Introduction
Lung cancer is a major cause of cancer-related
deaths worldwide and has limited treatment options. The numbers of
patients with lung cancer has been increased by 51% in the world
since 1985(1). Recent advances in
the treatment of lung cancer have increased our understanding of
tumor progression and disease biology. However, its cure and
survival rates remain poor, especially in advanced lung cancer.
Therefore, strategies to overcome drug resistance and develop new
drugs are required to increase the applicability of current
treatments to a larger lung cancer population (2,3).
TNF-related apoptosis-inducing ligand (TRAIL) induces apoptosis via
interaction with the death receptors TRAILR1/death receptor (DR)4
and TRAILR2/DR5 in several types of cancer, but not in normal
cells. Despite the significant potential for use in cancer
treatment, the translation of TRAIL into clinical use has been
limited because of TRAIL resistance (4).
Oncogenic drivers in lung cancer are directed
against a large proportion of targeted therapies, which are more
prevalent in patients with only light exposure to tobacco smoke
(5). A large proportion of
glioblastoma patients continue to smoke or use e-cigarettes during
treatment, which may affect the efficacy of treatment (6). Nicotine is the primary component of
tobacco. Tobacco nicotine can lead to drug resistance during
glioblastoma chemotherapy (6).
Tobacco smoke induces LKB1/AMPK pathway deficiency by reducing
epithelial growth factor receptor-tyrosine kinase inhibitor
sensitivity in lung cancer (7).
Some studies show that lncRNAs might be useful drug
targets (8). A number of lncRNAs
play pivotal roles in lung cancer drug resistance (9). The lncRNA SNHGS is also involved in
cancers (10). To study whether
nicotine influences TRAIL resistance, the dysregulation of lncRNA
SNHGs in lung cancer tissues from smokers and nonsmokers was
screened. It was discovered that lncRNA SNHG5 was upregulated by
nicotine in lung cancer cells and that SNHG5 overexpression could
promote TRAIL resistance in lung cancer. The recruitment of
X-linked inhibitor of apoptosis protein (XIAP) by overabundant
cytoplasm SNHG5 regulates cleaved caspase-3 expression to influence
cell apoptosis. The results showed that SNHG5/XIAP can promote
TRAIL resistance in lung cancer by altering cleaved caspase-3
expression, which is pivotal for lung cancer treatment.
Materials and methods
Patients and samples
Lung cancer tissues were collected from the Chaohu
Hospital affiliated to Anhui Medical University. All patients
signed informed consent for using samples. Inclusion criteria were
primary patients with lung cancer receiving surgery as initial
treatment and all clinicopathologic data were collected. Smoking
patients had been smoking for at least 20 years and smoked 20
cigarettes a day before surgical operation. The human ethics and
research ethics committees of Anhui Medical University approved the
study (approval no. 202104001).
Cells culture and transfection
Lung cancer cell lines of A549, HCC827 were obtained
from Cell Culture Center, Chinese Academy of Medical Sciences. The
characteristics of A549 and HCC827 cell lines were identified by
STR analysis in 2020 and 2021 respectively. All experiments were
performed with mycoplasma-free cells. A549 and HCC827 cell lines
were cultured in RPMI 1640 (Invitrogen; Thermo Fisher Scientific,
Inc.) with 10% fetal calf serum (HyClone; Cytiva), streptomycin
(100 µg/ml) and penicillin (100 U/ml). SNHG5 siRNAs, XIAP siRNAs
and control siRNAs were purchased from Shanghai GenePharma Co.,
Ltd. The sequences of siRNA are in Table SI. A549 and HCC827 cells were
cultured in 6-well plates with 1x106 cells in 2 ml
complete medium for 24 h until they were 90% confluent. Cells were
transiently transfected with 50 nM SNHG5 siRNAs, XIAP siRNAs and
control siRNAs, respectively, using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's instructions. Following ~6 h of transfection at room
temperature, the medium was refreshed. The subsequent experiments
were performed after 48 h. Nicotine was obtained from Sigma-Aldrich
(cat. no. N3876). Nicotine was added to lung cancer cells with the
concentration 5 µg/ml or 10 µg/ml for an additional 24 h.
RNA isolation and reverse
transcription-quantitative (RT-q) PCR
SNHG5 was identified by RT-qPCR. Cells were seeded
at a density of 1 million cells per ml for each subculture and a
minimum of 2 million cells were used for each RNA extraction. As
following the manufacturer's protocols, total cells or tissues RNAs
were extracted with TRIzol® (Thermo Fisher Scientific,
Inc.). The quantity and purity of RNA were measured with a Thermo
Scientific NanoDrop 2000 spectrophotometer (Thermo Fisher
Scientific Inc.). Regarding the cDNA synthesis, 1 µg total RNA was
reverse transcribed with the oligo(dT)18 primer and Reverse
Transcriptase M-MLV (Takara Bio Inc.) following the manufacturer's
instructions. qPCR was conducted using QuantiTect SYBR Green PCR
kit (Takara Bio, Inc.) according to the manufacturer's protocol on
a StepOne Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The reaction conditions were as follows for Cq
values: 95˚C for 30 sec, followed with 40 cycles of 95˚C for 5 sec,
60˚C for 34 sec; and for melting curves: 95˚C for 15 sec, 60˚C for
1 min and 95˚C for 15 sec. All primers were designed using Primer
Premier 5.0 (Premier, Inc.). The primers were obtained from Sangon
Biotech Co., Ltd. (Table SII).
RNA expression was normalized to GAPDH or U6. RNA relative
expression levels were calculated by using the 2-ΔΔCq
method (Bio-Rad CFX manager software 3.1; Bio-Rad Laboratories,
Inc.) (11). The whole experiment
was repeated with two different sets of biological samples.
