Introduction
Endometrial carcinoma/cancer (EC) is a common
reproductive system malignancy in menopausal and perimenopausal
women, ranking fourth for incidence among female malignant tumors
in developed countries (1). The
incidence of EC has increased in recent years, threatening the
health of women globally (2,3).
Once EC has progressed to an advanced stage, the 5-year survival
rate is low (4-6).
Early-stage EC is rarely found by physical examination, as once the
uterus or its appendages are palpated with abnormal masses, they
are generally in the advanced stage of EC (7,8). The
occurrence and development of EC is a result of the interaction of
environmental factors and genetic variation, involving a series of
components such as differential expression of various genes and
transcription factors, abnormal regulation of cell signal
transduction pathways and imbalance of the cellular
microenvironment homeostasis (9,10).
Different pathological changes and molecular features determine the
risk level and prognosis of patients with EC (11). To date, the molecular mechanisms
regulating the tumorigenesis, development and metastasis of EC have
not been elucidated fully. Identifying these mechanisms is
important for the identification of key therapeutic targets.
Centrosome amplification, resulting in multiple
centrosomes, occurs frequently in EC cells (12). This process can promote the growth
of several tumors by inducing chromosome mis-segregation and
regulation of the microtubule cytoskeleton, resulting in enhanced
directed migration and invasion of malignant cells (13). Centromere coiled-coil protein 110
(CCP110, also known as CP110) is an evolutionarily conserved
protein that is important for centrosome function (14,15).
Overexpression of CCP110 leads to centrosome expansion and
increases the invasive phenotype of cells (14,16).
CCP110 is involved in docetaxel-induced drug resistance of breast
cancer cells (17) and mediates
metastasis in prostate cancer (18). In addition, inhibition of
cyclin-dependent kinase 2 can downregulate CCP110 expression,
causing anaphase catastrophe and inhibiting lung cancer cell growth
(19). However, to the best of our
knowledge, the role of CCP110 in the tumorigenesis and development
of EC has not yet been reported.
MicroRNAs (miRNAs/miRs) are a class of
evolutionarily conserved, non-coding, single-stranded small RNAs
(~22 nucleotides) that can degrade or inhibit the translation of
target mRNAs via specific binding (20,21).
MiRNAs regulate cell proliferation, differentiation and apoptosis
by controlling the expression of certain signaling molecules,
including transcription factors, cytokines, growth factors and
pro-apoptotic and anti-apoptotic genes (22). Increasing numbers of miRNAs have
been shown to play important roles in EC (23-25).
Next-generation sequencing and miRNA screening technologies have
revealed that several miRNAs are differentially expressed between
EC and normal endometrial cells (25). For example, miR-103, -106a, -107,
-185 and -429 are upregulated in EC and are involved in
tumorigenesis, invasion and metastasis, whereas miR-30c, -152, -193
and -221 are downregulated in EC (26,27).
These miRNAs participate in various cell signaling pathways by
targeting downstream genes and play a regulatory role in the
proliferation, migration, invasion and apoptosis of EC cells
(28,29). MiR-129 also reportedly plays a role
in EC (30,31).
Long non-coding RNAs (lncRNAs) are defined as RNA
molecules that exceed 200 nucleotides and are not translated into
proteins (32). These RNAs can
regulate the expression of target genes by participating in
epigenetic, transcriptional and post-transcriptional pathways, and
are related to the proliferation, migration and invasion of various
tumors, such as female reproductive or gynecological cancers
(33,34). Notably, abnormally expressed
lncRNAs play an important role in the development of EC (35-37).
X-inactive-specific transcript (XIST), one of the first lncRNAs to
be discovered, is located at the X inactivation center of
chromosome Xq13.2 and plays a prominent role in X inactivation
(38). XIST is involved in the
occurrence and development of several disorders, including
pulmonary fibrosis, inflammation, neuropathic pain, cardiomyocyte
hypertrophy and osteoarthritic chondrocyte differentiation
(39). It also plays an important
regulatory function in various types of cancer, and can be used as
a diagnostic. It can also function as a prognostic biomarker and
potential therapeutic target for brain tumors, leukemia and lung,
breast and liver cancer (40).
There are few reports of the involvement of XIST in EC and the
detailed mechanism remains to be determined, and thus, this was
investigated in the current study.
The present study examined the expression levels of
CCP110, XIST and miR-129-2-3p in human EC tissues and cell lines,
and explored the targeting relationship between these species and
their functional roles in the proliferation, migration and invasion
of EC cells.
Materials and methods
Ethical statements and tissue
samples
A total of 19 female patients (age range, 37-77
years; mean age, 55.58±10.26 years) with EC who underwent surgical
excision at the First Affiliated Hospital of Jinan University
(Guangzhou, China) were enrolled in the study between March 2006
and May 2008. Inclusion criteria: Patients had been admitted to the
hospital for the first time and did not receive any malignant tumor
treatments, such as chemotherapy or radiotherapy, before surgery;
postoperative tissues were diagnosed by pathologists as EC with
parallel International Federation of Gynecology and Obstetrics
staging; patients were aware of and agreed to the whole process of
the study; and patient clinical data and follow-up data were
complete and available. Exclusion criteria: Patients with other
malignant tumors; patients with immune system diseases or
infectious diseases; and patients with severe impairment of the
heart, liver, kidney and other organ functions. EC tumor samples
and adjacent non-tumor tissues (distance from cancer tissue >2
cm) were collected immediately after the excision and were frozen
quickly in liquid nitrogen (-320˚F). For the analysis of overall
survival, follow-up of all patients was carried out for at least 8
years.
Immunohistochemistry (IHC)
For IHC assay, tissue sections (5-µm) were
deparaffined, rehydrated, and incubated for 1 h at 80˚C in citrate
buffer (10 mM; pH 6.0) with 0.05% Tween 20 for antigen retrieval.
After saturation for 30 min with 1% BSA (cat. no. #ST023-50g;
Beyotime Institute of Biotechnology) in PBS at room temperature,
the sections were incubated with CCP110 antibody (1:1,000; cat. no.
ab99337; Abcam) overnight at 4˚C, followed by incubation with
biotin-linked anti-rabbit IgG (cat. no. #E043201-6; 1:1,000;
Agilent Technologies, Inc.) for 1 h at room temperature and then
with the ABC complex (cat. no. #ab8647; Abcam) at 37˚C for 20 min.
IHC scores were determined as follows: Cells with <10% staining
were scored as negative staining (-, 1); 10-49% staining as (+, 2);
50-74% staining as (++, 3); 75-100% staining as (+++, 4). The
staining color was scored as light-yellow particle (score 1),
brown-yellow particle (score 2) and brown particle (score 3).
Number score times the color score was used as the final score
(41). A score value ≤5 was
grouped as low expression, while a score >5 was grouped as high
expression.
Cell culture and transfection
The HEC-1B and Ishikawa human EC cell lines were
procured from the Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences. Both cell lines were cultured in RPMI
1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.),
supplemented with 10% FBS (cat. no. #10270-106; Gibco; Thermo
Fisher Scientific, Inc.) and 100 U/ml penicillin and streptomycin
(cat. no. #15140122; Gibco; Thermo Fisher Scientific, Inc.) and
were maintained at 37˚C in a humid atmosphere containing 5%
CO2.
