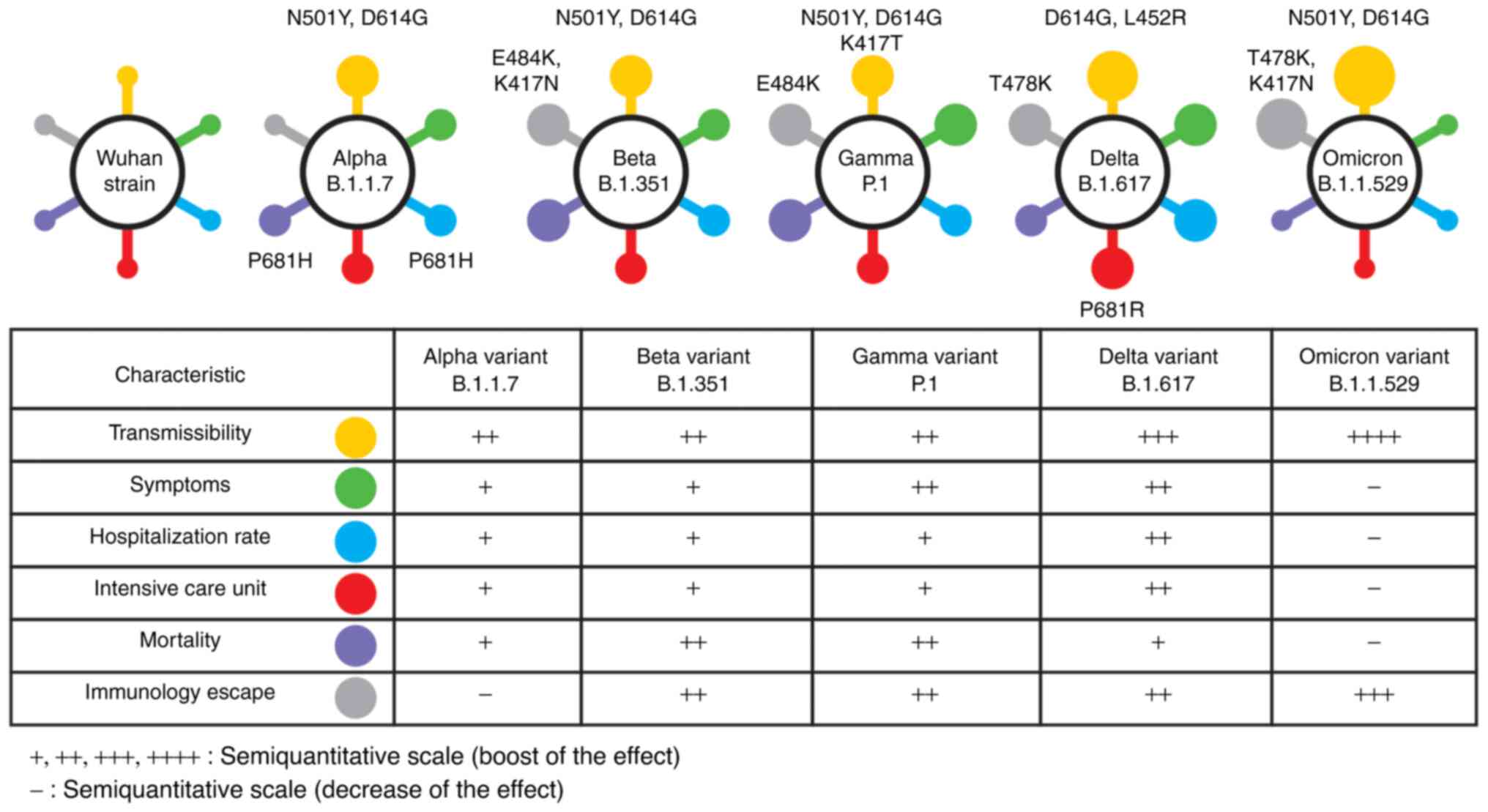
1. Introduction
Coronaviruses are a large virus family that cause a
number of diseases in mammals and birds, such as Middle East
respiratory syndrome (MERS) and severe acute respiratory syndrome
(SARS). Phylogenetic evidence suggests that RNA-dependent RNA
polymerase sequence divergence occurred 7,000-8,000 years ago in
mammals, showing that this type of virus is prone to interspecies
transmission and pathogenicity (1).
There are currently seven types of coronavirus with
human tropism: Human coronavirus (HCoV)-HKU1, HCoV-OC43, HCoV-229E,
HCoV-NL63 (2003), MERS-CoV, SARS-CoV and SARS-CoV-2(2). Among the Coronaviridae family, the
β-coronavirus genus (which includes SARS-CoV and SARS-CoV-2) has
the highest human pathogenic potential.
Coronaviruses are named for the crown pattern formed
by surface spikes. Their genomes are large RNA-type structures,
26-32 kb in length, with single-stranded positive-sense RNA
(3). Viruses such as SARS-CoV-2
acquire mutations due to rapid replication and error-prone viral
polymerase (4).
The open reading frame (ORF)1a/b region of
coronaviruses encodes 16 non-structural proteins, while ORFs region
encodes structural proteins such as spike (S), envelope, membrane
and nucleocapsid (N) protein. Surface antigenic structures are
specific and mutations result in viral variants with unique
features (5).
Viruses such as SARS-CoV-2 adapt to the immune
response in different species various tissues, leading to mutation.
Most mutations, however, do not lead to a change in phenotype or
infectivity (6).
All seven types of coronavirus in humans have
different genetic mutations. MERS-CoV exhibits mutant S, ORF4b and
ORF3 genes, while SARS-CoV exhibits mutant S and ORF8 genes
(4). The primary genetic anomalies
in SARS-CoV-2 are located in the S gene (4). Natural selection produces variants
that promote virus survival via more efficient inter-human
transmissibility, replication or intracellular penetration
capacity. Additionally, the frequency of random events serves a key
role in the creation of novel variants; evolution of more
transmissible forms is more likely than evolution of more
pathogenic variants (4).
A nomenclature to define novel emerging viruses was
developed to facilitate surveillance of epidemiological events.
Variant refers to changes in genomic structures; strain denotes
changes in terms of virulence or transmissibility (7).