SNHG5 expression vector construction
and transfection
According to the instructions of the manufacturer,
total RNA in the cells was extracted by using Rneasy Mini kit
(Invitrogen; Thermo Fisher Scientific, Inc.). The first-strand
complementary DNA (cDNA) of SNHG5 was obtained by using Maxima TM
First Strand cDNA Synthesis kit (Fermentas; Thermo Fisher
Scientific, Inc.) and DNA was amplified using the primers (Table SII). RT-qPCR products and pcDNA3.1
vector were digested. The full-length sequence of SNHG5 was
inserted into pcDNA3.1 vector to obtain pcDNA3.1-SNHG5.
pcDNA3.1-SNHG5 and pcDNA3.1 empty vector were transfected in A549
and HCC827 cells as the overexpression group and control group,
respectively, using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) following the manufacturer's
instructions. Cells were plated at a density of 500,000 cells/10-cm
plate 2 days prior to transfection. A total of 1 µg
SNHG5-overexpression plasmid and vector were transfected into the 6
well-plate cells, respectively. Following ~6 h of transfection, the
medium was refreshed. After 48 h, the cells were used for the
subsequent experiments.
Cellular fractionation
Norgen's cytoplasmic and nuclear RNA purification
kits (Norgen Biotek Corp.) were used to extract nuclear and
cytoplasmic RNA.
Apoptosis detection
The cells were washed three times using
phosphate-buffered saline (PBS). The cells were stained using the
Annexin V/fluorescein isothiocyanate (FITC). The single cell
suspension was first incubated with 500 µl of 1X buffer, 5 µl of
Annexin V and 5 µl of Propidium Iodide (PI; Beyotime Institute of
Biotechnology) at 37˚C for 35 min in the dark. The cells were
detected by a FACSAria III flow cytometer (BD Biosciences) and
analyzed by Win MDI. 2.9 software (TSRI Flow Cytometry Core
Facility). The rate of apoptosis was defined as the total
percentage of early and late apoptotic cells.
Western blotting
Using RIPA lysis buffer (RIPA; Beyotime Institute of
Biotechnology) total protein was extracted from the cells. Using
bicinchoninic acid (BCA) method (Pierce; Thermo Fisher Scientific,
Inc.) the protein was quantified, and each sample (2 µg) was loaded
on 10% gels for SDS-PAGE. The protein was transferred on a
polyvinylidene difluoride membrane (Millipore Sigma). Then the
membranes were blocked with 5% skimmed milk for ~2 h at room
temperature and the membrane was soaked in primary antibodies
against pro Caspase-3 (1:1,000; cat. no. ab32150; Abcam), cleaved
Caspase-3 (1:500; cat. no. ab32042; Abcam) and GAPDH (1:1,000; cat.
no. bs-0755R; BIOSS) at 4˚C for 12 h and secondary antibodies (goat
anti-rabbit IgG H&L; 1:1,000; cat. no. bs-0295G; BIOSS) for 2 h
at room temperature. Bands were visualized by enhanced
chemiluminescence (Amersham ECL; cat. no. RPN3243; Cytiva) and
analyzed by ImageJ Software v1.53 (National Institutes of
Health).
RNA pull-down assay
SNHG5 and SNHG5-antisense were transcribed using the
templates of pcDNA3.1-SNHG5 and pcDNA3.1-SNHG5-antisense plasmids.
The plasmids were amplified. The primers are shown in the Table SI. Using Biotin-RNA Labeling Mix
(Roche Diagnostics) biotin-labeled RNAs were transcribed in vitro.
Then biotin-labeled RNAs were treated with RNase-free DNase I
(Takara Bio, Inc.) and purified with an RNeasy Mini kit (Roche
Diagnostics). RNA structure buffer [10 mM Tris (pH 7), 0.1 M KCl,
and 10 mM MgCl2] included biotinylated RNA was in 98˚C
for ~2 min, on ice for 5 min, and then kept at room temperature for
~35 min. Total protein lysates of A549 cells were mixed with
biotinylated RNA and incubated at room temperature. Streptavidin
agarose beads (5 µl; Cytiva; Thermo Fisher Scientific, Inc.) were
added to each binding reaction and further incubated at room
temperature with rotation. The RNA-protein-beads compound were then
centrifuged at 1,000 x g for 1 min at room temperature and washed
three times with Wash buffer I and dissolved into RNase-free water.
After washes, the pull-down complexes were eluted by denaturation
in 1X protein loading buffer for 10 min at 100˚C. The samples were
then assessed by western blotting or proceeded to mass spectrometry
analysis. The captures were given electrophoresis with 12% sodium
dodecyl sulfate polyacrylamide gel and silver stained. Mass
spectrometry (LC-MS/MS, A TripleTOF; AB Sciex Pte. Ltd.) identified
the protein bands after in-gel trypsin digestion.
RNA binding protein
immunoprecipitation (RIP) assay
Magna RIP RNA-binding protein immunoprecipitation
kit (Millipore Sigma) was used to perform RIP according to the
manufacturer's instructions in A549 cells induced by nicotine for
36 h. Antibody against XIAP obtained from Cell Signaling
Technology, Inc. The coprecipitated RNAs were adsorbed with
magnetic beads and detected by qPCR.
UV-RIP
A549 cells induced by nicotine for 36 h applied 100
mM 4-thiouridine (4-SU) were cultured for ~12 h. Then cells were
cross-linked by 365 nm UV light at a dose of 400 mJ/cm2
in UV-RIP assay. The other UV-RIP protocols were the same as
RIP.
