Introduction
Astrocyte elevated gene-1 (AEG-1), which is also
known as metadherin and lysine-rich carcinoembryonic
antigen-related cell adhesion molecule 1 co-isolated, was first
observed to be upregulated in brain astrocytes from patients with
HIV-induced Alzheimer's disease (1). AEG-1 has been shown to localize to
the cell membrane, cytoplasm, endoplasmic reticulum, nucleolus and
nucleus in cancer cells (2). At
present, the majority of previous studies have focused on the
function of AEG-1 in the development and progression of various
malignancies such as gastric cancer, breast cancer and malignant
glioma (3,4). As an oncogene, AEG-1 has been found
to facilitate tumor growth, metastasis, angiogenesis and drug
resistance (5,6). In addition, AEG-1 as a key modulator
regulates aberrant cellular processes within the central nervous
system (CNS), where it is involved in neurological diseases such as
Huntington's chorea, migraine and HIV-induced neurological
disorders (7-9).
AEG-1 has differential regulatory effects on
astrocytes (AST) and neurons. In AST, AEG-1 causes glutamatergic
excitotoxicity by downregulating the activity of the excitatory
amino acid transporter 2 (EAAT2) promoter, leading to neuronal cell
death in glioma-induced neurodegenerative disease (10). Furthermore, AEG-1 can promote AST
activation and hyperplasia induced by brain injury in a mouse model
of reactive astrogliosis (11).
Silencing AEG-1 expression has also been shown to suppress AST
migration and proliferation to wounded areas (11). By contrast, in neurons,
downregulation of AEG-1 expression has been shown to reduce the
viability of motor neurons in models of amyotrophic lateral
sclerosis (ALS), both in vivo and in vitro (12). In addition, upregulation of AEG-1
expression can protect nigral dopaminergic neurons from injury
caused by aberrant apoptotic signaling pathways (13). AEG-1 has also been reported to
regulate embryonic neural development (14). Although AEG-1 is evolutionarily
conserved in vertebrates, the expression of AEG-1 in CNS cells,
including neurons and astrocytes (9,15),
has not been previously studied in depth to the best of our
knowledge. Therefore, further investigation into the potential role
of AEG-1 in neurons and its physiological mechanisms is
warranted.
CRISPR/Cas9 technology is a state-of-the-art gene
editing method that was originally derived from the prokaryotic
adaptive immune system (16). The
CRISPR/Cas9 system contains a single guide RNA (sgRNA) molecule and
a nucleic acid endonuclease, Cas9. These two components
cooperatively cleave the desired sequence in the target DNA at
specific locations by forming complementary base pairings. A
double-stranded break is subsequently formed (17). In the present study, an
AEG-1-deficient neuronal HT22 cell line was constructed using such
CRISPR/Cas9 technology. Cell morphology was subsequently observed
in AEG-1-deficient cells using light microscope. A list of
differentially expressed genes (DEGs) were obtained using
RNA-sequencing (RNA-seq) analysis. Functional enrichment analysis
on these DEGs was then performed to investigate neuronal AEG-1
function.
Materials and methods
Cell culture
The mouse hippocampal neuronal cells (HT22) were
purchased from Jennio Biotech Co., Ltd. The cells were cultured in
DMEM (Biological Industries) supplemented with 10% FBS (Biological
Industries) and 1% penicillin streptomycin (Beijing Solarbio
Science & Technology Co., Ltd.) at 37˚C in an incubator with 5%
CO2 for 2 days.
GV392-AEG-1-sgRNA plasmid generation
and lentivirus packaging
HT22 cells are highly sensitive to glutamate, have
cholinergic neuronal properties and have therefore been proposed to
be viable models for studying neurodegenerative diseases (18,19).
The present study was designed and performed based on the mouse
genome (GRCm39/m29). The full-length cDNA sequence
(ATGGCTGCACGAAGCTGGCAGGACGAGCTGGCCCAGCAGGCCGAGGAGGGCTCTGCCCGGCTGCGGGAGTTGCTCTCGGTCGGCCTAGGTTTTCTGCGCACGGAGTTGGGCCTCGACCTGGGGCTAGAGCCGAAGCGGTACCCGGGCTGGGTGATCCTGGTGGGCACCGGCGCTCTCGGGCTGCTCCTGCTCTTCCTTCTAGGTTACGGCTGGGCCGCGGCTTGCGCCGGCGCCCGCAAGAAGCGAAGGAGCCCGCCCCGCAAACGGGAGGAGGCGGCCCCGCCGACTCCGGCCCCCGACGACCTAGCCCAGCTGAAGAATCTCAGAAGCGAGGAGCAAAAGAAGAAGAACCGGAAGAAGCTTCCTGAAAAGCCCAAACCAAATGGACGGACTGTTGAAGTACCCGAGGATGAAGTTGTTAGAAATCCCCGAAGTATAACTGCAAAACAAGCACCAGAGACAGACAAGAAAAATGAAAAGTCAAAGAAAAATAAGAAGAAATCAAAGTCAGATGCTAAAGCAGTGCAAAACAGTTCACGCCATGATGGAAAGGAAGTTGATGAAGGAGCCTGGGAAACTAAAATTAGTCACAGAGAGAAACGACAACAGCGTAAACGTGATAAAGTGCTGACTGATTCTGGTTCATTGGATTCAACTATCCCTGGGATAGAAAATATCATCACAGTTACCACCGAGCAACTTACAACTGCATCATTTCCTGTTGGTTCCAAGAAGAATAAAGGTGATTCTCATCTAAATGTTCAAGTTAGCAACTTTAAGTCTGGAAAAGGAGATTCTACACTGCAGGTTTCTTCAAGGCTGAATGAAAATCTTACTGTCAATGGAGGAGGCTGGAGTGAAAAGTCTGTAAAACTCTCCTCACAATTGAGTGAGGAGAAGTGGAACTCTGTCCCACCTGCTTCTGCAGGCAAGAGGAAAACAGAGCCATCGGCTTGGACTCAAGACACTGGTGACACTAATGCAAATGGGAAAGACTGGGGAAGGAATTGGAGTGATCGCTCAATATTTTCTGGCATTGGATCTACTGCTGAGCCAGTTTCTCAGTCTACCACTTCTGATTATCAGTGGGATGTTAGCCGTAATCAACCTTATATCGATGATGAATGGTCTGGGTTAAATGGTTTGTCTTCTGCTGACCCTAGCTCAGACTGGAATGCACCAGCAGAGGAGTGGGGGAACTGGGTAGATGAAGATAGAGCTTCACTTCTGAAGTCCCAGGAACCAATTTCTAATGATCAAAAGGTTTCAGATGATGATAAAGAAAAAGGGGAGGGAGCTCTTCCAACTGGAAAATCTAAAAAGAAAAAGAAGAAAAAGAAGAAGCAAGGGGAAGATAACTCTCACACACAGGACACAGAAGACCTAGAAAAGGACACTAGAGAAGAGCTTCCAGTGAATACCTCAAAAGCCCGACCAAAACAGGAGAAAGCTTGTTCCCTGAAGACCATGAGCACTAGTGACCCAGCTGAAGTACTCATCAAAAATAGCCAGCCTGTCAAGACTCTTCCTCCTGCTATCTCTGCCGAGCCATCTATTACCTTATCAAAAGGTGACTCTGACAACAGCTCTTCCCAAGTGCCACCGATGTTACAAGACACAGACAAGCCCAAGTCAAATGCTAAGCAAAACAGTGTGCCTCCCTCACAGACCAAGTCTGAAACTAACTGGGAATCTCCAAAACAAATAAAAAAGAAGAAAAAGGCCAGACGGGAAACGTGA)
of mouse AEG-1 gene was 3,611 bp and contained 12 transcripts and
12 exons. Exon regions corresponding to the conserved protein
sequences were selected for target screening from Feng Zhang Lab
Library version 2 (http://www.addgene.org/pooled-library/zhang-mouse-gecko-v2/).
Sequences with higher CFD specificity scores and lower number of
off-target sites according to the Feng Zhang Lab website
(https://zlab.bio/guide-design-resources), were used as
main sequences.
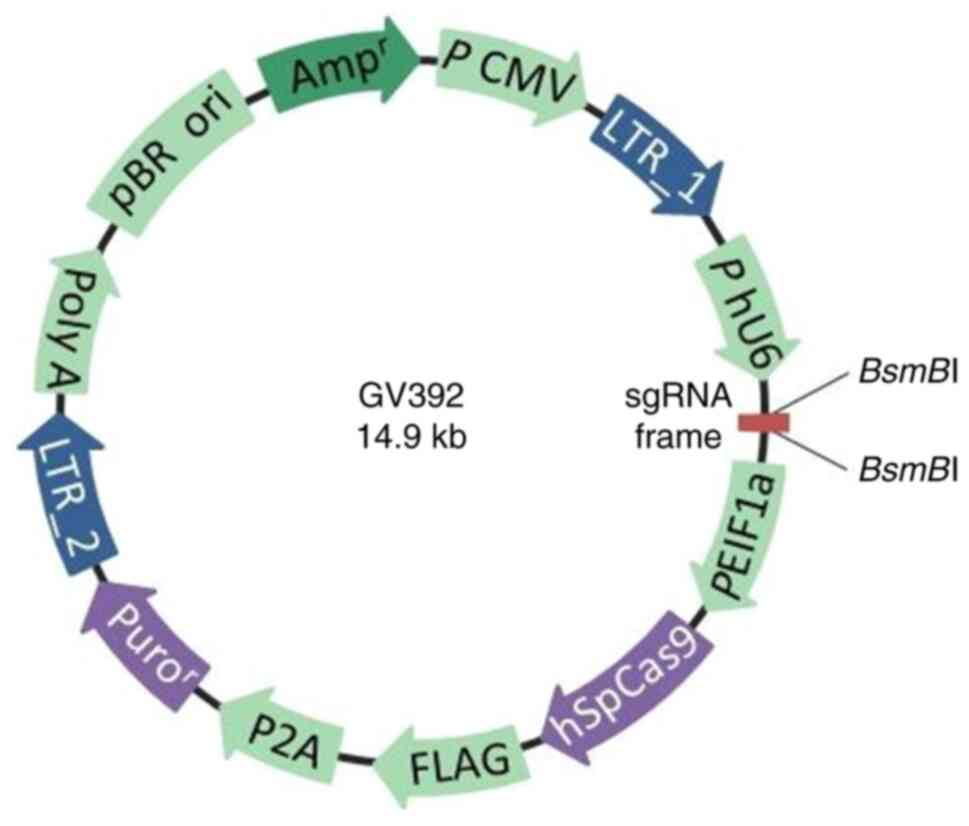
The GV392 plasmid (Fig.