Viral genotype changes are classified as follows:
Mutation, single viral genomic mutation; lineage, a group of
associated viruses with common origin and variant, viral genomes
that contain one or more mutations. To guide implementation of
measures to protect the population, public health organizations
introduced the terms variant of concern (VOC), variant of interest
(VOI) and variant under monitoring based on the impact on the
population (disease severity and transmissibility). The United
States also uses the terms variant being monitored and variant of
high consequence, although no variant of SARS-CoV-2 has been
included in the latter category so far.
The Global Initiative on Sharing All Influenza Data,
Nextstrain, and Pango systems have specified SARS-CoV-2 lineage
nomenclature. Furthermore, the World Health Organisation (WHO)
founded the Technical Advisory Group on Virus Evolution to define
key genetic lines emerging from SARS-CoV-2 mutations and classified
these as VOC or VOI. VOCs include Alpha, Beta, Delta, Gama and
Omicron and VOIs include Mu and Lambda variants (Table I) (8).
 | Table IVariants of concern. |
Table I
Variants of concern.
| WHO
nomenclature | Pango lineage | GISAID clade | Nextstrain
clade | Emergence |
|---|
| Alpha | B.1.1.7. | GRY | 20I (V1) | United Kingdom, Sep
2020 |
| Beta | B.1.351 | GH/501Y.V2 | 20H (V2) | South Africa, May
2020 |
| Delta | B.1.617.2 | G/478K.V1 | 21A, 21I, 21J | India, Oct
2020 |
| Gamma | P.1 | GR/501Y.V3 | 20J (V3) | Brazil, Nov
2020 |
| Omicron | B.1.1.529 | GRA | 21K, 21L, 21M | South Africa, 24
Nov 2021 |
The present review investigated SARS-CoV-2 variants
and their clinical features. The present study aimed to support
healthcare practitioners and public health policymakers in treating
patients and minimizing the outbreaks.
2. SARS-CoV-2 variant gene mutations and
impact of clinical features on disease evolution
Numerous mutations, including five VOCs have been
identified as a result of the global effort to detect mutations in
the SARS-CoV-2 genome (Table
I).
Alpha variant (Lineage B.1.1.7)
The first notable variant, Alpha, was detected in
southeast England in September 2020 and caused concern due to its
increased transmissibility and number of viral replications
compared with its predecessors. SARS-CoV-2 Alpha variant was
reported to have a transmissibility of 1.3-1.5 times that of the
original strain (9).
The Alpha variant has 10 mutations in the S protein
that led to structural changes. The N501Y mutation appears in the S
glycoprotein receptor-binding domain (RBD) at position 501 and
results in amino acid asparagine (N) being replaced with tyrosine
(Y). The N501Y mutation increases the binding affinity via an
additional binding site for angiotensin-converting enzyme (ACE)2
SARS-CoV-2 entry receptor (10,11).
H69del/V70del mutation is potentially associated with immune
evasion and is not detected by S-gene PCR assays, resulting in S
gene target failure. The P681H mutation near the S1/S2 furin
cleavage site facilitates epithelial cell entry (10,12).
As a result of these mutations, there has been an increase in the
percentage of hospitalizations and mortality. The D614G mutation
(also registered in all subsequent VOCs) is hypothesized to enhance
viral replication (10).
A model was created to evaluate the relative change
in transmissibility and level of immune evasion for distinct
SARS-CoV-2 variants while accounting for false negatives, reporting
delays, disease seasonality, non-pharmaceutical intervention (such
as self-isolation and social distancing) and vaccination (13). The study estimated that Alpha
variant exhibits a 46.6% increase in transmissibility but no
immunological escape from protection conferred by previous
wild-type infection.
Monel et al (14) analysed 426 nasopharyngeal swabs
from patients who had microbiologically confirmed SARS-CoV-2
infection in a retrospective study: 200 samples were from the
pre-Alpha dominance period (before September 2020) and 226 were
from the dominant Alpha surge. The aforementioned study found a
strong correlation between viral antigen detection and viable viral
shedding, as well as an association between infectious titre and
rapid diagnostic test positivity, low cycle threshold (Cq) value,
early symptom onset and the absence of nasopharyngeal IgG or IgA.
Alpha variant exhibits higher nasopharyngeal viral load and longer
viral shedding, as well as a stronger affinity for ACE2 receptor
binding and higher fusogenicity (15,16).
A study of 381,773 participants used the national
Covid-19 Infection Survey, a representative, longitudinal household
sample, to investigate the spread of Alpha variant in the United
Kingdom (17). A total of 9,032
(50.3%) positive results were triple-gene-positive, indicating
detection of all three regions of the SARS-CoV-2 genome tested
(ORF1ab region and N and S gene); 5,258 (29.3%) had S gene target
failure (indicative of Alpha variant) and 3,673 (20.4%) exhibited
other gene combinations. Although infection with S gene target
failure was more common than triple-gene-positive infection in
symptomatic infection, absolute increases in confirmed diagnosis
were similar regardless of whether people reported symptoms,
suggesting that asymptomatic infection may serve a key role in the
spread of Alpha variant. No notable change was observed in the
efficacy of natural or vaccine-induced antibodies, potentially due
to the lack of pressure induced by introduction of large-scale
vaccination at that time (December 2020-August 2021) (18).
According to Vassallo et al (19), patients infected with the Alpha
variant of SARS-CoV-2 exhibit a more unfavourable evolution than
those infected with previous strains. A total of 65 patients
infected with Alpha variant participated in the aforementioned
study, which compared patients who had never been immunized against
COVID-19 with a control group of patients infected with a previous
strain. The non-immunized mortality rate was 15.4 compared with
12.9% in the control group. There were also significant differences
in the percentage of patients admitted to intensive care unit: 27.7
in the study group vs. 8.6% for control group. The severity of
pneumonia was associated with intensive care admission and
mortality rate. In patients with Alpha infection, the severity of
the disease was also associated with increased viral load, as
assessed by Cq value in reverse transcription-quantitative PCR
diagnostic testing in the aforementioned study. By contrast,
mortality was lower during the second wave of disease (March-May
2020), which was characterised by the spread of Alpha infection,
according to a retrospective investigation in the United Kingdom
(20).
SARS-CoV-2 PCR positivity was predicted by seven
symptoms in a study of >1 million people in England: Loss or
change of smell, loss or change of taste, fever, new persistent
cough, chills, appetite loss and muscle ache (21).
