Introduction
Diabetic nephropathy (DN), one of the most common
and severe complications of diabetes mellitus, has developed in
40-50% of patients who are diabetic worldwide in the 2015-2020 and
become the leading cause of end-stage renal disease (1,2),
aggravating the global health and economic burden.
Glomerulosclerosis, tubulointerstitial fibrosis (TIF) and chronic
inflammation are characteristic pathological manifestations of DN
(3). Increasing evidence indicates
that renal TIF is the final common pathway and outcome of DN,
accompanied by tubular atrophy and extracellular matrix (ECM)
accumulation (4,5). However, the intrinsic mechanisms
underlying TIF remain unclear. Thus, it is important to explore the
mechanisms of TIF in DN.
Transfer RNA-derived fragments (tRFs), special types
of small non-coding RNA (sncRNA), are more highly conserved than
other sncRNAs and have attracted wide attention (6,7).
tRFs are produced by specific cleavage of precursor or mature tRNA,
and are classified into five subtypes: i) tRF-1; ii) tRF-2; iii)
tRF-3; iv) tRF-5; and v) internal tRF (i-tRF) (8). Previous studies reported that tRFs
can play crucial roles in biological processes in various types of
disease, including neuronal degeneration, tumor cell proliferation
and gene expression (9).
Additionally, tRFs may participate in the early diagnosis of cancer
by serving as prognostic biomarkers (10). A growing number of studies have
demonstrated that tRFs may be associated with chronic kidney
disease (CKD) progression (11-13).
Khurana et al (11)
identified that tRFVal and tRFLeu, derived
from urinary exosomes, are suitable biomarkers for the early
diagnosis of CKD. Our previous study also showed that tRFs
contribute to podocyte differentiation to regulate CKD and revealed
the potential mechanism of idiopathic nephrotic syndrome (12,13).
To the best of our knowledge, however, little is known about the
role of tRFs in TIF.
To explore the role of tRFs in TIF during DN
progression, the present study performed high-throughput sequencing
to detect differential expression profiles of tRFs in high glucose
(HG)-treated tubular epithelial cells. Bioinformatics analyses were
then conducted on these differentially regulated tRFs (fold change
>2, P<0.05). The potential effect of specific tRF was
investigated on HG-induced ECM accumulation of tubular epithelial
cells. Therefore, the present study attempted to reveal the
underlying mechanism of TIF from the novel perspective of tRFs and
provide a promising therapeutic target for DN.
Materials and methods
Cell culture
A total of 10 male 8-week-old C57BL/6 mice weighing
22-25 g (Vital River Laboratory Animal Technology Company; Beijing,
China) were placed a standard environment on a 12-h light/dark
cycle and suitable temperature (22-25˚C) and humidity (40-70%) with
free access to water and food. Primary mouse tubular epithelial
cells were isolated from the renal cortex as previously described
(14). All mice were euthanised by
intraperitoneal injection of pentobarbital sodium (100 mg/kg) and
the kidney was harvested. The cortical tissue was cut into 2-4 mm
pieces and digested in 0.75 mg/ml collagenase for 1 h at 37˚C.
Subsequently, the digested tissues were filtered through 80- and
100-mesh steel sieves. Tubules in the 100-mesh steel sieve were
collected and centrifuged at 3,000 x g for 20 min at 25˚C. Finally,
5x106 cells were suspended in Dulbecco's Modified Eagle
Medium (DMEM) supplemented with 10% foetal bovine serum and 1%
penicillin-streptomycin (all Gibco; Thermo Fisher Scientific,
Inc.). 2x105 cells (per well) were seeded into 12-well
plates and treated with HG (35 mM) or normal glucose (control; 5
mM). D-mannitol (5 mM glucose + 30 mM D-mannitol) was used as the
third group for balancing osmolality. Following 48 h HG treatment
at 37˚C, cells were harvested for subsequent experiments. The
animal experiment was performed in accordance with Animal Research:
Reporting of In Vivo Experiments guidelines (15), as well as the National Institutes
of Health Guide for the Care and Use of Laboratory Animals
(16), and was approved by the
Institutional Animal Care and Use Committee of Nanjing Medical
University (approval no. 2206034).
Human serum samples
Human serum samples from patients with DN were
obtained from the Department of Endocrinology, The Second
Affiliated Hospital of Nanjing Medical University, between 21
January 2021 and 17 September 2021. This study was approved by the
Ethics Committee of Nanjing Medical University [approval no.
(2022)-KY-037-01] and written informed consent was obtained from
all patients. The information about the human subjects is
summarized in Table SI.
Immunofluorescence
Cultured primary tubular epithelial cells were fixed
with 4% paraformaldehyde for 15 min at room temperature and then
incubated with primary antibody cytokeratin 18 (CK18; 1:200; cat.
no. sc-32329; Santa Cruz Biotechnology, Inc.) at 4˚C overnight.
Subsequently, cells were incubated with Alexa Fluor®
647-conjugated secondary antibody (goat anti-mouse IgG H&L;
1:400; cat. no. ab150115; Abcam) in the dark at room temperature
for 2 h. After washing with phosphate-buffered saline, 0.02% DAPI
staining was performed for 10 min at room temperature.
Immuno-stained cells were observed under a fluorescence microscope
(magnification, x100; Nikon Corporation).
High-throughput sequencing
High-throughput sequencing was performed as
described previously (12).
Briefly, total RNA samples were pretreated to remove RNA
modifications that influence small RNA-sequencing (RNA-seq) library
construction with the rtStar™ tRF&tiRNA Pretreatment kit
(Arraystar, Inc.). The total RNA of each sample was sequentially
ligated to 3' and 5' small RNA adapters. cDNA was synthesised and
amplified using proprietary reverse transcription (RT) primers and
amplification primers (NEBNext® Multiplex Small RNA
Library Prep Set for Illumina; Illumina, Inc.). Subsequently,
134-160 bp PCR-amplified fragments were extracted and purified via
8% PAGE. Subsequently, completed libraries were quantified using
the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.). The
libraries were denatured and diluted to a loading volume of 1.3 ml
and loading concentration of 1.8 pM following the manufacturer's
protocol. Subsequently, according to the manufacturer's
instructions, diluted libraries were loaded onto a reagent
cartridge and forwarded to the sequencing run on Illumina NextSeq
500 system using NextSeq 500/550 V2 kit (cat. no. FC-404-2005;
Illumina, Inc.).
RNA extraction and RT-quantitative
(q)PCR
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and RNA concentration and purity were detected using a
NanoDrop instrument (Thermo Fisher Scientific, Inc.). RT-qPCR was
performed using the HiScript III RT SuperMix and ChamQ™
SYBR® qPCR Master Mix (Vazyme Biotech Co., Ltd.) using
the StepOne Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
GAPDH was used as an internal control for normalisation. For
expression of tRFs, total RNA samples were pretreated to remove RNA
modifications with rtStar™ tRF&tiRNA Pretreatment kit
(Arraystar, Inc.). PCR amplification was performed using the
Bulge-Loop™ miRNA RT-qPCR Primer Sets (one RT primer and a pair of
PCR primers for each set), which were designed specifically for
each tRF by Guangzhou RiboBio Co., Ltd. After adding forward primer
and universal reverse primer, the reaction mixtures were incubated
at 95˚C for 10 min, followed by 95˚C for 2 sec, 60˚C for 20 sec and
70˚C for 10 sec, for 40 PCR cycles in the StepOne Real-Time PCR
System (Applied Biosystems; Thermo Fisher Scientific, Inc.). U6
small nuclear RNA (snRNA) was used as the internal reference. All
tRF primers were designed specifically by Guangzhou RiboBio Co.,
Ltd. and all mRNA primers were obtained from Generay Biotech Co.