1) was purchased from Shanghai GeneChem Co., Ltd. The
BsmBI site in this GV392 plasmid was selected to be the
sgRNA insertion point. The three sets of designed oligonucleotide
sequences are listed in Table I.
For each of the sgRNA sequences used, ‘CACCG’ was added to the 5'
ends whereas ‘AAAC’ was added to the 3' end. The GV392 plasmid was
subsequently cleaved using the restriction endonuclease
BsmBI (Shanghai GeneChem Co., Ltd.) to obtain the linearized
vector. The synthesized sgRNA oligonucleotides (Shanghai GeneChem
Co., Ltd.) were then phosphorylated, and annealed at 95˚C for 15
min (Shanghai Yuanye Biotech Co., Ltd.) to form double-stranded DNA
according to the manufacturer's instructions. T4 DNA Ligase High
Concentration Rapid Kit (Fermentas; Thermo Fisher Scientific, Inc.)
ligated the linearized GV392 plasmid with the double-stranded sgRNA
at 22˚C for 3 h, which were then amplified using DH5α-competent E.
Coli (Beijing Mei5 Biotech Co., Ltd.).
 | Table IAEG-1 sgRNA oligonucleotides. |
Table I
AEG-1 sgRNA oligonucleotides.
| Name | Sequence | Exon |
|---|
|
M-AEG-1-sgRNA-1 |
5'-CACCGACTTCAACAGTCCGTCCATT-3' | 2 |
| |
3'-CTGAAGTTGTCAGGCAGGTAACAAA-5' | |
|
M-AEG-1-sgRNA-2 |
5'-CACCGTCATTGGATTCAACTATCCC-3' | 3 |
| |
3'-CAGTAACCTAAGTTGATAGGGCAAA-5' | |
|
M-AEG-1-sgRNA-3 |
5'-CACCGCAAAACAGTTCACGCCATGA-3' | 4 |
| |
3'-CGTTTTGTCAAGTGCGGTACTCAAA-5' | |
| Negative control
sgRNA |
5'-CACCGCGCTTCCGCGGCCCGTTCAA-3' | |
| |
3'-CGCGAAGGCGCCGGGCAAGTTCAAA-5' | |
The GV392-AEG-1 sgRNA lentiviral expression vector
was co-transfected with lentivirus packaging helper plasmids (2nd
generation; 10 µg lentivirus plasmid used for transfection: 5 µg
lentivirus, 3.75 µg packaging and 1.25 µg envelope plasmids) into
293T cells (high sugar DMEM containing 10% FBS, 37˚C, 5%
CO2; Cell bank of Chinese Academy of Sciences) for
lentivirus packaging. Before the virus titer was determined,
lentivirus (M-AEG-1-sgRNA-1/M-AEG-1-sgRNA-2/M-AEG-1-sgRNA-3) stock
solutions were collected and concentrated by ultracentrifugation at
4,000 x g (4˚C) after 48 h at 37˚C of transfection.
Generation of AEG-1-deficient HT22
cell lines
The proliferation of the HT22 cells was observed in
the presence of 6 and 4 µg/ml puromycin (Fermentas; Thermo Fisher
Scientific, Inc.) before the cells were infected with the
lentivirus. Cells at logarithmic growth stage were digested with
0.25% trypsin-EDTA solution and inoculated into six-well plates at
a density of 1x105 cells per well. The cells were
infected at 37˚C with 50 µl virus M-AEG-1-sgRNA-1 (titer
2x108/ml), 50 µl virus M-AEG-1-sgRNA-2 (titer
2x108/ml), 50 µl negative control virus (titer
2x108/ml) and 33 µl M-AEG-1-sgRNA-3 (titer
3x108/ml) at a multiplicity of infection value 50 at the
same time, to achieve the optimum infection by each virus. After 3
days of culture in DMEM containing 6 µg/ml puromycin (Beijing
Solarbio Science & Technology Co., Ltd.), the puromycin
concentration was reduced to 4 µg/ml. Stably infected cells were
then inoculated into 96-well plates by trypsin digestion. After 6 h
at 37˚C, the cells were observed under a light microscope to
establish monoclonal cell lines.
Verification of sgRNA activity
To analyze the mutations caused by CRISPR/Cas9,
whole genomic DNA was extracted from AEG-1-deficient HT22 cells
using PureLink™ Genomic DNA Mini kit (Omega Bio-Tek,
Inc.). The DNA fragment was amplified by TB Green®
Premix Ex Taq™ II (Tli RNaseH Plus) kit (Takara Bio,
Inc.) using AEG-1-sgRNA-specific primers. The forward and reverse
primers of AEG-1-sgRNAs are provided in Table II. The PCR cycle was as follows:
Initial denaturation at 94˚C for 4 min, followed by 35 cycles of
denaturation at 94˚C for 30 sec, annealing at 48˚C for 30 sec,
elongation at 72˚C for 30 sec and a final extension at 72˚C for 12
min. The annealed PCR products were digested with T7 Endonuclease I
(New England BioLabs, Inc.), which specifically cleaves mismatched
DNA, at 37˚C for 1 h. The cleaved DNA fragment was then analyzed
using 1.5% agarose gel electrophoresis and visualized by ethidium
bromide staining.
 | Table IIPrimer list. |
Table II
Primer list.
| Name | Direction | Sequence |
|---|
| Primers of
M-AEG-1-sgRNA-1 | Forward |
5'-TTTCATTGTTGTATATGTTATTTCC-3' |
| | Reverse |
5'-AGACTACACTCTTATTAACCATGAA-3' |
| Primers of
M-AEG-1-sgRNA-2 | Forward |
5'-CTTTTATACCACACACCTCAGTTTA-3' |
| | Reverse |
5'-AGTTGTCAGTTTCATACATTTCATT-3' |
| Primers of
M-AEG-1-sgRNA-3 | Forward |
5'-TCAGCACAAAGTTAGCAGTTCAAAA-3' |
| | Reverse |
5'-TCAGGAGCCTGGGAAACTAAAATTA-3' |
| AEG-1 qPCR
primer | Forward |
5'-CCGCAAGAAGCGAAGGAGC-3' |
| | Reverse |
5'-CCGTCCATTTGGTTTGGGCT-3' |
| GAPDH qPCR
primer | Forward |
5'-AGGCCGGTGCTGAGTATGTC-3' |
| | Reverse |
5'-TGCCTGCTTCACCACCTTCT-3' |
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was collected from normal HT22 cells,
negative control virus cells and cells infected with the three
M-AEG-1-sgRNA viruses using the TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The normal HT22 cells were used as the
blank control group, whilst the HT22 cells infected with viruses
encoding control gRNA were used as the negative control group. RNA
purity and concentration were measured using a
NanoPhotometer® Spectrophotometer (Implen GmbH). Total
RNA was used for the reverse transcriptase reaction with FastKing
gDNA Dispelling RT SuperMix kit (Tiangen Biotech Co., Ltd.), the
reaction conditions are as follows: Genome removal and reverse
transcriptional reaction at 42˚C for 15 min, enzyme inactivation
process at 95˚C for 3 min. To evaluate the expression of blank
control group, negative control group and groups infected with the
three M-AEG-1-sgRNA viruses at the mRNA level, TB Green®
Premix Ex Taq™ II (Tli RNaseH Plus) kit (Takara Bio,
Inc.) and CFX96 Real-Time PCR Detection System (Bio-Rad
Laboratories, Inc.) were used for PCR amplification. The
thermocycling profiles incorporated an initial denaturation at 95˚C
for 30 sec, followed by 40 cycles of denaturation at 95˚C for 5 sec
and annealing at 60˚C for 30 sec. The sequences of the primers used
for qPCR are provided in Table
II. The Cq values of qPCR results in each group were converted
to 2-ΔΔCq values for comparison (20). The expression levels of each mRNA
in the cells were normalized to the level of GAPDH mRNA
expression.
Western blotting
The normal HT22 cells, negative control virus cells
and cells infected with the three M-AEG-1-sgRNA viruses were lysed
using Radio-Immunoprecipitation Assay buffer (100 ml; Jiangsu
Keygen Biotech Co., Ltd.) containing phosphatase and proteinase
inhibitors at 4˚C. The supernatant was then collected from the
lysate by centrifugation at 300 x g for 15 min at 4˚C. The
harvested protein concentrations were measured using the BCA
protein quantitation reagent (Nanjing KeyGen Biotech Co., Ltd.).
After boiling for 5 min at 95˚C, the total protein (50 µg) was
resolved using 10% SDS-PAGE gels and transferred onto
polyvinylidene difluoride membranes (MilliporeSigma). The membranes
were blocked in 5% skimmed milk for 1 h at room temperature and
then probed with the anti-AEG-1 antibody (cat. no. 13860-1-AP;
1:1,000; ProteinTech Group, Inc.) or the anti-GAPDH antibody (cat.
no. GTX100118; 1:1,000; GeneTex, Inc.) overnight at 4˚C, before
being incubated with the secondary antibody (Dylight 800; goat
anti-rabbit IgG; 1:2,000; cat. no. A23920; Abbkine Scientific Co.,
Ltd.) for 1 h at room temperature. Finally, the membranes were
developed using ECL reagents (Pierce; Thermo Fisher Scientific,
Inc.) and visualized using the full-automatic chemiluminescence
imaging analysis system (Bio-Rad Laboratories, Inc.). The
integrated band densities were measured using ImageJ v1.8.0.
software (National Institutes of Health).
The group of cells (those transfected with the
M-AEG-1-sgRNA-3) with the highest knockdown effect were screened
based on the western blotting and qPCR results. Genomic DNA of the
AEG-1-KO monoclonal cell line was extracted and amplified by AEG-1
specific primers (methods as aforementioned). The PCR products were
then purified using MiniBEST DNA Fragment Purification Kit Ver.4.0
(Takara Bio, Inc.) and ligated to a pMD19-T vector using
pMD™ 19-T Vector Cloning Kit (Takara Bio, Inc.). The
product was transformed into TOP10 competent cell (Shanghai
GeneChem Co., Ltd.) and positive clones were obtained by colony
PCR. The positive bacterial colonies were selected to extract
plasmids for Sanger sequencing analysis.