Beta variant (B.1.351)
In December 2020, South Africa announced the
discovery of a novel variant with an E484K mutation. Three
subgroups of this variant with a total of 12 mutations and one
depletion were discovered: K417N, E484K and N501Y mutations led to
an increase in S protein affinity for ACE2(7).
Certain evidence suggests that one S protein
mutation, E484K, may suppress the efficacy of neutralizing
antibodies (22,23).
Studies have investigated the effect of three amino
acid changes found in numerous VOCs (including Alpha, Beta and
Gamma) on the structure and function of SARS-CoV-2 S glycoprotein
RBD (24,25). The aforementioned studies
discovered that these alterations change the structure, stability
and ability of the RBD to bind to ACE2 in an unpredictable manner.
Thus, RBD VOC substitutions change the structure and stability of
the RBD, with K417N and N501Y increasing stability and E484K
decreasing stability. These substitutions result in stability
similar to the wild-type/Wuhan strain RBD, but with a more open
conformation and higher ACE2 binding affinity.
The aforementioned mutations caused contagiousness
of the novel variants to increase by up to 52% (7). The E484K mutation causes
conformational changes that put pressure on immunological evasion
(7). These RBD substitutions in
Beta S protein decrease the binding and neutralization of both mRNA
vaccine-induced antibodies and potent human monoclonal antibodies
(24).
According to one study, Beta exhibits increased
rates of transmissibility (32.4%) and immune escape (61.3%)
(13).
To the best of our knowledge, there are few studies
on Beta variant and with most reports focus on increased viral load
and immune response evasion (7,26). A
study evaluating the impact of Beta variant in South Africa during
the second wave of infection found an increase in hospitalization
rate and in-hospital mortality (26). The increased mortality was due to a
higher percentage of elderly people being admitted to hospitals, as
well greater demand on the healthcare system.
Gamma variant (P.1. or 20J/501Y.V3,
Lineage B.1.1.28)
Due to three changes in the RBD domain (K417T, E484K
and N501Y), the WHO included Gamma variant in the VOC group
simultaneously with Beta variant. This novel variant has 17
mutations, 11 of which are in the S protein. These mutations have
consequences in terms of infectivity, risk of reinfection and
immune evasion: Compared with wild-type, Gamma variant exhibits a
161% increase in infection rate and 50% increase in mortality
(7).
A model simulation indicated that Gamma variant has
a 43.3% increase in transmissibility rate and 52.5% increase in
immune escape (13). Gamma variant
shows an improved ability to resist the immune response acquired
during infection with previous variants (13).
The most common symptoms identified in 423 people
infected with Gamma variant of SARS-CoV-2 who worked in the health
system in Sao Paulo, Brazil, were coryza, headache, cough, sore
throat, myalgia, and asthenia (27).
SARS-CoV-2 caused by Gamma variant is more likely
than wild-type to cause cold-like symptoms. Hyposmia/anosmia and
dysgeusia are more common in younger and female patients according
to one study (7). Coryza (73%) and
headache (72%) are among the most common symptoms (27).
Delta variant (1.2.7., Lineage
B.1.617. or B.1.617.2)
In October 2020, India announced the discovery of a
novel variant including three key S protein mutations (L452R, E484Q
and P681R) that increased rate of transmission (8). This variant caused concern due to
increased household transmissibility (+64%) and doubled risk of
hospitalization compared with Alpha (28). Because the genomic changes
discovered in this variant include mutations found in both Alpha
and Beta variants, it was considered to be an epidemic variant with
significant risk (29). The strain
subsequently became dominant in multiple countries (including
Denmark, Germany and the Netherlands), suggesting that it had a
competitive advantage over previously identified strains (30). A Delta variant outbreak was
discovered in Guangzhou, China, in May 2021, with rapid spread
(four transmission generations within 10 days) (31).
To understand the links between viral variants,
disease severity and viral shedding kinetics, a retrospective study
evaluated the outcomes of individuals infected with Alpha, Beta and
Delta variant (32). Compared with
wild-type, Delta was associated with greater risk of ventilation,
intensive care unit admission or mortality and the OR odds ratio
for pneumonia was 1.88 (95% CI, 0.95-3.76). Alpha and Beta did not
display these associations. Vaccination status was linked to
decreased severity. Delta was associated with lower PCR Cq values
and increased duration of Cq value ≤30 (median duration, 18 days
for Delta, 13 days for wild-type). The aforementioned study
revealed a potential link between infection with Delta variant and
risk of pneumonia or severe COVID-19. In respiratory samples, Delta
was associated with higher viral load and longer viral shedding
(32).
Twohig et al (33) found that patients infected with
Delta variant had more than double the risk of hospital admission
and increased risk of hospital attendance (emergency care
attendance or hospital admission) compared with those infected with
Alpha variant. The aforementioned study also discovered that
non-vaccinated patients infected with Delta were more than twice as
likely to be admitted to hospital as those infected with Alpha
variant. Compared with wild-type and Alpha variant, Delta variant
causes more severe illness and poorer clinical outcomes (33).
In a study comparing Delta and Alpha infection
traits in southern Italy, researchers discovered a decrease in the
proportion of subjects under the age of 36 years with Delta
infection, as well as a higher risk of hospitalization; for the two
groups, risk of death was similar (34).
A study involving 1,915 patients in South Korea
found that individuals diagnosed during community-based spread of
Delta were more likely to exhibit symptomatic and severe
SARS-CoV-2(35). Symptoms such as
fever, chills, fatigue, cough, sputum production and dyspnea were
more common in the Delta-dominant group compared with the
Delta-minor group. Moreover, compared with the incidence of
asymptomatic cases during the isolation period, pneumonia was more
common in the Delta-dominant group. Delta-dominant infection was an
independent risk factor for all severity factors (oxygen saturation
<95%, progression of dyspnoea, increased pneumonic infiltration)
as well as for the probability of hospital transfer in multivariate
analysis (35).
The clinical characteristics of the Delta variant
include a shorter incubation time, shorter period of evolution
towards critical forms of the disease (associated with increase
mortality and intensive care admission rate) and a higher frequency
of critical forms (31).
A national cohort study in Qatar compared patients
infected with Beta with those infected with Delta variant (36). The study revealed that patients
with Beta variant were more likely to be hospitalized (27.3 vs.
