|
1
|
Su ZZ, Kang DC, Chen Y, Pekarskaya O, Chao
W, Volsky DJ and Fisher PB: Identification and cloning of human
astrocyte genes displaying elevated expression after infection with
HIV-1 or exposure to HIV-1 envelope glycoprotein by rapid
subtraction hybridization, RaSH. Oncogene. 21:3592–3602.
2002.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Thirkettle HJ, Girling J, Warren AY, Mills
IG, Sahadevan K, Leung H, Hamdy F, Whitaker HC and Neal DE:
LYRIC/AEG-1 is targeted to different subcellular compartments by
ubiquitinylation and intrinsic nuclear localization signals. Clin
Cancer Res. 15:3003–3013. 2009.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Yoo BK, Emdad L, Lee SG, Su ZZ,
Santhekadur P, Chen D, Gredler R, Fisher PB and Sarkar D: Astrocyte
elevated gene-1 (AEG-1): A multifunctional regulator of normal and
abnormal physiology. Pharmacol Ther. 130:1–8. 2011.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Emdad L, Sarksr D, Su ZZ, Randolph A,
Boukerche H, Valerie K and Fisher PB: Activation of the nuclear
factor kappaB pathway by astrocyte elevated gene-1: Implications
for tumor progression and metastasis. Cancer Res. 66:1509–1516.
2006.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Emdad L, Sarkar D, Su ZZ, Lee SG, Kang DC,
Bruce JN, Volsky DJ and Fisher PB: Astrocyte elevated gene-1:
Recent insights into a novel gene involved in tumor progression,
metastasis and neurodegeneration. Pharmacol Ther. 114:155–170.
2007.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhang C, Li HZ, Qian BJ, Liu CM, Guo F and
Lin MC: MTDH/AEG-1-based DNA vaccine suppresses metastasis and
enhances chemosensitivity to paclitaxel in pelvic lymph node
metastasis. Biomed Pharmacother. 70:217–226. 2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Roussel BD, Kruppa AJ, Miranda E, Crowther
DC, Lomas DA and Marciniak SJ: Endoplasmic reticulum dysfunction in
neurological disease. Lancet Neuro1. 12:105–118. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Anttila V, Stefansson H, Kallela M, Todt
U, Terwindt GM, Calafato MS, Nyholt DR, Dimas AS, Freilinger T,
Müller-Myhsok B, et al: Genome-wide association study of migraine
implicates a common susceptibility variant on 8q22.1. Nat Genet.
42:869–873. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Kang DC, Su ZZ, Sarkar D, Emdad L, Volsky
DJ and Fisher PB: Cloning and characterization of HIV-1-inducible
astrocyte elevated gene-1, AEG-1. Gene. 353:8–15. 2005.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Lee SG, Kim K, Kegelman TP, Dash R, Das
SK, Choi JK, Emdad L, Howlett EL, Jeon HY, Su ZZ, et al: Oncogene
AEG-1 promotes glioma-induced neurodegeneration by increasing
glutamate excitotoxicity. Cancer Res. 71:6514–6523. 2011.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Vartak-Sharma N and Ghorpade A: Astrocyte
elevated gene-1 regulates astrocyte responses to neural injury:
Implications for reactive astrogliosis and neurodegeneration. J
Neuroinflammation. 9(195)2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yin X, Ren M, Jiang H, Cui S, Wang S,
Jiang H, Qi Y, Wang J, Wang X, Dong G, et al: Downregulated AEG-1
together with inhibited PI3K/Akt pathway is associated with reduced
viability of motor neurons in an ALS model. Mol Cell Neurosci.
68:303–313. 2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Leem E, Kim HJ, Choi M, Kim S, Oh YS, Lee
KJ, Choe YS, Um JY, Shin WH, Jeong JY, et al: Upregulation of
neuronal astrocyte elevated gene-1 protects nigral dopaminergic
neurons in vivo. Cell Death Dis. 9(449)2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Jeon HY, Choi M, Howlett EL, Vozhilla N,
Yoo BK, Lloyd JA, Sarkar D, Lee SG and Fisher PB: Expression
patterns of astrocyte elevated gene-1 (AEG-1) during development of
the mouse embryo. Gene Expr Patterns. 10:361–367. 2010.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Carnemolla A, Fossale E, Agostoni E,
Michelazzi S, Calligaris R, De Maso L, Del Sal G, MacDonald ME and
Persichetti F: Rrs1 is involved in endoplasmic reticulum stress
response in Huntington disease. J Biol Chem. 284:18167–18173.