RNA quantification and library
construction
RNA purity and concentration was checked using a
NanoPhotometer spectrophotometer (Implen, Inc.) and
Qubit® RNA Assay kit (cat. no. Q10210) in combination
with the Qubit 2.0 Fluorometer (both Thermo Fisher Scientific,
Inc.). RNA integrity was assessed using an RNA 6000 Nano Kit with a
Bioanalyzer 2100 system (both Agilent Technologies, Inc.).
For the RNA sample preparations, sequencing
libraries were generated with a total amount of 993 mole RNA per
sample using the NEBNext® Ultra™ RNA Library
Prep Kit for Illumina® (cat. no. E7530L; New England
BioLabs, Inc.). Briefly, the mRNA was separated from total RNA
samples using magnetic beads coupled with oligo (dT) and fragmented
using divalent cations in 5X NEBNext First Strand Synthesis
Reaction Buffer (New England Biolabs, Inc.) according to the
manufacturer's protocol. Next, the mRNA was reverse transcribed to
synthesize double-stranded complementary DNA (cDNA) using Moloney
Murine Leukemia Virus Reverse Transcriptase (New England BioLabs,
Inc.) with random hexamer primers. The 3' ends of the cDNA were
added to the base A and an NEBNext Adaptor (New England BioLabs,
Inc.) with a hairpin loop structure to prepare for hybridization.
The cDNA target fragments were purified with the AMPure XP system
(cat. no. A63381; Beckman Coulter, Inc.) to select fragments 200 bp
in length with the direction of 5'-3'. Subsequently, 3 µl USER
Enzyme (New England Biolabs, Inc.) was used with size-selected,
adaptor-ligated cDNA at 37˚C for 15 min followed by 5 min at 95˚C
before PCR. PCR was performed with Phusion High-Fidelity DNA
polymerase, Universal PCR primers and Index (X) Primer. Finally,
the PCR products were purified also using the AMPure XP system and
used to obtain a sequencing library. The library quality was
assessed using the Bioanalyzer 2100 system (Agilent Technologies,
Inc.) and the effective concentration (>2 nM) of the library was
accurately quantified using RT-qPCR.
Clustering and sequencing
Indicator-coded samples were clustered on the cBot
Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS
(cat. no. PE-401-3001; Illumina, Inc.). Sequencing was then
performed via the HiSeq X Ten System (cat. no. SY-412-1001;
Illumina, Inc.).
Data analysis
Clean data (clean reads) of fastq format were first
obtained by excluding adapters, ploy-N and low-quality reads (reads
for which >50% of the read length had a Phred quality value ≤20)
from the original data (raw reads) through in-house Perl Script
(fastq v1.0, the parameter was fastq-g-q 5-u 50-n 15-l 150;
https://www.novogene.cn/) programming. The Q20,
Q30 and guanine-cytosine content of the clean data were calculated
simultaneously using fastp (v0.19.4) software (Shenzhen Haplox
Biotech Co., Ltd.). High-quality clean reads which passed through
the described above filtering steps were used for downstream
analysis and mapped to the mouse reference genome (accession no.
GRCm39) using HISAT2 v2.0.5 (http://daehwankimlab.github.io/hisat2/). The clean
reads were compared with reference genes (accession no. GRCm39)
(https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.27/)
using TopHat2 v2.0.0 (http://ccb.jhu.edu/software/tophat/index.shtml).
Gene expression characterization and
differential gene expression analysis
Gene expression levels were estimated using
featureCounts v1.5.0-p3 (http://subread.sourceforge.net/). The expected number
of fragments per kilobase of transcript per million fragments
mapped (21) was calculated using
the Cufflinks software v2.0.1 (http://cufflinks.cbcb.umd.edu/) based on the length of
each gene. EdgeR R package v3.18.1 (http://www.bioconductor.org/) (22) was employed to evaluate differential
gene expression analysis between different groups (two biological
replicates per condition) (23).
The DESeq2 package v1.16.1 (http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html)
was used to identify differentially expressed genes. The method of
Benjamini and Hochberg (24) was
used to adjust for the resulting P-values to exclude false positive
results. Genes were assigned to be differentially expressed by
DESeq2 with adjusted P-values <0.05 and log2 fold
change >0.
Gene ontology (GO) analysis and Kyoto
encyclopedia of genes and genomes (KEGG) pathway analysis
GO (25) analysis
used |log2 fold change|>0 and padj <0.05 as
threshold (padj stands for corrected P-value), and performed on the
DEGs using the clusterProfiler R package (V3.4.4) (26). KEGG (27) is a database resource for
understanding the advanced functions and utilities of biological
systems at the molecular level. The clusterProfiler R package was
used to analyze the statistical enrichment of DEGs in KEGG pathways
(http://www.genome.jp/kegg/), with a
Benjamini-Hochberg-adjusted P-value cutoff of <0.05.
Protein-protein interaction (PPI)
network analysis
The PPI network of DEGs was analyzed using the
STRING database (v11.5; ELIXIR; https://string-db.org) (28), a pre-computed database of known and
predicted protein interactions, including direct (physical) and
indirect (functional) associations derived from co-expression,
co-occurrence, genomic context, gene fusion, high-throughput
experiments and text mining. The protein networks were then
presented using Cytoscape software v3.7.1 (http://www.cytoscape.org/) with a confidence level of
0.7 and calculation conditions were P<0.05 and no non-coding
RNAs allowed. The ‘Molecular Complex Detection (MCODE)’ plugin for
Cytoscape was used to analyze network modules (29). The parameters of densely connected
regions or clusters in the co-expression network were set as
follows: ‘Degree cut-off’=2, ‘k-core’=2 and ‘max. depth’=100. The
‘cytoHubba’ plugin for Cytoscape was used to screen for hub genes
(30) under the following
conditions: ‘Top10 node(s)’ ranked by ‘Degree’.
Statistical analysis
Values were presented as the mean ± standard error
of the mean (n≥3). All statistical analysis and graph illustrations
were performed using Prism V8.0 (GraphPad Software, Inc.). One-way
ANOVA analysis was assessed the differences between multiple
groups. Dunnett's test was used for all comparisons with a negative
control group. P<0.05 was considered to indicate a statistically
significant difference.
Results
GV392-AEG-1-sgRNA plasmid construction
and sgRNA activity validation
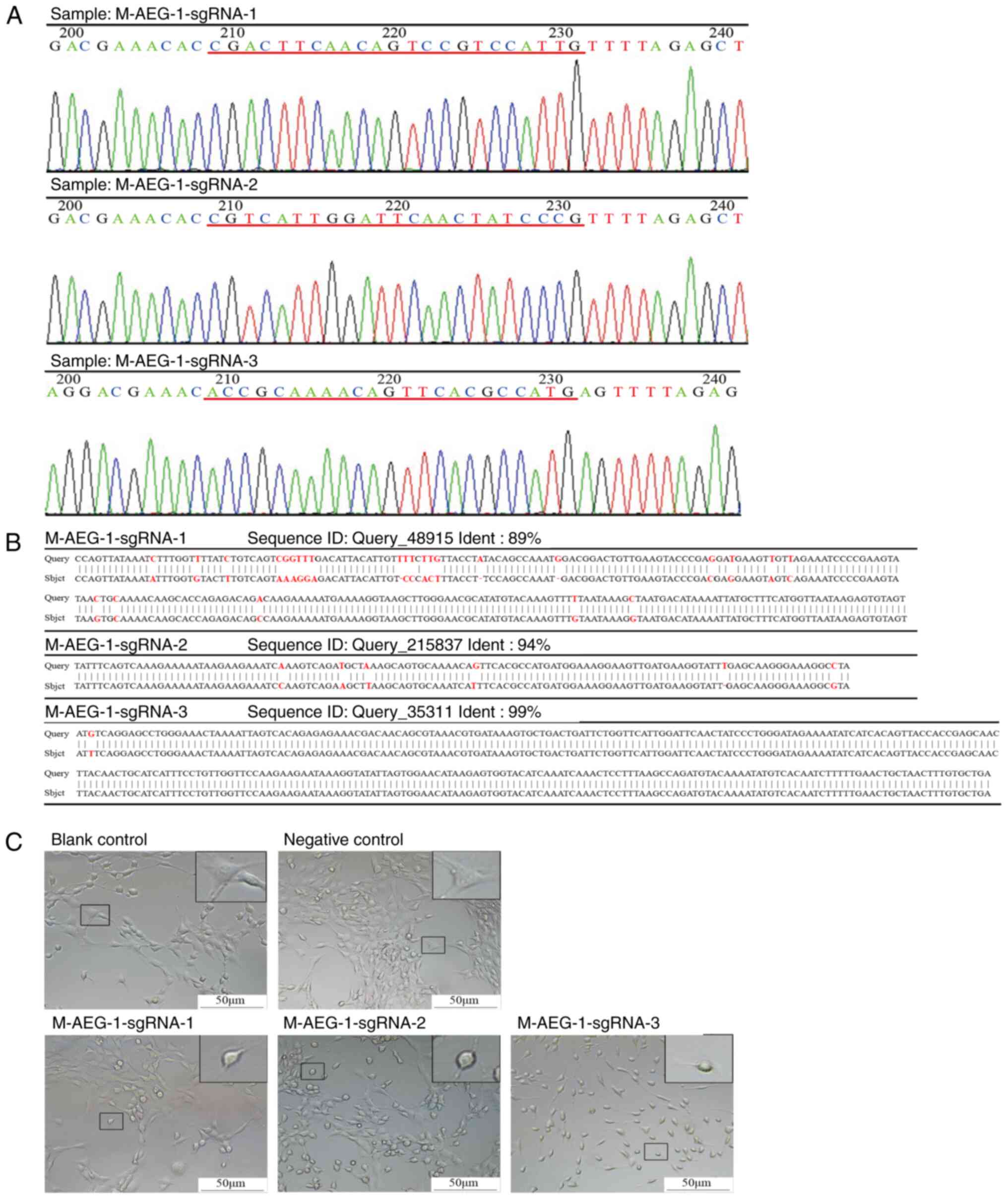
The inserts of the GV392-AEG-1-sgRNA plasmid with
the three AEG-1-sgRNA sites were validated by Sanger sequencing
(Fig. 2A). To verify the
efficiency of AEG-1-sgRNAs, both insertion and deletion mutations
were evaluated in the genomic DNA sequence of AEG-deficient
neuronal HT22 cells. SgRNA directs Cas9 protein to bind to the
target sequence, Cas9 protein is a nuclease that creates a cut at
the target sequence site and DNA repair occurs by mutation
(31). All three AEG-1-sgRNAs
effectively caused mutations in the specific sites with 89, 94 and
99% efficiency for M-AEG-1-sgRNA-1, M-AEG-1-sgRNA-2 and
M-AEG-1-sgRNA-3, respectively (Fig.