The CCP10 and XIST genes were cloned
and inserted into the pcDNA3.1 vector (cat. no. XY8014; Xinyu
Biology) The small interfering RNA (siRNA) targeting CCP110
(siCCP110), non-targeting negative control (NC) siRNA, miR-129-2-3p
mimics, miR-129-2-3p inhibitors, NC mimic, and NC inhibitor were
synthesized by Shanghai GenePharma Co., Ltd. For transfection,
Lipofectamine® 2000 (cat. no. #11668027; Invitrogen;
Thermo Fisher Scientific, Inc.) was used according to the
manufacturer's instructions with >1.3 µg nucleic acid. Cells
were harvested 48 h after transfection for total RNA or protein
isolation. The oligonucleotide sequences were as follows: NC,
5'-CCCACCAGUUUGAGACUCCACAAAU-3'; siCCP110,
5'-AAGACGUUCCAGGACAUCAUC-3'; miR-NC, 5'-UUGUCCGAACGUGUCACGUTT-3';
miR-129-2-3p mimics, 5'-AAGCCCUUACCCCAAAAAGCAU-3'; miR-NC
inhibitor, 5'-CACUACUUUUGUGUAGUACAA-3'; miR-129-2-3p inhibitors,
5'-AUGCUUUUUGGGGUAAGGGCUU-3'.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from tissues or cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's instructions. EasyScript
cDNA Synthesis SuperMix (cat. no. #AE301-02; Beijing Transgen
Biotech Co., Ltd.) was used for reverse transcription of miRNA,
according to the manufacturer's instructions. The RT-qPCR analysis
was performed using the LightCycler FastStart DNA MasterPlus SYBR
Green I kit (Roche Diagnostics), as per the manufacturer's
instructions. The following primers were used: Human CCP110
(NM_001323570.2; forward, 5'-AGAGGTGGAGTCGGGGTGGCAG-3'; reverse,
5'-CCCGCCATATTACGGATCTCAGGCT-3'); actin (forward,
5'-TCACCCACACTGTGCCCATCTACGA-3'; reverse,
5'-CAGCGGAACCGCTCATTGCCAATGG-3'), miR-129-2-3p (MIMAT0004605;
forward, 5'-TTCCAAGCCCTTACCCCA-3'; reverse,
5'-CACTTCCTCAGCACTTGTTCCTAT-3'), XIST (NR_001564; forward,
5'-TCAGCCCATCAGTCCAAGATC-3'; reverse,
5'-CCTAGTTCAGGCCTGCTTTTCAT-3') and U6 (forward,
5'-CTCGCTTCGGCAGCACA-3'; reverse, 5'-AACGCTTCACGAATTTGCGT-3').
Conditions for PCR were as follows: 1 cycle of 94˚C for 3 min and
38 cycles of 94˚C for 20 sec, 60˚C for 30 sec and 68˚C for 30 sec.
The expression level of miR-129-2-3p was normalized to that of
U6, whereas those of XIST and CCP110 were normalized
to the expression level of actin, using the 2-ΔΔCq
method (42). All PCRs were
repeated at least three times.
Cell Counting Kit-8 (CCK-8) assay
Adherent cells in the logarithmic growth phase were
trypsinized and counted, and an appropriate number of cells were
seeded into 96-well plates (in triplicate) and cultured at 37˚C for
24 h. After treatment with miR-129-2-3p mimics and inhibitors (or
the negative controls) for various time points (0, 24, 48, 72 and
96 h), the cells were incubated with 10 µl of CCK-8 reagent
(Dojindo Molecular Technologies, Inc.) for 1 h, and then the
absorbance was measured at 450 nm to evaluate cell
proliferation.
Transwell cell migration and invasion
assay
Cell migration and invasion were examined via a
Transwell assay (Corning Inc.). Briefly, cells were trypsinized and
counted, and then 1x105 cells were resuspended in
serum-free culture medium (RPMI 1640 medium) and seeded into the
upper chamber of a Transwell plate (6.5-mm insert with a 8-µm pore
size polycarbonate membrane). The lower chamber was loaded with
culture medium (RPMI 1640 medium) containing 20% FBS. After 24 h of
culture at 37˚C, cells that had migrated into the lower chamber
were fixed with 100% methanol for 10 min at room temperature and
stained with 0.1% crystal violet for 10 min at room temperature.
The cells were then observed using an inverted microscope with
bright field and five random fields were counted. For the invasion
assay, the chamber was paved with a layer of Matrigel (BD
Biosciences) at 2.5 mg/ml at room temperature for 4 h before cell
culture.
Cell colony formation assay
Ishikawa and HEC-1B cells were trypsinized and
resuspended, then 2,000 cells were seeded into six-well plates in
triplicate and cultured in a humidified atmosphere containing 5%
CO2 at 37˚C for 14 days. Subsequently, the cell colonies
were washed with PBS, fixed for 30 min with 100% methanol, and
stained for 20 min with 0.1% crystal violet (1 mg/ml; Beyotime
Institute of Biotechnology) at room temperature. One colony means a
visible staining spot counted by the naked eye. The mean colony
numbers were then calculated. A colony was a staining spot visible
to the naked eye.
Flow cytometry assay
To detect apoptosis, EC cells were stained with the
Annexin V-FITC/PI Dual Staining kit (cat. no. #640914; Biolegend,
Inc.) and then subjected to flow cytometric analysis. Briefly,
cells were digested by 0.5% trypsin at 37˚C for 2 min and then a
100 µl aliquot of the cell suspension was transferred to a
microcentrifuge tube and mixed with 5 µl of Annexin V-FITC (1
mg/ml) and 5 µl of propidium iodide (2.5 mg/ml). The sample was
then vortexed for 10 sec at room temperature and incubated for 15
min at room temperature in the dark. Subsequently, binding buffer
(400 µl) was added and the samples were analyzed using BD FACSDiva
software 7.0 (BD Biosciences).
Western blotting
Tissues or cells were lysed with RIPA lysis buffer
(cat. no. #P0013C; Beyotime Institute of Biotechnology). The
concentration of proteins was determined by BCA kit (cat. no.
#P0012; Beyotime Institute of Biotechnology). The proteins (30 µg
in each lane) were separated using 10% SDS-PAGE and transferred to
PVDF membranes (cat. no. #IPVH00010; MilliporeSigma). The membranes
were blocked with 5% skimmed milk solution at room temperature for
1 h and then incubated with the following primary antibodies at 4˚C
overnight: Anti-CCP110 (1:1,000; cat. no. ab99337; Abcam) and
anti-GAPDH (1:2,000; cat. no. ab8245; Abcam). Subsequently, the
membranes were incubated with goat anti-rabbit (cat. no. #A0208)
and anti-mouse (cat. no. #A0216) HRP-conjugated secondary
antibodies (1:1,000; Beyotime Institute of Biotechnology) at room
temperature for 1 h, and blots were visualized using an ECL kit
(cat. no. #P0018S; Beyotime Institute of Biotechnology). The gray
density was determined by ImageJ Version 1.50 (National Institutes
of Health) and the CCP110 signal was normalized to that of the
loading control (GAPDH). Each experiment was performed at least
three times.
Bioinformatics analysis and
dual-luciferase reporter assay
Potential binding sites for miR-129-2-3p and
CCP110/XIST were predicted using the TargetScan (43), miRDB (44), StarBase (45) and picTar (46) online software packages. The 3' UTR
fragments of CCP110 and XIST containing potential binding
sites for miR-129-2-3p were cloned into the pGL3 vector (Promega
Corporation) to construct wild-type (WT) and mutant (MUT) reporter
vectors. The vectors were then co-transfected by Lipofectamine 2000
(Thermo Fisher Scientific, Inc.) with the miR-129-2-3p mimics or
miR-NCs (as aforementioned) into EC cells. Cells were collected 48
h later and the luciferase activity was determined using a
dual-luciferase reporter assay system (Promega Corporation). The
activity of firefly luciferase was normalized to that of
Renilla luciferase.
Statistical analysis
The data are presented as the mean ± standard
deviation and the statistical analysis was performed using SPSS
version 16 (SPSS Inc.). Kaplan-Meier survival curves were used to
analyze the relationship between the expression levels of
miRNA-129-2-3p and the overall survival of the patients.
Differences between two groups or among multiple groups were
analyzed using Student's t-test (paired for tissue analysis and
unpaired for the others) or one-way ANOVA (with Tukey's post hoc
test where appropriate), respectively. The expression between
cancer tissue and adjacent normal tissue was compared using a
non-parametric Wilcoxon signed rank test. Spearman's rank
correlation coefficient was used to measure the statistical
dependence between two variables. Kaplan-Meier and log rank tests
were used to analyze the survival curve. P<0.05 was considered
to indicate a statistically significant difference.