20.0%) and exhibit mild-moderate or severe-critical disease (27.9
vs. 20.2%). There were no significant differences in the need for
supplemental or high-flow oxygen, mechanical ventilation or death
between the two groups (36). Old
age and comorbidities in patients infected with the Delta variant
were associated with higher risk of poor outcomes compared with
patients infected with Beta variant (36).
Following infection with Delta, double-vaccinated
patients exhibit a considerably decreased risk of intermediate or
severe outcomes. Vaccination is associated with decreased peak
levels of systemic inflammation, fewer symptoms, fewer instances of
asymptomatic infection and improved clinical outcomes (37).
Omicron variant (B.1.1.529)
Omicron variant emerged in South Africa and Botswana
~12 months after the previous VOC. This variant contains >60
substitutions, deletions and insertions, including 39 on S protein,
of which 15 occur in the RBD and confer a considerable increase in
morbidity (10,38,39).
Multiple Omicron mutations reported to be identical in the Alpha
and Delta variants result in increased transmissibility rate of
+105% compared with Delta variant (40). Omicron variant has the ability to
avoid infection-blocking antibodies and causes less severe
symptoms, exerting less impact on the lungs (39). Tropism for the upper respiratory
tract is associated with milder clinical manifestation and
decreased mortality. However, the increased presence of the virus
in the upper respiratory tract may result in easier spread
(39).
One Omicron variant mutation is associated with S
gene target failure (or S gene dropout), which means that one
region of the gene targeted by PCR testing will result in a false
negative (39).
The aforementioned studies indicated that Omicron
variant has unique characteristics, such as altered
transmissibility and severity of disease, as a result of its dozens
of mutations, resulting in altered rates of infection
dissemination, morbidity and mortality.
According to preliminary data from South Africa,
there are no unique symptoms associated with Omicron variant
infection, although more patients are asymptomatic compared with
other variants (10).
A study showed that the Omicron-driven fourth wave
had a lower severity of disease, with fewer deaths, ICU admissions
and length of hospital stay in its first global epicentre in South
Africa. This clinical profile is also likely to have been
influenced by younger patient age as this age group was less
affected by previous variants of the virus (41).
The wave grew faster than preceding waves, replacing
the Delta variant within weeks and starting to diminish in both
cases and hospital admissions in the fifth week since its start
(41).
Mild symptoms were commonly observed in Omicron
variant infection. Mild cough, fever, generalized myalgia, malaise,
scratchy but not painful throat, headache, body discomfort and
moderate to severe fatigue are among symptoms reported by patients
(39).
3. Discussion
The proportion of individuals with asymptomatic
disease did not significantly change as the incidence of Alpha
increased (42). A study of 165
people found that those infected with Alpha (50%) and Beta (90%)
variants are mostly asymptomatic, but those infected with Delta
(17%) variant exhibit severe clinical symptoms (43). It is important to identify the
asymptomatic cases in order to decrease virus transmission, however
this is difficult to do in practice and consumes a lot of
resources.
With the emergence of multiple variants of
SARS-CoV-2, identifying infected patients may become difficult.
Identification is accomplished using viral whole-genome sequencing,
which is not globally available and has a high cost in terms of
both material and human resources (44). Although a number of cheaper tests
have been proposed (such as detection of multiple variant lineages
with >20 key SARS-CoV-2 S mutations), efforts to adapt
diagnostic techniques to rapid viral evolution are ongoing
(45). Rapid antigen tests are a
less expensive alternative to molecular assay for diagnosis and are
sensitive to Alpha, Beta and Gamma variants (46,47).
Serological tests determine prior infection based on the detection
of antibodies but there is no data on the ability to discriminate
between different viral strains (48).
SARS-CoV-2 has a range of clinical features and
undergoes continuous evolution (Fig.
1).
Although respiratory symptoms (assessed by lung
damage) are the most common, the mechanisms triggered by infection
are various and dynamic; these include cytokine storm, coagulation
disorders, as evaluated by the appearance of thrombosis, oxidative
stress that causes ferroptosis, changes in immune cells and
neurological impairment (49-53).
Typical clinical manifestations in the first reports of SARS-CoV-2
(including fever, fatigue, loss of smell, myalgia, headache, cough
and shortness of breath), have either become less significant or
associated with specific variants (49-53).
During SARS-CoV-2 infection, interferon pathways become impaired,
causing higher mortality and longer disease course (54).
Alpha, Beta, Gamma, and Delta variants were
discovered in 2020. All of these variants were isolated before
SARS-CoV-2 vaccination became widely available (8,55).
As a result, their emergence was more likely due to spontaneous
mutations arising during gene transcription instead of the vaccine
or convalescent plasma treatment. Omicron variant emerged following
immunisation efforts, especially in developed countries. However,
only ~25% of the population in South Africa (in which Omicron
emerged) was vaccinated at the end of November 2021, which suggests
that there was not enough pressure from immunological escape to
lead to novel variants (8). Based
on these findings, it was hypothesized that the occurrence of
mutations in SARS-CoV-2 is the result of natural processes rather
than human intervention.
From the perspective of the impact of each variant
on public health, as assessed by indicators such as mortality,
hospitalization risk, and admission in intensive care units,
variations may arise from differences between national health
systems. On the other hand, concerns about the severity of variants
were not always realised, based on data from retrospective studies.
The better preparation of health systems from one wave to the next
and early introduction of antiviral therapy (such as remdesivir),
thromboprophylaxis, high flow oxygen therapy or non-pharmacological
treatments (placing patients in a prone position) had a
considerable impact (56-59).
Changes in clinical features such as anosmia or
dysgenesis occurred during the emergence of Gamma infection and
were notable following emergence of the Omicron variant. Clinical
signs have changed, suggesting a progression towards a cold-like
character with a self-limiting evolution (39).