Statistical analysis
The statistical analysis was performed with SPSS
software (version 15.0; SPSS, Inc.). The results were analyzed by
t-test or ANOVA followed by Bonferroni's test. Categorical
variables were analyzed by χ2 test. Data are presented
as the mean ± SEM. P<0.05 was considered to indicate a
statistically significant difference.
Results
SNHG5 expression is increased in lung
cancer tissues from smokers
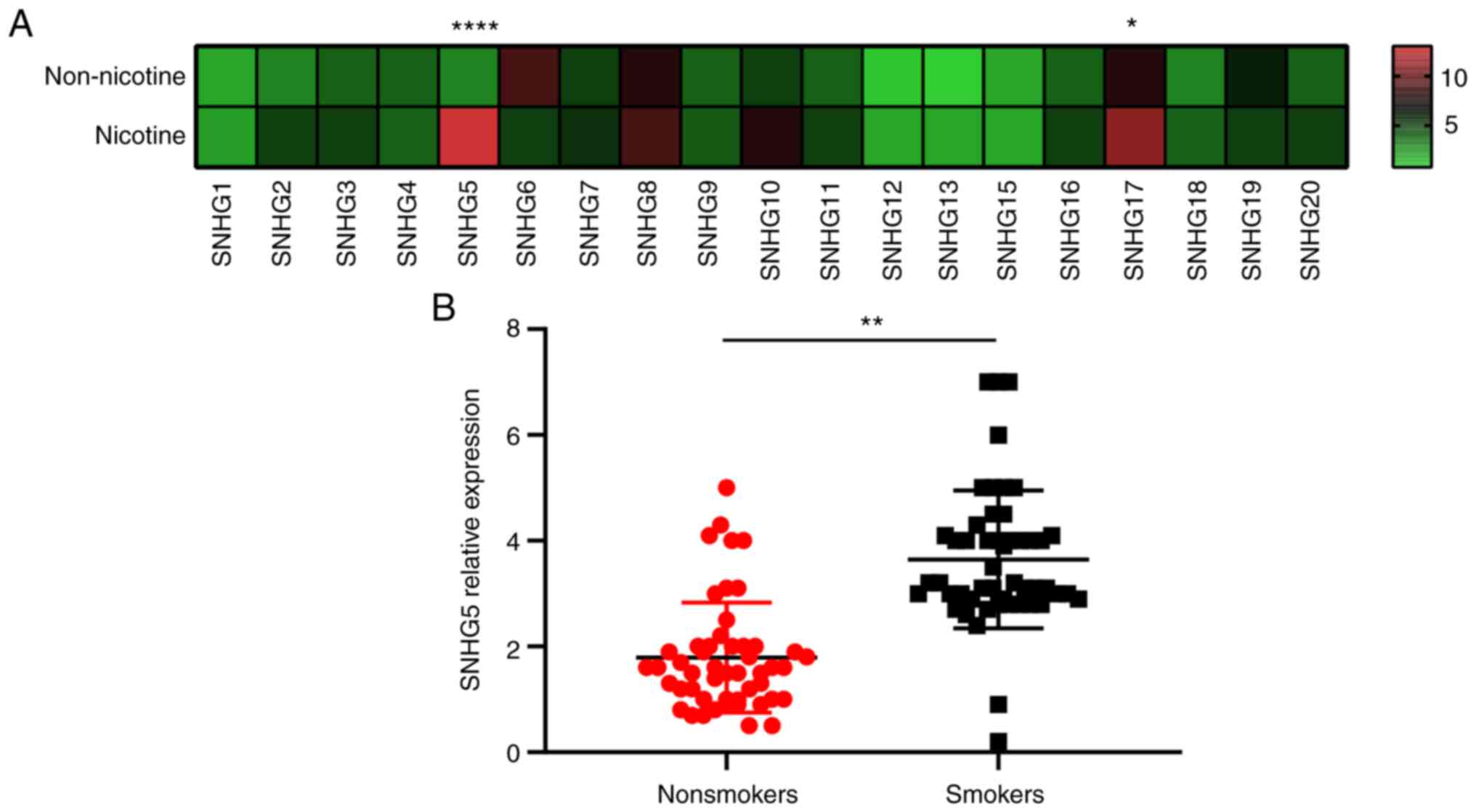
RT-qPCR was performed to identify the panel of SNHGs
in A549 cells. One group of cells was induced by nicotine and the
other group of cells was not induced. Among all the SNHGs tested,
nicotine induced increased expression of SNHG5 and SNHG17 in A549
cells, indicating that they might be important in lung cancer
(Fig. 1A). RT-qPCR was performed
on lung cancer samples from smokers (n=50) and nonsmokers (n=50). A
comparison between tissues from nonsmokers and smokers showed a
marked enhancement of SNHG5 expression and no increase in SNHG17
expression (Fig. 1B). There were
no significant correlations of SNHG5 expression with age, sex, or
the lung cancer tissue differentiation level of patients. However,
SNHG5 expression level was correlated with tumor node metastasis
stage (P=0.039), distant metastasis (P=0.040), lymph node
metastasis (P=0.002), and smoking (P=0.0004; Table SIII). These results suggested that
SNHG5 might play an important role in smokers with lung cancer.