Results
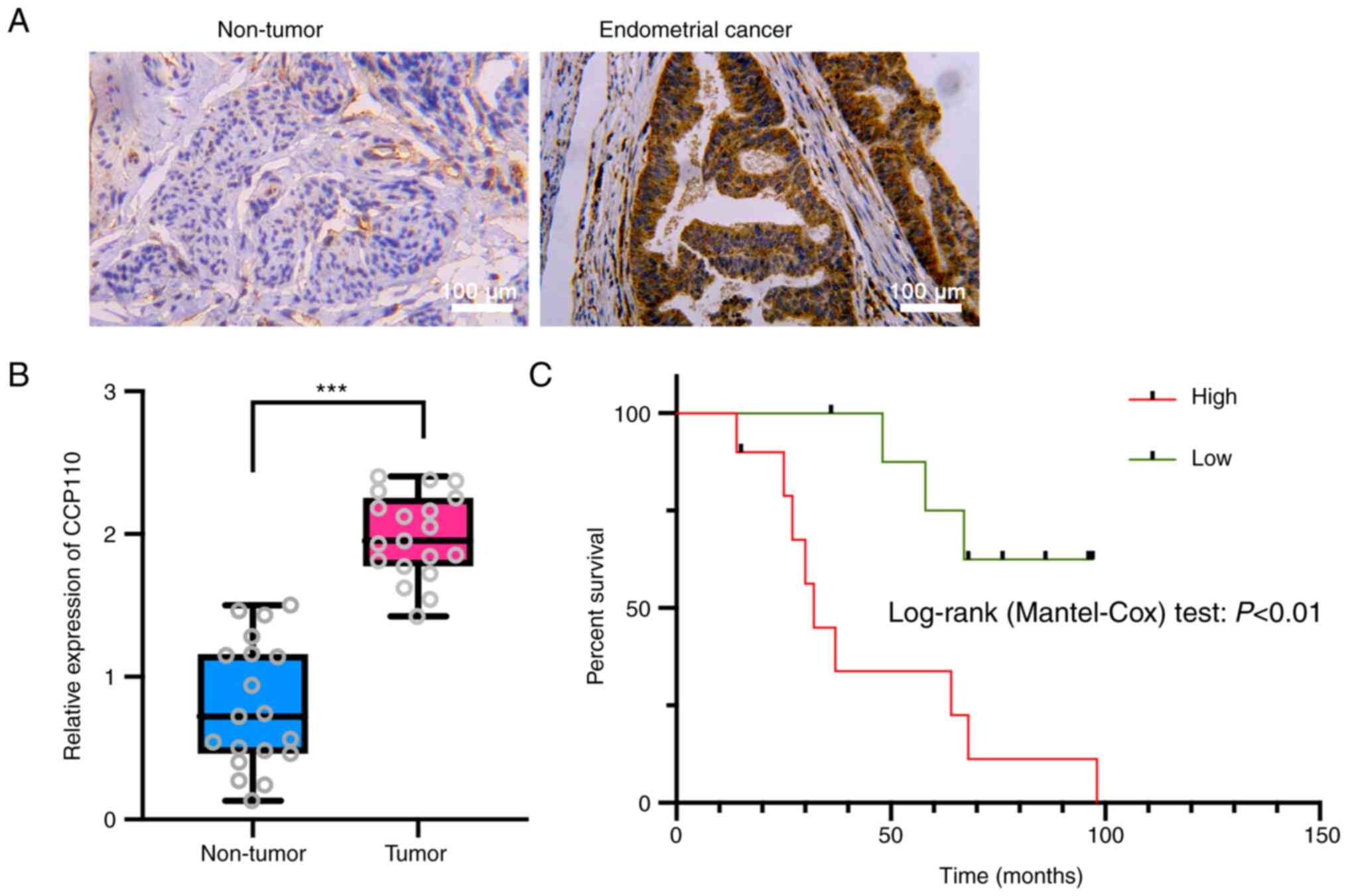
CCP110 is upregulated in EC
Cancerous and paracancerous tissues were collected
from 19 patients with EC and the expression level of CCP110 was
detected using immunohistochemistry. CCP110 expression was
significantly upregulated in cancer tissues compared with adjacent
tissues (Fig. 1A and B), and the overall survival of patients
with high CCP110 expression was significantly shorter compared with
that of patients with low CCP110 expression (Fig. 1C). These findings suggested that
CCP110 was a carcinogenic factor that affected the prognosis of
patients with EC.
Genetic knockdown of CCP110 induces
apoptosis and suppresses the development of EC cells
To examine the role of CCP110 in EC further, the
Ishikawa and HEC-1B EC cell lines were transfected with a NC siRNA
or a siRNA targeting CCP110 (siCCP110). The expression of the
protein was measured using western blotting. Compared with the NC,
treatment of both cell lines with siCCP110 inhibited expression of
the CCP110 protein significantly (Fig.
2A and B and Fig. S1). CCK-8 and Transwell assays
showed that knockdown of CCP110 significantly suppressed the
proliferation, migration and invasion of both cell lines (Fig. 2C-G). In addition, a colony
formation assay revealed that treatment with siCCP110 significantly
inhibited the colony formation ability of both cell lines (Fig. 2H and I). Next, flow cytometry assay was used to
examine apoptosis, which revealed that silencing of CCP110
significantly upregulated the apoptosis rate in both cell lines
(Fig. 2J and K). Overall, these results indicated that
genetic knockdown of CCP110 induced apoptosis and suppressed the
growth, migration and invasion of EC cells.
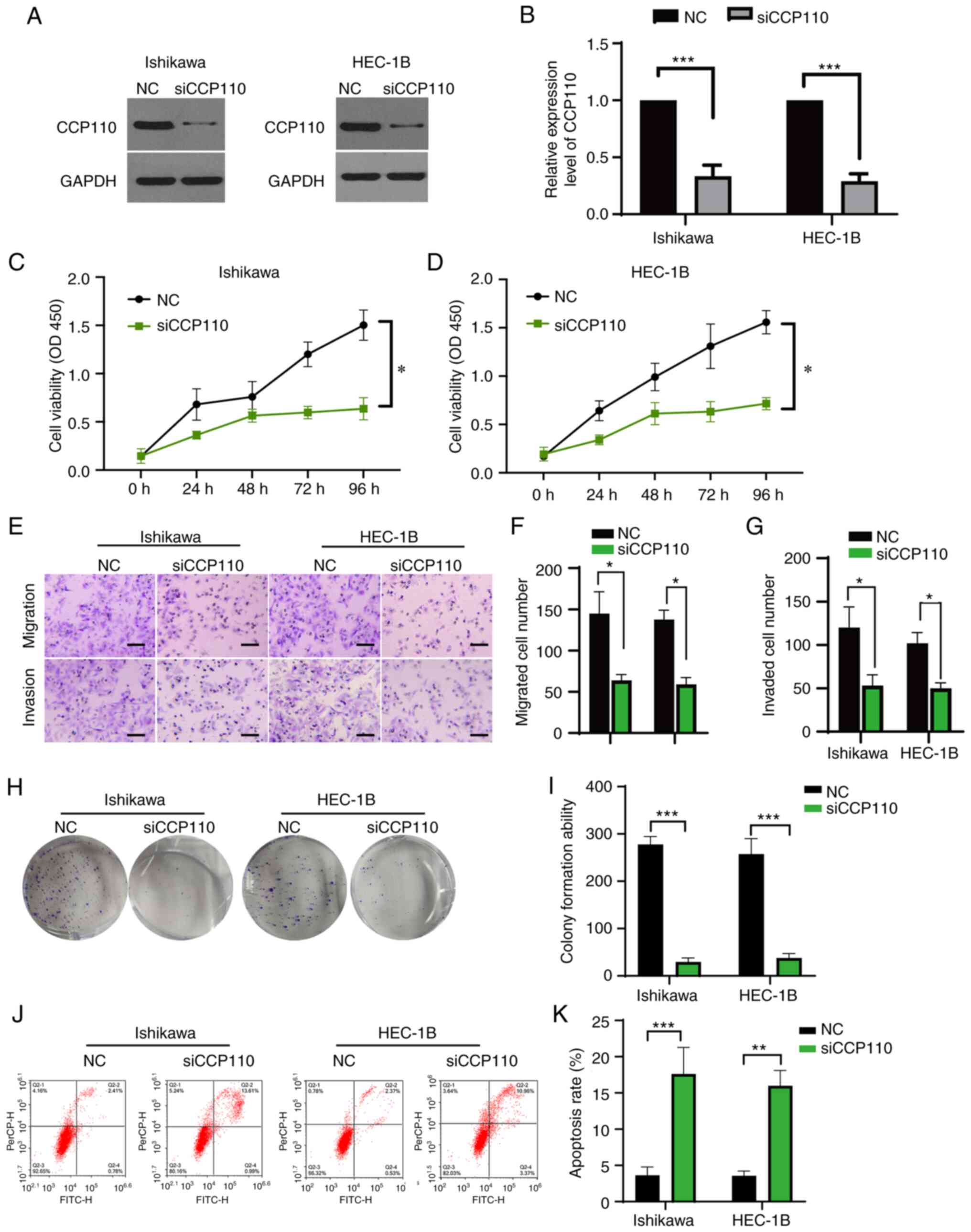 | Figure 2Inhibition of CCP110 suppresses the
development of EC cells. Ishikawa and HEC-1B EC cell lines were
transfected with a NC siRNA or a CCP110-specific siRNA (siCCP110)
and lysed 24 h later. (A) Expression levels of CCP110 and GAPDH
were detected using western blotting and then (B) quantified.