Ltd. The mRNA and tRF forward primer sequences are listed in
Tables SII and SIII. The Bulge-Loop™ U6 snRNA qPCR
Primer Set (cat. no. MQP-0201) and universal reverse primer (cat.
no. ssD089261711) cannot be provided due to the patent of Guangzhou
RiboBio Co., Ltd. The relative expression levels were normalized to
endogenous controls and were expressed as
2-ΔΔCq (17).
Western blotting
Western blotting was performed as described by Ji
et al (18). Briefly, cells
were harvested using RIPA Lysis Buffer supplemented with 1%
protease inhibitor phenylmethylsulfonyl fluoride (both Beyotime
Institute of Biotechnology). Total protein (25 ug) loaded per lane
were separated by 10% SDS-PAGE and were transferred onto PVDF
membranes (MilliporeSigma; cat. no. HATF09025). Then, membranes
were blocked with 5% skimmed milk at room temperature for 2 h.
Subsequently, membranes were incubated with primary antibodies
against α-smooth muscle actin (α-SMA; 1:1,000; cat. no. BF9212;
Affinity Biosciences, Ltd.), collagen I (1:1,000; cat. no. ab34710;
Abcam), fibronectin (1:1,000; cat. no. ab2413; Abcam) and GAPDH
(1:1,000; cat. no. AF7021; Affinity Biosciences, Ltd.) at 4˚C
overnight. Next day, membranes were washed using 1x TBST
supplemented with 0.1% Tween 20, and then incubated with the
secondary horseradish peroxidase (HRP)-conjugated antibodies (goat
anti-mouse IgG; 1:2,000; cat. no. S002; Affinity Biosciences, Ltd;
or goat anti-rabbit IgG; 1:2,000; cat. no. S001; Affinity
Biosciences, Ltd.) at room temperature for 2 h. Finally, the
membranes were incubated with ECL Developer (Biosharp) Intensity
values expressed as relative protein expression were analysed using
ImageJ software (version 1.8.0; National Institutes of Health) and
normalised to the expression of GAPDH.
Gene Ontology (GO) and pathway
analysis
To explore the potential functions of differentially
expressed tRFs, target genes of differentially expressed tRFs (fold
change >2 and P<0.05 for significantly differentially
expressed tRFs) were selected. DAVID (david.ncifcrf.gov/) website was used for GO and Kyoto
Encyclopedia of Genes and Genomes (KEGG; kegg.jp/) was used to
perform pathway analysis.
Transfection
Mouse renal tubular epithelial cells were seeded in
12-well plates and grown to ~70% confluence (2x105 cells
per well) at 37˚C. The tRF-1:30-Gln-CTG-4 mimic (tRF mimic;
5'-GGTTCCATGGTGTAATGGTGAGCACTCTGG-3'; Guangzhou RiboBio Co., Ltd.)
or negative control (NC mimic;
5'-UUAUAGUCGUGGGAGCAGGGAUCGGCUUCUNC-3'; Guangzhou RiboBio Co.,
Ltd.) were transfected into cells at a concentration of 30 nM with
RiboFECT™ CP Transfection Reagent (Guangzhou RiboBio Co., Ltd.) for
6 h at 37˚C according to the manufacturer's instructions. Then, the
medium was replaced with complete DMEM and cells were stimulated
with HG (35 mM) at 37˚C for 48 h.
Statistical analysis
Data are expressed as the mean ± standard error of
the mean. All experiments were repeated three times. Statistical
analyses were performed using Statistical Package for the Social
Sciences version 22.0 (IBM Corp.). Student's t test was used for
comparisons between two groups (unpaired). To compare >2 groups,
one-way ANOVA followed by a Bonferroni's correction was used to
analyse differences. P<0.05 was considered to indicate a
statistically significant difference.
Results
tRF expression profiles in HG-treated
tubular epithelial cells
First, the purity of primary tubular epithelial
cells was identified using immunofluorescence staining with an
epithelial-specific marker (CK18). The present results showed that
>95% of cells expressed CK18 (Fig.
S1). Subsequently, tubular epithelial cell ECM accumulation was
induced by HG treatment. Meanwhile, D-mannitol was used to examine
the effects of osmotic pressure on cells; osmotic pressure had no
effect on the production of ECM in tubular epithelial cells
(Fig. S2), which is consistent
with previous studies (19,20).
Thereafter, cells with a 5 mM glucose culture were selected as the
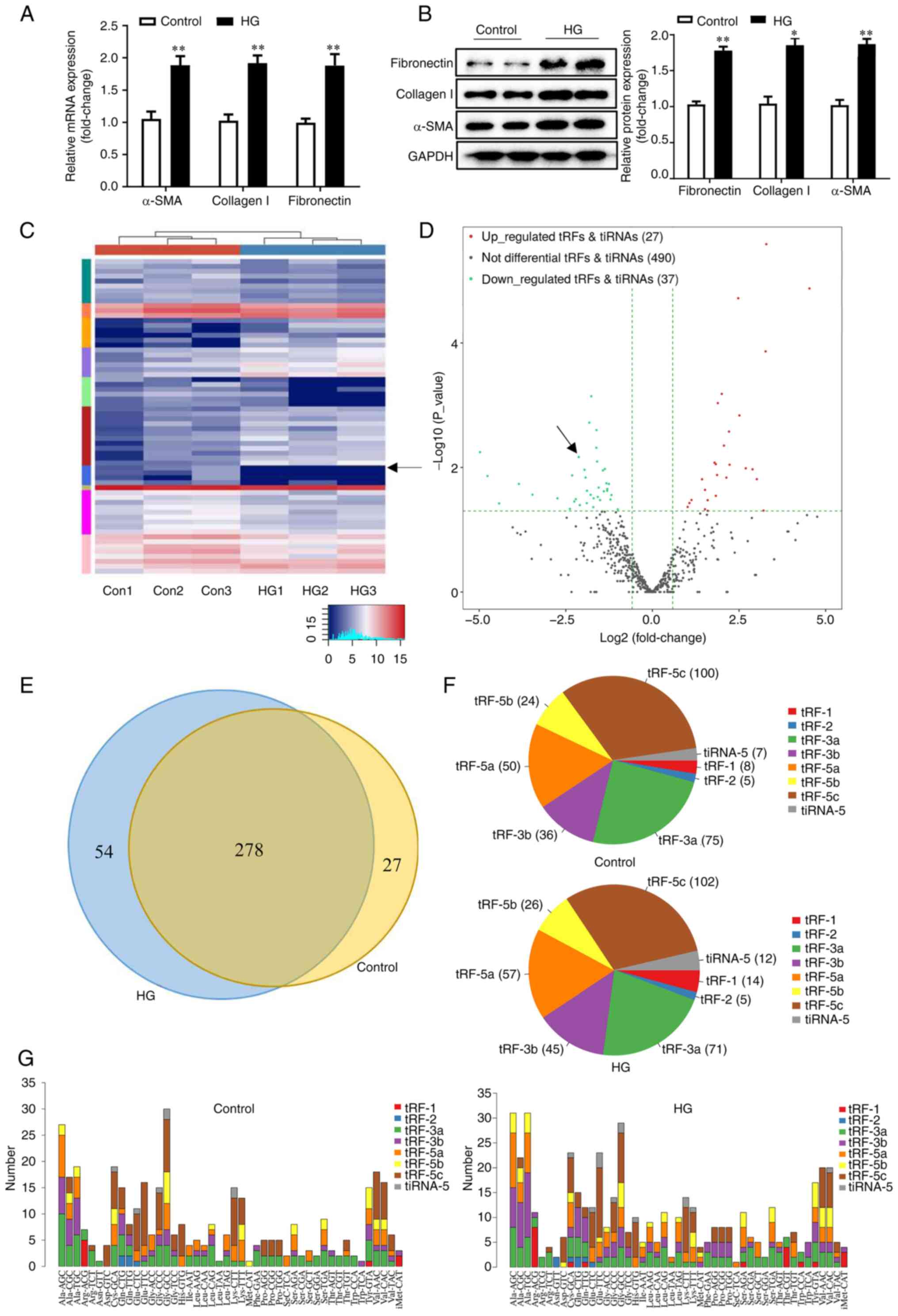
control group. mRNA and protein expression of TIF-associated
markers α-SMA, collagen I and fibronectin (21,22)
were significantly induced in the HG-treated group (Fig. 1A and B). Differentially expressed tRFs were
analysed with high-throughput sequencing. Of 554 distinct tRFs
detected, 64 differentially expressed tRFs (fold change >2;
P<0.05) were identified. Of these, 27 tRFs were upregulated and
37 tRFs were downregulated, as displayed by hierarchical clustering
heatmap and the volcano plot (Fig.
