Introduction
Thyroid cancer (THCA) is one of the commonest
malignancies associated with increased recurrence (1). Generally, THCA shows a good overall
prognosis and low fatality rate in most cases; however, due to its
aggressive characteristics and metastasis, and poor prognosis can
be seen in some patients with THCA (2). Identifying available molecular
markers and therapeutic targets is urgently required to improve the
treatment outcome.
The tripartite motif (TRIM) family of proteins
belongs to the subfamily of E3 ubiquitin ligases and participates
in various biological and pathophysiological processes, including
tumor progression (3-5).
TRIMs share similar domains in their protein structure, including
the N-terminal RING domain with E3 ubiquitin ligase activity, the
B-box domain, and the coiled-coil domain (6). Several members of the TRIM family are
associated with tumorigenesis and disease progression of THCA.
TRIM14 has been reported as an oncogene in THCA (7). TRIM44 knockdown suppresses the tumor
progression of THCA by inhibiting the Wnt/β-catenin signaling
pathway (8). TRIM8 serves as a
target for miR-182 in promoting tumor growth and increasing
chemoresistance in human THCA (9).
However, the roles of other TRIMs in THCA remain to be elucidated.
The authors of the present study aim to investigate the roles of
other TRIMs in THCA and so far, TRIM21 is the one which has been
elucidated. Therefore, the present study reported TRIM21. TRIM21 is
a member of the TRIM family involved in innate immunity and the
development of diseases such as systemic lupus erythematosus, and
Sjögren's syndrome (10). TRIM21
may play opposing roles in tumorigenesis and its progression
(6,11). TRIM21 suppresses hepatocellular
carcinoma cell invasion (12); it
inhibits renal cancer tumorigenesis and metastasis by mediating
hypoxia-inducible factor-1 alpha (HIF-1α) degradation (13). TRIM21 promotes glioma progression
by regulating cell proliferation and migration (14). By contrast, TRIM21 is related to
the therapeutic sensitivity of several tumors: By suppressing EZH1
stability, TRIM21 improves the sensitivity of gastric cancer to
apatinib (15). In squamous cell
carcinoma of the head and neck, high TRIM21 expression is
associated with a shorter overall survival rate (16). However, the participation of TRIM21
in THCA regulation remains unelucidated.
The present study aimed to identify the role of
TRIM21 in THCA and to analyze the functional networks related to
TRIM21 using public databases such as The Cancer Genome Atlas
(TCGA) database. The function of TRIM21 in the proliferation,
migration and invasion of THCA cells was evaluated.
Materials and methods
Clinical sample collection
Paraffin tumor tissue samples and paraffin
para-tumor normal tissues 1 cm away from the tumor tissues were
collected from 120 patients diagnosed with papillary thyroid
carcinoma and who underwent surgical resection in Liaocheng
People's Hospital between 2018 and 2020. The patients were aged
from 21-70. The clinical information of all cases was collected and
is given in Table I. All patients
were free of other malignancies or a history of chemoradiotherapy.
Written informed consent was obtained from all the participants.
The experiment was approved by the Ethics committee of Liaocheng
People's Hospital (approval no. LC2021059).
 | Table IRelationship between TRIM21 expression
and clinicopathologic features of patients with thyroid papillary
carcinoma. |
Table I
Relationship between TRIM21 expression
and clinicopathologic features of patients with thyroid papillary
carcinoma.
| | Expression of TRIM21
protein in cancer tissue | |
|---|
| Pathological clinical
data | High-expression (89
cases) | Low-expression (31
cases) | Statistical
quantity | P-value |
|---|
| Sex | | | | |
|
Male | 35 | 10 |
χ2=0.23 | 0.62 |
|
Female | 54 | 21 | | |
| Age (years) | | | | |
|
<55 | 46 | 19 |
χ2=0.51 | 0.47 |
|
≥55 | 43 | 12 | | |
| Tumor diameter
(cm) | | | | |
|
~1.0-2.0 | 36 | 11 |
χ2=0.55 | 0.75 |
|
~2.1-3.0 | 28 | 12 | | |
|
~3.1-4.0 | 25 | 8 | | |
| Extragranular
invasion | | | | |
|
Yes | 60 | 11 |
χ2=2.78 | 0.09 |
|
No | 29 | 21 | | |
| Lymph node
metastasis (pieces) | | | | |
|
No | 11 | 10 |
χ2=12.63 | 0.002 |
|
~1-3 | 30 | 15 | | |
|
≥4 | 48 | 6 | | |
Immunohistochemical staining and
scoring
Immunohistochemical staining (IHC) was performed to
detect the expression of TRIM21 in papillary thyroid carcinoma
tissues. Endogenous peroxidase activity was blocked using 3%
hydrogen peroxide (cat. no. 88597; Merck KGaA) after routine
dewaxing, hydration, and antigen retrieval. Permeabilization of
samples was performed using 0.1% Triton X-100 (cat. no. ST797;
Beyotime Institute of Biotechnology) and blocked with 5% bovine
serum albumin (BSA) (cat. no. ST025; Beyotime Institute of
Biotechnology). Tissue sections (10 µm thick) were incubated with
TRIM21 antibodies (1:200 dilution; ProteinTech; cat. no.
12108-1-AP) at 4˚C for 12 h. After washing with phosphate-buffered
saline (PBS), the sections were incubated with horseradish
peroxidase (HRP)-conjugated goat anti-rabbit IgG (1:1,000 dilution;
cat. no. A0208; Beyotime Institute of Biotechnology) for 1 h at
20˚C. TRIM21 expression was visualized using 3,3'-diaminobenzidine
(DAB; cat. no. P0202; Beyotime Institute of Biotechnology) staining
at 20˚C for 1 min. A blind evaluation was performed by two
pathologists. Brown-yellow staining indicated a positive
expression. Staining score=staining intensity score x
staining-positive area score. The staining intensity was scored as
0 (negative), 1 (weakly positive), 2 (moderately positive) and 3
(strongly positive). The score of the staining-positive area was
recorded according to the proportion of positive cells: 0 (<5%),
1 (5-25%), 2 (26-50%), 3 (51-75%) and 4 points (>75%). A
staining score of <3 was classified as low TRIM21 expression,
and those ≥3 were classified as high TRIM21 expression.
Reverse-transcription quantitative
(RT-q) PCR
TRIzol® reagent (cat. no. 15596-026;
Thermo Fisher Scientific, Inc) was used for total RNA isolation
according to the manufacturer's protocols. To quantify TRIM21
expression, the total RNA was reverse-transcribed into cDNA using a
PrimeScript RT Reagent kit (cat. no. RR037A; Takara Bio, Inc.)
according to the manufacturer's protocols, which, in turn, was
subjected to qPCR analysis with a SYBR Premix Ex Taq kit (cat. no.