Given the large number of symptomatic people and
increased demand for SARS-CoV-2 infection tests, it is unclear
whether extensive testing is still feasible and necessary (Cojocaru
et al, unpublished data). Analysis of the average number of
tests reported by certain European countries, the United States and
Canada in the first 7 days of 2022 revealed significant variation
in the number of diagnostic tests performed for SARS-CoV-2
infection (Table II) (60). The positive rate is higher where
the number of tests is lower, suggesting a large number of
undiagnosed positive cases. In these cases, it seems logical to
change the approach to symptomatic infections by focusing resources
on cases with a high risk of adverse effects and expanding hotline
networks to assist these patients (Cojocaru et al,
unpublished data).
 | Table IINumber of severe acute respiratory
syndrome coronavirus 2 tests in different countries. |
Table II
Number of severe acute respiratory
syndrome coronavirus 2 tests in different countries.
| Country,
publication date | Mean number of
tests (7-day average) | Number of positive
tests (7-day average) | Proportion of
positive tests, % | Population,
100,000 | Number of
tests/100,000 inhabitants |
|---|
| France,
07.01.2022 | 1,371.513 | 272.931 | 19.9 | 673.9 | 2,035.2 |
| Germany,
09.01.2022 | 212.644 | 48.483 | 22.8 | 841.9 | 252.5 |
| Italy,
07.01.2022 | 829.723 | 136.904 | 16.5 | 603.2 | 1,375.5 |
| Spain,
06.01.2022 | 295.479 | 89.678 | 30.3 | 467.8 | 631.6 |
| UK, 07.01.2022 | 1,800.852 | 180.085 | 10.0 | 684.3 | 2,631.6 |
| US, 07.01.2022 | 2,304.498 | 665.539 | 28.9 | 3,324.0 | 693.2 |
| Canada,
07.01.2022 | 166.253 | 40.133 | 24.1 | 382.5 | 434.6 |
A study has revealed an average of 7.23 mutations
per sample and a significant frequency of single nucleotide
transitions (61). Shen et
al (62) found SARS-CoV-2
genetic variation in certain infected patients, indicating rapid
viral evolution. Although the appearance of novel variants is
directly associated with the number of existing viral units, the
emergence of novel mutations with negative effects is possible even
in areas where viral transmission is low. However, it is predicted
that mutations will be promote an increase in contagiousness rather
than severity. Therefore, an increasing number of people will
develop immune responses following vaccination or infection.
SARS-CoV-2 infection is predicted to form a pattern of
influenza-like outbreaks of varying magnitude. However,
reproduction of the seasonal pattern of the infections is less
likely as SARS-CoV-2 is not associated with periodicity in viral
transmission and outbreaks (Cojocaru et al, unpublished
data).
Immune evasion as a way of avoiding the host immune
response creates a risk of emergence of a novel variant (Cojocaru
et al, unpublished data). An in vitro study has shown
that substitution of the E484K gene, which is found in Beta and
Gamma variants, is responsible for immune escape from convalescent
serum or vaccine-induced antibodies (22). A high degree of immune escape has
also been reported for the Delta and Omicron variants and may be
associated with mutation on T478K gene (63).
Another approach that may be considered is based on
the spread of infection in unimmunized people, which is supported
by the fact that partial protection generated by vaccinations that
only protect against severe forms of disease. SARS-CoV-2 infection
may be more common among young children, especially since
vaccination for children aged 5-12 years has been introduced in few
countries (Cojocaru et al, unpublished data).
As SARS-CoV-2 is expanding faster than previously
known seasonal coronaviruses or influenza viruses, an
epidemiological model predicts that risk of infection will persist
for a long period (6). This will
have an impact on travel, event participation and public health.
Although the general principles of combating the spread have been
known since the sixteenth century, based on quarantine and virus
transmission prevention, these measures are difficult to implement
in the modern era due to high population mobility, urban sprawl and
the easy spread of misinformation through communication channels
(64). The international rapid
response in creating vaccines and treatments that lower viral
replication and risk of multisystem impairment is unprecedented.
Societal attitudes do not fully keep up with rapid progress of
medical research and pose a barrier to current challenges.
The incidence of cancer in post-COVID patients is
not yet known. Currently, it is only known that COVID-19 pandemic
delayed cancer screening and late therapy resulted in more
aggressive cancer behaviour. This may be due to difficulties in
scheduling physician visits and delayed cancer diagnosis.
The emergence of SARS-CoV-2 variants has raised
issues about vaccination efficacy. Multiple studies have been
performed to identify mutations that may be responsible for the
decline in vaccine effectiveness (65,66).
Current data suggest that protection for the Alpha variant induced
by Pfizer-BioNTech vaccine is similar to that for wild-type
SARS-CoV-2 virus, but is significantly lower for Beta, Gamma and
Delta variants (65,66). Preliminary data on Pfizer-BioNTech
vaccine indicated that a third dose is required for adequate
protection against Omicron variant infection (67).
Inequalities in the ability to respond to viral
spread remain primarily due to global social, economic, and
cultural differences; this may provide a viral reservoir that will
trigger outbreaks. SARS-CoV-2 will continue to be a major challenge
spread of viral infection is controlled (68). In the past two years, SARS-CoV-2
has resulted in >313 million cases and 5.5 million deaths, as
well as a global economic crisis, which has affected the $90
trillion global economy (69).
4. Limitations
Although the present systematic review adds to
knowledge of SARS-CoV-2 variants and their associated clinical
symptomatology, it has certain limitations. The present review
analysed only SARS-CoV-2 VOC mutations and clinical signs. The
number of reports which describe SARS-CoV-2 symptoms is still
limited. The knowledge of various strains and variants, as well as
their effect on symptoms, is incomplete. Researchers may also
struggle to discover new mutations and describe new symptoms given
the capacity of the virus to develop new variants.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
CC and EC conceptualized the study. EC designed the
methodology. CC, EC, DZ and AMT performed the literature review.
AMT constructed the figure. DZ edited the manuscript. EC wrote the
manuscript and supervised the project. Data authentication is not
applicable. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chan JF, To KK, Tse H, Jin DY and Yuen KY:
Interspecies transmission and emergence of novel viruses: Lessons
from bats and birds. Trends Microbiol. 21:544–555. 2013.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Zhu N, Zhang D, Wang W, Li X, Yang B, Song
J, Zhao X, Huang B, Shi W, Lu R, et al: A novel coronavirus from
patients with Pneumonia in China. N Engl J Med. 382:727–733.
2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou
J, Liu W, Bi Y and Gao G: Epidemiology, genetic recombination, and
pathogenesis of coronaviruses. Trends Microbiol. 24:490–502.
2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Zhou P, Yang XL, Wang XG, Hu B, Zhang L,
Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al: A pneumonia outbreak
associated with a new coronavirus of probable bat origin. Nature.