Growth rates of the transfected (C) Ishikawa and (D) HEC-1B cells
were determined using a Cell Counting Kit-8 assay. (E) A Transwell
assay to detect migration and invasion of the transfected cells
(scale bar, 20 µm); the results of the Transwell assay (F)
migration and (G) invasion were then quantified. (H) Colony
formation assay of the transfected cells, which was then (I)
quantified. (J) Flow cytometry analysis to detect apoptosis of the
transfected cells, which was then (K) quantified.
*P<0.05, **P<0.01,
***P<0.001. CCP110, centromere coiled-coil protein
110; EC, endometrial carcinoma/cancer; NC, negative control; si-,
short interfering; OD, optical density. |
MiR129-2-3p attenuates EC cell growth
and migration by targeting CCP110
Next, the present study attempted to elucidate the
molecular mechanism underlying the role of CCP110 in EC. A
bioinformatics analysis identified two potential binding sites for
miR-129-2-3p in the 3' UTR region of CCP110 (Fig. 3A). Consequently, CCP110 luciferase
reporter plasmids containing WT or MUT versions of the miR-129-2-3p
binding sequences were constructed (Fig. 3A). The WT and MUT reporter
constructs were co-transfected into Ishikawa and HEC-1B cells with
a miR-129-2-3p mimic or NC mimic. The expression levels of
miR-129-2-3p were first verified using RT-qPCR (Fig. S2). In both cell lines, compared
with the NC mimic group, the relative luciferase activity of the WT
CCP110 reporter plasmid was significantly reduced after
co-transfection with the miR-129-2-3p mimic compared with the mimic
NC, whereas co-transfection with the miR-129-2-3p mimic had no
significant effect on the relative luciferase activity of the MUT
reporter (Fig. 3B and C).
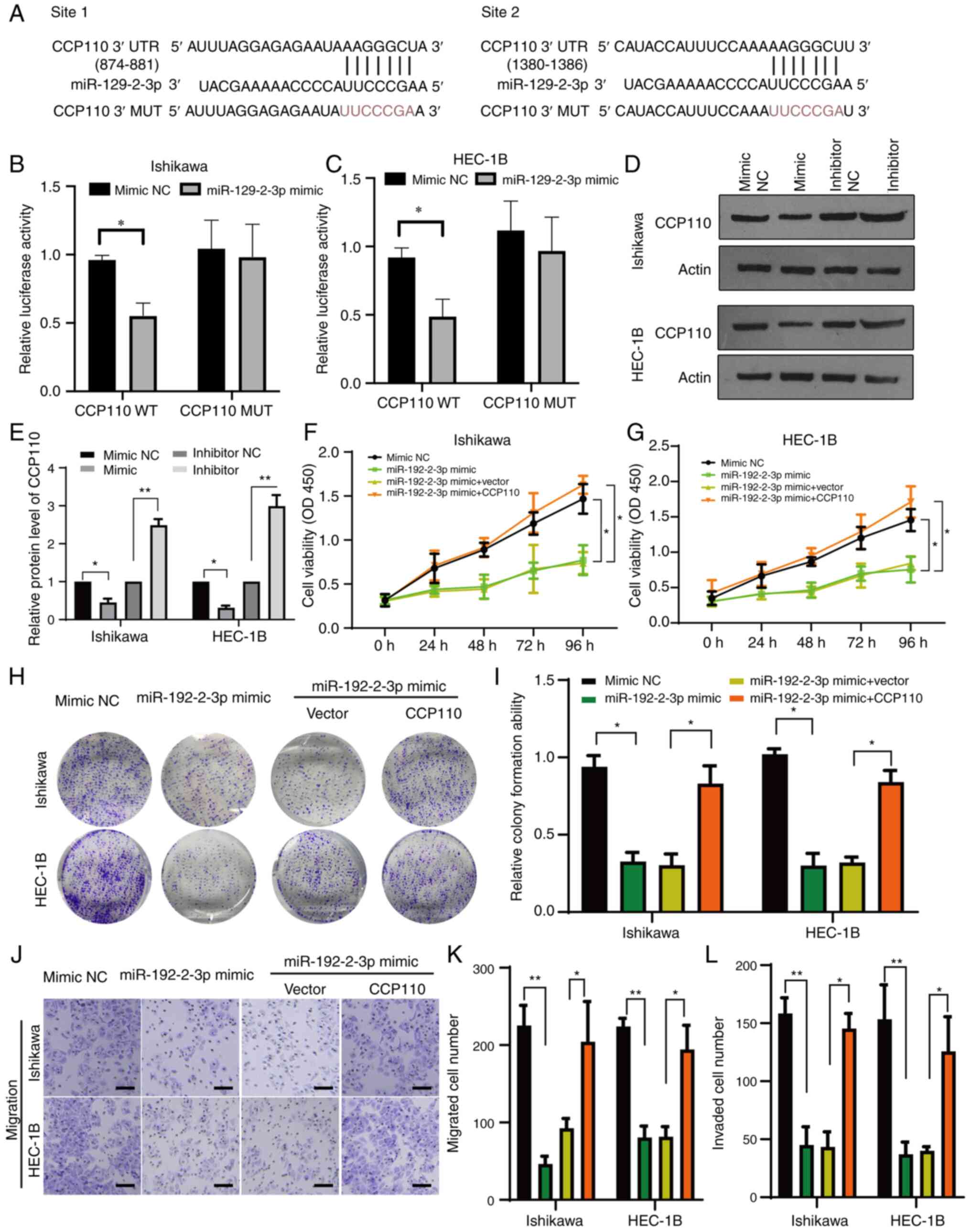 | Figure 3MiR-129-2-3p targets CCP110 and
regulates EC cell development. (A) Bioinformatic prediction of
miR-129-2-3p binding sites in the CCP110 3’ UTR. Luciferase
reporter gene analysis of (B) Ishiwaka and (C) HEC-1B EC cell lines
expressing a WT or MUT CCP110 reporter plasmid and a miR-129-2-3p
mimic or NC mimic. (D) Western blotting analysis of CCP110
expression in EC cell lines transfected with a miR-129-2-3p mimic
or inhibitor, (E) which was then quantified. A Cell Counting Kit-8
assay to measure the growth rates of (F) Ishiwaka and (G) HEC-1B EC
cell lines transfected with a NC mimic, miR-129-2-3p mimic,
miR-129-2-3p mimic + empty vector or miR-129-2-3p mimic +
CCP110-expressing vector. (H) Colony formation and (I)
quantification; (J) Transwell (scale bar, 20 µm) (K) migration and
(L) invasion assays using the cells described in panels F and G.
*P<0.05, **P<0.01. miR, microRNA;
CCP110, centromere coiled-coil protein 110; EC, endometrial
carcinoma/cancer; UTR, untranslated region; WT, wild-type; MUT,
mutant; NC, negative control; OD, optical density. |
To examine the regulatory relationship between
miR-129-2-3p and CCP110 further, both EC cell lines were
transfected with a NC mimic, miR-129-2-3p mimic, NC inhibitor or a
miR-129-2-3p inhibitor. Cells were collected after 24 h, lysed and
subjected to western blotting to detect CCP110 expression. The
miR-129-2-3p mimic inhibited the expression of CCP110 significantly
in both Ishikawa and HEC-1B cells compared with the mimic NC,
whereas the miR-129-2-3p inhibitor significantly increased the
protein expression compared with the inhibitor NC (Fig. 3D and E).