DRR041A; Takara Bio, Inc.) according to the manufacturer's
protocols. The PCR conditions were as follows: 95˚C, 10 min; (95˚C,
15 sec; 60˚C, 60 sec) x 40 cycles. The primer sequences were as
follows: TRIM21, 5'-CCATGTGCCAGGGCTGAAGAAG-3' (forward),
5'-AGGTATGCTCTGCTGGGTGTCTC-3' (reverse); β-actin,
5'-CATGGAGTCCTGTGGCATC-3' (forward), 5'-CAGGGCAGTGATCTCCTTCT-3'
(reverse). All the primers were synthesized in Sangon Biotech Co.,
Ltd. Relative gene expression was calculated using the
2-ΔΔCq method (17). These experiments were replicated
three times.
Public database data sources
THCA transcriptome data were downloaded from The
Cancer Genome Atlas (TCGA) database using the UCSC Xena tool
(https://xena.ucsc.edu/). TRIM21 expression levels
in all types of cancer obtained from the cBioPortal database
(https://www.cbioportal.org/) (18) were analyzed using R software
version 3.6.1 (http://www.R-project.org/) (19). An unpaired Student's t-test was
applied to compare TRIM21 expression in various cancers with that
in normal tissues.
Gene set enrichment analysis (GSEA) of
TRIM21-related cancer pathways
THCA transcriptome expression profiles were obtained
from the TCGA database (https://gdc-portal.nci.nih.gov/). A total of 510 THCA
samples and 59 normal tissues derived from healthy individuals were
included in the present study. The correlations between the Kyoto
Encyclopedia Of Genes And Genomes (KEGG) signaling pathways of
TRIM21 and co-expressed genes were explored using GSEA (http://software.broadinstitute.org/gsea/index.jsp)
(20). The latter was performed
using three enrichment statistics: Enrichment scores, normalized
enrichment scores and nominal P-values. The enrichment score
indicates the degree of enrichment of a functional gene set before
or after a given sequence. The normalized enrichment score is the
major parameter in enrichment analyses of functional gene sets. The
nominal P-value indicates the statistical significance of the
enrichment score of a given functional gene subset with lower
P-values. KEGG enrichment was reported at a significance threshold
of P<0.05.
Cell culture and lentivirus
infection
FTC-133 cell line (cat. no. 1101HUM-PUMC000687) was
purchased from the National Infrastructure of Cell Line Resource of
China. The cells were cultured in Dulbecco's modified eagle medium
F-12 (DMEM-F12, cat. no. 11320-033, Gibco; Thermo Fisher
Scientific, Inc.) containing 10% fetal bovine serum (FBS, cat. no.
10100147, Gibco; Thermo Fisher Scientific, Inc.) in an incubator
with 5% CO2 at 37˚C.
Cultured cells were seeded into 24-well plates at
30,000 cells/well. Once the cells reached 90% confluence,
lentivirus (Lv-shCon or Lv-shTRIM21) was added to the wells at a
multiplicity of infection (MOI) of 10. At 48 h later, infected
cells were selected using 10 µg/ml puromycin (cat. no. ST551;
Beyotime Institute of Biotechnology). Virally infected cells were
observed under a fluorescence microscope (cat. no. IX73; Olympus
Corporation). TRIM21 expression was determined using RT-qPCR
analysis. Lv-shCon and Lv-shTRIM21 were designed and constructed at
Shanghai GeneChem Co., Ltd. The targeting sequence of Lv-shCon was:
5'-AACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTG-3'. The
targeting sequence of Lv-shTRIM21 was:
5'-GGAAGTCACTTCACCATCACTCGAGTGATGGTGAAGTGACTTCCTTTTTT-3'.
Transwell assay
Migration: Cells were seeded into the upper chambers
of a Transwell plate. The upper chambers contained DMEM media
without FBS, whereas the lower chambers contained DMEM media with
10% FBS. The cells were cultured for 24 h, and then the remaining
cells in the upper chamber were wiped away with a swab. The cells
passed through the membrane were stained with crystal violet (cat.
no. C0121; Beyotime Institute of Biotechnology). The cells in three
randomly selected visual fields were counted under a fluorescence
microscope (cat. no. IX73; Olympus Corporation).
Invasion: The invasion assay was performed with the
similar procedures as the migration assay, excepting that the wells
were pre-coated with 20 µg Matrigel at 37˚C for 2 h
(MilliporeSigma).
CCK-8 assay
Cells were seeded into a 96 well plate at 5,000
cell/well. After the cells were cultured for 0, 24 and 48 h in an
incubator with 5% CO2 at 37˚C, 10 µl of CCK-8 reagent
(cat. no. C0037; Beyotime Institute of Biotechnology) was added.
The cells were then incubated for 1 h at 37˚C and the absorbance at
450 nm wavelength was measured using a Multiskan GO microplate
reader (Thermo Fisher Scientific, Inc.).
Statistical analysis
All pathological and experimental data were analyzed
using SPSS software (version 25.0; IBM Corp.). The measurement data
conformed to a normal distribution and are presented as mean ±
standard deviation (SD). The χ2 test was performed to
analyze the association between TRIM21 expression and
clinicopathologic features of patients with thyroid papillary
carcinoma. Considering the normal tissues derived from different
patients to those who donated the cancer tissues, the expression of
TRIM21 in 14 different types of tumors was evaluated using an
unpaired Student's t-test. TRIM21 expression in 120 pairs THCA and
the corresponding adjacent normal tissues was evaluated using
paired Student's t-test. P-value was obtained from a two-tailed
Student's t-test. The survival time of the patients was calculated
using the Kaplan-Meier method. Log-rank test was performed to
analyze the Kaplan-Meier survival curves. P<0.05 was considered
to indicate a statistically significant difference.
Results
TRIM21 expression in multiple tumor
sites
TRIM21 expression was assessed in various cancers
based on TCGA and GTEX databases. TRIM21 expression was
dysregulated in the 14 tumor types compared with the corresponding
normal tissues, including kidney renal clear cell carcinoma (KIRC),
lung squamous cell carcinoma (LUSC), head and neck squamous
carcinoma (HNSC), lung adenocarcinoma (LUAD), cholangiocarcinoma
(CHOL), liver hepatocellular carcinoma (LIHC), uterine corpus
endometrial carcinoma (UCEC), esophageal carcinoma (ESCA),
glioblastoma multiforme (GBM), stomach adenocarcinoma (STAD),
kidney chromophobe (KICH), skin cutaneous melanoma (SKCM) tumor and
THCA. (Fig. 1 and Table II).