1C and D). All upregulated and
downregulated tRFs in the two groups are listed in Table SIV. Commonly and specifically
expressed tRFs are displayed in a Venn diagram (Fig. 1E). In total, 278 commonly expressed
tRFs overlapped between the two groups, indicating non-zero counts
per million mapped reads (CPM) values in both groups. In addition,
54 specifically expressed tRFs were found in the HG-treated group,
whereas 27 specifically expressed tRFs were only detected in the
control group. The tRF-5c, formed by cleavage at the D- and
anticodon stems, was the most abundant tRF subtype in both the
HG-treated and control groups, with expression in the HG-treated
group higher than that in the control group (Fig. 1F and G), suggesting that tRF-5c may be
associated with DN progression. Collectively, these results
revealed that tRFs might play a role in HG-induced tubular ECM
accumulation.
Verification of expression levels of
selected tRFs
To verify the RNA-seq results, 10 tRFs with the
greatest difference (fold change >2; P<0.05) were selected as
candidate tRFs after excluding tRFs with zero CPM values. RT-qPCR
analysis revealed that tRF-53:70-chrM.Trp-TCA,
tRF-49:70-chrM.Trp-TCA, tRF-1:32-Glu-TTC-2 and tRF-1:32-Glu-TTC-1
were significantly upregulated in HG-treated tubular epithelial
cells (Fig. 2B-E).
tRF-1:30-Gln-CTG-4, tRF-1:28-Gly-CCC-1, tRF-1:29-Val-AAC-5 and
tRF-1:30-Val-AAC-5 were significantly downregulated (Fig. 2F, G, I and
J). However, there were no
significant differences in the expression of tRF-1:32-Cys-GCA-1-M3
and tRF-1:29-Val-CAC-1 between the two groups (Fig. 2A and H). Among these verified tRFs,
tRF-1:32-Glu-TTC-2, tRF-1:32-Glu-TTC-1, tRF-1:30-Gln-CTG-4,
tRF-1:28-Gly-CCC-1, tRF-1:29-Val-AAC-5, tRF-1:32-Cys-GCA-1-M3,
tRF-1:29-Val-CAC-1 and tRF-1:30-Val-AAC-5 belong to the tRF-5c
subtype, tRF-53:70-chrM.Trp-TCA belongs to the tRF-3a subtype and
tRF-49:70-chrM.Trp-TCA belongs to the tRF-3b subtype (Table SIV).
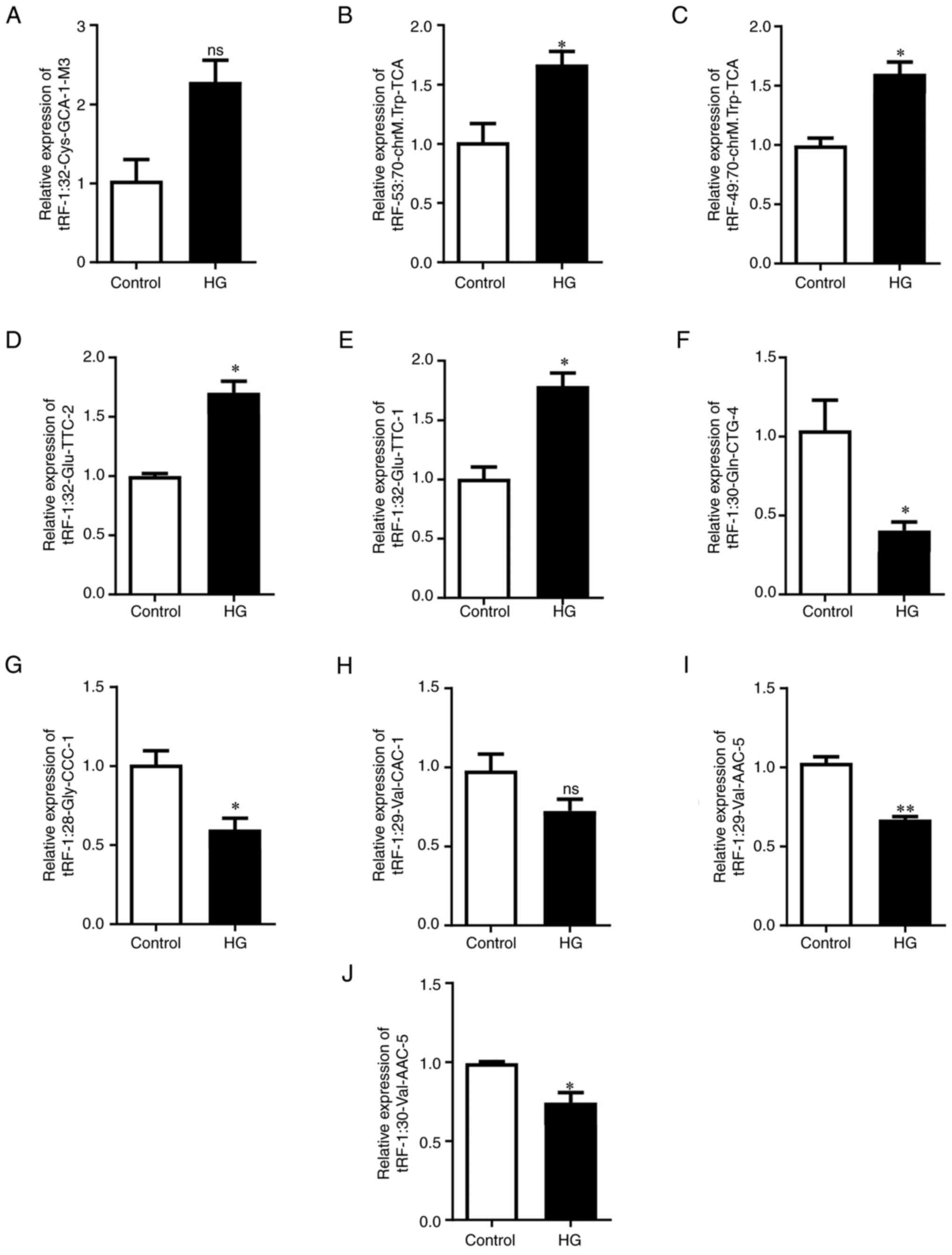 | Figure 2Verification of expression levels of
selected tRFs by reverse transcription-quantitative PCR. Expression
of (A) tRF-1:32-Cys-GCA-1-M3, (B) tRF-53:70-chrM.Trp-TCA, (C)
tRF-49:70-chrM.Trp-TCA, (D) tRF-1:32-Glu-TTC-2, (E)
tRF-1:32-Glu-TTC-1, (F) tRF-1:30-Gln-CTG-4, (G) tRF-1:28-Gly-CCC-1,
(H) tRF-1:29-Val-CAC-1, (I) tRF-1:29-Val-AAC-5 and (J)
tRF-1:30-Val-AAC-5. U6 was used as the internal reference for
normalisation. *P<0.05 and **P<0.01.
ns, non-significant; tRF, transfer RNA-derived fragment. |
GO and KEGG analysis of differentially
expressed tRF targets
tRFs may play a key role in post-transcriptional
regulation via substantial tRF-target gene interactions (23). In the present study, target genes
of the eight significantly differentially expressed tRFs were
predicted to reveal potential underlying mechanisms of tRFs
(Table SV). GO and KEGG analyses
of these target genes evaluated the function of the differentially
expressed tRFs. First, the potential function of downregulated tRF
targets were assessed. The most highly enriched biological
processes (BPs) were ‘cellular process’, ‘biological regulation’
and ‘metabolic process’ (Fig. 3A).