Next, the impact of miR-129-2-3p-regulated CCP110
expression was examined in endometrial cell function. The
expression levels of CCP110 by overexpression of pcDNA3.1-CCP110
encoding plasmids in two cell lines were verified and shown in
Fig. S3A and B. A CCK-8 assay revealed that
transfection with the miR-129-2-3p mimic suppressed the growth of
both EC cell lines significantly compared with the mimic NC, and
co-overexpression of CCP110 rescued this inhibitory effect
(Fig. 3F and G). Moreover, the colony formation
abilities (Fig. 3H and I) and Transwell migratory activities
(Figs. 3J-L and S3C) of both cell lines were suppressed
by transfection with the miR-129-2-3p mimic, and these effects were
neutralized by co-overexpression of CCP110. Taken together, these
data suggested that miR-129-2-3p regulated the development of EC by
modulating CCP110 expression.
LncRNA XIST targets miR-129-2-3p in EC
cells
Given its role in controlling CCP110 expression and
the development of EC, the present study examined the mechanism by
which miR-129-2-3p was regulated. lncRNAs can act as miRNA sponges,
reducing their regulatory effect on mRNAs (47). XIST, the first lncRNA identified in
mammals (48), was investigated
because, to the best of our knowledge, there are few reports of its
involvement in EC and the detailed mechanism remains to be
determined. Thus, RT-qPCR was used to examine the expression levels
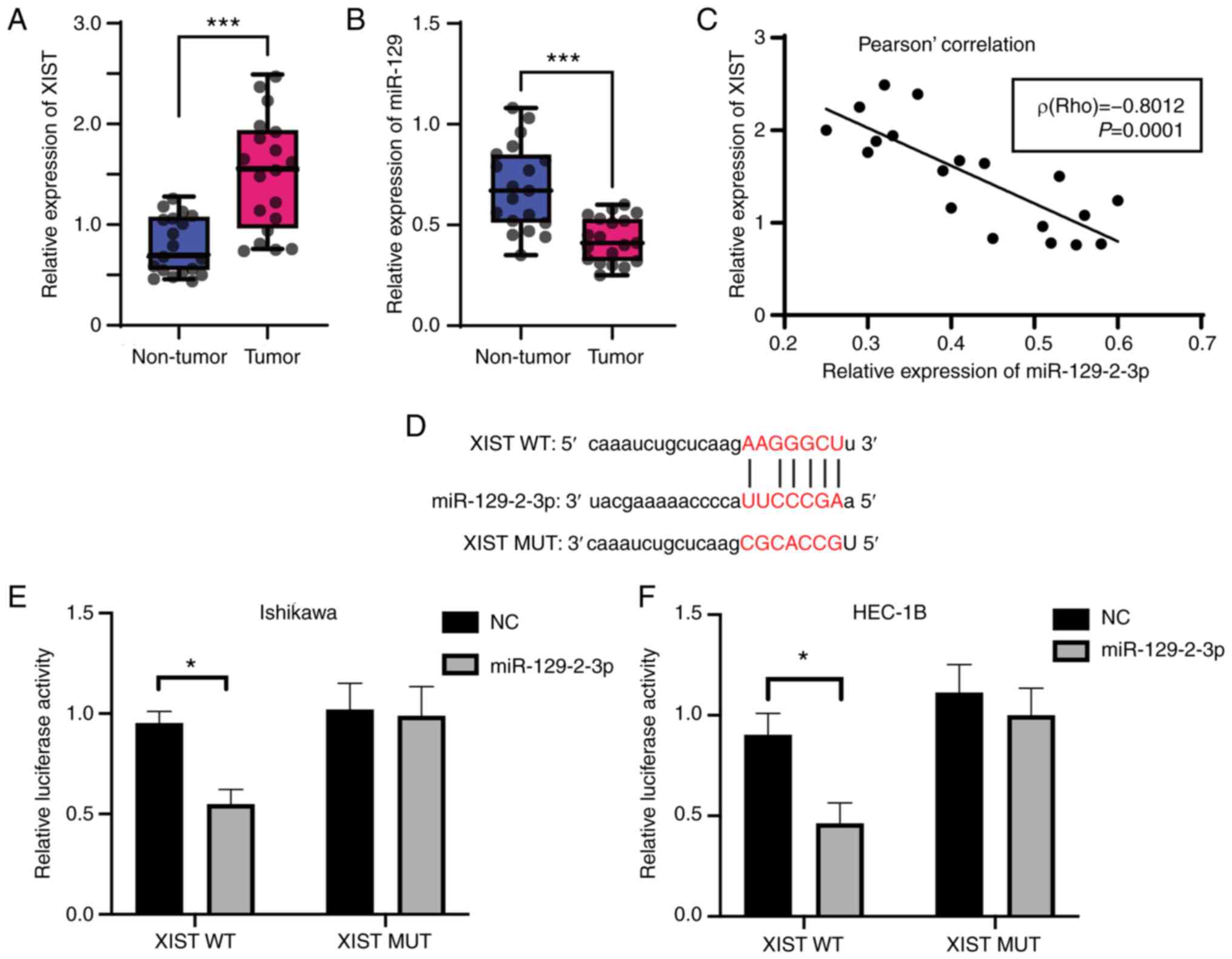
of XIST and miR-129-2-3p in cancerous and paracancerous tissues
collected from 19 patients with EC. Compared with those in
paracancerous tissues, the expression levels of XIST (Fig. 4A) and miR-129-2-3p (Fig. 4B) were, significantly upregulated
and downregulated, respectively, in cancer tissues. In addition,
the expression levels of XIST and miR-129-2-3p were negatively
correlated [Rho value (r)=0.608; Fig.
4C]. A bioinformatics prediction revealed the existence of
interaction sites between XIST and miR-129-2-3p (Fig. 4D), suggesting a potential targeting
relationship between the two.
Next, XIST luciferase reporter plasmids containing a
WT or MUT version of the predicted miR-129-2-3p binding site were
created and co-transfected into Ishikawa and HEC-1B cells with a
miR-129-2-3p mimic or NC mimic. Co-transfection of the miR-129-2-3p
mimic significantly reduced the relative luciferase activity of the
XIST WT reporter compared with the NC group, but had no such effect
on that of the XIST MUT reporter (Fig.
4E and F). Overall, these data
indicated the lncRNA XIST targeted miR-129-2-3p in EC cells.