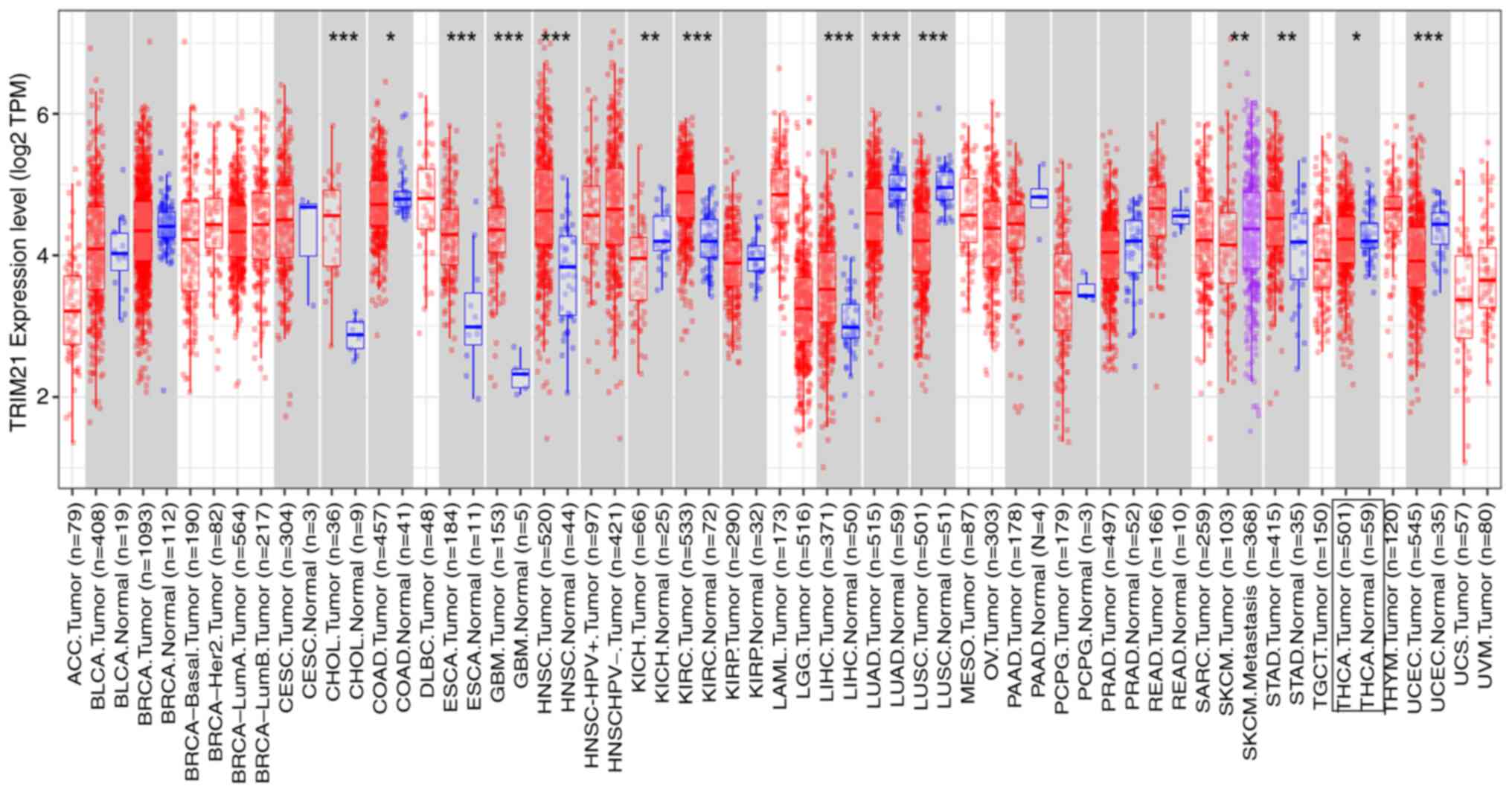 | Figure 1TRIM21 expression levels in different
types of tumors. *P<0.05; **P<0.01;
***P<0.001. ACC, adrenocortical carcinoma; BLCA,
bladder carcinoma; BRCA, breast carcinoma; CESC, cervical squamous
cell carcinoma; CHOL, cholangiocarcinoma; COAD, colon
adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell
lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme;
HNSC, head and neck squamous carcinoma; KICH, kidney chromophobe;
KIRC, kidney renal clear cell carcinoma; KIRC, kidney renal clear
cell carcinoma; LAML, acute myeloid leukemia; LIHC, liver
hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung
squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous
cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG,
pheochromocytoma and paraganglioma; PRAD, rectum adenocarcinoma;
READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous
melanoma; STAD, stomach adenocarcinoma; TGCT, testis germ cell
tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine
corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM,
uveal melanoma. |
 | Table IIExpression levels of TRIM in various
cancers vs. normal tissue included in TCGA. |
Table II
Expression levels of TRIM in various
cancers vs. normal tissue included in TCGA.
| Tumor | Normal | P-value |
|---|
| KIRC.Tumor
(n=533) | KIRC.Normal
(n=72) |
2.08x10-20 |
| LUSC.Tumor
(n=501) | LUSC.Normal
(n=51) |
1.22x10-18 |
| HNSC.Tumor
(n=520) | HNSC.Normal
(n=44) |
2.37x10-11 |
| LUAD.Tumor
(n=515) | LUAD.Normal
(n=59) |
8.63x10-9 |
| CHOL.Tumor
(n=36) | CHOL.Normal
(n=9) |
6.77x10-8 |
| LIHC.Tumor
(n=371) | LIHC.Normal