2B). The number of protrusions was reduced in AEG-1-deficient
neuronal HT22 cells compared with normal HT22 cells, especially in
the cell group transfected with the M-AEG-1-sgRNA-3 virus (Fig. 2C).
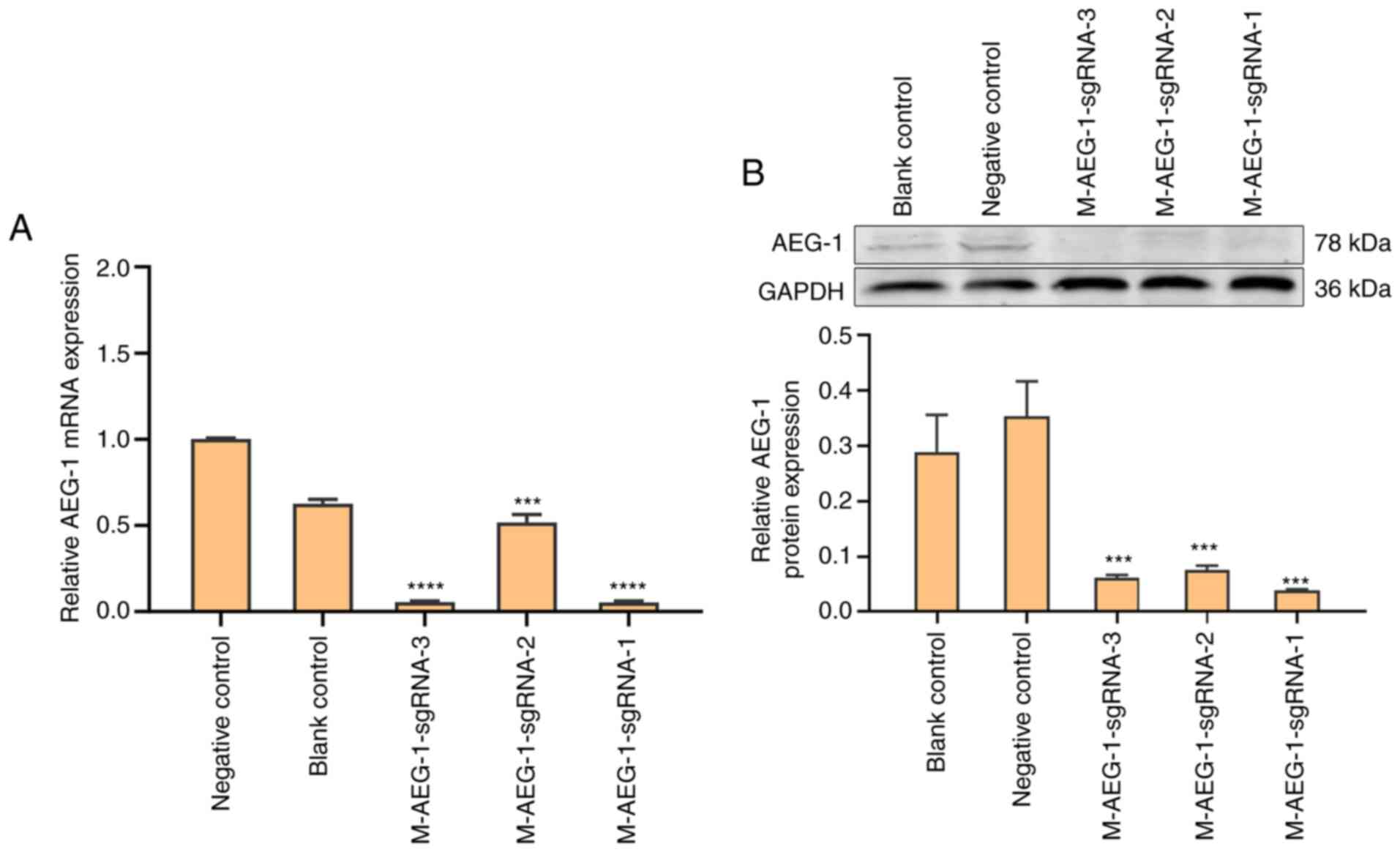
Verification of AEG-1 knockdown
qPCR and western blot analysis were performed to
select for AEG-1-deficient HT22 cell lines with the highest
knockdown efficiency. As mentioned above, SgRNA directs Cas9
protein to bind to the target sequence, Cas9 protein is a nuclease
that creates a cut at the target sequence site and DNA repair
occurs by mutation. The results suggest that all three
M-AEG-1-sgRNA cell lines showed significant reductions in AEG-1
expression compared with that in the negative control group, with
the highest knockdown efficiency in the M-AEG-1-sgRNA-1 cell line
(Figs. 3A and B and S1). Therefore, the M-AEG-1-sgRNA-1 group
was selected for subsequent transcriptomic analysis. The higher
AEG-1 expression in the negative control compared with the blank
control may have been the effect of the viral empty vector on the
AEG-1 virus.
RNA-seq analysis and reference gene
sequence comparison
An average of 58,514,448 clean reads of
differentially expressed mRNAs were obtained in the control group,
whilst an average of 64,377,723 clean reads were obtained in the
AEG-1-KO group. Sequencing quality information is listed in
Table III. The clean reads were
compared with reference genes using TopHat2 v2.0.0. The comparison
efficiency refers to the percentage of mapped reads in clean reads,
and the comparison efficiency was 95.91% in the control group and
96.14% in the AEG-1-KO group (Table
SI).
 | Table IIIRNA-sequencing data statistics. |
Table III
RNA-sequencing data statistics.
| Groups | Clean reads | Clean bases, G | GC content, % | % ≥ Q20 | % ≥ Q30 |
|---|
| N_con_1 | 58731670 | 8.81 | 53.91 | 97.91 | 94.15 |
| N_con_2 | 59762786 | 8.96 | 51.26 | 97.52 | 93.23 |
| N_con_3 | 57048890 | 8.56 | 50.90 | 97.84 | 93.93 |
| AEG-KO_1 | 70543436 | 10.58 | 50.94 | 97.86 | 94.01 |
| AEG-KO_2 | 66312428 | 9.95 | 51.86 | 97.81 | 93.89 |
| AEG-KO_3 | 56271306 | 8.44 | 51.61 | 97.85 | 93.97 |
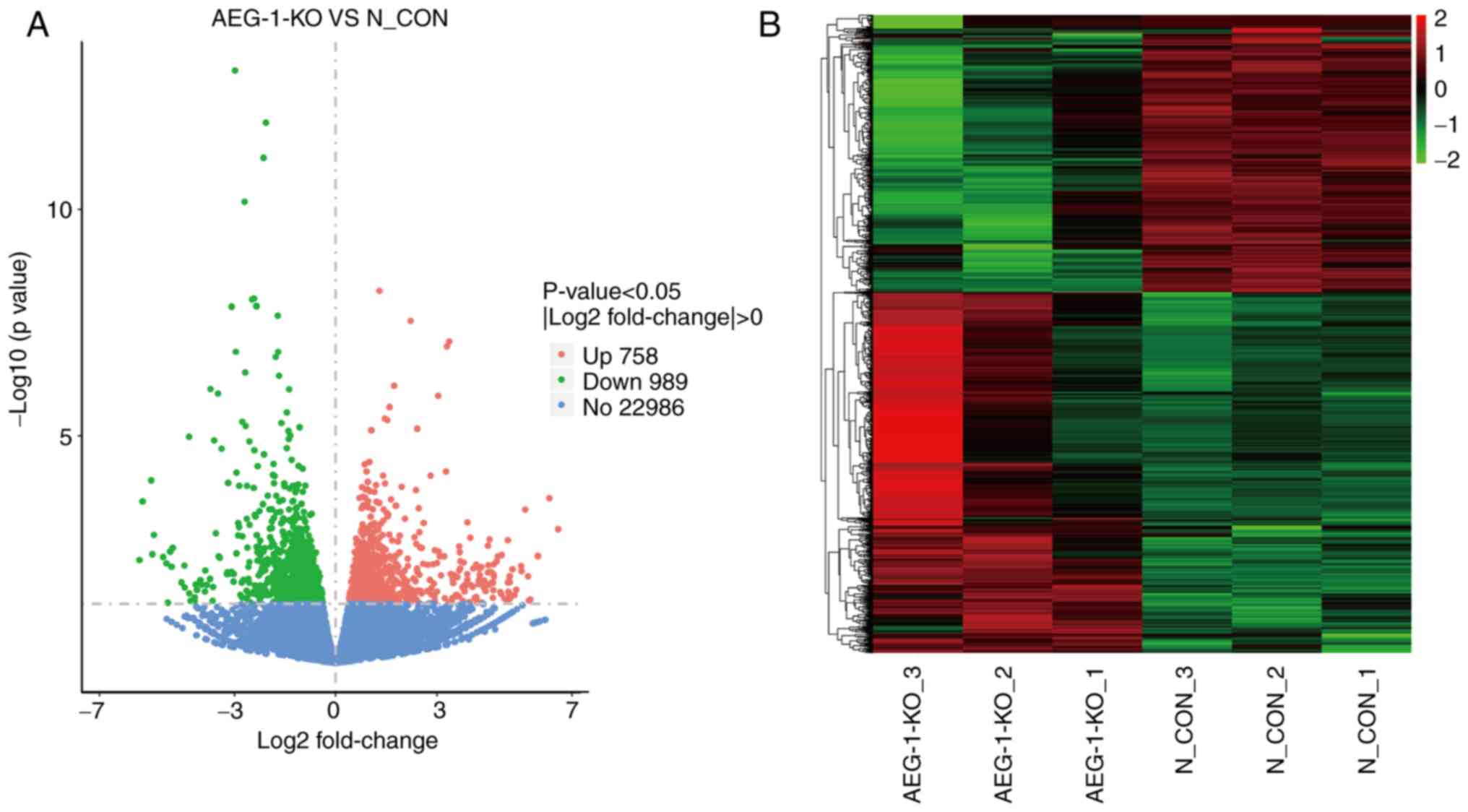
Gene expression identification and DEG
analysis
Variations in gene expression were presented in
volcano plots by comparing the control and AEG-1-KO groups. Genes
with similar expression patterns were categorized and clustered
into hierarchical clustering heat maps. A total of 1,747 DEGs were
screened based on the selection criteria of |log2 fold change|>0
and P<0.05. Among the 1,747 DEGs, 758 genes were upregulated and
989 genes were downregulated in the AEG1-KO group compared with the
control group (Fig. 4A and
B).