2009.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Garneau JE, Dupuis MÈ, Villion M, Romero
DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH and
Moineau S: The CRISPR/Cas bacterial immune system cleaves
bacteriophage and plasmid DNA. Nature. 468:67–71. 2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Doudna JA and Charpentier E: Genome
editing. The new frontier of genome engineering with CRISPR-Cas9.
Science: Nov 28, 2014 (Epub ahead of print).
|
|
18
|
Caldwell JD, Shapiro RA, Jirikowski GF and
Suleman F: Internalization of sex hormone-binding globulin into
neurons and brain cells in vitro and in vivo. Neuroendocrinology.
86:84–93. 2007.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Murphy TH, Miyamoto M, Sastre A, Schnaar
RL and Coyle JT: Glutamate toxicity in a neuronal cell line
involves inhibition of cystine transport leading to oxidative
stress. Neuron. 2:1547–1558. 1989.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Florea L, Song L and Salzberg SL:
Thousands of exon skipping events differentiate among splicing
patterns in sixteen human tissues. F1000Res. 2(188)2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140.
2010.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Anders S and Huber W: Differential
expression analysis for sequence count data. Genome Biol.
11(R106)2010.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc B (Methodol). 57:289–300. 1995.
|
|
25
|
Harris MA, Clark J, Ireland A, Lomax J,
Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C,
et al: The gene ontology (GO) database and informatics resource.
Nucleic Acids Res. 32:D258–D261. 2004.PubMed/NCBI View Article : Google Scholar
|
|
26
|
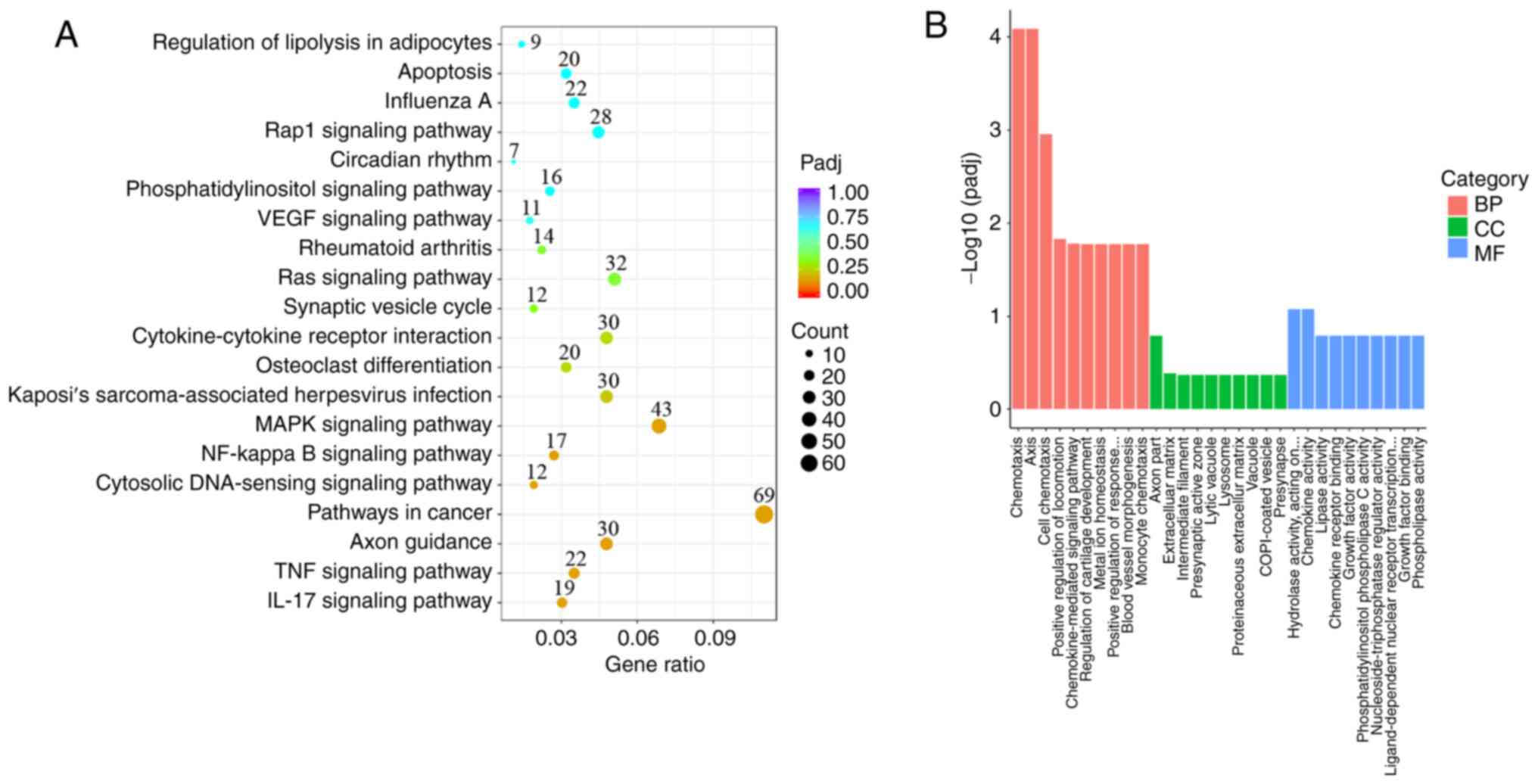
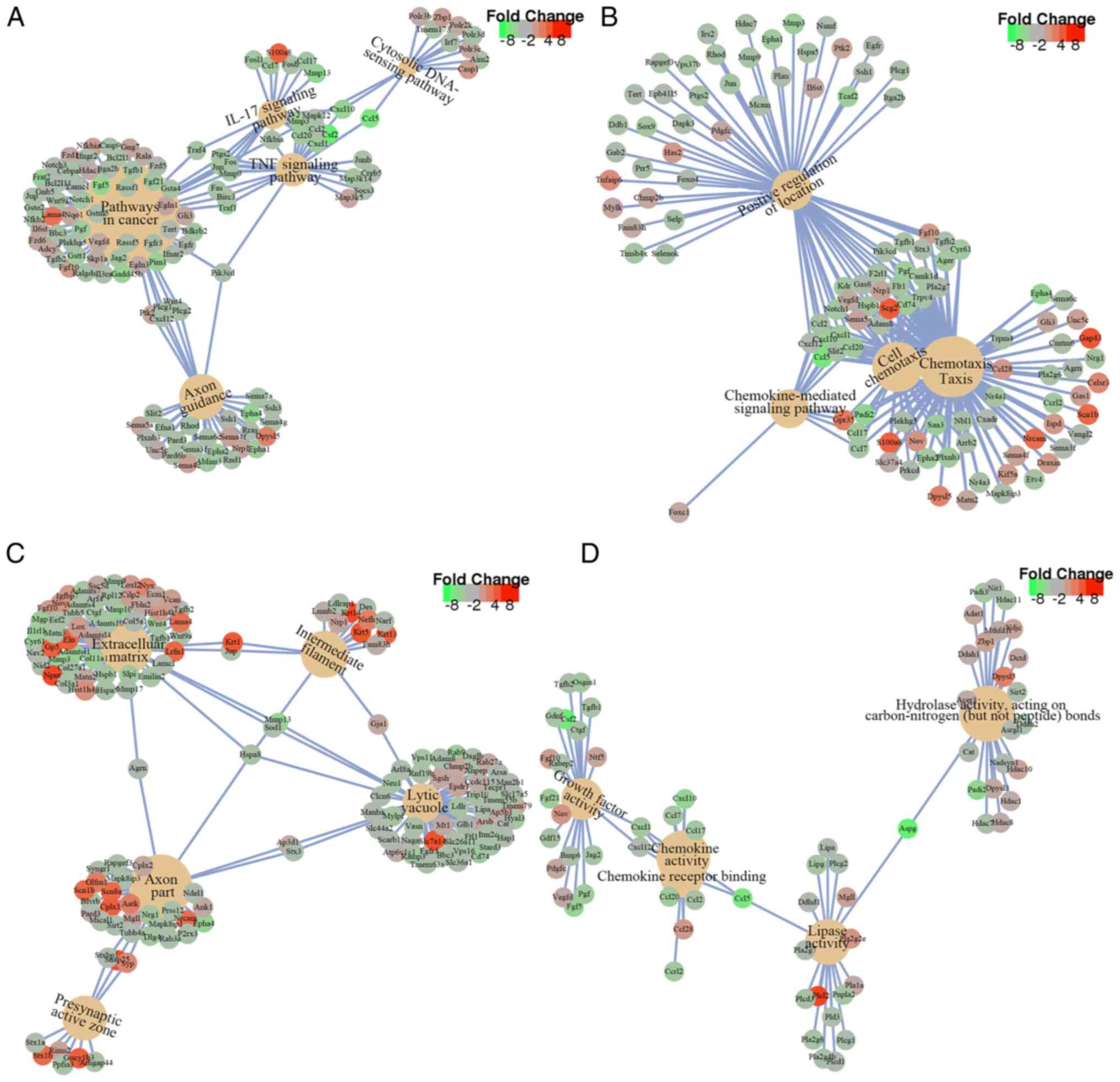