SNHG5 is associated with TRAIL
resistance in lung cancer
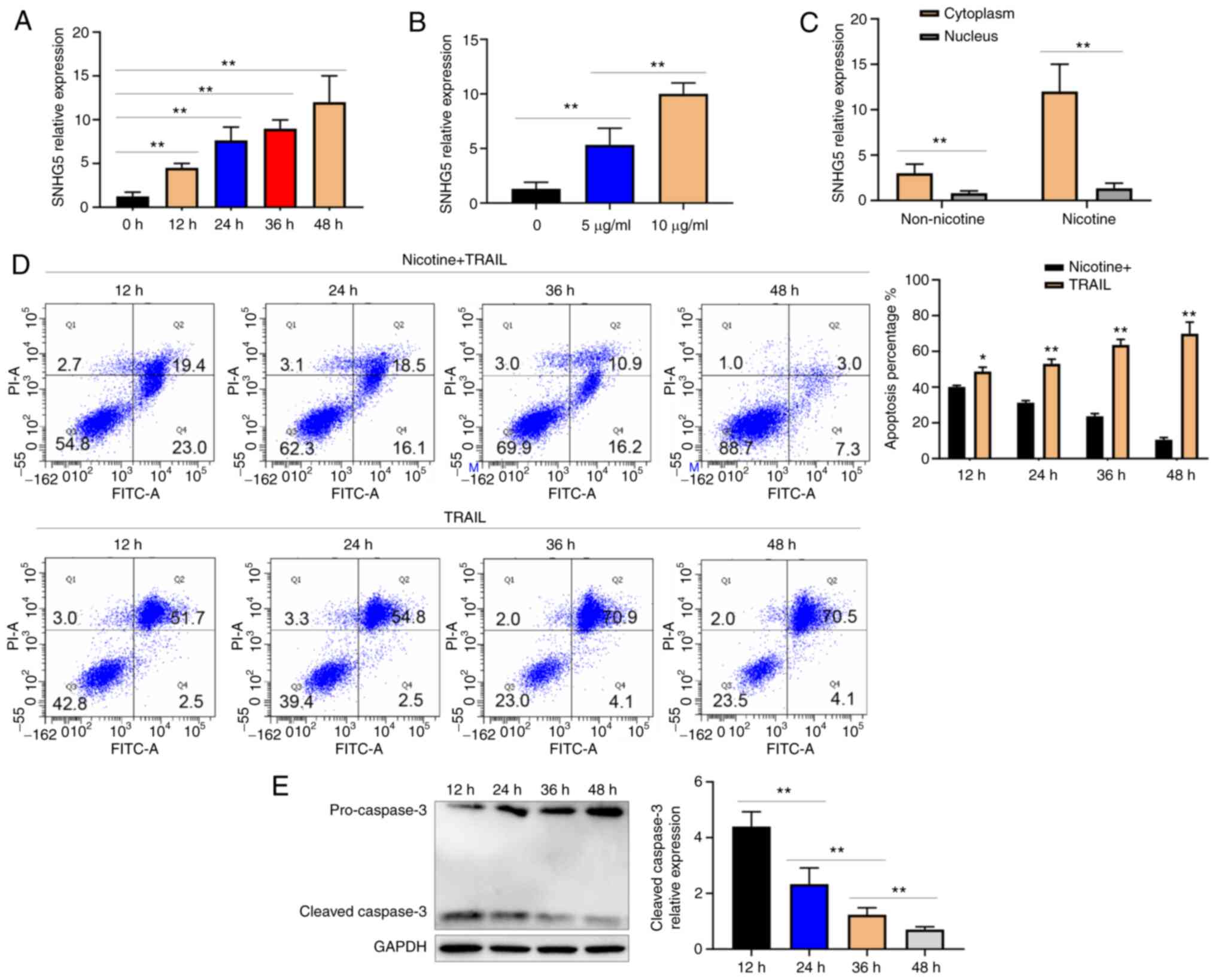
To further determine whether nicotine alters the
SNHG5 expression in vitro, the present study measured the SNHG5
expression following nicotine (10 µg/ml) induction in A549 cells at
different times. The cells were in good condition after nicotine
was applied. The results showed that SNHG5 expression increased
with nicotine induction in a time-dependent manner (Fig. 2A). The A549 cells were exposed to
different doses of nicotine (5 and 10 µg/ml) for 48 h. The SNHG5
expression increased in a dose-dependent manner (Fig. 2B). Next, the SNHG5 expression in
the cytoplasmic and nuclear fractions of RNA of A549 cells exposed
to nicotine (10 µg/ml) for 48 h was quantified. It was found that
the SNHG5 upregulation was greater among cytoplasmic RNA than
nuclear RNA (Fig. 2C). Nicotine
(10 µg/ml) and TRAIL (1.6 µg/ml) were then added to A549 cells, and
flow cytometry was performed. Flow cytometry showed that the
apoptosis rate gradually decreased with increasing SNHG5
expression. The addition of TRAIL (1.6 µg/ml) alone to A549 cells
did not decrease the apoptosis rate (Fig. 2D). It was also found that the
expression of cleaved caspase-3 decreased with increasing SNHG5
expression, as detected by western blotting (Fig. 2E). The results showed that nicotine
promoted the expression of SNHG5 and SNHG5-induced TRAIL resistance
in lung cancer.