KEGG pathway analysis and GO analysis
on DEGs
KEGG pathway analysis and GO analysis were performed
to uncover the potential roles of the selected DEGs following AEG-1
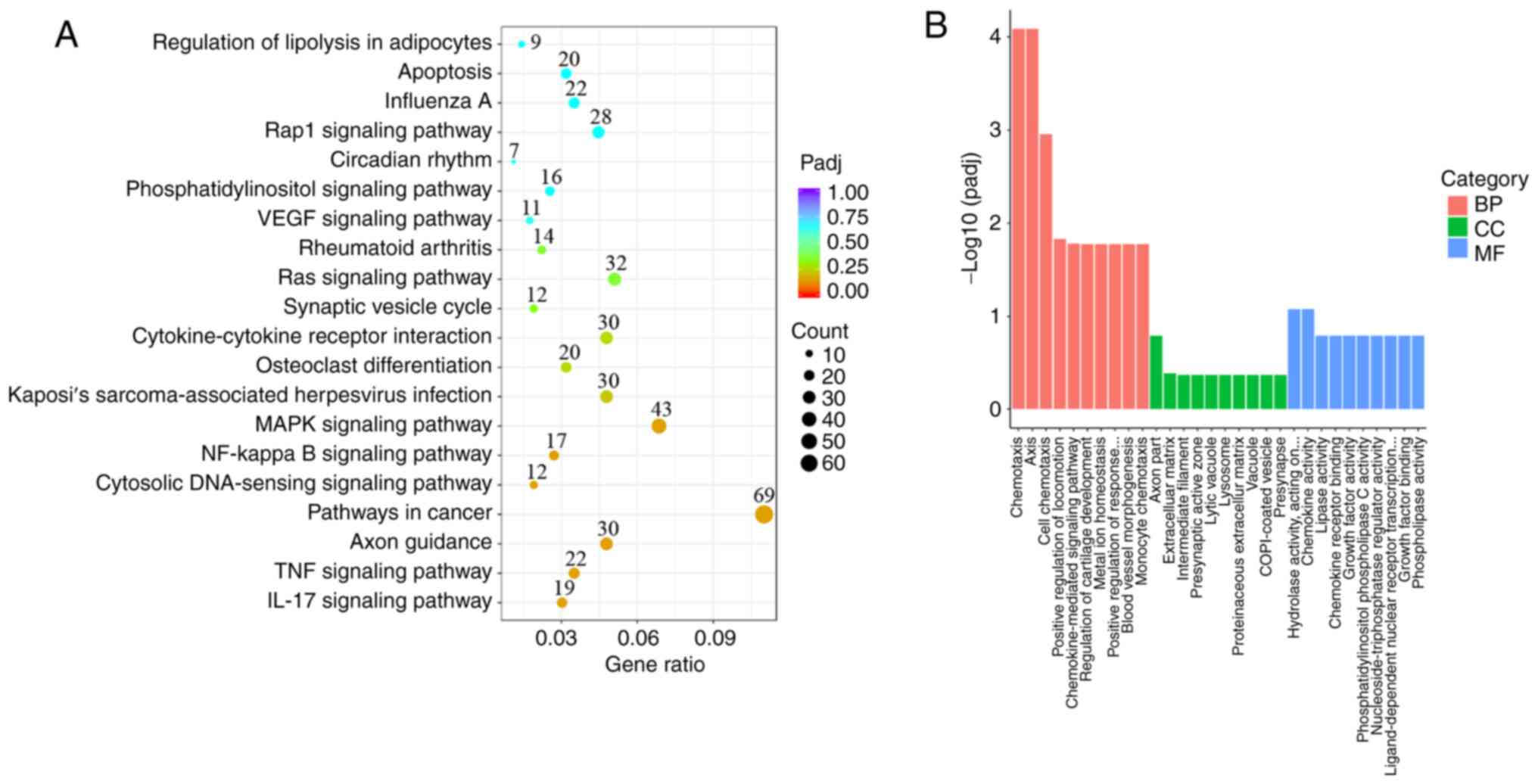
knockdown. The selected DEGs were particularly enriched in 20
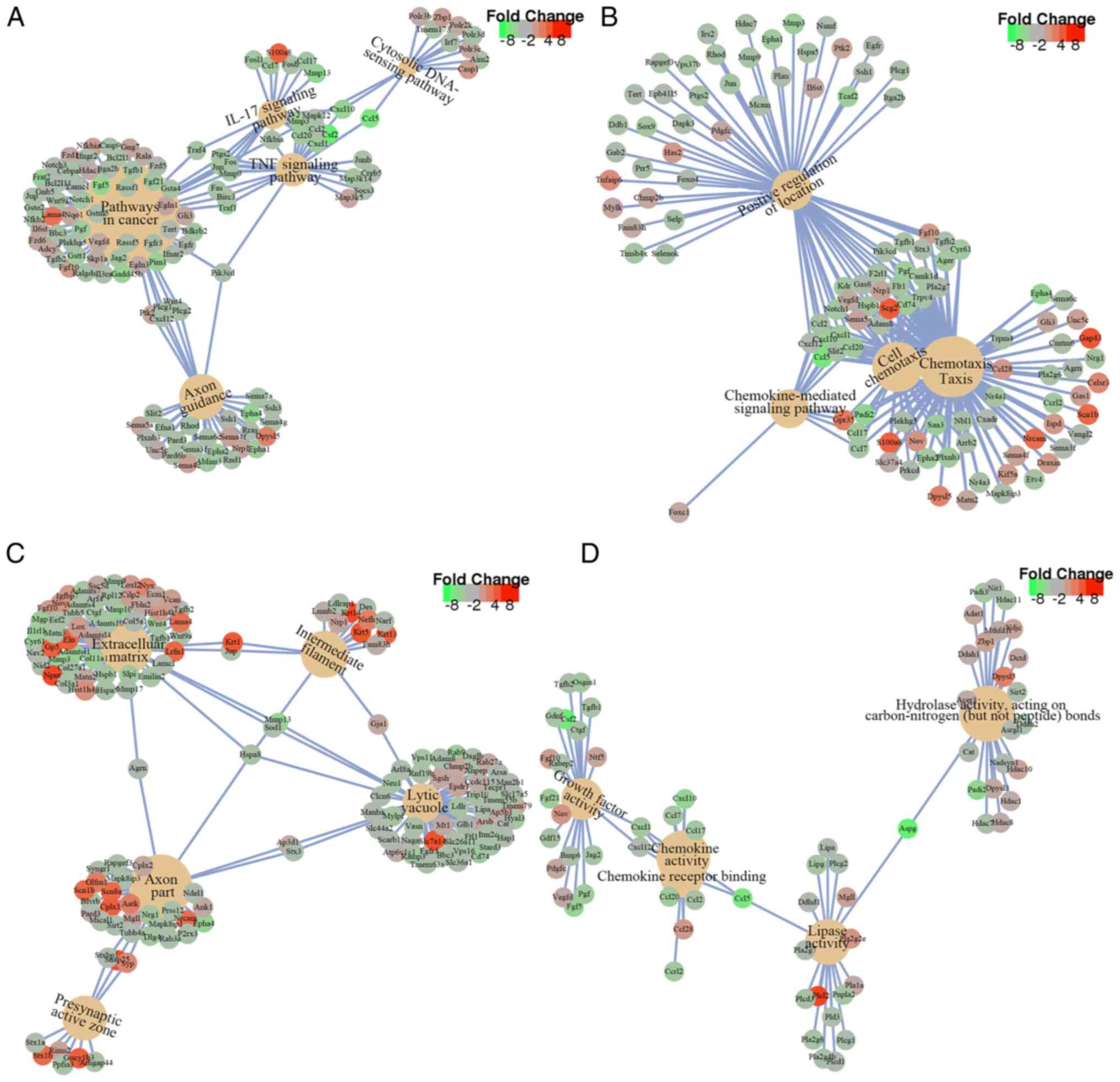
signaling pathways (Fig. 5A). The
top five enriched pathways were ‘Axon guidance’, ‘Pathways in
cancer’, ‘TNF signaling pathway’, ‘IL-17 signaling pathway’ and
‘Cytosolic DNA-sensing signaling pathway’, which are closely
associated with inflammation, cell proliferation and
differentiation (Figs. 5A and
6A) (32-34).
GO analysis showed that the DEGs were mainly enriched in terms of
biological processes (BP), cell composition (CC) and molecular
function (MF) (Fig. 5B). Within
BP, DEGs were particularly enriched in ‘chemotaxis’, ‘taxis’, ‘cell
chemotaxis’, ‘positive regulation of locomotion’ and
‘chemokine-mediated signaling pathway’ (Figs. 5B and 6B). Within CC, DEGs were refined in ‘axon
part’, ‘extracellular matrix’, ‘intermediate filament’,
‘presynaptic active zone’ and lytic vacuole’ (Figs. 5B and 6C). Within MF, DEGs were primarily
enriched in ‘hydrolase activity’, ‘carbon-nitrogen (except peptide)
bonds, ‘chemokine activity’, ‘lipase activity’, ‘chemokine receptor
binding’ and ‘growth factor activity’ (Figs. 5B and 6D).