(n=50) |
8.37x10-6 |
| UCEC.Tumor
(n=545) | UCEC.Normal
(n=35) |
1.55x10-5 |
| ESCA.Tumor
(n=184) | ESCA.Normal
(n=11) |
6.16x10-5 |
| GBM.Tumor
(n=153) | GBM.Normal
(n=5) |
1.96x10-4 |
| STAD.Tumor
(n=415) | STAD.Normal
(n=35) |
2.54x10-3 |
| KICH.Tumor
(n=66) | KICH.Normal
(n=25) |
2.94x10-3 |
| THCA.Tumor
(n=501) | THCA.Normal
(n=59) |
1.55x10-2 |
| COAD.Tumor
(n=457) | COAD.Normal
(n=41) |
4.65x10-2 |
| PRAD.Tumor
(n=497) | PRAD.Normal
(n=52) |
9.42x10-2 |
| PAAD.Tumor
(n=178) | PAAD.Normal
(n=4) |
9.59x10-2 |
| BRCA.Tumor
(n=1093) | BRCA.Normal
(n=112) |
1.59x10-1 |
| KIRP.Tumor
(n=290) | KIRP.Normal
(n=32) |
3.10x10-1 |
| READ.Tumor
(n=166) | READ.Normal
(n=10) |
3.69x10-1 |
| BLCA.Tumor
(n=408) | BLCA.Normal
(n=19) |
5.75x10-1 |
| CESC.Tumor
(n=304) | CESC.Normal
(n=3) |
6.83x10-1 |
| PCPG.Tumor
(n=179) | PCPG.Normal
(n=3) |
9.38x10-1 |
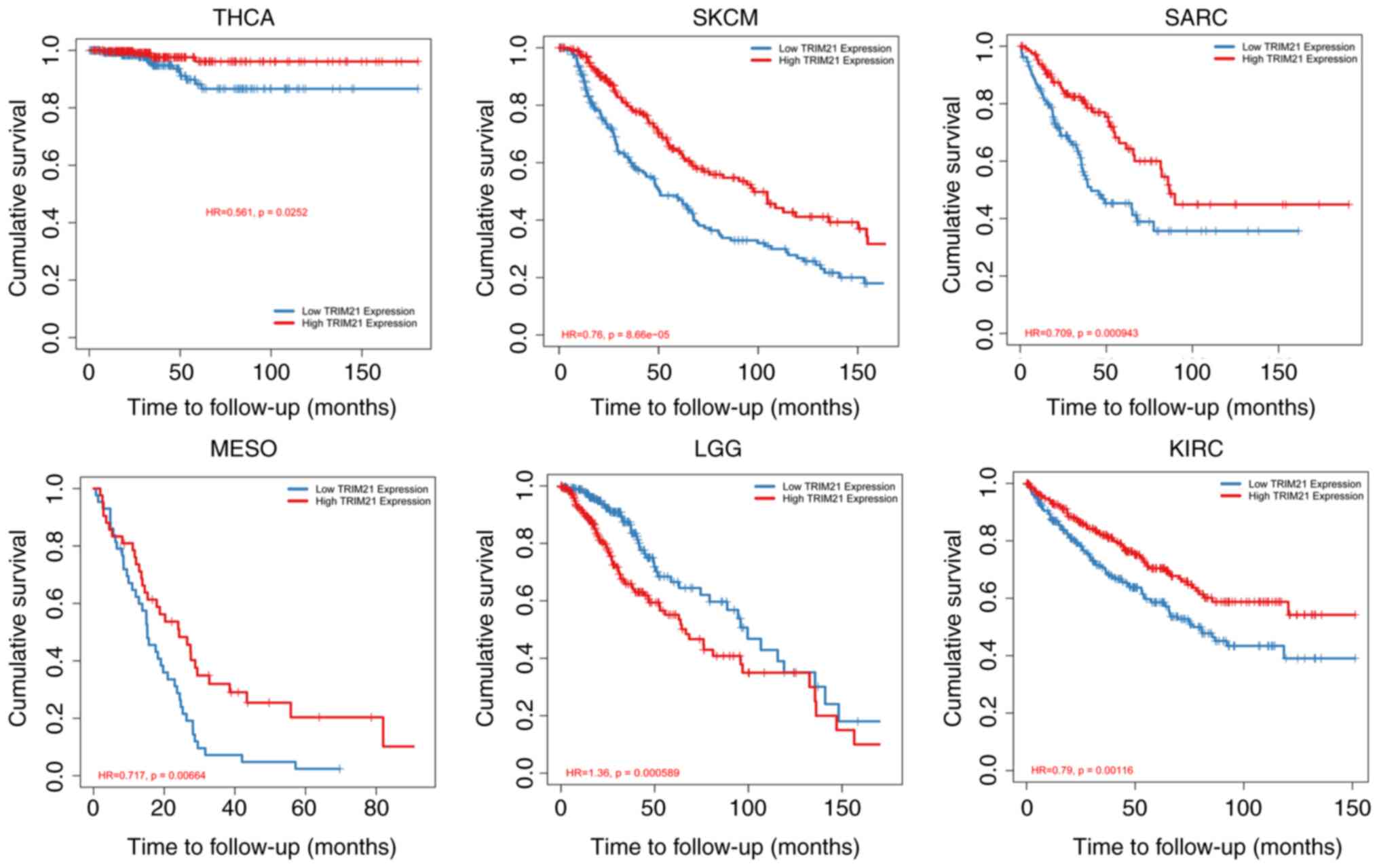
Association of TRIM21 with the
survival of patients with THCA
In the TCGA data set, according to TRIM21
expression, tumors were divided into Low TRIM21 group and High
TRIM21 group. Fig. 2 and Table I show the survival analysis of the
two groups. In low grade gliomas, the low TRIM21 group showed a
better prognosis than that in the High TRIM21 group. In contrast,
in SKCM, mesothelioma (MESO), sarcoma (SARC), KIRC, and THCA, the
patients in the high TRIM21 group showed a better prognosis
compared with that in the low TRIM21 group.
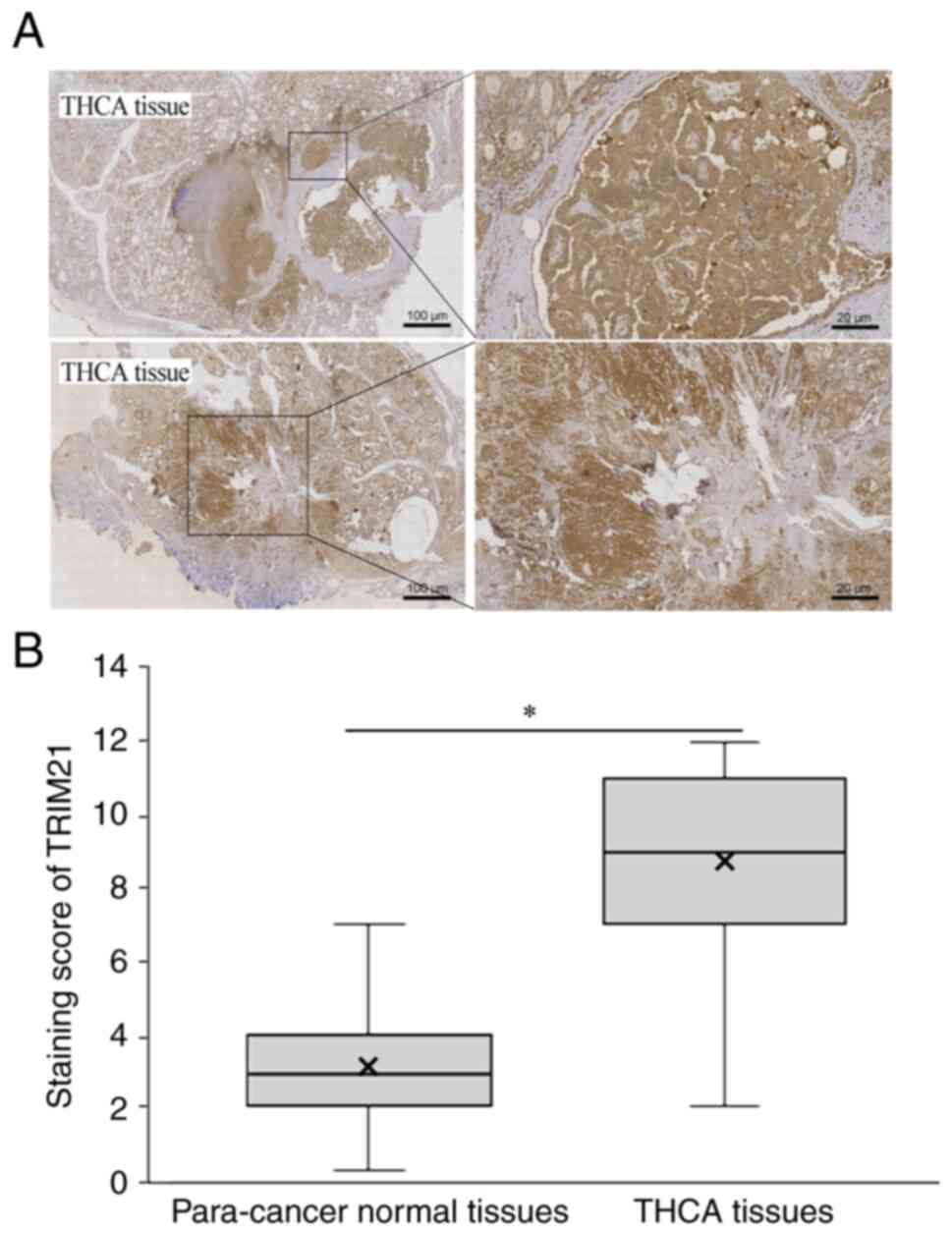
TRIM21 expression in THCA and normal
tissues
The expression of TRIM21 in 120 THCA and
corresponding para-cancer normal tissues was evaluated using IHC.