579:270–273. 2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Wu F, Zhao S, Yu B, Chen YM, Wang W, Song
ZG, Hu Y, Tao ZW, Tian JH, Pei YY, et al: A new coronavirus
associated with human respiratory disease in China. Nature.
579:265–269. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kistler K, Huddleston J and Bedford T:
Rapid and parallel adaptive mutations in spike S1 drive clade
success in SARS-CoV-2. Cell Host Microbe. 30:545–555.e4.
2022.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Khan A, Khan T, Ali S, Aftab S, Wang Y,
Qiankun W, Khan M, Suleman M, Ali S, Heng W, et al: SARS-CoV-2 new
variants: Characteristic features and impact on the efficacy of
different vaccines. Biomed Pharmacother. 143(112176)2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
World Health Organization (WHO): Tracking
SARS-CoV-2 variants. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/.
Accessed January 19, 2022.
|
|
9
|
Davies NG, Abbott S, Barnard RC, Jarvis
CI, Kucharski AJ, Munday JD, Pearson CAB, Russell TW, Tully DC,
Washburne AD, et al: Estimated transmissibility and impact of
SARS-CoV-2 lineage B.1.1.7 in England. Science.
372(eabg3055)2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Centers for Disease Control and Prevention
(CDC): Science Brief: Emerging SARS-CoV-2 Variants. CDC, Atlanta,
GA, 2021. https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/scientific-brief-emerging-variants.html.
Updated January 2021.
|
|
11
|
Letko M, Marzi A and Munster V: Functional
assessment of cell entry and receptor usage for SARS-CoV-2 and
other lineage B betacoronaviruses. Nat Microbiol. 5:562–569.
2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hoffmann M, Kleine-Weber H and Pöhlmann S:
A Multibasic cleavage site in the spike protein of SARS-CoV-2 is
essential for infection of human lung cells. Mol Cell.
78:779–784.e5. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Yang W and Shaman J: Development of a
model-inference system for estimating epidemiological
characteristics of SARS-CoV-2 variants of concern. Nat Commun.
12(5573)2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Monel B, Planas D, Grzelak L, Smith N,
Robillard N, Staropoli I, Goncalves P, Porrot F, Guivel-Benhassine
F, Guinet ND, et al: Release of infectious virus and cytokines in
nasopharyngeal swabs from individuals infected with non-alpha or
alpha SARS-CoV-2 variants: An observational retrospective study.
EBioMedicine. 73(103637)2021.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kidd M, Richter A, Best A, Cumley N, Mirza
J, Percival B, Mayhew M, Megram O, Ashford F, White T, et al:
S-variant SARS-CoV-2 lineage B1.1.7 is associated with
significantly higher viral loads in samples tested by TaqPath
Polymerase Chain Reaction. J Infect Dis. 223:1666–1670.
2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Calistri P, Amato L, Puglia I, Cito F, Di
Giuseppe A, Danzetta ML, Morelli D, Di Domenico M, Caporale M,
Scialabba S, et al: Infection sustained by lineage B.1.1.7 of
SARS-CoV-2 is characterised by longer persistence and higher viral
RNA loads in nasopharyngeal swabs. Int J Infect Dis. 105:753–755.
2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Walker AS, Vihta KD, Gethings O, Pritchard
E, Jones J, House T, Bell I, Bell JI, Newton JN, Farrar J, et al:
Tracking the emergence of SARSCoV-2 alpha variant in the United
Kingdom. N Engl J Med. 385:2582–2585. 2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Shenai MB, Rahme R and Noorchashm H:
Equivalency of protection from natural immunity in COVID-19
recovered versus fully vaccinated persons: A systematic review and
pooled analysise. Cureus. 13(e19102)2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Vassallo M, Manni S, Klotz C, Fabre R,
Pini P, Blanchouin E, Sindt A, Lotte L, Dubertrand JM, Liguori S,
et al: Patients admitted for variant alpha COVID-19 have poorer
outcomes than those infected with the old strain. J Clin Med.
10(3550)2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Turnbull CD, Porter BML, Evans SB, Smith
O, Lardner R, Hallifax R, Bettinson HV, Talbot NP, Bafadhel M,
Rahman NM, et al: Improved COVID-19 outcomes in a large
non-invasive respiratory support cohort despite emergence of the
alpha variant. BMJ open Respir Res. 8(e001044)2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Elliott J, Whitaker M, Bodinier B, Eales
O, Riley S, Ward H, Cooke G, Darzi A, Chadeau-Hyam M and Elliott P:
Predictive symptoms for COVID-19 in the community: REACT-1 study of
over 1 million people. PLoS Med. 18(e1003777)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Weisblum Y, Schmidt F, Zhang F, DaSilva J,
Poston D, Lorenzi JC, Muecksch F, Rutkowska M, Hoffmann HH,
Michailidis E, et al: Escape from neutralizing antibodies by
SARS-CoV-2 spike protein variants. Elife. 9(e61312)2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Resende P, Bezerra J, Teixeira Vasconcelos
RH, Arantes I, Appolinario L, Mendonça AC, Paixao AC, Duarte
Rodrigues AC, Silva T, Sampaio Rocha A, et al: Severe Acute
Respiratory Syndrome Coronavirus 2 P.2 Lineage Associated with
Reinfection Case, Brazil, June-October 2020. Emerg Infect Dis.
27:1789–1794. 2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wang Z, Schmidt F, Weisblum Y, Muecksch F,
Barnes CO, Finkin S, Schaefer-Babajew D, Cipolla M, Gaebler C,
Lieberman JA, et al: mRNA vaccine-elicited antibodies to SARS-CoV-2
and circulating variants. Nature. 592:616–622. 2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Moss DL and Rappaport J: SARS-CoV-2 beta
variant substitutions alter spike glycoprotein receptor binding
domain structure and stability. J Biol Chem.
297(101371)2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Jassat W, Mudara C, Ozougwu L, Tempia S,
Blumberg L, Davies MA, Pillay Y, Carter T, Morewane R, Wolmarans M,
et al: Difference in mortality among individuals admitted to
hospital with COVID-19 during the first and second waves in South
Africa: A cohort study. Lancet Glob Heal. 9:e1216–e1225.
2021.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Luna-Muschi A, Borges IC, de Faria E,
Barboza AS, Maia FL, Leme MD, Guedes AR, Mendes-Correa MC, Kallas
EG, Segurado AC, et al: Clinical features of COVID-19 by SARS-CoV-2
Gamma variant: A prospective cohort study of vaccinated and
unvaccinated healthcare workers. J Infect. 84:248–288.