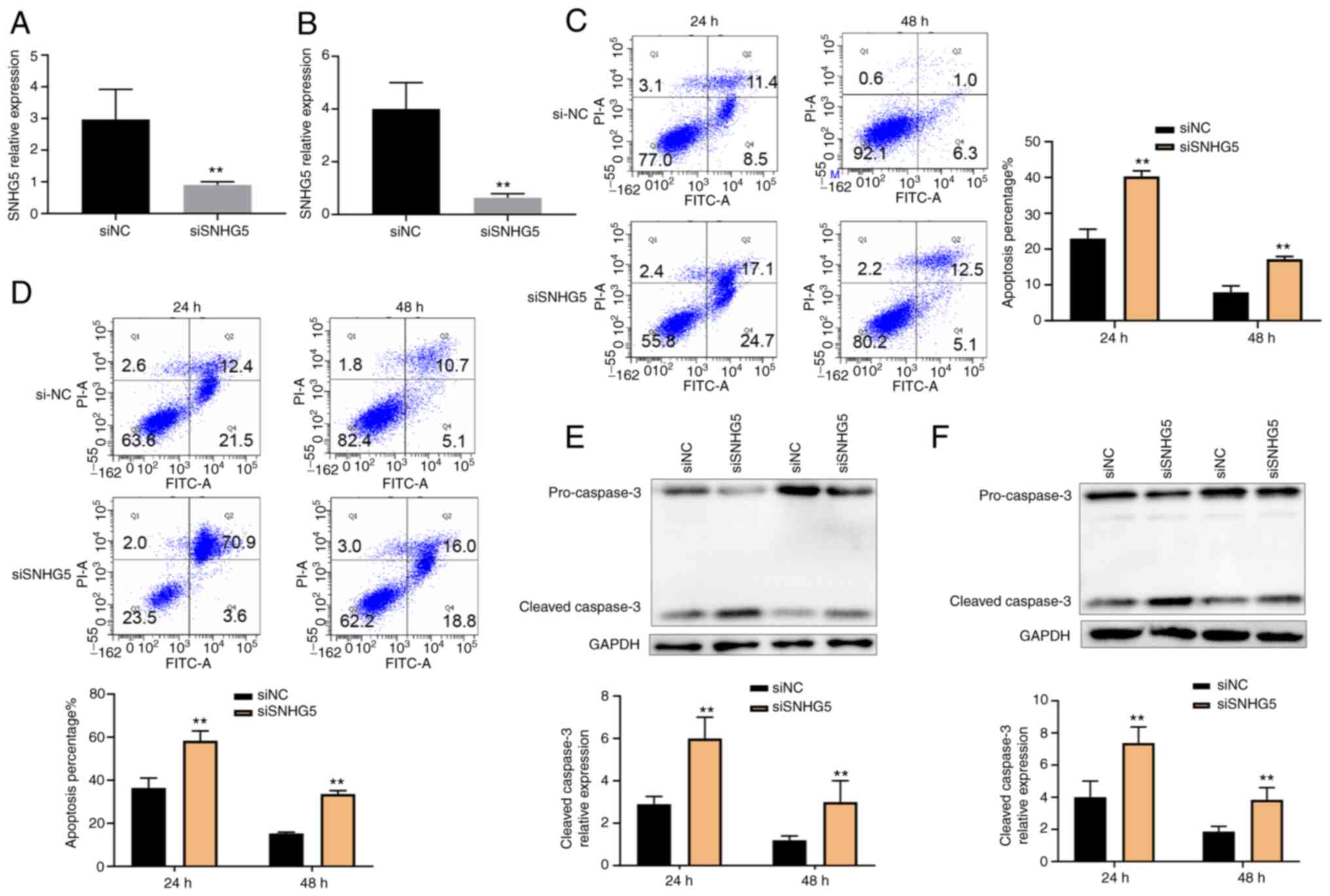
Knockdown of SNHG5 expression
decreases the TRAIL resistance in A549 and HCC827 cells
A549 and HCC827 cells were exposed to nicotine (10
µg/ml) and TRAIL (1.6 µg/ml). As shown in Fig. 3A and B, SNHG5 expression was significantly
knocked down in A549 and HCC827 cells by RT-qPCR of siRNA. The
siRNA sequences are presented in Table SI. SNHG5 knockdown resulted in a
markedly increased apoptosis rate in A549 and HCC827 cells as
measured by flow cytometry (Fig.
3C and D). Moreover, the
expression of cleaved caspase-3 was significantly increased with
decreasing SNHG5 expression (Fig.
3E and F). Collectively, these
data revealed a functional role of SNHG5 in TRAIL resistance.
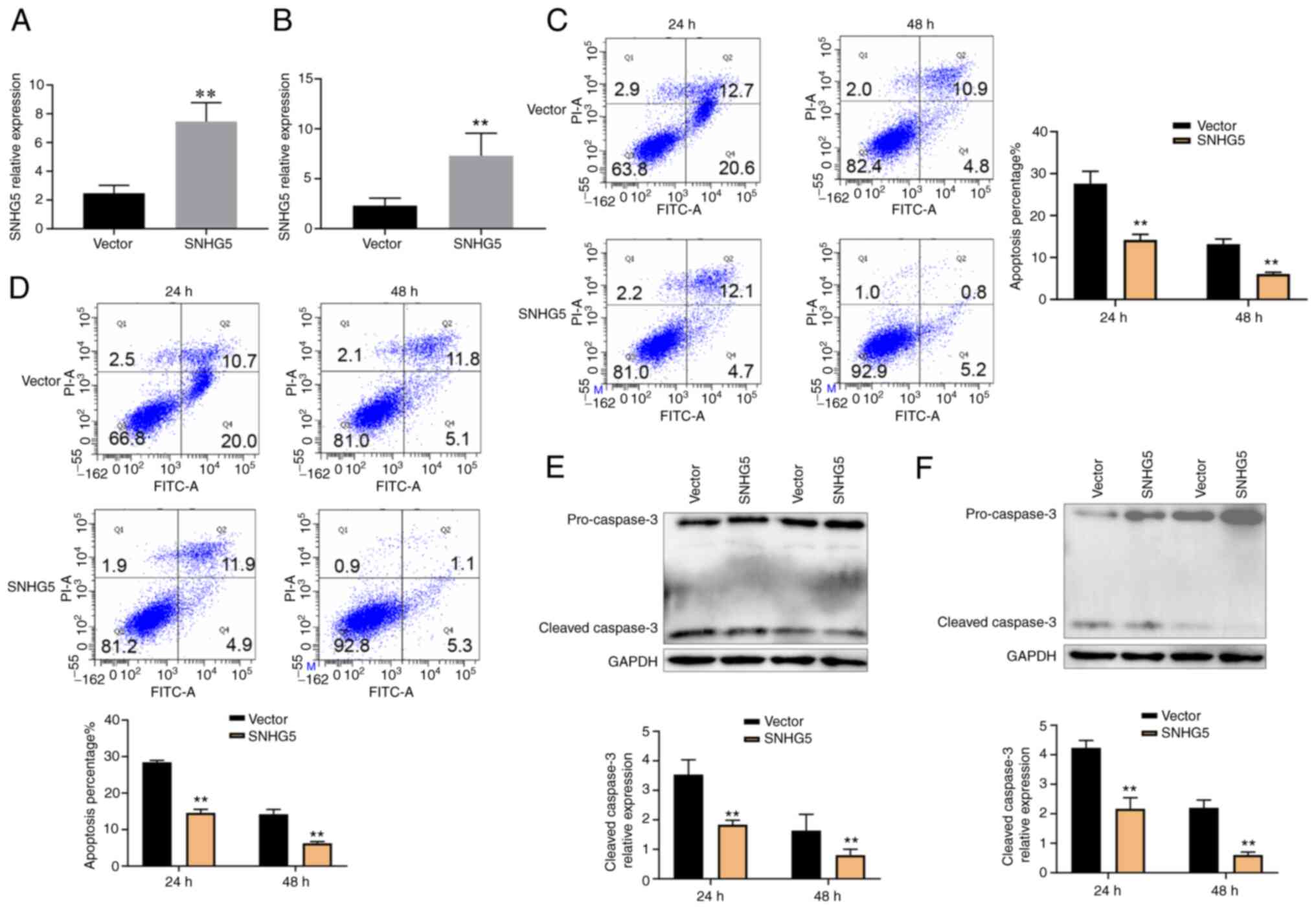
SNHG5 overexpression increases TRAIL
resistance in A549 and HCC827 cells
SNHG5 was overexpressed in A549 (Fig. 4A) and HCC827 (Fig. 4B) cells as revealed by RT-qPCR. The
cells were exposed to nicotine (10 µg/ml) and TRAIL (1.6 µg/ml) for
48 h. SNHG5 overexpression resulted in a significantly decreased
apoptosis rate in A549 and HCC827 cells by flow cytometry (Fig. 4C and D). The expression of cleaved caspase-3
decreased with increasing SNHG5 expression by western blotting
(Fig. 4E and F). Therefore, the results showed that
nicotine might promote SNHG5 overexpression, as well as suggested
an important role of SNHG5 in TRAIL resistance.
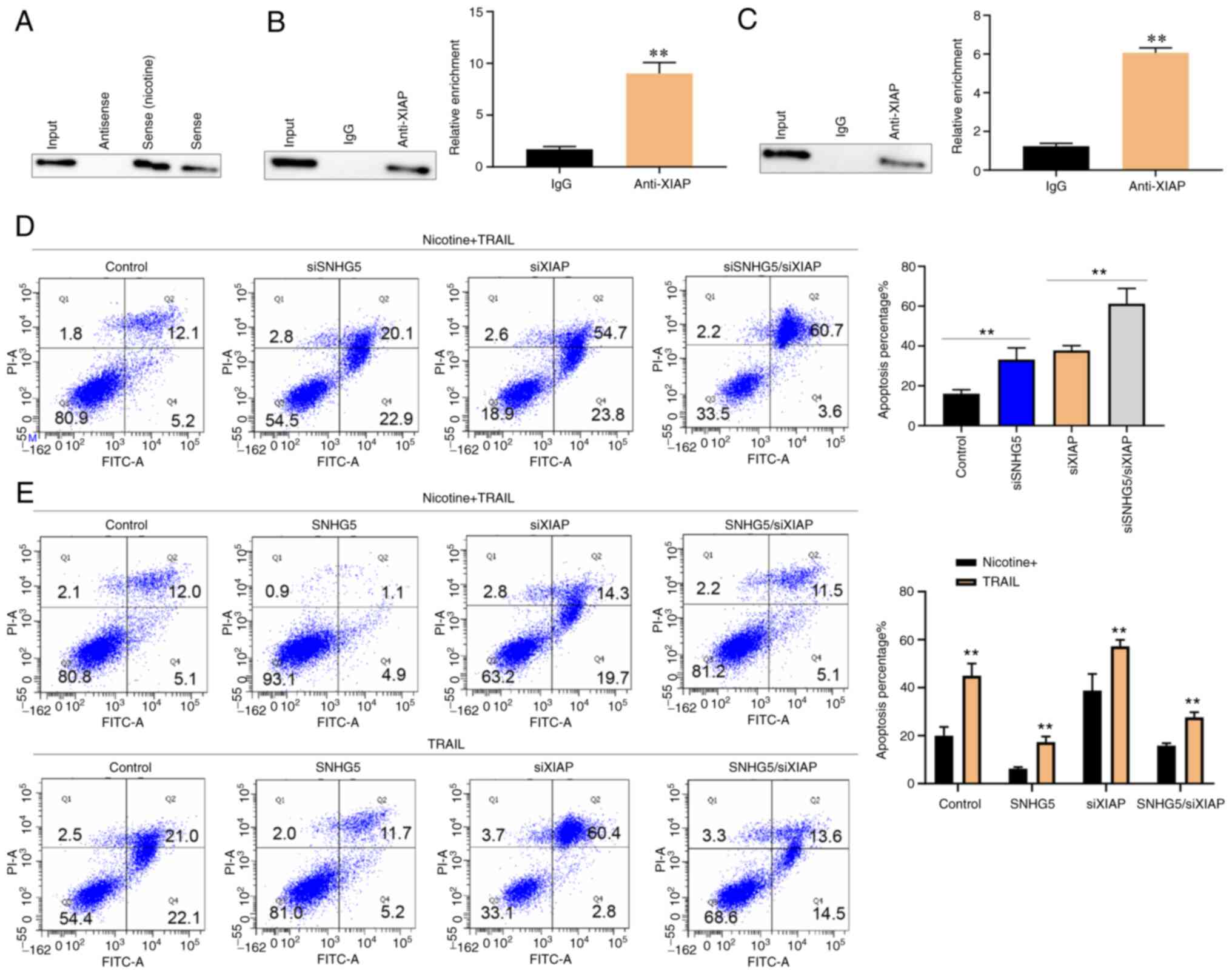
Cytoplasm SNHG5 directly interacts
with XIAP to regulate TRAIL resistance
The mechanistic insights into the mechanism and
interactions of SNHG5 were explored. As SNHG5 was simultaneously
located in the nucleus and cytoplasm, the possibility that SNHG5
functions by physically interacting with proteins was explored. A
biotinylated SNHG5 RNA pull-down assay and subsequent mass
spectrometry analysis of the differentially displayed bands
revealed that XIAP was the main protein bound to SNHG5. A number of
studies show that XIAP is involved in the apoptosis pathway