As shown in Fig. 3A, the staining
intensity of TRIM21 in THCA tissues was significantly higher than
that in para-cancer normal tissues. Simultaneously, the staining
score of TRIM21 protein was calculated, and the score in THCA
tissues was significantly higher than that in para-cancer normal
tissues (Fig. 3B). These results
indicate that TRIM21 is overexpressed in THCA.
Relevance of TRIM21 in clinicopathological features
of THCA cases. According to TRIM21 expression, 120 THCA cases were
divided into TRIM21 high- and low-expression groups. Subsequently,
the relevance of TRIM21 to the clinicopathologic features of THCA
was evaluated. As shown in Table
I, the expression of TRIM21 showed no relevance to patients'
gender, age, tumor diameter, and extra-granular invasion; however,
TRIM21 was significantly associated with lymph node metastasis.
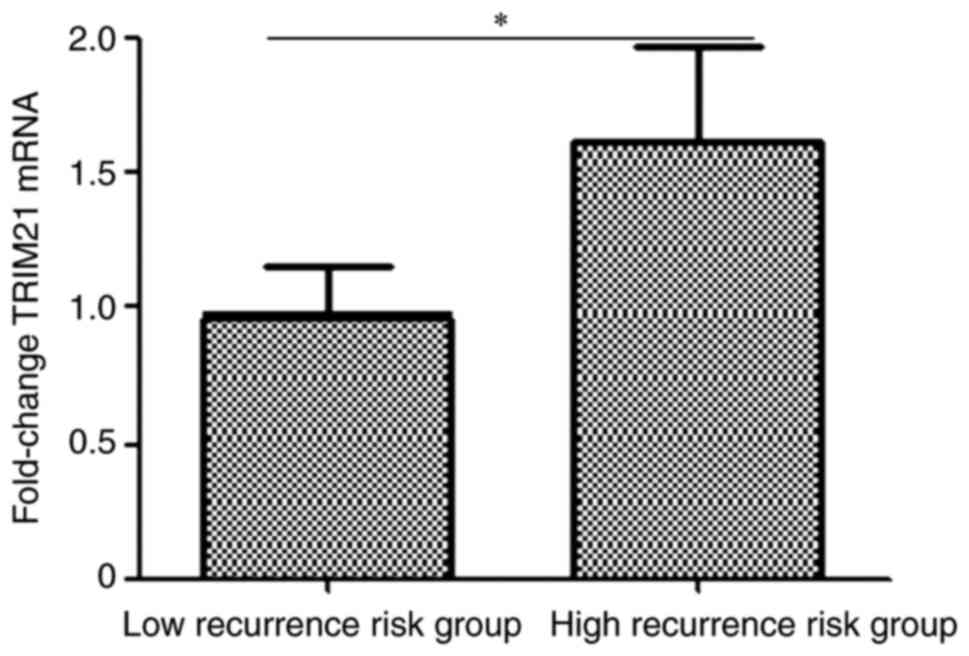
According to the recurrence risk based on the American Thyroid
Association (ATA) guidelines 2021(21), 120 patients were divided into high-
and low recurrence-risk groups. As shown in Fig. 4, the expression of TRIM21 was
measured using RT-qPCR and the expression of TRIM21 in the high
recurrence-risk group was 1.69 folds of that in the low
recurrence-risk group (P=0.0242).
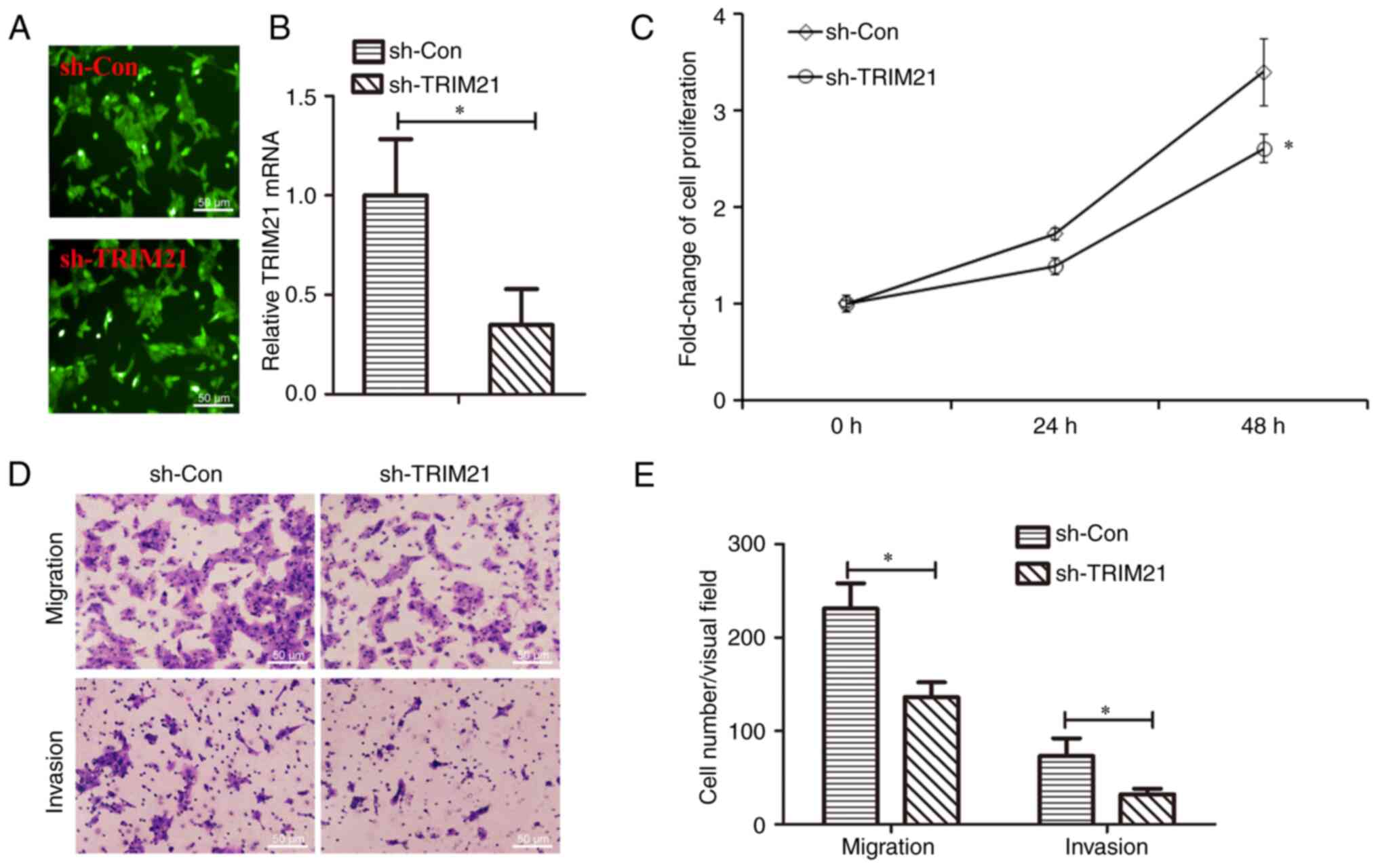