2022.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Allen H, Vusirikala A, Flannagan J, Twohig
KA, Zaidi A, Chudasama D, Lamagni T, Groves N, Turner C, Rawlinson
C, et al: Household transmission of COVID-19 cases associated with
SARS-CoV-2 delta variant (B.1.617.2): National casecontrol study.
Lancet Reg Health Eur. 12(100252)2022.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Cherian S, Potdar V, Jadhav S, Yadav P,
Gupta N, Das M, Rakshit P, Singh S, Abraham P, Panda S and Team N:
SARS-CoV-2 Spike Mutations, L452R, T478K, E484Q and P681R, in the
Second Wave of COVID-19 in Maharashtra, India. Microorganisms.
9(1542)2021.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Challen R, Dyson L, Overton CE,
Guzman-Rincon LM, Hill EM, Stage HB, Brooks-Pollock E, Pellis L,
Scarabel F, Pascall DJ, et al: Early epidemiological signatures of
novel SARS-CoV-2 variants: Establishment of B.1.617.2 in England.
medRxiv: doi: https://doi.org/10.1101/2021.06.05.21258365.
|
|
31
|
Wang Y, Chen R, Hu F, Lan Y, Yang Z, Zhan
C, Shi J, Deng X, Jiang M, Zhong S, et al: Transmission, viral
kinetics and clinical characteristics of the emergent SARS-CoV-2
Delta VOC in Guangzhou, China. EClinicalMedicine.
40(101129)2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ong SWX, Chiew CJ, Ang LW, Mak TM, Cui L,
Toh MPHS, Lim YD, Lee PH, Lee TH, Chia PY, et al: Clinical and
virological features of severe acute respiratory syndrome
coronavirus 2 (SARSCoV-2) variants of concern: A retrospective
cohort study comparing B.1.1.7 (Alpha), B.1.351 (Beta), and
B.1.617.2 (Delta). Clin Infect Dis: Aug 23, 2021 (Epub ahead of
print).
|
|
33
|
Twohig KA, Nyberg T, Zaidi A, Thelwall S,
Sinnathamby MA, Aliabadi S, Seaman SR, Harris RJ, Hope R,
Lopez-Bernal J, et al: Hospital admission and emergency care
attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with
alpha (B.1.1.7) variants of concern: A cohort study. Lancet Infect
Dis. 22:35–42. 2022.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Loconsole D, Centrone F, Morcavallo C,
Campanella S, Accogli M, Sallustio A, Peccarisi D, Stufano A,
Lovreglio P and Chironna M: Changing Features of COVID-19:
Characteristics of Infections with the SARS-CoV-2 Delta (B.1.617.2)
and Alpha (B.1.1.7) Variants in Southern Italy. Vaccines (Basel).
9(1354)2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Ryu BH, Hong SI, Lim SJ, Cho Y, Hwang C,
Kang H, Kim SH, Wi YM, Hong KW, Bae IG and Cho OH: Clinical
Features of Adult COVID-19 Patients without Risk Factors before and
after the Nationwide SARS-CoV-2 B.1.617.2 (Delta)-variant Outbreak
in Korea: Experience from Gyeongsangnam-do. J Korean Med Sci.
36(e341)2021.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Butt AA, Dargham SR, Chemaitelly H, Al
Khal A, Tang P, Hasan MR, Coyle PV, Thomas AG, Borham AM,
Concepcion EG, et al: Severity of Illness in Persons Infected With
the SARS-CoV-2 Delta Variant vs Beta Variant in Qatar. JAMA Intern
Med. 182:197–205. 2022.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Chia PY, Ong SWX, Chiew CJ, Ang LW,
Chavatte JM, Mak TM, Cui L, Kalimuddin S, Chia WN, Tan CW, et al:
Virological and serological kinetics of SARS-CoV-2 Delta variant
vaccine breakthrough infections: A multicentre cohort study. Clin
Microbiol Infect. 28:612.e1–612.e7. 2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kim S, Nguyen TT, Taitt AS, Jhun H, Park
HY, Kim SH, Kim YG, Song EY, Lee Y, Yum H, et al: SARSCoV-2 omicron
mutation is faster than the chase: Multiple mutations on spike/ACE2
interaction residues. Immune Netw. 21(e38)2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Meo SA, Meo AS, Al-Jassir FF and Klonoff
DC: Omicron SARS-CoV-2 new variant: Global prevalence and
biological and clinical characteristics. Eur Rev Med Pharmacol Sci.
25:8012–8018. 2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Sofonea MT, Roquebert B, Foulongne V,
Verdurme L, Trombert-Paolantoni S, Roussel M, Haim-Boukobza S and
Alizonet S: From Delta to Omicron: Analysing the SARS-CoV-2
epidemic in France using variant-specific screening tests
(September 1 to December 18, 2021). medRxiv: doi: https://doi.org/10.1101/2021.12.31.21268583.
|
|
41
|
Abdullah F, Myers J, Basu D, Tintinger G,
Ueckermann V, Mathebula M, Ramlall R, Spoor S, de Villiers T, Van
der Walt Z, et al: Decreased severity of disease during the first
global omicron variant covid-19 outbreak in a large hospital in
tshwane, South Africa. Int J Infect Dis. 116:38–42. 2022.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Graham MS, Sudre CH, May A, Antonelli M,
Murray B, Varsavsky T, Kläser K, Canas LS, Molteni E, Modat M, et
al: Changes in symptomatology, reinfection, and transmissibility
associated with the SARS-CoV-2 variant B.1.1.7: An ecological
study. Lancet Public Health. 6:e335–e345. 2021.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Wang S, Zou X, Li Z, Fu J, Fan H, Yu H,
Deng F, Huang H, Peng J, Zhao K, et al: Analysis of clinical
characteristics and virus strains variation of patients infected
with SARS-CoV-2 in Jiangsu Province-A retrospective study. Front
Public Health. 9(791600)2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Neagu M, Constantin C and Surcel M:
Testing antigens, antibodies, and immune cells in COVID-19 as a
public health topic-experience and outlines. Int J Environ Res
Public Health. 18(13173)2021.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Lownik JC, Farrar JS, Way GW, McKay A,
Roychoudhury P, Greninger AL and Martin RK: Fast SARS-CoV-2 variant
detection using snapback primer high-resolution melting.