‘Cellular anatomical entity’, ‘intracellular’, ‘organelle’,
‘cytoplasm’ and ‘membrane’ were the most highly enriched cellular
components (CCs; Fig. 3B).
Molecular functions (MFs) primarily included ‘protein binding’,
‘ion binding’, ‘organic cyclic compound binding’, ‘heterocyclic
compound binding’ and ‘DNA binding’ (Fig. 3C). The potential functions of
upregulated tRF targets were consistent with those of downregulated
tRF targets (Fig. 3D-F). In KEGG
pathway enrichment analysis, forkhead box O (‘FoxO signaling
pathway’), ‘PD-L1 expression and PD-1 checkpoint pathway in
cancer’, ‘proteoglycans in cancer’, ‘endocrine resistance’, ‘mTOR
signalling pathway’ and ‘MAPK signaling pathway’ were highly
enriched in the differentially upregulated tRFs (Fig. 3G). ‘GABAergic synapse’,
‘autophagy-animal’, ‘neurotrophin signalling pathway’, ‘Ras
signalling pathway’, helper T (‘Th1 and Th2 cell differentiation’),
‘glutamatergic synapse’, ‘T cell receptor signalling pathway’ and
‘insulin signalling pathway’ were highly enriched in the
differentially downregulated tRFs (Fig. 3H).
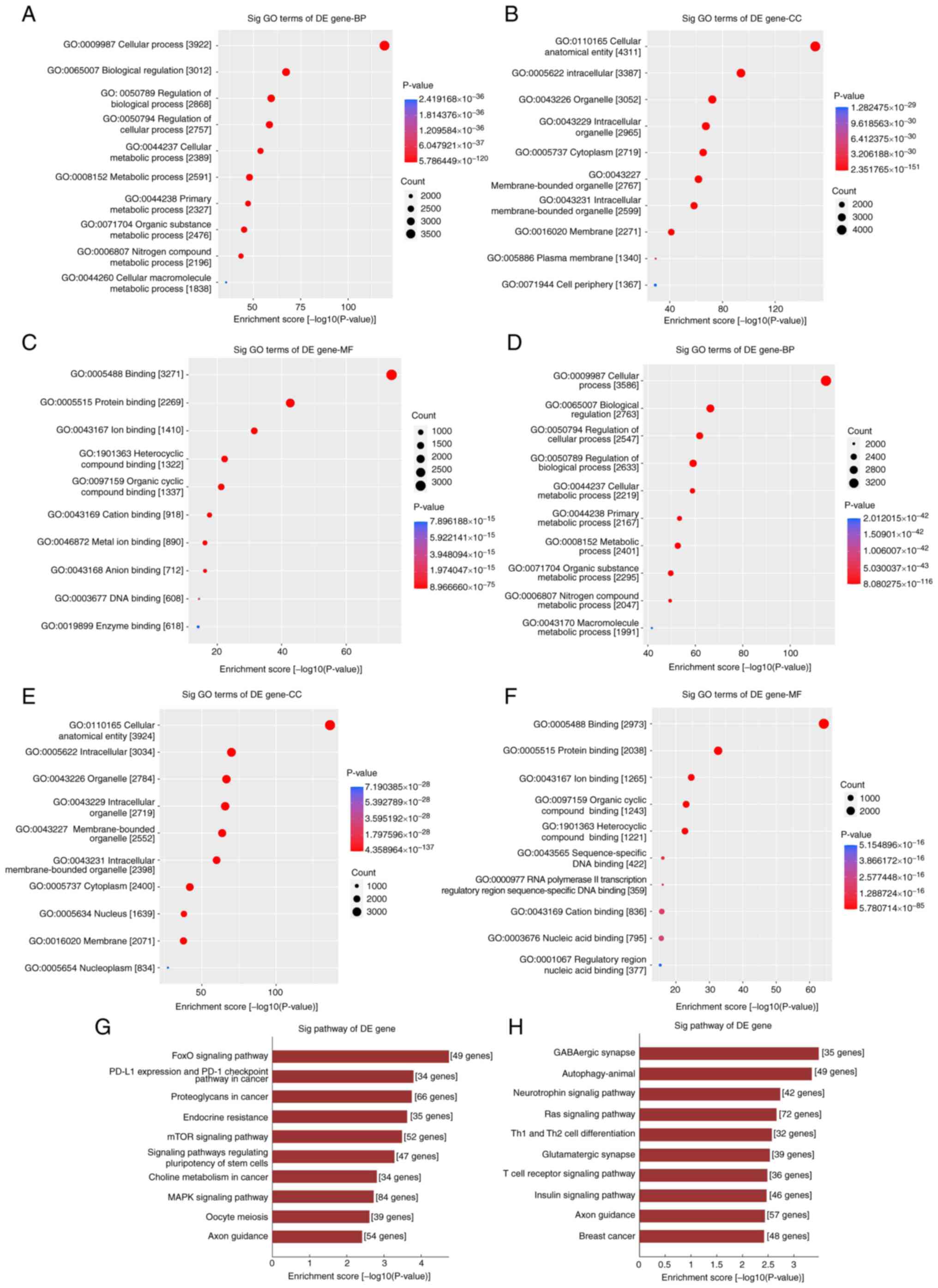 | Figure 3GO and KEGG analysis of potential
genes of selected differentially downregulated and upregulated
tRFs. GO analysis of the target genes of downregulated tRFs
including (A) BP, (B) CC and (C) MF. GO analysis of the target
genes of upregulated tRFs including (D) BP, (E) CC and (F) MF. (G)
Analysis of potential target genes of selected differentially
upregulated tRFs by KEGG pathway clustering. (H) KEGG pathway
clustering analysis of potential target genes of the selected
differentially downregulated tRFs. GO, Gene Ontology; KEGG, Kyoto
Encyclopedia of Genes and Genomes; tRF, transfer RNA-derived
fragment; BP, biological process; CC, cellular component; MF,
molecular function; Sig, significant; DE, differentially
expressed. |
Overexpression of tRF-1:30-Gln-CTG-4
attenuates ECM deposition
tRF-1:30-Gln-CTG-4 showed the most pronounced
difference in expression and was significantly decreased in
response to HG (Fig. 2). Moreover,
tRF-1:30-Gln-CTG-4 expression in the serum of patients with DN was
decreased by 30% (Fig. 4A). To
explore the effects of tRF-1:30-Gln-CTG-4 on ECM deposition,
tRF-1:30-Gln-CTG-4 was overexpressed using tRF-1:30-Gln-CTG-4
mimic, followed by HG treatment (Fig.