(12,13). The relationship between SNHG5 and
XIAP was studied using an immunoblot assay (Fig. 5A). RNA immunoprecipitation (UV-RIP)
assays with an XIAP-specific antibody confirmed a direct
interaction between SNHG5 and XIAP (Fig. 5B). Then, native RIP was performed
to confirm the binding between SHNG5 and XIAP (Fig. 5C). The interactive effects of SNHG5
and XIAP on the TRAIL resistance were evaluated using XIAP
knockdown in A549 cells. The cells were exposed to nicotine (10
µg/ml) and TRAIL (1.6 µg/ml) for 48 h. As shown in Fig. 5D, XIAP knockdown promoted A549 cell
apoptosis. In addition, apoptosis of SNHG5-knockdown A549 cells was
enhanced by XIAP knockdown. The TRAIL resistance in
SNHG5-overexpression A549 cells was rescued by XIAP siRNA with or
without nicotine treatment (Fig.
5E).
Discussion
In humans and other mammals, most small nucleolar
RNAs (snoRNAs) are located in the introns of protein-coding and
non-coding genes. Some previous studies assume that the host snoRNA
genes have no function and only carried the snoRNA-encoding
sequences in their introns. However, a number of SNHGs have been
found to play important roles in cancers (13-15).
The present study detected the SNHG expression in A549 cells
exposed to nicotine and found that SNHG5 expression was
significantly increased by nicotine treatment. The alterations
suggested that SNHG5 overexpression might be associated with
nicotine. The present study also found that SNHG5 expression was
significantly higher in lung cancer tissues from smokers compared
with those from nonsmokers. The results suggested that SNHG5
expression was related to important cell behaviors in lung cancer.
The present study also found that SNHG5 overexpression due to
nicotine treatment was associated with TRAIL resistance in lung
cancer. Tobacco nicotine plays a role in cancer drug resistance and
can inhibit cancer cell apoptosis induced by chemotherapeutic drugs
via increasing XIAP expression (16). Nicotine can suppress apoptosis by