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Kanehisa M, Sato Y, Kawashima M, Furumichi
M and Tanabe M: KEGG as a reference resource for gene and protein
annotation. Nucleic Acids Res. 44:D457–D462. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
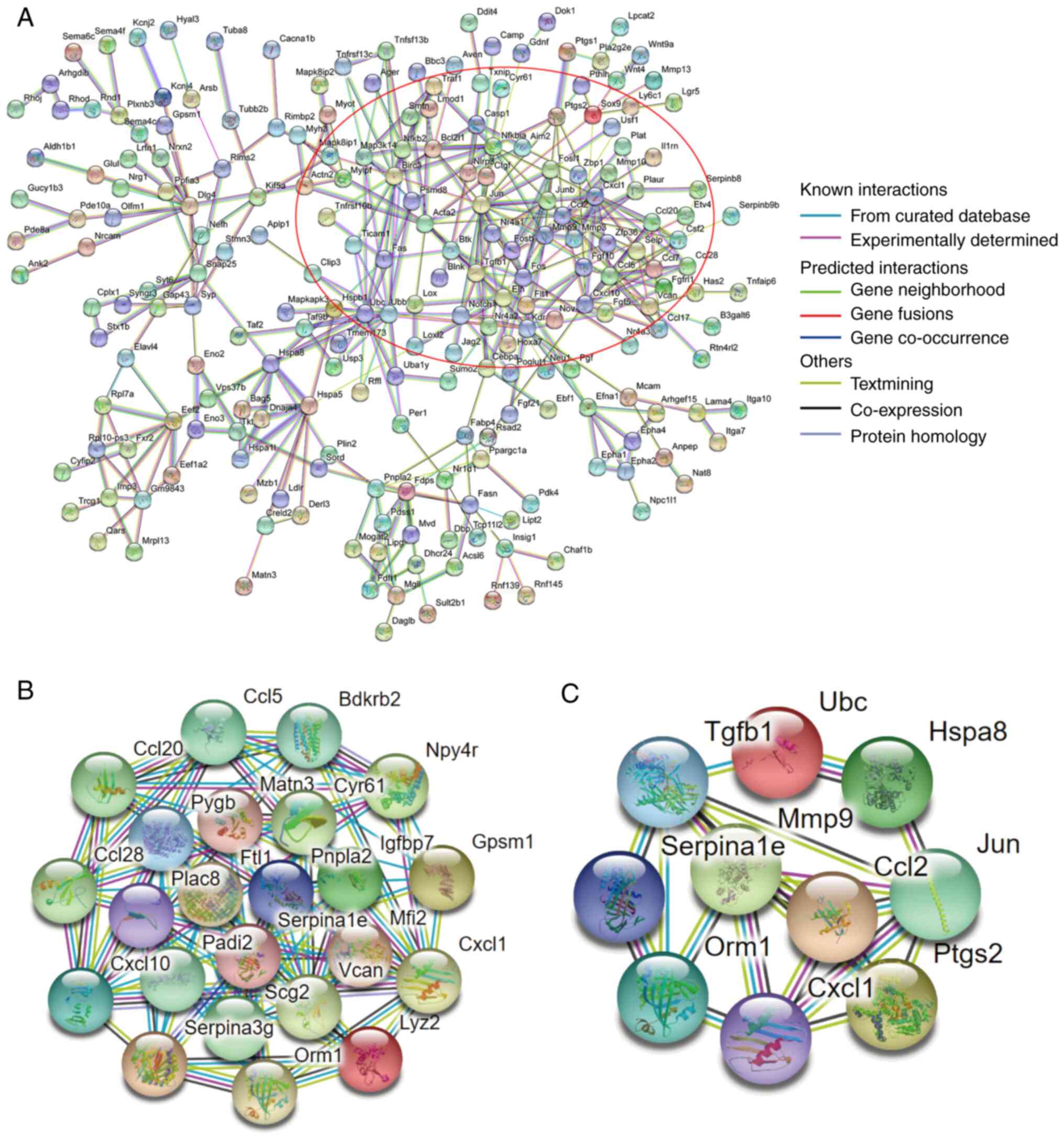
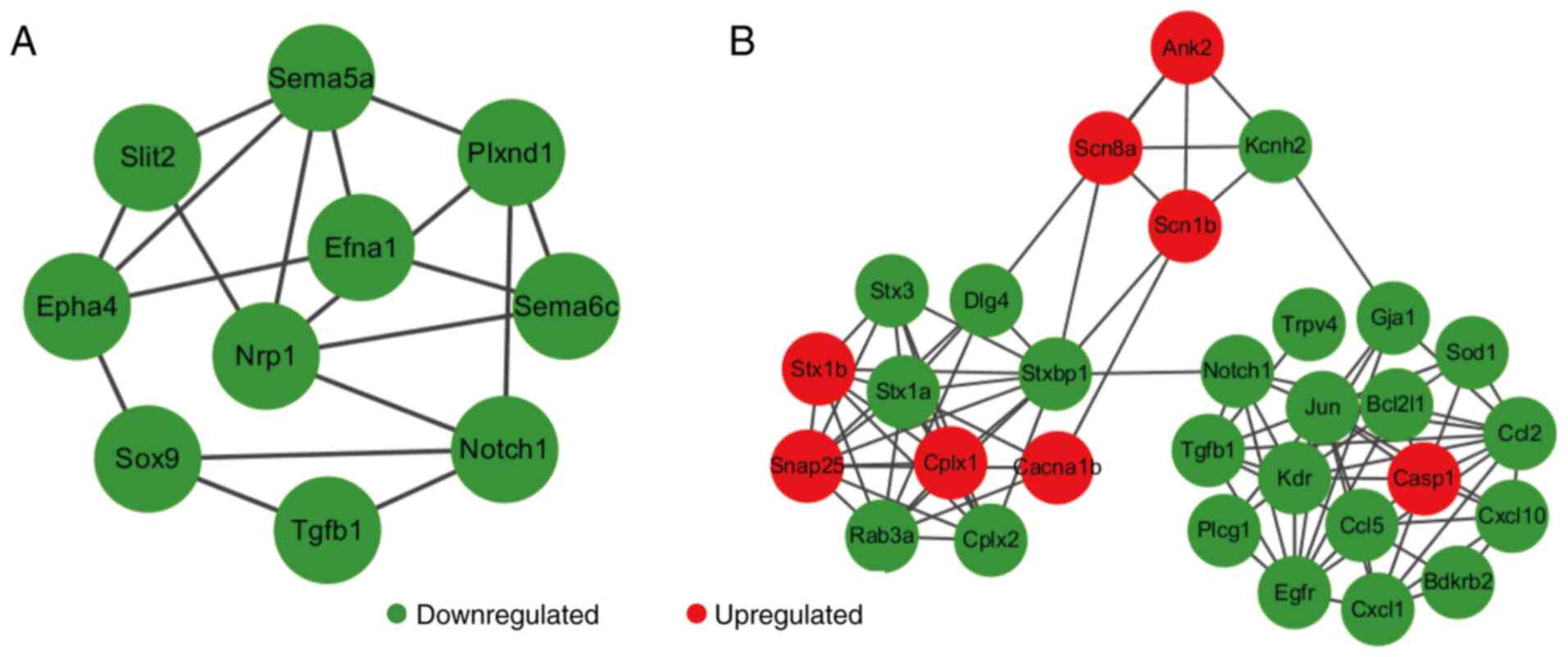
von Mering C, Jensen LJ, Snel B, Hooper
SD, Krupp M, Foglierini M, Jouffre N, Huynen MA and Bork P: STRING:
Known and predicted protein-protein associations, integrated and
transferred across organisms. Nucleic Acids Res. 33:D433–D437.