4B). There was a marked decrease in α-SMA, collagen I and
fibronectin levels (Fig. 4C and
D), indicating an attenuated ECM
deposition in tubular epithelial cells. These results demonstrated
that tRF-1:30-Gln-CTG-4 was involved in DN progression and may be a
novel biomarker for DN progression.
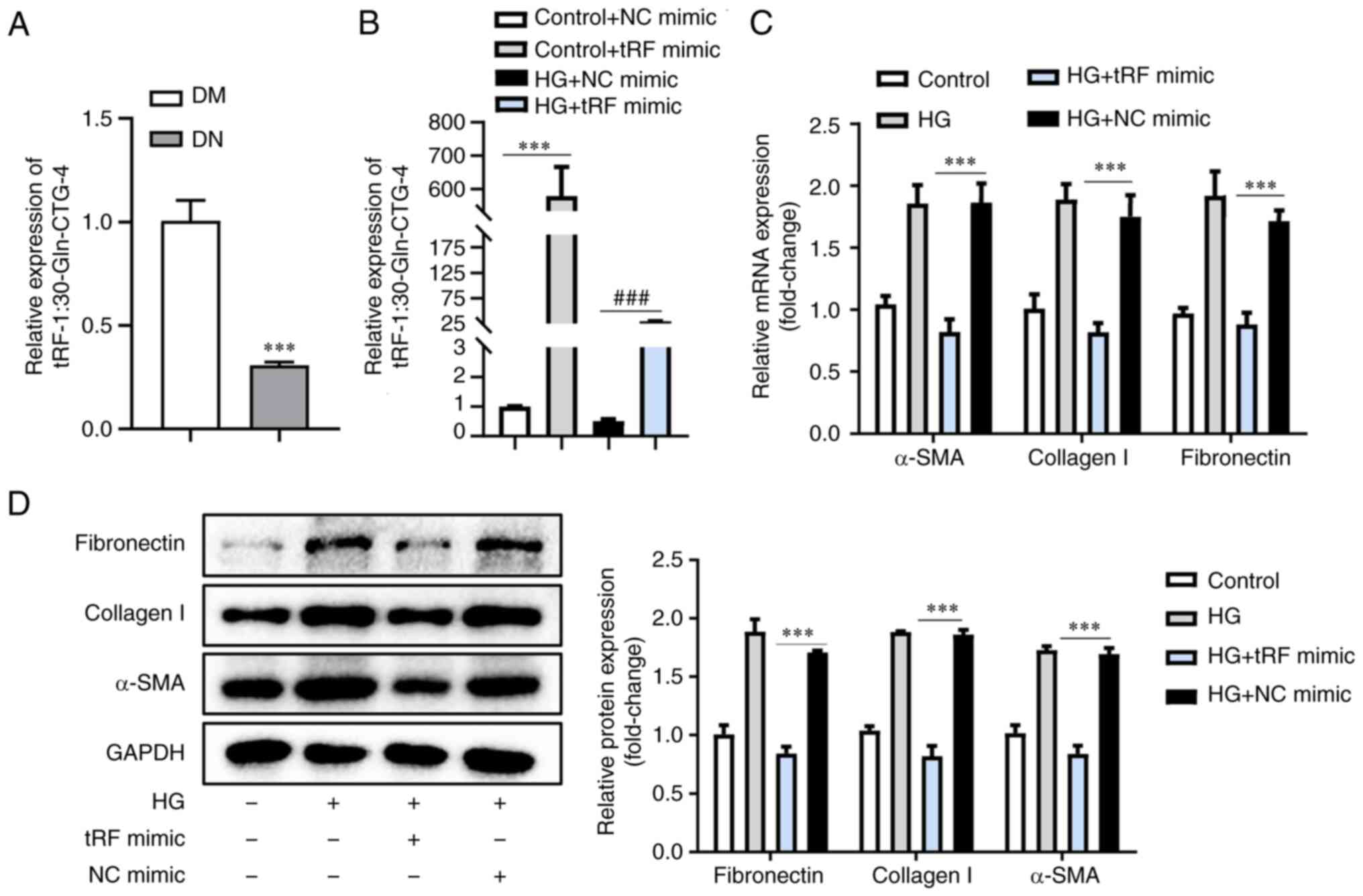 | Figure 4tRF-1:30-Gln-CTG-4 overexpression
contributes to inhibition of extracellular matrix accumulation. (A)
tRF-1:30-Gln-CTG-4 was validated by RT-qPCR, with U6 as a
normalisation control. (B) RT-qPCR analysis of tRF-1:30-Gln-CTG-4
expression in mouse tubular epithelial cells transfected with
tRF-1:30-Gln-CTG-4 mimic with or without HG treatment. (C) mRNA
expression levels of α-SMA, collagen-1 and fibronectin by RT-qPCR
analysis. (D) Western blot analysis showing protein expression
levels of α-SMA, collagen-1 and fibronectin. The relative levels
were normalised to those of GAPDH. ***P<0.001 and
###P<0.001. RT-qPCR, reverse
transcription-quantitative PCR; tRF, transfer RNA-derived fragment;
α-SMA, α-smooth muscle actin; HG, high glucose; DM, diabetes
mellitus; DN, diabetic nephropathy; NC, negative control. |
Discussion
Despite technological advances, the pathogenesis of
DN remains unclear and, to the best of our knowledge, there are no
valid therapeutic strategies (24,25).
TIF is the key factor in DN progression, which is characterised by
the deposition of ECM, including α-SMA, fibronectin and collagen I
(26). The present study found
that tRFs, a special kind of sncRNA, are associated with TIF and
involved in DN. These findings not only provide novel insights into
the pathogenesis of DN but may also help to screen effective
therapies for DN.
With advances in next-generation sequencing
technology, numerous sncRNAs, including microRNAs, circular,
p-element-induced wimpy testis-interacting and small nucleolar RNAs
and tRNAs, have been confirmed to serve significant roles in a
variety of diseases (27).
Recently, tRNA-derived small fragments, called tsRNAs, have
attracted considerable attention (28-30).
The two types of tsRNA, tRNA-derived stress-induced RNA (tiRNA) and
tRFs, are classified according to the different cleavage positions
of the precursor or mature tRNA transcript (31). An increasing number of studies have
demonstrated that tRFs serve a variety of biological functions in
regulating cell proliferation, interacting with proteins or mRNA
and regulating the cell cycle, DNA damage response and epigenetic
modifications (32-34).
In addition, previous studies showed that tRFs may be novel
diagnostic and therapeutic targets in kidney disease (11,35).
Therefore, it was hypothesised that tRFs serve a key role in DN. To
understand the potential effects of tRFs on DN, small RNA-seq was
performed in the present study and 27 upregulated and 37
downregulated differentially expressed tRFs were found. These
results suggest that tRFs may serve a crucial role in DN
development and are worthy of further study.
To date, five types of tRFs have been identified and
characterised by their provenance on tRNAs: tRF-1, tRF-2, tRF-3,
tRF-5 and i-tRF. The tRF-1 subtype is derived from the 3' trailer
of primary tRNA and is formed during the tRNA precursor sequence
maturation. The tRF-2 subtype is derived from tRNAGlu,
tRNAGly and tRNATyr. The tRF-3 subtype that
results from cleavage of the T-loop by the Dicer enzyme and
ribonuclease angiogenin is further classified into two subgroups:
tRF-3a; and tRF-3b. The tRF-5 subtype that is generated from the
cleavage of the D-loop of tRNAs by the Dicer enzyme is further
divided into three subtypes: tRF-5a, tRF-5b and tRF-5c. The i-tRF
subtype originates from the internal bodies of mature tRNA
(36-38).
According to the present tRF high-throughput sequencing data,
tRF-5c was the most abundant subtype in both the HG-treated and
control groups. Furthermore, expression of tRF-5c in the HG-treated
group was higher than that in the control group, suggesting that
tRF-5c may be associated with DN progression.