Knockdown of TRIM21 induced inhibition
of cell proliferation, migration and invasion of THCA cells
As TRIM21 was overexpressed in THCA and was
associated with lymph node metastasis, the role of TRIM21 in THCA
cell migration and invasion was further examined. TRIM21 was
knocked down in lentivirus-infected FTC-133 cells. As shown in
Fig. 5A, green fluorescent
labeling indicated that the cells were infected with lentivirus.
Fig. 5B demonstrated that TRIM21
expression was reduced ~62.88% in Lv-shTRIM21 infected cells,
indicating that the efficiency of TRIM21 knockdown was 37.12%. Cell
migration and invasion capacities were measured using Transwell
assays. The migration and invasion capacities of
Lv-shTRIM21-infected cells were inhibited compared to those
infected with Lv-shCon (Fig. 5C,
D and E). These results indicate that TRIM21
knockdown inhibits proliferation, migration and invasion of THCA
cells.
KEGG analysis of TRIM21 and TRIM21
co-expression genes in THCA
The potential biological functions of TRIM21 with
high or low expression in THCA were investigated using GSEA, and
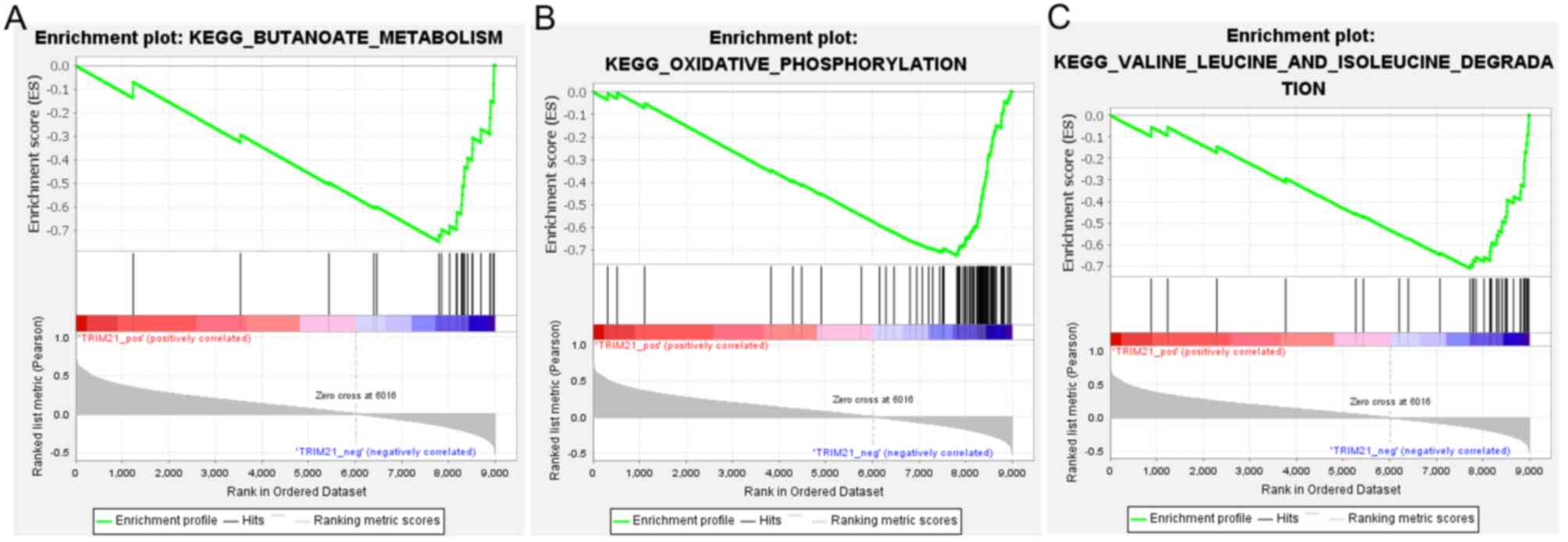
the genes were significantly enriched in 38 KEGG pathways,
including ‘butanoate metabolism’, ‘oxidative phosphorylation’, and
‘valine leucine and isoleucine degradation’ (Fig. 6).