XIST/miR-129-2-3p axis regulates the
development of EC cells
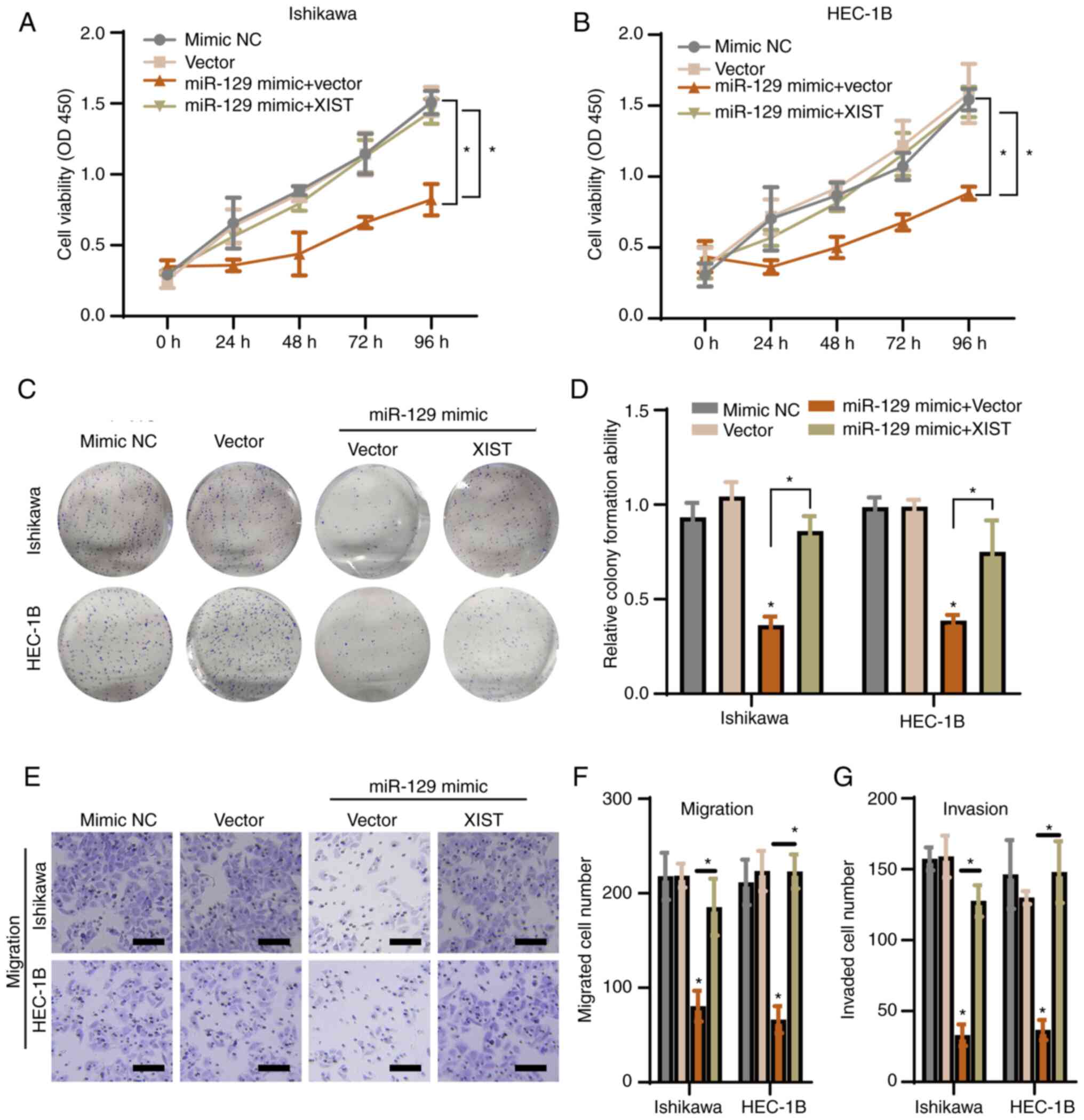
To examine the role of the lncRNA XIST/miR-129-2-3p
axis in EC further, Ishikawa and HEC-1B cells were transfected with
vector only, a NC mimic, a miR-129-2-3p mimic + vector or a
miR-129-2-3p mimic + XIST vector. The expression levels of XIST by
overexpression of pcDNA3.1-XIST encoding plasmids in two cell lines
were verified using RT-qPCR and shown in Fig. S4A. A CCK-8 assay showed that the
miR-129-2-3p mimic inhibited EC cell proliferation (compared to the
vector group), while overexpression of XIST antagonized this effect
in both cell lines (Fig. 5A and
B). Furthermore, in both cell
lines, overexpression of XIST significantly antagonized the
miR-129-2-3p mimic-induced inhibition of cell colony formation
(Fig. 5C and D) and migration and invasion ability
(Figs. 5E-G and S4). These data suggested that the lncRNA
XIST functions via miR-129-2-3p to regulate the proliferation,
migration and invasion of EC cells.
XIST/miR-129-2-3p axis modulates the
expression of CCP110
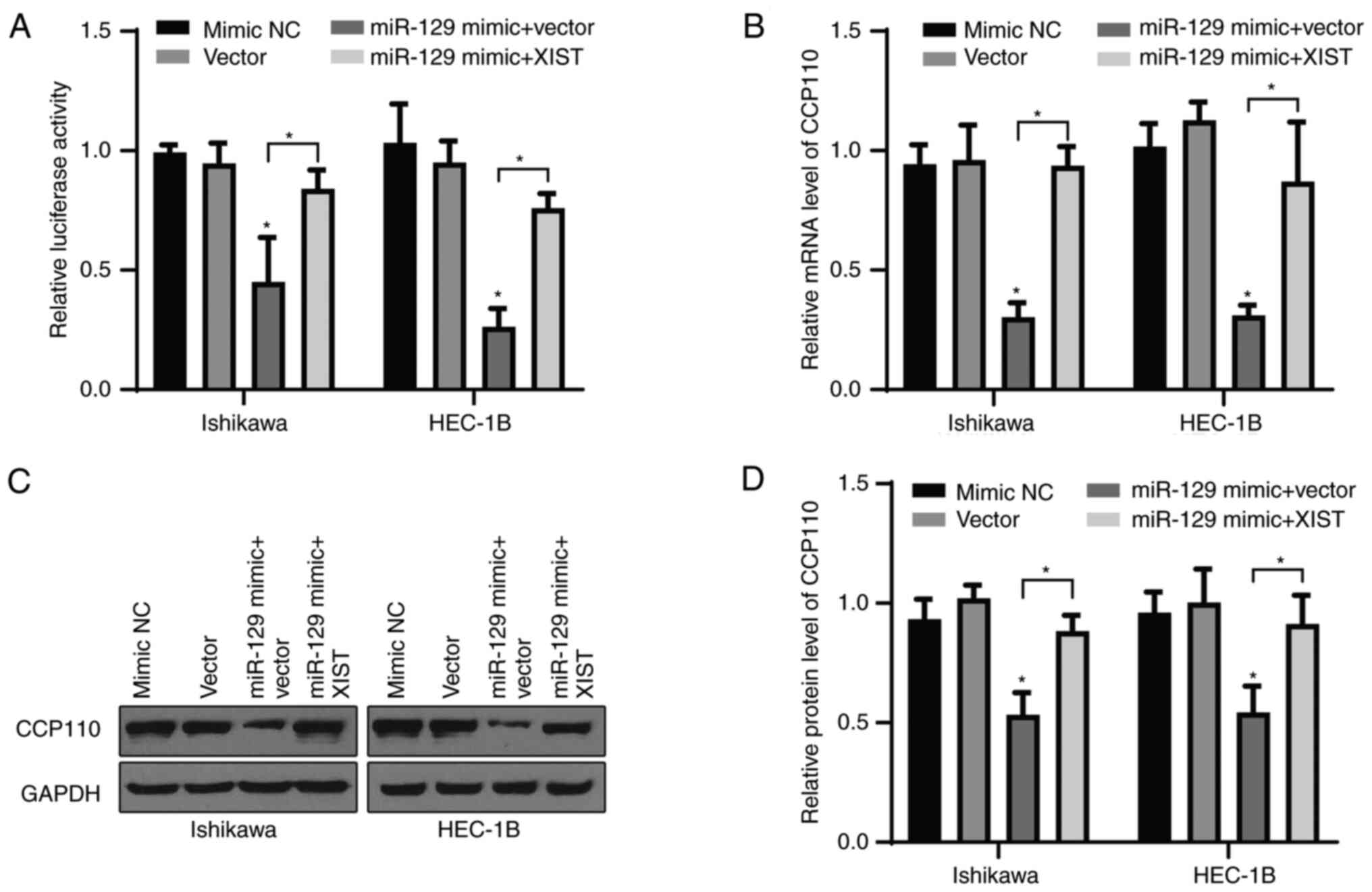
Given its targeting relationship with miR-129-2-3p,
the present study examined whether XIST could regulate the
expression of CCP110 through miR-129-2-3p. Ishikawa and HEC-1B
cells were co-transfected with a WT CCP110 reporter and either
vector only, a NC mimic, a miR-129-2-3p mimic + vector or a
miR-129-2-3p mimic + XIST vector, and then subjected to a
luciferase reporter assay. Compared with the control group, the
miR-129-2-3p mimic + vector group displayed a significantly
decreased relative luciferase activity in both cell lines, while
overexpression of XIST abolished the inhibitory effect of
miR-129-2-3p (Fig. 6A).
Furthermore, the miR-129-2-3p mimic suppressed the mRNA (Fig. 6B) and protein (Fig. 6C and D) levels of CCP110 significantly in both
cell lines, and these effects were alleviated by overexpression of
XIST. Overall, these data suggested that the XIST/miR-129-2-3p axis
regulated the development of EC cells by modulating CCP110
expression.
Discussion
EC endangers the physical and mental health of women
and occurs most frequently in menopausal women (49). At present, there are no specific
markers of EC in laboratory tests, resulting in delayed detection
of the disease (50). CCP110 is
involved in the interaction of several protein complexes that
regulate centrosome replication and segregation, chromosome
segregation and cilia formation, and it structurally regulates
microtubule growth and centriole length (51,52).
High expression levels of CCP110 induce centrosome expansion, which
subsequently delays centrosome segregation and promotes centrosome
aggregation (14). Thus, CCP110 is
closely associated with tumor development and its inhibition
suppresses the development of several types of cancer (53,54).