Although there are some studies on the involvement
of tRFs in pathogenesis of disease (39,40),
to the best of our knowledge, the mechanisms by which tRFs regulate
occurrence and development of diseases remain unclear. Previous
studies have revealed that tRFs not only participate in
posttranscriptional regulation via miRNA-like actions but also
stabilise the mRNA by binding to RNA-binding proteins (41,42).
For example, tRF3008A suppresses progression and metastasis of
colorectal cancer by destabilising FOXK1 in an Argonaute
protein-dependent manner (43).
Goodarzi et al (44) showed
that tRFs suppress breast cancer progression by competitively
binding YBX1 to inhibit the stability of multiple oncogenic
transcripts. Potential target genes of tRF-53:70-chrM.Trp-TCA,
tRF-1:32-Glu-TTC-2, tRF-1:32-Glu-TTC-1, tRF-49:70-chrM.Trp-TCA,
tRF-1:30-Gln-CTG-4, tRF-1:28-Gly-CCC-1, tRF-1:30-Val-AAC-5 and
tRF-1:29-Val-AAC-5 were predicted in the present study. Analysis of
tRF target genes revealed that multiple tRFs are associated with
renal fibrosis. For example, tRF-1:28-Gly-CCC-1 is involved in
regulating the fibrotic process by targeting transforming growth
factor β receptor type 2, a type II serine/threonine kinase
receptor for transforming growth factor β-1 (TGF-β1) that induces
fibrosis by binding TGF-β1 and acting as the initiator and key
component of canonical TGF-β/SMAD signalling (45). tRF-1:30-Val-AAC-5 may function as
an anti-fibrosis agent by targeting kelch-like protein 42 (KLHL42).
Lear et al (46) showed
that KLHL42 impairs TGF-β-dependent profibrotic signalling and
KLHL42 knockdown decreases fibrotic tissue production. In addition,
in the present study FoxO3 was predicted as a target gene of
tRF-1:30-Gln-CTG-4. Evidence has indicated that FoxO3 is an
important player in fibrogenesis and a novel treatment strategy in
renal fibrosis (47). Hence,
tRF-1:30-Gln-CTG-4 may serve a role in renal fibrosis by targeting
FoxO3 expression. Further analysis of tRF target genes should be
performed to explore the mechanisms of tRFs in TIF.
GO clustering was performed in the present study to
investigate the potential functions of tRFs. For BP, most targets
of differentially expressed tRFs were associated with cellular
metabolic processes. In a pathology study of 34 cases of human
kidneys diagnosed with DN, lipid accumulation was found in renal
tubules (48). Moreover, in
vitro studies showed lipid droplets in cultured renal tubular
epithelial cells following HG treatment (49,50);
these data demonstrate that tRF-associated metabolic disorders of
renal epithelial cells may result in progressive fibrosis in DN. In
addition, the most highly enriched MF subcategory in the present
study was ‘protein binding’. tRFs competitively bind RNA-binding
proteins (RBPs) to regulate oncogenic mRNA expression. Falconi
et al (51) indicated that
a novel tRF derived from mature tRNAGlu was able to bind
and displace the 3' untranslated region of specific RBPs to
suppress breast cancer progression. The present GO analysis results
provide novel ideas for exploring the role of tRFs in DN.
In KEGG analysis, the pathways of autophagy, FoxO,
mTOR, MAPK and Ras signalling were highly enriched. Previously,
studies have demonstrated that these pathways were closely
associated with ECM (52,53). Dysregulation of autophagy under
stress conditions plays a crucial role in progressive renal and
hepatic fibrosis (54,55). The FoxO signalling pathway exerts
key effects on renal fibrosis (56,57).
Additionally, mTOR serves an essential role in cell proliferation
and metabolism. The dysfunction of the mTOR pathway is involved in
progression of renal fibrosis in various kidney diseases (58) and mediates fibrogenesis by
regulating ECM accumulation (59).
MAPK signalling is strongly active in ECM accumulation during the
progression of DN (60). RAS
effector RREB1 is considered to be a key partner of TGF-β-activated
SMAD transcription factors in ECM accumulation and
epithelial-to-mesenchymal transitions (61). These results indicate that the
differentially expressed tRFs may be associated with several
signalling pathways and play important regulatory roles in the
development of DN.
Finally, considering their potential
pathophysiological mechanisms, the present study focused on
downregulated tRFs. In particular, tRF-1:30-Gln-CTG-4 was further
investigated since it was the most downregulated tRFs. Expression
of tRF-1:30-Gln-CTG-4 decreased in the HG-treated group. ECM
accumulation induced by HG treatment was reversed by the
overexpression of tRF-1:30-Gln-CTG-4, which indicated that
tRF-1:30-Gln-CTG-4 may have an antifibrotic role in DN progression.
However, the mechanism by which tRF-1:30-Gln-CTG-4 regulates ECM
secretion in DN remains unclear and warrants further investigation.
Moreover, the effects of upregulated tRFs on HG-induced ECM
accumulation were not investigated in the present study.
Upregulated tRFs may also serve a critical role in promoting renal
tubular injury and ECM accumulation. Thus, the exact effects of
upregulated tRFs should be explored in a future study. It remains
to be established whether the present results may also be observed
in human kidney tissues.
In conclusion, the present study discovered that
tRFs, which are novel types of sncRNAs, are associated with TIF and
involved in DN. The underlying mechanism may involve the
dysregulation of autophagy and the FoxO, mTOR and MAPK signalling
pathways. The present findings advance understanding of the
pathophysiology of DN and may reveal promising therapeutic targets
for the treatment of DN.
Supplementary Material
Identification of primary mouse renal
tubular epithelial cells. Immunofluorescence detection of
epithelial cell marker (CK18). Scale bar, 50 μm. CK,
cytokeratin.
Osmotic pressure has no effect on
tubular epithelial cells. (A) mRNA and (B) protein expression
levels of fibronectin, collagen I and α-SMA. The relative levels
were normalized to GAPDH. **P<0.01 and
***P<0.001. ns, non-significant; M, D-mannitol; HG,
high glucose; α-SMA, α-smooth muscle actin.
Patient characteristics in serum
study.
Primers for α-SMA, collagen I,
fibronectin and GAPDH.
Primers for tRFs.
All differentially expressed
tRFs.
Target genes of seclected tRFs.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81970664), Natural Science
Foundation of Jiangsu Province (grants nos. BK20191082 and
BK20211385) and 789 Outstanding Talent Program of SAHNMU (grants
nos. 789ZYRC202080119 and 789ZYRC202090251).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Sequence Read Archive under
BioProject repository, https://www.ncbi.nlm.nih.gov/sra/PRJNA878883.
Authors' contributions
AZ and WG designed the study. JJ performed the
experiments and wrote the manuscript. JR, HZ, HS, GQ and SL
contributed to the data analysis. All authors have read and
approved the final manuscript. JJ and AZ confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
Animal experiments were approved by the
Institutional Animal Care and Use Committee of Nanjing Medical
University (approval no. 2206034). The human experimental
procedures in the present study were approved by Nanjing Medical
University [Nanjing, China; approval no. (2022)-KY-037-01] and
written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Xue R, Gui D, Zheng L, Zhai R, Wang F and
Wang N: Mechanistic insight and management of diabetic nephropathy:
Recent progress and future perspective. J Diabetes Res.
2017(1839809)2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Tuttle KR, Jones CR, Daratha KB, Koyama
AK, Nicholas SB, Alicic RZ, Duru OK, Neumiller JJ, Norris KC, Ríos
Burrows N and Pavkov ME: Incidence of Chronic Kidney Disease among
Adults with Diabetes, 2015-2020. N Engl J Med. 387:1430–1431.