After excluding the genes that met the condition of
false discovery rate <0.05 and with a correlation coefficient
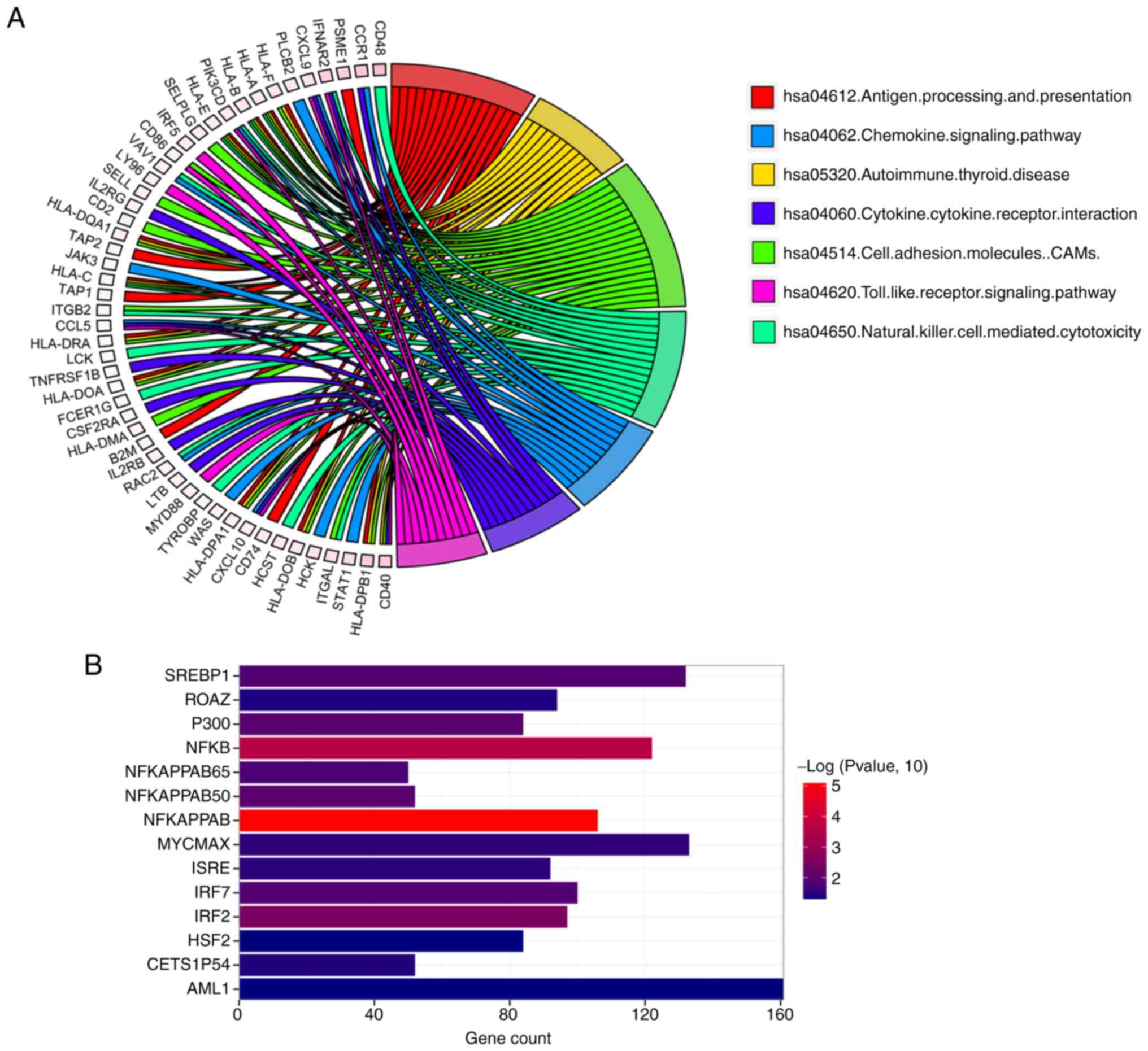
>0.6 were included. A total of 252 TRIM21 co-expression genes
were identified, including SP110, APOL2, and UBE2L6. Furthermore,
the KEGG database was used to screen for TRIM21 co-expression gene
enrichment pathways in thyroid carcinoma. As shown in Fig. 7A, the genes were significantly
enriched in seven KEGG pathways. The top three were ‘antigen
processing and presentation’, ‘autoimmune thyroid disease’, and
‘cell adhesion molecules’. The enrichment of co-expression genes in
the transcription factor (TF) and kinase datasets were further
investigated. As shown in Fig. 7B,
a total of 14 TFs of TRIM21 co-expressed genes were identified,
including NFKAPPAB, NFKB, and IRF2.
Discussion
THCA is one of the most common endocrine
malignancies worldwide. Dysregulation of TRIM21 is responsible for
the progression of various diseases, including tumors. However,
limited information is available regarding the potential
contribution of TRIM21 to THCA.
The current study found, using bioinformatics
analysis, that TRIM21 was upregulated in THCA. In addition, the
results of the bioinformatics analyses were verified by measuring
TRIM21 expression in THCA and matched adjacent normal tissues.
Higher TRIM21 expression was observed in THCA tissues compared with
matched adjacent normal tissues. Furthermore, a high TRIM21 level
was associated with a high risk of recurrence and lymph node
metastasis. The results indicated that TRIM21 may be a potential
biological marker to distinguish tumor recurrence rates.
TRIM21 expression and its role in various cancers
have been previously investigated. The effects of TRIM21 on tumor
progression differed in different types of cancer. Zhao et
al (14) observed TRIM21
upregulation in gliomas and confirmed its role in tumor
proliferation, migration, and drug resistance. By contrast, TRIM21
is downregulated in breast cancer, associated with tumor size and
clinical stage, and is considered an important factor for overall
survival (22). In patients with
colitis-associated colorectal cancer, decreased TRIM21 expression
causes dysregulation of epithelial cell proliferation, angiogenesis
and pro-inflammatory responses, resulting in intestinal epithelial
carcinogenesis (23). The present
study investigated the role of TRIM21 in THCA progression in
vitro. It was observed that TRIM21 knockdown inhibited THCA
cell proliferation, migration and invasion. This may be one of the
biological involvements of TRIM21 in the high recurrence risk and
lymph node metastasis of THCA.
TRIM21 may destabilize the tumor suppressor protein
p53, the disruption of which often leads to cancer development
(22). TRIM21 can degrade p27 and
enable cells to enter the S phase, leading to tumor progression
(24). By contrast, TRIM21
negatively regulates anti-apoptotic proteins and inactivates the
glycogen synthase kinase-3 β (GSK3β)-NF-κB pathway to initiate cell
apoptosis (25). In addition,
increased TRIM21 expression increases the activation of caspase-8
and enhances the death receptor-mediated apoptosis (26). In the present study, 252 TRIM21
co-expressed genes, including SP110, APOL2, and UBE2L6, were
identified. These significantly enriched genes were associated with
the ‘antigen processing and presentation’, ‘autoimmune thyroid
disease’ and ‘cell adhesion molecules’ pathways. The expression of
TRIM21 co-expression genes in THCA may be influenced by 14 TFs,
including NFKAPPAB, NF-κB, and IRF2. However, the mechanism by
which TRIM21 regulates THCA progression remains to be
elucidated.
There are several members of the TRIM family. TRIM14
has been reported as an oncogene in THCA (7). TRIM44 knockdown suppresses the tumor
progression of THCA by inhibiting the Wnt/β-catenin signaling
pathway (8). Although others are
also of concern, the current study focused on TRIM21. Further
studies on the expression and function of other members of the TRIM
family in THCA progression are required to establish the regulation
network of the TRIM family in THCA.
In conclusion, using bioinformatics analysis and an
in vitro study, the present study revealed that TRIM21
promoted tumor progression, indicating that TRIM21 may be a
potential biomarker and therapeutic target for THCA. In the future,
the mechanism by which TRIM21 regulates THCA progression will be
further investigated.