inducing multi-site Bad phosphorylation (17); it can also activate Akt in a dose-
and time-dependent manner. Akt inhibitors can lead to a mild
reduction in chemotherapy-induced apoptosis, and the effects from
nicotine are blocked in A549 cells (18).
In the present study, SNHG5 directly interacted with
XIAP, which promoted TRAIL resistance by inhibiting the cleaved
caspase-3 expression in A549 and HCC827 cells. XIAP can bind
caspase-3 to suppress apoptosis (19-21).
The activity of processed caspases can be inhibited by XIAP, which
increases the threshold of apoptosis activation. Proapoptotic
factors, which are released from mitochondria, can inhibit XIAP.
The proapoptotic factors SMAC/DIABLO can bind and prevent XIAP
activity (22). High levels of
XIAP can explain inducible melanoma TRAIL resistance. SMAC is
released from the mitochondria and inhibits apoptosis by XIAP
(23). XIAP inhibitors promote
TRAIL-induced apoptosis and counter tumors in pancreatic cancer
(24,25). The present study showed that SNHG5
promoted TRAIL resistance, which explains the association with XIAP
in lung cancer. The results have significant implications regarding
our understanding of the roles of SNHG5.
SNHG5 was evidently increased in lung cancer cells
treated with nicotine. Tobacco nicotine induced SNHG5 expression
and promoted TRAIL resistance through SNHG5/XIAP. How SNHG5 and
XIAP to combine needs further detection. The present study provided
a new strategy to allow the use of TRAIL as a cancer treatment. A
small clinic sample size is the limitation of the present study.
Animal experiments are also required.
Supplementary Material
siRNA sequences.
Primers sequences.
Association between SNHG5 expression
and pathological features in 100 patients with lung cancer.
Acknowledgements
Not applicable.
Funding
Funding: This study was supported by a grant from the Anhui
Medical University Project Fund (grant no. 2021xkj188).
Availability of data and materials
All data generated and/or analyzed in the present
study are included in this published article.