Diagnostics (Basel). 11(1788)2021.PubMed/NCBI View Article : Google Scholar
|
|
46
|
de Puig H, Lee RA, Najjar D, Tan X,
Soekensen LR, Angenent-Mari NM, Donghia NM, Weckman NE, Ory A, Ng
CF, et al: Minimally instrumented SHERLOCK (miSHERLOCK) for
CRISPR-based point-of-care diagnosis of SARS-CoV-2 and emerging
variants. Sci Adv. 7(eabh2944)2021.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Lunca C, Cojocaru C, Gurzu IL, Petrariu FD
and Cojocaru E: Performance of antigenic detection of SARS-CoV-2 in
nasopharyngeal samples. medRxiv: doi: https://doi.org/10.1101/2021.07.12.21260263.
|
|
48
|
Buchta C, Camp JV, Jovanovic J, Radler U,
Benka B, Puchhammer-Stöckl E, Müller MM, Griesmacher A, Aberle SW
and Görzer I: Inadequate design of mutation detection panels
prevents interpretation of variants of concern: Results of an
external quality assessment for SARS-CoV-2 variant detection. Clin
Chem Lab Med. 60:291–298. 2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Ruan Q, Yang K, Wang W, Jiang L and Song
J: Clinical predictors of mortality due to COVID-19 based on an
analysis of data of 150 patients from Wuhan, China. Intensive Care
Med. 46:846–848. 2020.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Ghebrehiwet B and Peerschke EI: Complement
and coagulation: Key triggers of COVID-19-induced multiorgan
pathology. J Clin Invest. 130:5674–5676. 2020.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Jacobs W, Lammens M, Kerckhofs A, Voets E,
Van San E, Van Coillie S, Peleman C, Mergeay M, Sirimsi S,
Matheeussen V, et al: Fatal lymphocytic cardiac damage in
coronavirus disease 2019 (COVID-19): autopsy reveals a ferroptosis
signature. ESC Hear Fail. 7:3772–3781. 2020.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Timpani CA and Rybalka E: Calming the
(Cytokine) Storm: Dimethyl fumarate as a therapeutic candidate for
COVID-19. Pharmaceuticals (Basel). 14(15)2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Wood H: New insights into the neurological
effects of COVID-19. Nat Rev Neurol. 16(403)2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Cojocaru E, Cojocaru C, Antoniu SA, Stafie
CS, Rajnoveanu A and Rajnoveanu RM: Inhaled interferons beta and
SARS-COV2 infection: A preliminary therapeutic perspective. Expert
Rev Respir Med. 16:257–261. 2022.
|
|
55
|
US Food and Drug (FDA): FDA Approves First
COVID-19 Vaccine. FDA, Silver Spring, MD, 2021. https://www.fda.gov/news-events/press-announcements/fda-approves-first-covid-19-vaccine.
Accessed August 23, 2021.
|
|
56
|
Beigel JH, Tomashek KM, Dodd LE, Mehta AK,
Zingman BS, Kalil AC, Hohmann E, Chu HY, Luetkemeyer A, Kline S, et
al: Remdesivir for the treatment of Covid-19-Final Report. N Engl J
Med. 383:1813–1826. 2020.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Lavinio A, Ercole A, Battaglini D, Magnoni
S, Badenes R, Taccone FS, Helbok R, Thomas W, Pelosi P and Robba C:
collaborators. Safety profile of enhanced thromboprophylaxis
strategies for critically ill COVID-19 patients during the first
wave of the pandemic: Observational report from 28 European
intensive care units. Crit Care. 25(155)2021.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Guy T, Créac'hcadec A, Ricordel C, Salé A,
Arnouat B, Bizec JL, Langelot M, Lineau C, Marquette D, Martin F,
et al: High-flow nasal oxygen: A safe, efficient treatment for
COVID-19 patients not in an ICU. Eur Respir J.
56(2001154)2020.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Horwitz LI, Jones SA, Cerfolio RJ,
Francois F, Greco J, Rudy B and Petrilli CM: Trends in COVID-19
Risk-Adjusted Mortality Rates. J Hosp Med. 16:90–92.
2021.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Our World in Data: COVID-19 Data Explorer.
https://ourworldindata.org/explorers/coronavirus-data-explorer.
Accessed March 12, 2022.
|
|
61
|
Mercatelli D and Giorgi FM: Geographic and
genomic distribution of SARS-CoV-2 mutations. Front Microbiol.
11(1800)2020.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Shen Z, Xiao Y, Kang L, Ma W, Shi L, Zhang
L, Zhou Z, Yang J, Zhong J, Yang D, et al: Genomic diversity of
severe acute respiratory syndrome-coronavirus 2 in patients with
coronavirus disease 2019. Clin Infect Dis. 71:713–720.
2020.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Mlcochova P, Kemp SA, Dhar MS, Papa G,
Meng B, Ferreira IATM, Datir R, Collier DA, Albecka A, Singh S, et
al: SARS-CoV-2 B.1.617.2 Delta variant replication and immune
evasion. Nature. 599:114–119. 2021.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Long MJC and Aye Y: Science's Response to
CoVID-19. ChemMedChem. 16:2288–2314. 2021.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Muik A, Wallisch AK, Sänger B, Swanson KA,
Mühl J, Chen W, Cai H, Maurus D, Sarkar R, Türeci O, et al:
Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by
BNT162b2 vaccine-elicited human sera. Science. 371:1152–1153.
2021.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Abdool Karim SS and de Oliveira T: New
SARS-CoV-2 variants-clinical, public health, and vaccine
implications. N Engl J Med. 384:1866–1868. 2021.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Pfizer: Pfizer and BioNTech Provide Update
on Omicron Variant. Pfizer, New York, NY 2021. https://www.pfizer.com/news/press-release/press-release-detail/pfizer-and-biontech-provide-update-omicron-variant.
Accessed December 08, 2021.
|
|
68
|
Cojocaru C, Cojocaru E, Radu S and Gurzu
B: Perception and attitude of the general population on the risk of
infection with SARS-CoV-2. J Biosci Med. 9:1–10. 2021.
|
|
69
|
Congressional Research Service: Global
Economic Effects of COVID-19, 2021. https://crsreports.congress.gov/. Updated November
10, 2021.
|