Protein-protein interaction network
analysis and hub gene selection
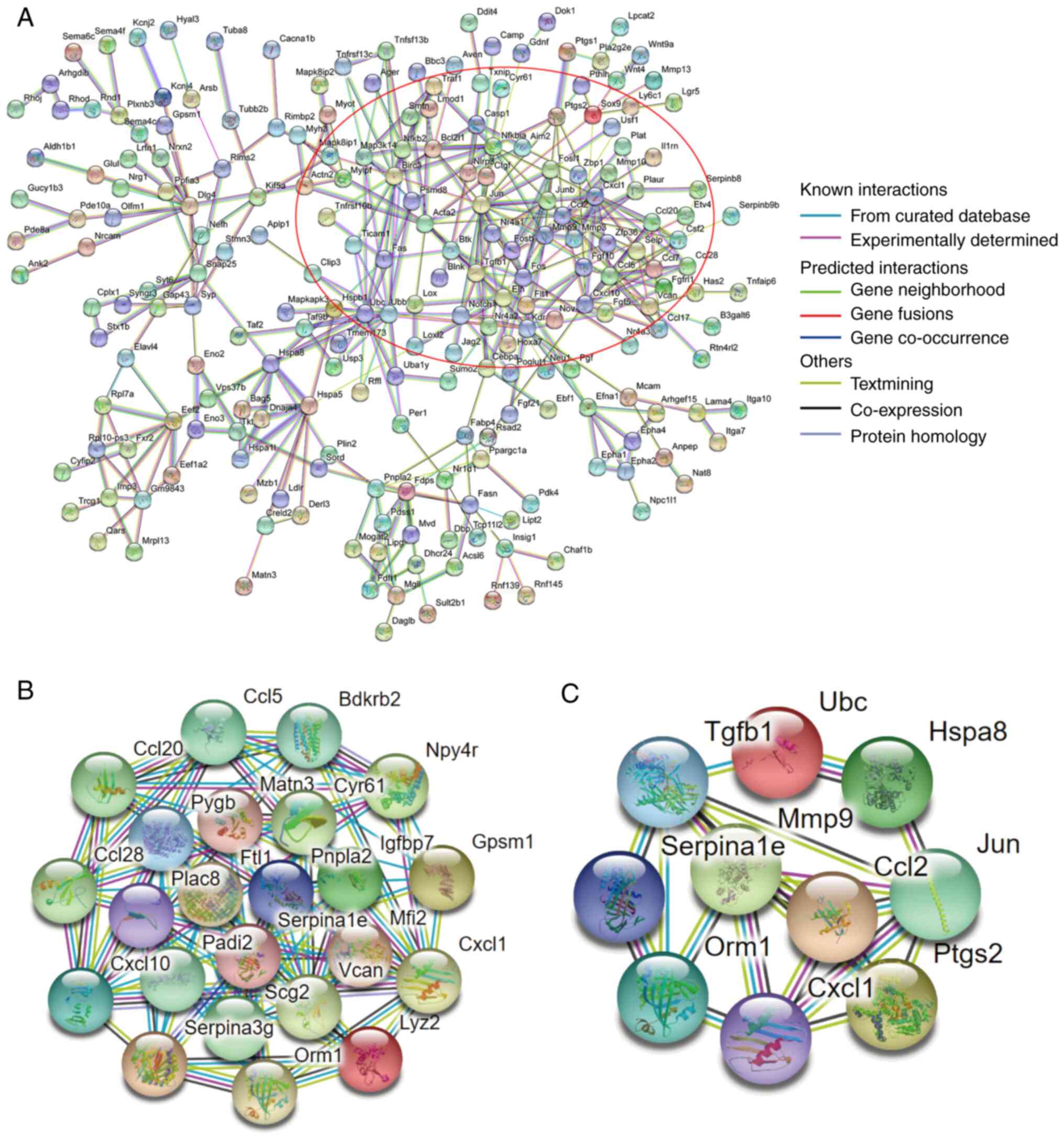
Protein interactions networks of DEGs were analyzed
using the STRING database. DEGs formed a complex network containing
221 nodes and 395 degrees with a higher degree of connectivity
(Fig. 7A). By using the MCODE
application within the Cytoscape software, significant modules in
the PPI network were detected. There was a top module in this
network, which contained 23 node genes and 81 interaction pathways
(Fig. 7B). Filtered by degree
calculation, 10 hub genes were obtained: Ubiquitin C (UBC), C-X-C
motif chemokine ligand 1 (CXCL1), matrix metallopeptidase 9 (MMP9),
orosomucoid 1, JUN, TGFβ1, SERPINA1E, heat shock protein family A
(Hsp70) member 8, CC motif chemokine ligand 2 and
prostaglandin-endoperoxide synthase 2 (Fig. 7C).
Characterization of genes associated
with neurogenesis and ion homeostasis
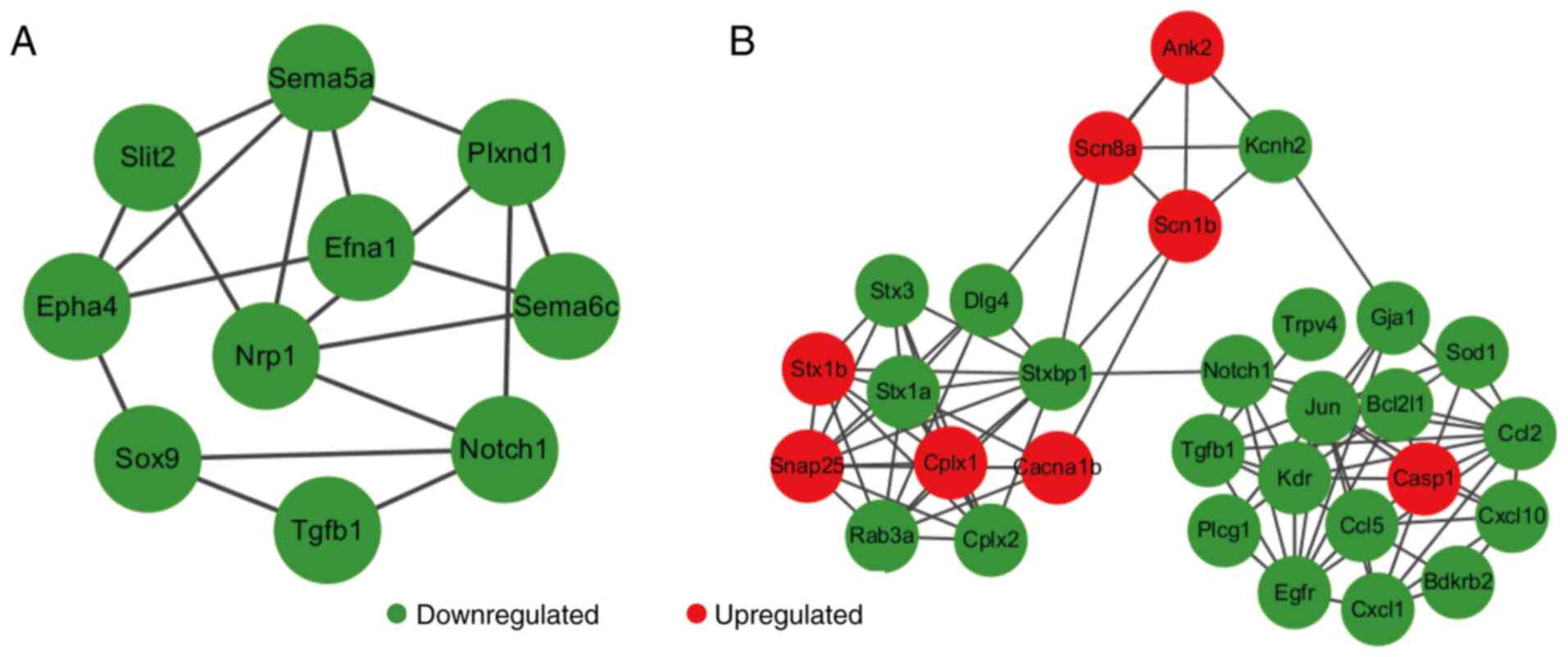
Certain DEGs such as neuropilin 1 (NRP-1) and Notch1
were involved in axon guidance, cell migration, neuronal
differentiation, regulation of exocytosis and formation of certain
neuronal circuits (Fig. 8A),
suggesting that the reduced expression of these DEGs greatly
affected the development of the nervous system process. In
addition, 30 DEGS associated with ion homeostasis were categorized,
especially the genes of ion channels such as calcium, sodium and
potassium channels and neurotransmitter release, including calcium
voltage-gated channel subunit α1 B, sodium voltage-gated channel β
subunit 1, potassium voltage-gated channel subfamily h member 2,
synaptosome associated protein 25, syntaxin 3 and RAB3A (Fig. 8B). These results suggest that AEG-1
deficiency contributes to the regulation of neurotransmitter
release. Therefore, AEG-1 may serve a potential role in the
synaptic function of neuronal systems.
Discussion
In recent years, the CRISPR/Cas9 system has greatly
promoted site-specific mutagenesis and has broad applications such
as gene function identification, disease modeling, gene therapy and
immunotherapy (35-37).
The CRISPR/Cas9 system only requires the function of sgRNAs, which
is less complex and more cost-effective to design compared with
alternative methods of knocking out or editing genes, such as zinc
finger nuclease and transcription activator-like effector nuclease
(38). In particular, CRISPR/Cas9
is a reliable method for knocking down the expression of the
desired gene (39). HT22 is a
widely used hippocampal neuron cell line that has been extensively
studied in the context of a variety of nervous system diseases,
such as Alzheimer's disease and Parkinson's disease (18,19).
In the present study, although the viability of the HT22 cells was
not assessed prior to the experiments, the cells could reach 80%
confluence after 2 days of culture, which indirectly suggest that
the HT22 cell line had maintained their viability. However, testing
the viability of this cell line before viral infection would render
the present study more rigorous. Therefore, this part of the
experiment should be performed in further studies. In the present
study, AEG-1-deficient HT22 cell lines were generated using the
CRISPR/Cas9 technology. Validation using qPCR and western blotting
showed that M-AEG-1-sgRNA-3 achieved high efficiencies of knocking
down AEG-1 expression, which provided the opportunity to study
AEG-1 function in the neuronal cells in vitro.
AEG-1 is a potent oncogene that has been reported to
contribute to distinct processes during HIV-1 infection, glutamate
regulation and tumorigenesis at the time of its initial cloning and
identification (10). AEG-1 has
also been proposed to contribute significantly to neurodegenerative
diseases and is a potential therapeutic target (40,41).
Huntington's disease (HD) is a fatal progressive neurodegenerative
disorder. In an immortalized striatal cell model of HD and brain
tissues from individuals with HD, immunohistochemical analysis has
demonstrated that AEG-1 expression was upregulated (15), suggesting that there may be an
association between AEG-1 and the pathogenesis of HD. Furthermore,
the role of AEG-1 in another neurodegenerative disease, ALS, has
also been investigated (12).
Upregulation of neuronal AEG-1 can protect nigral dopaminergic
neurons from mSOD1-induced cell injury and has been associated with
improvements in the viability of motor neurons of the mutant SOD1
in an ALS model (12). In
addition, previous findings on the role of AEG-1 in oxidative
stress, a common link in multiple CNS pathologies, revealed that
AEG-1 likely regulates a number of neurodegenerative processes such
as atherosclerosis, stroke and aging (42,43).
Although AEG-1 remain poorly explored in the context of
neurodegenerative diseases, AEG-1 has been previously implicated in
endoplasmic reticulum/nucleus stress (7) and glutamate-mediated neurotoxicity,
highlighting its plausible role in other neurodegenerative
diseases. In the present study, the gene expression profile after
AEG-1 expression was knocked down was studied to elucidate the
possible regulatory mechanism of neuronal AEG-1 in
neurodegenerative diseases.