Authors' contributions
XX, JX and XS collected and analyzed the data and
drafted the manuscript. XX and XS designed the project. CW, JD and
XS revised the manuscript. XX, JX, XS, CW and JD confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
The research protocol was reviewed and approved by
the Ethics Committee of Anhui Medical University (approval no.
202104001).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Akhtar N and Bansal JG: Risk factors of
lung cancer in nonsmoker. Curr Probl Cancer. 41:328–339.
2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Hirsch FR, Scagliotti GV, Mulshine JL,
Kwon R, Curran WJ Jr, Wu YL and Paz-Ares L: Lung cancer: Current
therapies and new targeted treatments. Lancet. 389:299–311.
2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Deng D and Shah K: TRAIL of hope meeting
resistance in cancer. Trends Cancer. 6:989–1001. 2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Middleton G, Fletcher P, Popat S, Savage
J, Summers Y, Greystoke A, Gilligan D, Cave J, O'Rourke N, Brewster
A, et al: The National lung matrix trial of personalized therapy in
lung cancer. Nature. 583:807–812. 2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
McConnell DD, Carr SB and Litofsky NS:
Potential effects of nicotine on glioblastoma and
chemoradiotherapy: A review. Expert Rev Neurother. 19:545–555.
2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Cheng FJ, Chen CH, Tsai WC, Wang BW, Yu
MC, Hsia TC, Wei YL, Hsiao YC, Hu DW, Ho CY, et al: Cigarette
smoke-induced LKB1/AMPK pathway deficiency reduces EGFR TKI
sensitivity in NSCLC. Oncogene. 40:1162–1175. 2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Liang WC, Ren JL, Wong CW, Chan SO, Waye
MM, Fu WM and Zhang JF: LncRNA-NEF antagonized epithelial to
mesenchymal transition and cancer metastasis via cis-regulating
FOXA2 and inactivating Wnt/β-catenin signaling. Oncogene.
37:1445–1456. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Liu MY, Li XQ, Gao TH, Cui Y, Ma N, Zhou Y
and Zhang GJ: Elevated HOTAIR expression associated with cisplatin
resistance in non-small cell lung cancer patients. J Thorac Dis.
8:3314–3322. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Yang Z, Li Q, Zheng X and Xie L: Long
Noncoding RNA small nucleolar host gene: A potential therapeutic
target in urological cancers. Front Oncol.
11(638721)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Tu H and Costa M: XIAP's profile in human
cancer. Biomolecules. 10(1493)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zimta AA, Tigu AB, Braicu C, Stefan C,
Ionescu C and Berindan-Neagoe I: An emerging class of long
Non-coding RNA with oncogenic role arises from the snoRNA host
genes. Front Oncol. 10(389)2020.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Qin Y, Sun W, Wang Z, Dong W, He L, Zhang
T and Zhang H: Long Non-Coding small nucleolar RNA host genes
(SNHGs) in endocrine-related cancers. Onco Targets Ther.
13:7699–7717. 2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Yang T and Shen J: Small nucleolar RNAs
and SNHGs in the intestinal mucosal barrier: Emerging insights and
current roles. J Adv Res: Jun 11, 2022 (Epub ahead of print).
|
|
16
|
Dasgupta P, Kinkade R, Joshi B, Decook C,
Haura E and Chellappan S: Nicotine inhibits apoptosis induced by
chemotherapeutic drugs by up-regulating XIAP and survivin. Proc
Natl Acad Sci USA. 103:6332–6337. 2006.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Jin Z, Gao F, Flagg T and Deng X: Nicotine
induces multi-site phosphorylation of Bad in association with
suppression of apoptosis. J Biol Chem. 279:23837–23844.
2004.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhang J, Kamdar O, Le W, Rosen GD and
Upadhyay D: Nicotine induces resistance to chemotherapy by
modulating mitochondrial signaling in lung cancer. Am J Respir Cell
Mol Biol. 40:135–146. 2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Riedl SJ, Renatus M, Schwarzenbacher R,
Zhou Q, Sun C, Fesik SW, Liddington RC and Salvesen GS: Structural
basis for the inhibition of caspase-3 by XIAP. Cell. 104:791–800.
2001.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Von Karstedt S, Montinaro A and Walczak H:
Exploring the TRAILs less travelled: TRAIL in cancer biology and
therapy. Nat Rev Cancer. 17:352–366. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Jost PJ, Grabow S, Gray D, McKenzie MD,
Nachbur U, Huang DC, Bouillet P, Thomas HE, Borner C, Silke J, et
al: XIAP discriminates between type I and type II FAS-induced
apoptosis. Nature. 460:1035–1039. 2009.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Jost PJ and Vucic D: Regulation of cell
death and immunity by XIAP. Cold Spring Harb Perspect Biol.
12(a036426)2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Eberle J: Countering TRAIL resistance in
melanoma. Cancers (Basel). 11(656)2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Vogler M, Walczak H, Stadel D, Haas TL,
Genze F, Jovanovic M, Bhanot U, Hasel C, Möller P, Gschwend JE, et
al: Small molecule XIAP inhibitors enhance TRAIL-induced apoptosis
and antitumor activity in preclinical models of pancreatic
carcinoma. Cancer Res. 69:2425–2434. 2009.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Stadel D, Mohr A, Ref C, MacFarlane M,
Zhou S, Humphreys R, Bachem M, Cohen G, Möller P, Zwacka RM, et al:
TRAIL-induced apoptosis is preferentially mediated via TRAIL
receptor 1 in pancreatic carcinoma cells and profoundly enhanced by
XIAP inhibitors. Clin Cancer Res. 16:5734–5749. 2010.PubMed/NCBI View Article : Google Scholar
|