However, the role of CCP110 in EC has previously been unclear.
The current study investigated the role of CCP110 in
regulating the proliferation, migration, invasion and colony
formation ability of EC cells, and identified miR-129-2-3p and the
lncRNA XIST as upstream signals. CCP110 expression was upregulated
in EC tissues and was closely related to the prognosis of patients.
Genetic knockdown of CCP110 increased apoptosis and suppressed the
proliferation, migration, invasion and colony formation ability of
EC cells significantly. A bioinformatics analysis and luciferase
reporter assay identified CCP110 as a direct target of
miR-129-2-3p. Overexpression of miR-129-2-3p suppressed the
proliferation, migration, invasion and colony formation ability of
EC cells, and these effects were counteracted by overexpression of
CCP110. The expression level of the lncRNA XIST was upregulated in
EC tissues, while that of miR-129-2-3p was downregulated, and the
expression levels of these two species were negatively correlated.
Overall, the present study identified a novel regulatory pathway in
which XIST acted as a sponge to inhibit miR-129-2-3p, leading to
upregulation of CCP110 and a subsequent increase in the
proliferation, migration, invasion and colony formation ability of
EC cells. These findings identified CCP110, miR-129-2-3p and XIST
as potential targets for the clinical treatment of EC.
As a member of the miR-129 family, miR-129-2-3p
occupies an important position in cancer development and other
diseases (55). For example,
miR-129-3p expression is decreased in rheumatoid arthritis, and its
overexpression suppresses expression of the inflammatory cytokine
IL-17 to abolish rheumatoid arthritis (56). In one study, during the
pathogenesis of osteoarthritis, overexpression of miR-129-3p has
been shown to significantly improve articular chondrocyte viability
and reduce apoptosis (57). In the
cardiovascular system, overexpression of miR-129-3p ameliorates
cardiomyocyte inflammation and apoptosis (58). In the nervous system,
downregulation of miR-129-3p increases calcium overload, reactive
oxygen species production, MMP-2 expression and apoptosis, leading
to changes in neuronal function (59). In addition, miR-129-2-3p plays a
notable role in cancer development, with altered expression in
various tumors. For example, miR-129-3p expression is downregulated
in prostate cancer, and its overexpression inhibits the
proliferation and invasion of prostate cancer cells, promotes cell
regulation, increases expression of the pro-apoptotic protein Bax
and decreases expression of the anti-apoptotic protein
Bcl-2(60). Furthermore,
overexpression of miR-129-3p reduces CCP110 levels in prostate
cancer cells and prevents the production of excess centrosomes
(18), which is line with the
current findings in EC cells. MiR-129-3p expression is also reduced
in gastric cancer tissues and its overexpression inhibits the
proliferation, migration and invasion of gastric cancer cells
(54). The antitumor effects of
miR-129-3p in gastric cancer cells include repression of
SUMO-activating enzyme subunit 1 expression by direct targeting of
the 3'UTR and suppression of the SUMOylation of X-ray repair cross
complementing 4, which disrupts the nuclear localization of the
protein (61). Overexpression of
miR-129-3p also inhibits the metastasis and invasion of
hepatocellular carcinoma cells by inducing inactivation of the
PI3K/Akt and p38-MAPK signaling pathways (62-64).
Low expression of miR-129-2-3p is significantly correlated with the
malignant clinical features of patients with intrahepatic
cholangiocarcinoma (65).
The lncRNA XIST plays an important role in the
progression of cancer, acting as a sponge to target miRNAs and
affect the biological function of tumors (66). In non-small cell lung cancer,
overexpression of XIST promotes the proliferation, invasion and
metastasis of cancer cells, acting as an oncogene regulating a
variety of miRNAs and signaling pathways, such as miR-186-5p,
miR-374a, miR-744/RING and CXCR4 (67-70).
In esophageal cancer, XIST activates the miR-494/CDK6/JAK2/STAT3
signaling pathway to induce carcinogenesis (71), and knockdown of XIST inhibits the
malignant behavior of esophageal cancer cells by antagonizing
miR-129-5p/CCND1 signaling (72).
XIST also has an oncogenic role in lung cancer and affects tumor
progression by regulating the miR-140/iASPP axis and TCF-4
expression (73). XIST expression
also predicts poor prognosis and promotes malignant phenotypes in
osteosarcoma (74). Furthermore,
XIST regulates osteosarcoma cell proliferation, migration,
invasion, epithelial-mesenchymal transition and apoptosis,
participating in gene regulation through multiple mechanisms
(75-79).
In the present study, XIST was upregulated in EC tissues and its
expression level was negatively correlated with that of
miR-129-2-3p. The present study revealed that the inhibitory effect
of miR-129-2-3p on the proliferation, metastasis and invasion of EC
cells was abolished by overexpression of XIST, which suggested a
direct relationship between the two species. Thus, as in other
types of cancer, XIST appeared to act as an oncogene in EC by
modulating the activity of miR-129-2-3p, leading to upregulation of
CCP110. This proposed regulatory mechanism should be explored
further in nude mice or other animal models.
To summarize, the present study reported the
upregulation of CCP110 in EC tissues and characterized its
regulation by a novel signal pathway involving miR-129-2-3p and the
lncRNA XIST. The current study identified a novel role for the
XIST/miR-129-2-3p/CCP110 axis in regulating endometrial cell
growth, colony formation, migration and invasion, which could
provide new insights into the clinical management of EC.
Supplementary Material
Ishikawa and HEC-1B EC cell lines were
transfected with a NC siRNA or a CCP110-specific siRNA (siCCP110)
and lysed 24 h later. The expression levels of CCP110 and GAPDH
were detected using western blotting. NC, negative control; EC,
endometrial carcinoma/cancer; siRNA, short interfering; CCP110,
centromere coiled-coil protein 110.
Ishikawa and HEC-1B EC cell lines were
transfected with a miR-129-2-3p mimic or inhibitor or the NC
control fragments. Reverse transcription-quantitative PCR was
performed to determine the levels of miR-129-2-3p in these cell
lines. *P<0.05 compared with the Mimic/inhibitor NC.
EC, endometrial carcinoma/cancer; miR, microRNA; NC, negative
control.
(A) Ishikawa and HEC-1B EC cell lines
were transfected with pcDNA3.1 or pcDNA3.1-CCP110, the expression
level of CCP110 was detected by western blotting and the quantified
data were shown in (B). (C) Ishikawa and HEC-1B EC cell lines were
transfected with indicated NC mimic, miR-129-2-3p mimic,
miR-129-2-3p mimic + empty vector, or miR-129-2-3p mimic +
CCP110-expressing vector. The representative images of cell
invasion by Transwell assay are shown in each group (scale bar, 50
μm). ***P<0.001 compared with the vector. EC,
endometrial carcinoma/cancer; CCP110, centromere coiled-coil
protein 110; miR, microRNA; NC, negative control.
(A) Ishikawa and HEC-1B EC cell lines
were transfected with pcDNA3.1 vector or the encoding plasmids of
XIST, the expression level of XIST was verified using reverse
transcription-quantitative PCR. (B) Ishikawa and HEC-1B EC cell
lines were transfected with vector only, a NC mimic, a miR-129-2-3p
mimic + vector or a miR-129-2-3p mimic + XIST vector. The
representative images of cell invasion by Transwell assay were
shown in each group (scale bar, 50 μm).
***P<0.001 compared with the vector. EC, endometrial
carcinoma/cancer; XIST, X-inactive-specific transcript; NC,
negative control; miR, microRNA.
Acknowledgements
Not applicable.
Funding
Funding: This work was supported by the Affiliated Shunde
Hospital of Jinan University Research and cultivation special fund
project (Summit project) (grant no. 202101008) and the Fundamental
Research Funds for the Central Universities (grant no.
21620310).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SC and YS designed the experiments, drafted the
manuscript, performed the experiments and analyzed and interpreted
the data. YL helped with data collection and analysis. YS
coordinated the research and participated in the experimental
design. XW conceived the idea and conducted the project. All
authors were involved in critically revising the manuscript, and
have read and approved the final manuscript. SC and YS confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
Written consent was signed by the patients, and the
study was approved by the ethics committee of Jinan University
(approval no. 20210826-25; Guangzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Moore K and Brewer MA: Endometrial cancer:
Is this a new disease? Am Soc Clin Oncol Educ Book. 37:435–442.