According to the GO analysis results, AEG-1 was
mainly involved in neuronal morphology and synaptic development.
Subcellular compartments of neurons include dendrites, axons and
synapses, all of which can impact the formation of neuronal
circuits and functional transmission of electrical information
(44). Defects in the formation or
development of dendrites, axons and synapses can lead to a number
of diseases, such as Alzheimer's disease and epilepsy (45). KEGG pathway analysis indicated that
several pathways were impacted after AEG-1 knockdown. The IL-17 and
the NF-κB signaling pathways, which are involved in inflammation
(32-34),
were altered. In addition, the MAPK, RAS and phosphatidylinositol
signaling pathways were changed, which are involved in cell
proliferation and differentiation (46-48).
The other regulatory pathways, such as ‘axon guidance and synaptic
vesicle cycle’, were validated further to analyze the relationship
between AEG-1 knockdown and neuronal morphology development.
Numerous DEGs were identified that associated with
neurogenesis and axonogenesis. Bioinformatics analysis revealed
that NRP-1 is a key gene that relies on AEG-1 function (Fig. 8A). Chemorepulsant activity of
semaphorins is mediated by NRP-1(49). It is also important for the
pre-target sorting of axon fasciculation in the peripheral nervous
system (50). Therefore, AEG-1
potentially mediates axonogenesis by regulating NRP-1 function.
Notably, Notch1 was categorized in the DEGs that was
AEG-1-dependent (Fig. 8A).
As a member of the Notch signaling pathway, Notch1
is involved in the proliferation and apoptosis of different cell
types in a variety of organisms (51). In particular, neurogenesis and
development of the dentate gyrus have been associated with Notch1
expression (52). Loss of Notch1
expression in the dentate gyrus leads to cognitive and emotional
impairments; the present study revealed that deletion of AEG-1
downregulated Notch1 suggesting that AEG-1 can regulate
neurogenesis through regulating Notch1(53). Since ephrin-A5/EphA4 interaction is
considered to be associated with neuron generation de novo,
microvascular remodeling and spontaneous recurrent seizures
(54,55), EphA4 and slit guidance ligand 2 are
also potential genes regulated by AEG-1 in nervous system diseases.
In addition, 30 DEGs were involved in ion channels, including
calcium, sodium and potassium channels (Fig. 8B). Calcium efflux in neurons is
essential for synaptic signaling processes, neuronal energy
metabolism and neurotransmission (56). It was revealed that that AEG-1
deletion disturbed genes related to the calcium signaling pathway,
which may indirectly affect the development of the nervous
system.
Furthermore, 10 hub genes were screened from the
DEGs that can potentially serve important roles in biological
process of the neuron. UBC, CXCL1 and MMP9 were among the top 10
hub genes based on ‘Top10’ node(s) ranked by ‘Degree’. UBC
participates in ubiquitination through forming a polyubiquitin
chain and labeling proteins (57).
A variety of neurological, skeletal and muscular disorders have
been associated with the downregulation of UBC (58). By contrast, CXCL1 is a chemokine
that has been reported to improve both nociceptor and central
sensitization through biding with C-X-C motif chemokine receptor
2(59). CXCL1 is a potential
target for novel analgesic drugs (60). MMP9 is a member of the
endopeptidase family that promotes tissue remodeling by degrading
extracellular matrix components (61). Therefore, AEG-1 deficiency may be
able to regulate the development of neurological disorders through
multiple targets.
In summary, RNA-seq analysis was used to identify
potential genes and pathways that are regulated by AEG-1 expression
upstream of neuronal function. AEG-1 is considered to be a key
protein in nervous system development by regulating signaling
pathways and gene expression. The present study found that AEG-1
knockdown disrupted the function of proteins involved in
intracellular signal transduction, ion channels and
neurotransmitter release. There are two major limitations in the
present study that should be addressed in future studies. The
present study focused on the RNA-seq analysis of DEGs and
functional enrichment after AEG-1 knockdown. However, the DEGs were
not validated using qPCR. In addition, this neuronal cell line
cannot be viewed as a perfect representation of primary neurons.
Despite these limitations, the present study provided novel
findings into potential genes and pathways that were regulated by
AEG-1 expression upstream of neuronal function through RNA-seq
analysis. Validation of the DEGs by qPCR and continued exploration
of the exact functional mechanism of AEG-1 in HT22 cells should be
the aim of future studies.
Supplementary Material
Expression of AEG-1 in virus-infected
cells. The protein expression level of AEG-1 was markedly lower in
the AEG-1-sgRNA virus-infected cell groups compared with that in
the negative control group. GAPDH was used as an internal
reference. These blots were the uncropped and unedited blots of
those presented in Fig. 3B. AEG-1,
astrocyte elevated gene-1; sgRNA, single guide RNA.
Clean data and reference genome
comparison results statistics.
Acknowledgements
Not applicable.
Funding
Funding: This work was supported by the National Natural Science
Foundation of China (grant nos. 31660267, 32070930 and 82160497),
the National Natural Science Foundation of Ningxia (grant nos.
2022AAC02034 and 2020AAC03177), the Ningxia Hui Autonomous Region
‘13th Five-Year Plan’ Major Science and Technology Projects (grant
no. 2016BZ07), the Key R&D Plan Project of Ningxia Autonomous
Region (grant no. 2020BFG02012), the First-Class Discipline
Construction Founded Project of Ningxia Medical University and the
School of Clinical Medicine (grant no. NXYLXK2017A05), the Science
Research Project of Ningxia's Colleges (grant no. NGY2020043).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the NCBI repository (https://www.ncbi.nlm.nih.gov/sra/PRJNA836164).
Authors' contributions
KL, PW and YH analyzed the RNA-sequencing data and
wrote the manuscript. BW constructed the AEG-1 knockout cell line.
XW and RZ performed data analysis and manuscript review. LG
conducted the study design and supervised the study. All authors
have read and approved the final manuscript. KL and LG confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Su ZZ, Kang DC, Chen Y, Pekarskaya O, Chao
W, Volsky DJ and Fisher PB: Identification and cloning of human
astrocyte genes displaying elevated expression after infection with
HIV-1 or exposure to HIV-1 envelope glycoprotein by rapid
subtraction hybridization, RaSH. Oncogene. 21:3592–3602.
2002.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Thirkettle HJ, Girling J, Warren AY, Mills
IG, Sahadevan K, Leung H, Hamdy F, Whitaker HC and Neal DE:
LYRIC/AEG-1 is targeted to different subcellular compartments by
ubiquitinylation and intrinsic nuclear localization signals. Clin
Cancer Res. 15:3003–3013. 2009.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Yoo BK, Emdad L, Lee SG, Su ZZ,
Santhekadur P, Chen D, Gredler R, Fisher PB and Sarkar D: Astrocyte
elevated gene-1 (AEG-1): A multifunctional regulator of normal and
abnormal physiology. Pharmacol Ther. 130:1–8. 2011.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Emdad L, Sarksr D, Su ZZ, Randolph A,
Boukerche H, Valerie K and Fisher PB: Activation of the nuclear
factor kappaB pathway by astrocyte elevated gene-1: Implications
for tumor progression and metastasis. Cancer Res. 66:1509–1516.
2006.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Emdad L, Sarkar D, Su ZZ, Lee SG, Kang DC,
Bruce JN, Volsky DJ and Fisher PB: Astrocyte elevated gene-1:
Recent insights into a novel gene involved in tumor progression,
metastasis and neurodegeneration. Pharmacol Ther. 114:155–170.
2007.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zhang C, Li HZ, Qian BJ, Liu CM, Guo F and
Lin MC: MTDH/AEG-1-based DNA vaccine suppresses metastasis and
enhances chemosensitivity to paclitaxel in pelvic lymph node
metastasis. Biomed Pharmacother. 70:217–226. 2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Roussel BD, Kruppa AJ, Miranda E, Crowther
DC, Lomas DA and Marciniak SJ: Endoplasmic reticulum dysfunction in
neurological disease. Lancet Neuro1. 12:105–118. 2013.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Anttila V, Stefansson H, Kallela M, Todt
U, Terwindt GM, Calafato MS, Nyholt DR, Dimas AS, Freilinger T,
Müller-Myhsok B, et al: Genome-wide association study of migraine
implicates a common susceptibility variant on 8q22.1. Nat Genet.
42:869–873. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Kang DC, Su ZZ, Sarkar D, Emdad L, Volsky
DJ and Fisher PB: Cloning and characterization of HIV-1-inducible
astrocyte elevated gene-1, AEG-1. Gene. 353:8–15. 2005.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Lee SG, Kim K, Kegelman TP, Dash R, Das
SK, Choi JK, Emdad L, Howlett EL, Jeon HY, Su ZZ, et al: Oncogene
AEG-1 promotes glioma-induced neurodegeneration by increasing
glutamate excitotoxicity. Cancer Res. 71:6514–6523. 2011.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Vartak-Sharma N and Ghorpade A: Astrocyte
elevated gene-1 regulates astrocyte responses to neural injury:
Implications for reactive astrogliosis and neurodegeneration. J
Neuroinflammation. 9(195)2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Yin X, Ren M, Jiang H, Cui S, Wang S,
Jiang H, Qi Y, Wang J, Wang X, Dong G, et al: Downregulated AEG-1
together with inhibited PI3K/Akt pathway is associated with reduced
viability of motor neurons in an ALS model. Mol Cell Neurosci.
68:303–313. 2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Leem E, Kim HJ, Choi M, Kim S, Oh YS, Lee
KJ, Choe YS, Um JY, Shin WH, Jeong JY, et al: Upregulation of
neuronal astrocyte elevated gene-1 protects nigral dopaminergic
neurons in vivo. Cell Death Dis. 9(449)2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Jeon HY, Choi M, Howlett EL, Vozhilla N,
Yoo BK, Lloyd JA, Sarkar D, Lee SG and Fisher PB: Expression
patterns of astrocyte elevated gene-1 (AEG-1) during development of
the mouse embryo. Gene Expr Patterns. 10:361–367. 2010.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Carnemolla A, Fossale E, Agostoni E,
Michelazzi S, Calligaris R, De Maso L, Del Sal G, MacDonald ME and
Persichetti F: Rrs1 is involved in endoplasmic reticulum stress
response in Huntington disease. J Biol Chem. 284:18167–18173.