2022.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Alicic RZ, Rooney MT and Tuttle KR:
Diabetic kidney disease: Challenges, progress and possibilities.
Clin J Am Soc Nephrol. 12:2032–2045. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Qi R and Yang C: Renal tubular epithelial
cells: The neglected mediator of tubulointerstitial fibrosis after
injury. Cell Death Dis. 9(1126)2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Huang F, Wang Q, Guo F, Zhao Y, Ji L, An
T, Song Y, Liu Y, He Y and Qin G: FoxO1-mediated inhibition of
STAT1 alleviates tubulointerstitial fibrosis and tubule apoptosis
in diabetic kidney disease. EBioMedicine. 48:491–504.
2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Pandey KK, Madhry D, Ravi Kumar YS,
Malvankar S, Sapra L, Srivastava RK, Bhattacharyya S and Verma B:
Regulatory roles of tRNA-derived RNA fragments in human
pathophysiology. Mol Ther Nucleic Acids. 26:161–173.
2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Fagan SG, Helm M and Prehn JHM:
tRNA-derived fragments: A new class of non-coding RNA with key
roles in nervous system function and dysfunction. Prog Neurobiol.
205(102118)2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Zeng TY, Hua YJ, Sun CX, Zhang Y, Yang F,
Yang M, Yang Y, Li J, Huang X, Wu H, et al: Relationship between
tRNA-derived fragments and human cancers. Int J Cancer.
147:3007–3018. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Karaca E, Weitzer S, Pehlivan D, Shiraishi
H, Gogakos T, Hanada T, Jhangiani SN, Wiszniewski W, Withers M,
Campbell IM, et al: Human CLP1 mutations alter tRNA biogenesis,
affecting both peripheral and central nervous system function.
Cell. 157:636–650. 2014.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Oberbauer V and Schaefer MR: tRNA-derived
small RNAs: Biogenesis, modification, function and potential impact
on human disease development. Genes (Basel). 9(607)2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Khurana R, Ranches G, Schafferer S,
Lukasser M, Rudnicki M, Mayer G and Hüttenhofer A: Identification
of urinary exosomal noncoding RNAs as novel biomarkers in chronic
kidney disease. RNA. 23:142–152. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Shi H, Yu M, Wu Y, Cao Y, Li S, Qu G, Gong
J, Gan W and Zhang A: tRNA-derived fragments (tRFs) contribute to
podocyte differentiation. Biochem Biophys Res Commun. 521:1–8.
2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Li S, Liu Y, He X, Luo X, Shi H, Qu G, Wen
X, Gan W, Wang J and Zhang A: tRNA-Derived fragments in podocytes
with Adriamycin-induced injury reveal the potential mechanism of
idiopathic nephrotic syndrome. Biomed Res Int.
2020(7826763)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Luo C, Zhou S, Zhou Z, Liu Y, Yang L, Liu
J, Zhang Y, Li H, Liu Y, Hou FF and Zhou L: Wnt9a promotes renal
fibrosis by accelerating cellular senescence in tubular epithelial
cells. J Am Soc Nephrol. 29:1238–1256. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kilkenny C, Browne W, Cuthill IC, Emerson
M and Altman DG: NC3Rs Reporting Guidelines Working Group. Animal
Research: Reporting of in vivo experiments guidelines. Br J
Pharmacol. 160:1577–1579. 2010.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
National Institutes of Health (NIH): Guide
for the Care and Use of Laboratory Animals. In: National Research
Council (US) Committee for the Update of the Guide for the Care and
Use of Laboratory Animals. 7th Edition. National Academy Press,
Washington, DC, 1996.
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ji JL, Zhao YJ, Na C, Yang M, Zhu X, Shi
H, Gan W and Zhang A: Connexin 43-autophagy loop in the podocyte
injury of diabetic nephropathy. Int J Mol Med. 44:1781–1788.
2019.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Peng F, Gong W, Li S, Yin B, Zhao C, Liu
W, Chen X, Luo C, Huang Q, Chen T, et al: circRNA_010383 Acts as a
Sponge for miR-135a and its downregulated expression contributes to
renal fibrosis in diabetic nephropathy. Diabetes. 70:603–615.
2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Liu XQ, Jiang L, Lei L, Nie ZY, Zhu W,
Wang S, Zeng HX, Zhang SQ, Zhang Q, Yard B and Wu YG: Carnosine
alleviates diabetic nephropathy by targeting GNMT, a key enzyme
mediating renal inflammation and fibrosis. Clin Sci (Lond).
134:3175–3193. 2020.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang XX, Wang D, Luo Y, Myakala K,
Dobrinskikh E, Rosenberg AZ, Levi J, Kopp JB, Field A, Hill A, et
al: FXR/TGR5 dual agonist prevents progression of nephropathy in
diabetes and obesity. J Am Soc Nephrol. 29:118–137. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Liu Y: Cellular and molecular mechanisms
of renal fibrosis. Nat Rev Nephrol. 7:684–696. 2011.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Yu X, Xie Y, Zhang S, Song X, Xiao B and
Yan Z: tRNA-derived fragments: Mechanisms underlying their
regulation of gene expression and potential applications as
therapeutic targets in cancers and virus infections. Theranostics.
11:461–469. 2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Tuttle KR, Agarwal R, Alpers CE, Bakris
GL, Brosius FC, Kolkhof P and Uribarri J: Molecular mechanisms and
therapeutic targets for diabetic kidney disease. Kidney Int.
102:248–260. 2022.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Nellaiappan K, Preeti K, Khatri DK and
Singh SB: Diabetic complications: An update on pathobiology and
therapeutic strategies. Curr Diabetes Rev.
18(e030821192146)2022.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Xu Z, Zhang M, Wang Y, Chen R, Xu S, Sun
X, Yang Y, Lin Z, Wang S and Huang H: Gentiopicroside ameliorates
diabetic renal tubulointerstitial fibrosis via inhibiting the
AT1R/CK2/NF-κB pathway. Front Pharmacol. 13(848915)2022.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Martens-Uzunova ES, Olvedy M and Jenster
G: Beyond microRNA-novel RNAs derived from small non-coding RNA and
their implication in cancer. Cancer Lett. 340:201–211.
2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wang Y, Weng Q, Ge J, Zhang X, Guo J and
Ye G: tRNA-derived small RNAs: Mechanisms and potential roles in
cancers. Genes Dis. 9:1431–1442. 2022.PubMed/NCBI View Article : Google Scholar
|
|
29
|
George S, Rafi M, Aldarmaki M, ElSiddig M,
Al Nuaimi M and Amiri KMA: tRNA derived small RNAs-Small players
with big roles. Front Genet. 13(997780)2022.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Lu Z, Su K, Wang X, Zhang M, Ma S, Li H
and Qiu Y: Expression profiles of tRNA-derived small rnas and their
potential roles in primary nasopharyngeal carcinoma. Front Mol
Biosci. 8(780621)2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Li S, Xu Z and Sheng J: tRNA-derived small
RNA: A novel regulatory small non-coding RNA. Genes (Basel).