Acknowledgements
The authors would like to thank Dr. Jun Li
(Precision Biomedical Laboratory, Liaocheng People's Hospital,
Liaocheng, Shandong, China) for donating the primers of
β-actin.
Funding
Funding: The present study was supported by the Science and
Technology Project of Shandong Society of Geriatrics (grant no.
LKJGG2021W069), and the Medical and Health Science project of
Shandong Province (grant no. 202104010413).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Figshare repository, https://figshare.com/articles/figure/Untitled_Item/19501369.
Authors' contributions
ZW and JF designed the study and wrote the
manuscript. ZW and YW participated in performing the experiments.
ZM was responsible for data acquisition and the interpretation of
data. ZY was responsible for statistical analysis and the
literature search. WD participated in collecting the tissue
samples, performing the RT-qPCR experiments and revising the
manuscript. All authors read and approved the final manuscript. ZW
and ZM confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
This study has been checked and approved by the
Ethics committee of Liaocheng People's Hospital (approval no.
LC2021059).
Patient consent for publication
Not applicable.
Competing interests
All the authors declare that they have no competing
interests.
References
|
1
|
Du L, Zhao Z, Zheng R, Li H, Zhang S, Li
R, Wei W and He J: Epidemiology of thyroid cancer: Incidence and
mortality in China, 2015. Front Oncol. 10(1702)2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Takami H, Ito Y, Okamoto T and Yoshida A:
Therapeutic strategy for differentiated thyroid carcinoma in Japan
based on a newly established guideline managed by Japanese society
of thyroid surgeons and Japanese association of endocrine surgeons.
World J Surg. 35:111–121. 2011.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Hatakeyama S: TRIM family proteins: Roles
in autophagy, immunity, and carcinogenesis. Trends Biochem Sci.
42:297–311. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Vunjak M and Versteeg GA: TRIM proteins.
Curr Biol. 29:R42–R44. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Valletti A, Marzano F, Pesole G, Sbisa E
and Tullo A: Targeting chemoresistant tumors: Could TRIM
proteins-p53 axis be a possible answer? Int J Mol Sci.
20(1776)2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Alomari M: TRIM21-A potential novel
therapeutic target in cancer. Pharmacol Res.
165(105443)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Sun W, Wang Y, Li D, Wu Y, Ji Q and Sun T:
Tripartite motif containing 14: An oncogene in papillary thyroid
carcinoma. Biochem Biophys Res Commun. 521:360–367. 2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zhou Z, Liu Y, Ma M and Chang L: Knockdown
of TRIM44 inhibits the proliferation and invasion in papillary
thyroid cancer cells through suppressing the Wnt/β-catenin
signaling pathway. Biomed Pharmacother. 96:98–103. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Liu Y, Zhang B, Shi T and Qin H: miR-182
promotes tumor growth and increases chemoresistance of human
anaplastic thyroid cancer by targeting tripartite motif 8.
OncoTargets Ther. 10:1115–1122. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Oke V and Wahren-Herlenius M: The
immunobiology of Ro52 (TRIM21) in autoimmunity: A critical review.
J Autoimmun. 39:77–82. 2012.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Simoes Eugénio M, Faurez F, Kara-Ali GH,
Lagarrigue M, Uhart P, Bonnet MC, Gallais I, Com E, Pineau C,
Samson M, et al: TRIM21, a new component of the TRAIL-induced
endogenous necrosome complex. Front Mol Biosci.
8(645134)2021.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Zhang Z, Zhu Z, Sheng H, Sun J and Cao C:
TRIM21 suppresses invasion of hepatocellular carcinoma cells by
promoting beta-catenin ubiquitylation and degradation. Nan Fang Yi
Ke Da Xue Xue Bao. 42:55–62. 2022.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
13
|
Chen X, Li Z, Yong H, Wang W, Wang D, Chu
S, Li M, Hou P, Zheng J and Bai J: Trim21-mediated HIF-1alpha
degradation attenuates aerobic glycolysis to inhibit renal cancer
tumorigenesis and metastasis. Cancer Lett. 508:115–126.
2021.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhao Z, Wang Y, Yun D, Huang Q, Meng D, Li
Q, Zhang P, Wang C, Chen H and Lu D: TRIM21 overexpression promotes
tumor progression by regulating cell proliferation, cell migration
and cell senescence in human glioma. Am J Cancer Res. 10:114–130.
2020.PubMed/NCBI
|
|
15
|
Ping M, Wang S, Guo Y and Jia J: TRIM21
improves apatinib treatment in gastric cancer through suppressing
EZH1 stability. Biochem Biophys Res Commun. 586:177–184.
2022.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Chuang CY, Chien YC, Lin CW, Chou CH, Chen
SC, Liu CL, Bai LY, Yang SF and Yu YL: TRIM21 polymorphisms are
associated with susceptibility and clinical status of oral squamous
cell carcinoma patients. Int J Med Sci. 18:2997–3003.
2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6(pl1)2013.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Shim SR, Kim SJ, Lee J and Rucker G:
Network meta-analysis: Application and practice using R software.
Epidemiol Health. 41(e2019013)2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Bible KC, Kebebew E, Brierley J, Brito JP,
Cabanillas ME, Clark TJ Jr, Di Cristofano A, Foote R, Giordano T,
Kasperbauer J, et al: 2021 American thyroid association guidelines
for management of patients with anaplastic thyroid cancer. Thyroid.
31:337–386. 2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Guha A, Ahuja D, Das Mandal S, Parasar B,
Deyasi K, Roy D, Sharma V, Willard B, Ghosh A and Ray PS: .
Integrated regulation of HuR by translation repression and protein
degradation determines pulsatile expression of p53 under DNA
damage. iScience. 15:342–359. 2019.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhou G, Wu H, Lin J, Lin R, Feng B and Liu
Z: TRIM21 is decreased in Colitis-associated cancer and negatively
regulates epithelial carcinogenesis. Inflamm Bowel Dis. 27:458–468.
2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Sabile A, Meyer AM, Wirbelauer C, Hess D,
Kogel U, Scheffner M and Krek W: Regulation of p27 degradation and
S-phase progression by Ro52 RING finger protein. Mol Cell Biol.
26:5994–6004. 2006.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Gao X, Xu F, Zhang HT, Chen M, Huang W,
Zhang Q, Zeng Q and Liu L: PKCα-GSK3β-NF-κB signaling pathway and
the possible involvement of TRIM21 in TRAIL-induced apoptosis.
Biochem Cell Biol. 94:256–264. 2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhang J, Fang L, Zhu X, Qiao Y, Yu M, Wang
L, Chen Y, Yin W and Hua ZC: Ro52/SSA sensitizes cells to death
receptor-induced apoptosis by down-regulating c-FLIP(L). Cell Biol
Int. 36:463–468. 2012.PubMed/NCBI View Article : Google Scholar
|