2005.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Xu WH, Xu Y, Wang J, Wan FN, Wang HK, Cao
DL, Shi GH, Qu YY, Zhang HL and Ye DW: Prognostic value and immune
infiltration of novel signatures in clear cell renal cell carcinoma
microenvironment. Aging (Albany NY). 11:6999–7020. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Chen Q, Yu D, Zhao Y, Qiu J, Xie Y and Tao
M: Screening and identification of hub genes in pancreatic cancer
by integrated bioinformatics analysis. J Cell Biochem.
120:19496–19508. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Stephenson AA, Raper AT and Suo Z:
Bidirectional degradation of DNA cleavage products catalyzed by
CRISPR/Cas9. J Am Chem Soc. 140:3743–3750. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Bernardino L, Agasse F, Silva B, Ferreira
R, Grade S and Malva JO: Tumor necrosis factor-alpha modulates
survival, proliferation, and neuronal differentiation in neonatal
subventricular zone cell cultures. Stem Cells. 26:2361–2371.
2008.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Li X, Bechara R, Zhao J, McGeachy MJ and
Gaffen SL: IL-17 receptor-based signaling and implications for
disease. Nat Immunol. 20:1594–1602. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Liu W, Ye J and Yan H: Investigation of
key genes and pathways in inhibition of oxycodone on
vincristine-induced microglia activation by using bioinformatics
analysis. Dis Markers. 2019(3521746)2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Ng SR, Rideout WM III, Akama-Garren EH,
Bhutkar A, Mercer KL, Schenkel JM, Bronson RT and Jacks T:
CRISPR-mediated modeling and functional validation of candidate
tumor suppressor genes in small cell lung cancer. Proc Natl Acad
Sci USA. 117:513–521. 2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
He XY, Ren XH, Peng Y, Zhang JP, Ai SL,
Liu BY, Xu C and Cheng SX: Aptamer/peptide-functionalized
genome-editing system for effective immune restoration through
reversal of PD-L1-mediated cancer immunosuppression. Adv Mater.
32(2000208)2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Ling X, Xie B, Gao X, Chang L, Zheng W,
Chen H, Huang Y, Tan L, Li M and Liu T: Improving the efficiency of
precise genome editing with site-specific Cas9-oligonucleotide
conjugates. Sci Adv. 6(eaaz0051)2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
LaFountaine JS, Fathe K and Smyth HD:
Delivery and therapeutic applications of gene editing technologies
ZFNs, TALENs, and CRISPR/Cas9. Int J Pharm. 494:180–194.
2015.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhou Z, Tan H, Li Q, Chen J, Gao S, Wang
Y, Chen W and Zhang L: CRISPR/Cas9-mediated efficient targeted
mutagenesis of RAS in Salvia miltiorrhiza. Phytochemistry.
148:63–70. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Noch EK and Khalili K: The role of
AEG-1/MTDH/LYRIC in the pathogenesis of central nervous system
disease. Adv Cancer Res. 120:159–192. 2013.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Wang Y, Zhang W, Zhu X and Wang Y, Mao X,
Xu X and Wang Y: Upregulation of AEG-1 involves in schwann cell
proliferation and migration after sciatic nerve crush. J Mol
Neurosci. 60:248–257. 2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Vartak-Sharma N, Nooka S and Ghorpade A:
Astrocyte elevated gene-1 (AEG-1) and the A(E)Ging HIV/AIDS-HAND.
Prog Neurobiol. 157:133–157. 2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Serviddio G, Romano AD, Cassano T,
Bellanti F, Altomare E and Vendemiale G: Principles and therapeutic
relevance for targeting mitochondria in aging and neurodegenerative
diseases. Curr Pharm Des. 17:2036–2055. 2011.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Mohan H, Verhoog MB, Doreswamy KK, Eyal G,
Aardse R, Lodder BN, Goriounova NA, Asamoah B, B Brakspear AB,
Groot C, et al: Dendritic and axonal architecture of individual
pyramidal neurons across layers of adult human neocortex. Cereb
Cortex. 25:4839–4853. 2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Jan YN and Jan LY: Branching out:
Mechanisms of dendritic arborization. Nat Rev Neurosci. 11:316–328.
2010.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Yoo BK, Emdad L, Su ZZ, Villanueva A,
Chiang DY, Mukhopadhyay ND, Mills AS, Waxman S, Fisher RA, Llovet
JM, et al: Astrocyte elevated gene-1 regulates hepatocellular
carcinoma development and progression. J Clin Invest. 119:465–477.