9(246)2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yu M, Lu B, Zhang J, Ding J, Liu P and Lu
Y: tRNA-derived RNA fragments in cancer: Current status and future
perspectives. J Hematol Oncol. 13(121)2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Guzzi N and Bellodi C: Novel insights into
the emerging roles of tRNA-derived fragments in mammalian
development. RNA Biol. 17:1214–1222. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Zhang Y, Bi Z, Dong X, Yu M, Wang K and
Song X, Xie L and Song X: tRNA-derived fragments: tRF-Gly-CCC-046,
tRF-Tyr-GTA-010 and tRF-Pro-TGG-001 as novel diagnostic biomarkers
for breast cancer. Thorac Cancer. 12:2314–2323. 2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Li D, Zhang H, Wu X, Dai Q, Tang S, Liu Y,
Yang S and Zhang W: Role of tRNA derived fragments in renal
ischemia-reperfusion injury. Ren Fail. 44:815–825. 2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Lee YS, Shibata Y, Malhotra A and Dutta A:
A novel class of small RNAs: tRNA-derived RNA fragments (tRFs).
Genes Dev. 23:2639–2649. 2009.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Pan Q, Han T and Li G: Novel insights into
the roles of tRNA-derived small RNAs. RNA Biol. 18:2157–2167.
2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Soares AR and Santos M: Discovery and
function of transfer RNA-derived fragments and their role in
disease. Wiley Interdiscip Rev RNA. 8(e1423)2017.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Zhu P, Lu J, Zhi X, Zhou Y, Wang X, Wang
C, Gao Y, Zhang X, Yu J, Sun Y and Zhou P: tRNA-derived fragment
tRFLys-CTT-010 promotes triple-negative breast cancer progression
by regulating glucose metabolism via G6PC. Carcinogenesis.
42:1196–1207. 2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhong F, Hu Z, Jiang K, Lei B, Wu Z, Yuan
G, Luo H, Dong C, Tang B, Zheng C, et al: Complement C3 activation
regulates the production of tRNA-derived fragments Gly-tRFs and
promotes alcohol-induced liver injury and steatosis. Cell Res.
29:548–561. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Haussecker D, Huang Y, Lau A, Parameswaran
P, Fire AZ and Kay MA: Human tRNA-derived small RNAs in the global
regulation of RNA silencing. RNA. 16:673–695. 2010.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Shen Y, Yu X, Zhu L, Li T, Yan Z and Guo
J: Transfer RNA-derived fragments and tRNA halves: Biogenesis,
biological functions and their roles in diseases. J Mol Med (Berl).
96:1167–1176. 2018.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Han Y, Peng Y, Liu S, Wang X, Cai C, Guo
C, Chen Y, Gao L, Huang Q, He M, et al: tRF3008A suppresses the
progression and metastasis of colorectal cancer by destabilizing
FOXK1 in an AGO-dependent manner. J Exp Clin Cancer Res.
41(32)2022.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Goodarzi H, Liu X, Nguyen HC, Zhang S,
Fish L and Tavazoie SF: Endogenous tRNA-derived fragments suppress
breast cancer progression via YBX1 displacement. Cell. 161:790–802.
2015.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Fabregat I, Moreno-Caceres J, Sanchez A,
Dooley S, Dewidar B, Giannelli G and Ten Dijke P: IT-LIVER
Consortium. TGF-β signaling and liver disease. FEBS J.
283:2219–2232. 2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Lear TB, Lockwood KC, Larsen M, Tuncer F,
Kennerdell JR, Morse C, Valenzi E, Tabib T, Jurczak MJ, Kass DJ, et
al: Kelch-like protein 42 is a profibrotic ubiquitin E3 ligase
involved in systemic sclerosis. J Biol Chem. 295:4171–4180.
2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Li L, Kang H, Zhang Q, D'Agati VD,
Al-Awqati Q and Lin F: FoxO3 activation in hypoxic tubules prevents
chronic kidney disease. J Clin Invest. 129:2374–2389.
2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Herman-Edelstein M, Scherzer P, Tobar A,
Levi M and Gafter U: Altered renal lipid metabolism and renal lipid
accumulation in human diabetic nephropathy. J Lipid Res.
55:561–572. 2014.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Wang W, Luo Y, Yang S, Zeng M, Zhang S,
Liu J, Han Y, Liu Y, Zhu X, Wu H, et al: Ectopic lipid
accumulation: Potential role in tubular injury and inflammation in
diabetic kidney disease. Clin Sci (Lond). 132:2407–2422.
2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Chen X, Han Y, Gao P, Yang M, Xiao L,
Xiong X, Zhao H, Tang C, Chen G, Zhu X, et al: Disulfide-bond A
oxidoreductase-like protein protects against ectopic fat deposition
and lipid-related kidney damage in diabetic nephropathy. Kidney
Int. 95:880–895. 2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Falconi M, Giangrossi M, Zabaleta ME, Wang
J, Gambini V, Tilio M, Bencardino D, Occhipinti S, Belletti B,
Laudadio E, et al: A novel 3'-tRNAGlu-derived fragment
acts as a tumor suppressor in breast cancer by targeting nucleolin.
FASEB J. 33:13228–13240. 2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Livingston MJ, Shu S, Fan Y, Li Z, Jiao Q,
Yin XM, Venkatachalam MA and Dong Z: Tubular cells produce FGF2 via
autophagy after acute kidney injury leading to fibroblast
activation and renal fibrosis. Autophagy. 1-22:2022.PubMed/NCBI View Article : Google Scholar : (Epub ahead of
print).
|
|
53
|
Majumder S, Ren L, Pushpakumar S and Sen
U: Hydrogen sulphide mitigates homocysteine-induced apoptosis and
matrix remodelling in mesangial cells through Akt/FOXO1 signalling
cascade. Cell Signal. 61:66–77. 2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Tang C, Han H, Yan M, Zhu S, Liu J, Liu Z,
He L, Tan J, Liu Y, Liu H, et al: PINK1-PRKN/PARK2 pathway of
mitophagy is activated to protect against renal
ischemia-reperfusion injury. Autophagy. 14:880–897. 2018.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Kong D, Zhang Z, Chen L, Huang W, Zhang F,
Wang L, Wang Y, Cao P and Zheng S: Curcumin blunts
epithelial-mesenchymal transition of hepatocytes to alleviate
hepatic fibrosis through regulating oxidative stress and autophagy.
Redox Biol. 36(101600)2020.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Rao P, Qiao X, Hua W, Hu M, Tahan M, Chen
T, Yu H, Ren X, Cao Q, Wang Y, et al: Promotion of
β-catenin/forkhead box protein O signaling mediates epithelial
repair in kidney injury. Am J Pathol. 191:993–1009. 2021.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Qiao X, Rao P, Zhang Y, Liu L, Pang M,
Wang H, Hu M, Tian X, Zhang J, Zhao Y, et al: Redirecting TGF-β
Signaling through the β-Catenin/Foxo complex prevents kidney
fibrosis. J Am Soc Nephrol. 29:557–570. 2018.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Ma MKM, Yung S and Chan TM: mTOR
inhibition and kidney diseases. Transplantation. 102 (2S Suppl
1):S32–S40. 2018.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Jimenez-Uribe AP, Gomez-Sierra T,
Aparicio-Trejo OE, Orozco-Ibarra M and Pedraza-Chaverri J:
Backstage players of fibrosis: NOX4, mTOR, HDAC and S1P; companions
of TGF-β. Cell Signal. 87(110123)2021.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Malik S, Suchal K, Khan SI, Bhatia J,
Kishore K, Dinda AK and Arya DS: Apigenin ameliorates
streptozotocin-induced diabetic nephropathy in rats via
MAPK-NF-κB-TNF-α and TGF-β1-MAPK-fibronectin pathways. Am J Physiol
Renal Physiol. 313:F414–F422. 2017.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Su J, Morgani SM, David CJ, Wang Q, Er EE,
Huang YH, Basnet H, Zou Y, Shu W, Soni RK, et al: TGF-β
orchestrates fibrogenic and developmental EMTs via the RAS effector
RREB1. Nature. 577:566–571. 2020.PubMed/NCBI View Article : Google Scholar
|