2009.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Garneau JE, Dupuis MÈ, Villion M, Romero
DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH and
Moineau S: The CRISPR/Cas bacterial immune system cleaves
bacteriophage and plasmid DNA. Nature. 468:67–71. 2010.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Doudna JA and Charpentier E: Genome
editing. The new frontier of genome engineering with CRISPR-Cas9.
Science: Nov 28, 2014 (Epub ahead of print).
|
|
18
|
Caldwell JD, Shapiro RA, Jirikowski GF and
Suleman F: Internalization of sex hormone-binding globulin into
neurons and brain cells in vitro and in vivo. Neuroendocrinology.
86:84–93. 2007.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Murphy TH, Miyamoto M, Sastre A, Schnaar
RL and Coyle JT: Glutamate toxicity in a neuronal cell line
involves inhibition of cystine transport leading to oxidative
stress. Neuron. 2:1547–1558. 1989.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Florea L, Song L and Salzberg SL:
Thousands of exon skipping events differentiate among splicing
patterns in sixteen human tissues. F1000Res. 2(188)2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140.
2010.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Anders S and Huber W: Differential
expression analysis for sequence count data. Genome Biol.
11(R106)2010.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc B (Methodol). 57:289–300. 1995.
|
|
25
|
Harris MA, Clark J, Ireland A, Lomax J,
Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C,
et al: The gene ontology (GO) database and informatics resource.
Nucleic Acids Res. 32:D258–D261. 2004.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Kanehisa M, Sato Y, Kawashima M, Furumichi
M and Tanabe M: KEGG as a reference resource for gene and protein
annotation. Nucleic Acids Res. 44:D457–D462. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
von Mering C, Jensen LJ, Snel B, Hooper
SD, Krupp M, Foglierini M, Jouffre N, Huynen MA and Bork P: STRING:
Known and predicted protein-protein associations, integrated and
transferred across organisms. Nucleic Acids Res. 33:D433–D437.
2005.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Xu WH, Xu Y, Wang J, Wan FN, Wang HK, Cao
DL, Shi GH, Qu YY, Zhang HL and Ye DW: Prognostic value and immune
infiltration of novel signatures in clear cell renal cell carcinoma
microenvironment. Aging (Albany NY). 11:6999–7020. 2019.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Chen Q, Yu D, Zhao Y, Qiu J, Xie Y and Tao
M: Screening and identification of hub genes in pancreatic cancer
by integrated bioinformatics analysis. J Cell Biochem.
120:19496–19508. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Stephenson AA, Raper AT and Suo Z:
Bidirectional degradation of DNA cleavage products catalyzed by
CRISPR/Cas9. J Am Chem Soc. 140:3743–3750. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Bernardino L, Agasse F, Silva B, Ferreira
R, Grade S and Malva JO: Tumor necrosis factor-alpha modulates
survival, proliferation, and neuronal differentiation in neonatal
subventricular zone cell cultures. Stem Cells. 26:2361–2371.
2008.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Li X, Bechara R, Zhao J, McGeachy MJ and
Gaffen SL: IL-17 receptor-based signaling and implications for
disease. Nat Immunol. 20:1594–1602. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Liu W, Ye J and Yan H: Investigation of
key genes and pathways in inhibition of oxycodone on
vincristine-induced microglia activation by using bioinformatics
analysis. Dis Markers. 2019(3521746)2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Ng SR, Rideout WM III, Akama-Garren EH,
Bhutkar A, Mercer KL, Schenkel JM, Bronson RT and Jacks T:
CRISPR-mediated modeling and functional validation of candidate
tumor suppressor genes in small cell lung cancer. Proc Natl Acad
Sci USA. 117:513–521. 2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
He XY, Ren XH, Peng Y, Zhang JP, Ai SL,
Liu BY, Xu C and Cheng SX: Aptamer/peptide-functionalized
genome-editing system for effective immune restoration through
reversal of PD-L1-mediated cancer immunosuppression. Adv Mater.
32(2000208)2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Ling X, Xie B, Gao X, Chang L, Zheng W,
Chen H, Huang Y, Tan L, Li M and Liu T: Improving the efficiency of
precise genome editing with site-specific Cas9-oligonucleotide
conjugates. Sci Adv. 6(eaaz0051)2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
LaFountaine JS, Fathe K and Smyth HD:
Delivery and therapeutic applications of gene editing technologies
ZFNs, TALENs, and CRISPR/Cas9. Int J Pharm. 494:180–194.
2015.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhou Z, Tan H, Li Q, Chen J, Gao S, Wang
Y, Chen W and Zhang L: CRISPR/Cas9-mediated efficient targeted
mutagenesis of RAS in Salvia miltiorrhiza. Phytochemistry.
148:63–70. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Noch EK and Khalili K: The role of
AEG-1/MTDH/LYRIC in the pathogenesis of central nervous system
disease. Adv Cancer Res. 120:159–192. 2013.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Wang Y, Zhang W, Zhu X and Wang Y, Mao X,
Xu X and Wang Y: Upregulation of AEG-1 involves in schwann cell
proliferation and migration after sciatic nerve crush. J Mol
Neurosci. 60:248–257. 2016.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Vartak-Sharma N, Nooka S and Ghorpade A:
Astrocyte elevated gene-1 (AEG-1) and the A(E)Ging HIV/AIDS-HAND.
Prog Neurobiol. 157:133–157. 2017.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Serviddio G, Romano AD, Cassano T,
Bellanti F, Altomare E and Vendemiale G: Principles and therapeutic
relevance for targeting mitochondria in aging and neurodegenerative
diseases. Curr Pharm Des. 17:2036–2055. 2011.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Mohan H, Verhoog MB, Doreswamy KK, Eyal G,
Aardse R, Lodder BN, Goriounova NA, Asamoah B, B Brakspear AB,
Groot C, et al: Dendritic and axonal architecture of individual
pyramidal neurons across layers of adult human neocortex. Cereb
Cortex. 25:4839–4853. 2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Jan YN and Jan LY: Branching out:
Mechanisms of dendritic arborization. Nat Rev Neurosci. 11:316–328.
2010.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Yoo BK, Emdad L, Su ZZ, Villanueva A,
Chiang DY, Mukhopadhyay ND, Mills AS, Waxman S, Fisher RA, Llovet
JM, et al: Astrocyte elevated gene-1 regulates hepatocellular
carcinoma development and progression. J Clin Invest. 119:465–477.
2009.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Yu C, Chen K, Zheng H, Guo X, Jia W, Li M,
Zeng M, Li J and Song L: Overexpression of astrocyte elevated
gene-1 (AEG-1) is associated with esophageal squamous cell
carcinoma (ESCC) progression and pathogenesis. Carcinogenesis.
30:894–901. 2009.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Lee SG, Su ZZ, Emdad L, Sarkar D and
Fisher PB: Astrocyte elevated gene-1 (AEG-1) is a target gene of
oncogenic Ha-ras requiring phosphatidylinositol 3-kinase and c-Myc.
Proc Natl Acad Sci USA. 103:17390–17395. 2006.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Rohm B, Ottemeyer A, Lohrum M and Püschel
AW: Plexin/neuropilin complexes mediate repulsion by the axonal
guidance signal semaphorin 3A. Mech Dev. 93:95–104. 2000.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Rolny C, Capparuccia L, Casazza A, Mazzone
M, Vallario A, Cignetti A, Medico E, Carmeliet P, Comoglio PM and
Tamagnone L: The tumor suppressor semaphorin 3B triggers a
prometastatic program mediated by interleukin 8 and the tumor
microenvironment. J Exp Med. 205:1155–1171. 2008.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Mumm JS, Schroeter EH, Saxena MT,
Griesemer A, Tian X, Pan DJ, Ray WJ and Kopan R: A ligand-induced
extracellular cleavage regulates gamma-secretase-like proteolytic
activation of Notch1. Mol Cell. 5:197–206. 2000.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Liu X, Yang Z, Yin Y and Deng X: Increased
expression of Notch1 in temporal lobe epilepsy: Animal models and
clinical evidence. Neural Regen Res. 9:526–533. 2014.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Feng S, Shi T, Qiu J, Yang H, Wu Y, Zhou
W, Wang W and Wu H: Notch1 deficiency in postnatal neural
progenitor cells in the dentate gyrus leads to emotional and
cognitive impairment. FASEB J. 31:4347–4358. 2017.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Shu Y, Xiao B, Wu Q, Liu T, Du Y, Tang H,
Chen S, Feng L, Long L and Li Y: The ephrin-A5/EphA4 interaction
modulates neurogenesis and angiogenesis by the p-Akt and p-ERK
pathways in a mouse model of TLE. Mol Neurobiol. 53:561–576.
2016.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Feng L, Shu Y, Wu Q, Liu T, Long H, Yang
H, Li Y and Xiao B: EphA4 may contribute to microvessel remodeling
in the hippocampal CA1 and CA3 areas in a mouse model of temporal
lobe epilepsy. Mol Med Rep. 15:37–46. 2017.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Salińska E and Łazarewicz JW: Role of
calcium in physiology and pathology of neurons. Postepy Biochem.
58:403–417. 2012.PubMed/NCBI(In Polish).
|
|
57
|
Chadwick L, Gentle L, Strachan J and
Layfield R: Review: Unchained maladie-a reassessment of the role of
Ubb(+1)-capped polyubiquitin chains in Alzheimer's disease.
Neuropathol Appl Neurobiol. 38:118–131. 2012.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Manavalan A, Mishra M, Feng L, Sze SK,
Akatsu H and Heese K: Brain site-specific proteome changes in
aging-related dementia. Exp Mol Med. 45(e39)2013.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Kothur K, Bandodkar S, Wienholt L, Chu S,
Pope A, Gill D and Dale RC: Etiology is the key determinant of
neuroinflammation in epilepsy: Elevation of cerebrospinal fluid
cytokines and chemokines in febrile infection-related epilepsy
syndrome and febrile status epilepticus. Epilepsia. 60:1678–1688.
2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Silva RL, Lopes AH, Guimaraes RM and Cunha
TM: CXCL1/CXCR2 signaling in pathological pain: Role in peripheral
and central sensitization. Neurobiol Dis. 105:109–116.
2017.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Ringland C, Schweig JE, Eisenbaum M, Paris
D, Ait-Ghezala G, Mullan M, Crawford F, Abdullah L and Bachmeier C:
MMP9 modulation improves specific neurobehavioral deficits in a
mouse model of Alzheimer's disease. BMC Neurosci.
22(39)2021.PubMed/NCBI View Article : Google Scholar
|