2009.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Yu C, Chen K, Zheng H, Guo X, Jia W, Li M,
Zeng M, Li J and Song L: Overexpression of astrocyte elevated
gene-1 (AEG-1) is associated with esophageal squamous cell
carcinoma (ESCC) progression and pathogenesis. Carcinogenesis.
30:894–901. 2009.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Lee SG, Su ZZ, Emdad L, Sarkar D and
Fisher PB: Astrocyte elevated gene-1 (AEG-1) is a target gene of
oncogenic Ha-ras requiring phosphatidylinositol 3-kinase and c-Myc.
Proc Natl Acad Sci USA. 103:17390–17395. 2006.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Rohm B, Ottemeyer A, Lohrum M and Püschel
AW: Plexin/neuropilin complexes mediate repulsion by the axonal
guidance signal semaphorin 3A. Mech Dev. 93:95–104. 2000.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Rolny C, Capparuccia L, Casazza A, Mazzone
M, Vallario A, Cignetti A, Medico E, Carmeliet P, Comoglio PM and
Tamagnone L: The tumor suppressor semaphorin 3B triggers a
prometastatic program mediated by interleukin 8 and the tumor
microenvironment. J Exp Med. 205:1155–1171. 2008.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Mumm JS, Schroeter EH, Saxena MT,
Griesemer A, Tian X, Pan DJ, Ray WJ and Kopan R: A ligand-induced
extracellular cleavage regulates gamma-secretase-like proteolytic
activation of Notch1. Mol Cell. 5:197–206. 2000.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Liu X, Yang Z, Yin Y and Deng X: Increased
expression of Notch1 in temporal lobe epilepsy: Animal models and
clinical evidence. Neural Regen Res. 9:526–533. 2014.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Feng S, Shi T, Qiu J, Yang H, Wu Y, Zhou
W, Wang W and Wu H: Notch1 deficiency in postnatal neural
progenitor cells in the dentate gyrus leads to emotional and
cognitive impairment. FASEB J. 31:4347–4358. 2017.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Shu Y, Xiao B, Wu Q, Liu T, Du Y, Tang H,
Chen S, Feng L, Long L and Li Y: The ephrin-A5/EphA4 interaction
modulates neurogenesis and angiogenesis by the p-Akt and p-ERK
pathways in a mouse model of TLE. Mol Neurobiol. 53:561–576.
2016.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Feng L, Shu Y, Wu Q, Liu T, Long H, Yang
H, Li Y and Xiao B: EphA4 may contribute to microvessel remodeling
in the hippocampal CA1 and CA3 areas in a mouse model of temporal
lobe epilepsy. Mol Med Rep. 15:37–46. 2017.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Salińska E and Łazarewicz JW: Role of
calcium in physiology and pathology of neurons. Postepy Biochem.
58:403–417. 2012.PubMed/NCBI(In Polish).
|
|
57
|
Chadwick L, Gentle L, Strachan J and
Layfield R: Review: Unchained maladie-a reassessment of the role of
Ubb(+1)-capped polyubiquitin chains in Alzheimer's disease.
Neuropathol Appl Neurobiol. 38:118–131. 2012.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Manavalan A, Mishra M, Feng L, Sze SK,
Akatsu H and Heese K: Brain site-specific proteome changes in
aging-related dementia. Exp Mol Med. 45(e39)2013.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Kothur K, Bandodkar S, Wienholt L, Chu S,
Pope A, Gill D and Dale RC: Etiology is the key determinant of
neuroinflammation in epilepsy: Elevation of cerebrospinal fluid
cytokines and chemokines in febrile infection-related epilepsy
syndrome and febrile status epilepticus. Epilepsia. 60:1678–1688.
2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Silva RL, Lopes AH, Guimaraes RM and Cunha
TM: CXCL1/CXCR2 signaling in pathological pain: Role in peripheral
and central sensitization. Neurobiol Dis. 105:109–116.
2017.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Ringland C, Schweig JE, Eisenbaum M, Paris
D, Ait-Ghezala G, Mullan M, Crawford F, Abdullah L and Bachmeier C:
MMP9 modulation improves specific neurobehavioral deficits in a
mouse model of Alzheimer's disease. BMC Neurosci.
22(39)2021.PubMed/NCBI View Article : Google Scholar
|