2017.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Amant F, Mirza MR, Koskas M and Creutzberg
CL: Cancer of the corpus uteri. Int J Gynaecol Obstet. 143 (Suppl
2):S37–S50. 2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Urick ME and Bell DW: Clinical
actionability of molecular targets in endometrial cancer. Nat Rev
Cancer. 19:510–521. 2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sorosky JI: Endometrial cancer. Obstet
Gynecol. 120:383–397. 2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Practice Bulletin No. 149. Endometrial
cancer. Obstet Gynecol. 125:1006–1026. 2015.PubMed/NCBI View Article : Google Scholar
|
|
6
|
ACOG committee opinion no. 557. Management
of acute abnormal uterine bleeding in nonpregnant reproductive-aged
women. Obstet Gynecol. 121:891–896. 2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Morice P, Leary A, Creutzberg C,
Abu-Rustum N and Darai E: Endometrial cancer. Lancet.
387:1094–1108. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Tran AQ and Gehrig P: Recent advances in
endometrial cancer. F1000Res. 6(81)2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Tsikouras P, Bouchlariotou S, Vrachnis N,
Dafopoulos A, Galazios G, Csorba R and von Tempelhoff GF:
Endometrial cancer: Molecular and therapeutic aspects. Eur J Obstet
Gynecol Reprod Biol. 169:1–9. 2013.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Bell DW and Ellenson LH: Molecular
genetics of endometrial carcinoma. Annu Rev Pathol. 14:339–367.
2019.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Caponio MA, Addati T, Popescu O, Petroni
S, Rubini V, Centrone M, Trojano G and Simone G: P16(INK4a) protein
expression in endocervical, endometrial and metastatic
adenocarcinomas of extra-uterine origin: Diagnostic and clinical
considerations. Cancer Biomark. 14:169–175. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Fukasawa K: Centrosome amplification,
chromosome instability and cancer development. Cancer Lett.
230:6–19. 2005.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Sluder G and Nordberg JJ: The good, the
bad and the ugly: The practical consequences of centrosome
amplification. Curr Opin Cell Biol. 16:49–54. 2004.PubMed/NCBI View Article : Google Scholar
|
|
14
|
D'Angiolella V, Donato V, Vijayakumar S,
Saraf A, Florens L, Washburn MP, Dynlacht B and Pagano M:
SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity
through CP110 degradation. Nature. 466:138–142. 2010.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Tsang WY, Spektor A, Luciano DJ, Indjeian
VB, Chen Z, Salisbury JL, Sánchez I and Dynlacht BD: CP110
cooperates with two calcium-binding proteins to regulate
cytokinesis and genome stability. Mol Biol Cell. 17:3423–3434.
2006.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Li J, D'Angiolella V, Seeley ES, Kim S,
Kobayashi T, Fu W, Campos EI, Pagano M and Dynlacht BD: USP33
regulates centrosome biogenesis via deubiquitination of the
centriolar protein CP110. Nature. 495:255–259. 2013.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhang Y, Wang Y, Wei Y, Li M, Yu S, Ye M,
Zhang H, Chen S, Liu W and Zhang J: MiR-129-3p promotes docetaxel
resistance of breast cancer cells via CP110 inhibition. Sci Rep.
5(15424)2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Bijnsdorp IV, Hodzic J, Lagerweij T,
Westerman B, Krijgsman O, Broeke J, Verweij F, Nilsson RJA,
Rozendaal L, van Beusechem VW, et al: miR-129-3p controls
centrosome number in metastatic prostate cancer cells by repressing
CP110. Oncotarget. 7:16676–16687. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Hu S, Danilov AV, Godek K, Orr B, Tafe LJ,
Rodriguez-Canales J, Behrens C, Mino B, Moran CA, Memoli VA, et al:
CDK2 inhibition causes anaphase catastrophe in lung cancer through
the centrosomal protein CP110. Cancer Res. 75:2029–2038.
2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014.PubMed/NCBI View
Article : Google Scholar
|
|
21
|
Kwon SC, Baek SC, Choi YG, Yang J, Lee YS,
Woo JS and Kim VN: Molecular basis for the single-nucleotide
precision of primary microRNA processing. Mol Cell. 73:505–518
e505. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Lu TX and Rothenberg ME: MicroRNA. J
Allergy Clin Immunol. 141:1202–1207. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Widodo, Djati MS and Rifa'I M: Role of
MicroRNAs in carcinogenesis that potential for biomarker of
endometrial cancer. Ann Med Surg (Lond). 7:9–13. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Banno K, Kisu I, Yanokura M, Masuda K,
Ueki A, Kobayashi Y, Susumu N and Aoki D: Epigenetics and genetics
in endometrial cancer: New carcinogenic mechanisms and relationship
with clinical practice. Epigenomics. 4:147–162. 2012.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Yanokura M, Banno K, Iida M, Irie H, Umene
K, Masuda K, Kobayashi Y, Tominaga E and Aoki D: MicroRNAS in
endometrial cancer: Recent advances and potential clinical
applications. EXCLI J. 14:190–198. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Cohn DE, Fabbri M, Valeri N, Alder H,
Ivanov I, Liu CG, Croce CM and Resnick KE: Comprehensive miRNA
profiling of surgically staged endometrial cancer. Am J Obstet
Gynecol. 202:656 e651–e658. 2010.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Montagnana M, Benati M, Danese E, Giudici
S, Perfranceschi M, Ruzzenenete O, Salvagno GL, Bassi A, Gelati M,
Paviati E, et al: Aberrant microRNA expression in patients with
endometrial cancer. Int J Gynecol Cancer. 27:459–466.
2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Li L and Ma L: Upregulation of miR-582-5p
regulates cell proliferation and apoptosis by targeting AKT3 in
human endometrial carcinoma. Saudi J Biol Sci. 25:965–970.
2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Lu Z, Nian Z, Jingjing Z, Tao L and Quan
L: MicroRNA-424/E2F6 feedback loop modulates cell invasion,
migration and EMT in endometrial carcinoma. Oncotarget.
8:114281–114291. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Huang YW, Liu JC, Deatherage DE, Luo J,
Mutch DG, Goodfellow PJ, Miller DS and Huang THM: Epigenetic
repression of microRNA-129-2 leads to overexpression of SOX4
oncogene in endometrial cancer. Cancer Res. 69:9038–9046.
2009.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Shan T, Uyar DS, Wang LS, Mutch DG, Huang
THM, Rader JS, Sheng X and Huang YW: SOX11 hypermethylation as a
tumor biomarker in endometrial cancer. Biochimie. 162:8–14.
2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
He WP, Chen YY, Wu LX, Guo YY, You ZS and
Yang GF: A novel necroptosis-related lncRNA signature for
predicting prognosis and anti-cancer treatment response in
endometrial cancer. Front Immunol. 13(1018544)2022.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Ong MS, Cai W, Yuan Y, Leong HC, Tan TZ,
Mohammad A, You ML, Arfuso F, Goh BC, Warrier S, et al: ‘Lnc’-ing
Wnt in female reproductive cancers: therapeutic potential of long
non-coding RNAs in Wnt signalling. Br J Pharmacol. 174:4684–4700.
2017.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Hosseini ES, Meryet-Figuiere M,
Sabzalipoor H, Kashani HH, Nikzad H and Asemi Z: Dysregulated
expression of long noncoding RNAs in gynecologic cancers. Mol
Cancer. 16(107)2017.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Xie P, Cao H, Li Y, Wang J and Cui Z:
Knockdown of lncRNA CCAT2 inhibits endometrial cancer cells growth
and metastasis via sponging miR-216b. Cancer Biomark. 21:123–133.
2017.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Guo C, Song WQ, Sun P, Jin L and Dai HY:
LncRNA-GAS5 induces PTEN expression through inhibiting miR-103 in
endometrial cancer cells. J Biomed Sci. 22(100)2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Sun MY, Zhu JY, Zhang CY, Zhang M, Song
YN, Rahman K, Zhang LJB and Zhang H: Autophagy regulated by lncRNA
HOTAIR contributes to the cisplatin-induced resistance in
endometrial cancer cells. Biotechnol Lett. 39:1477–1484.
2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Brown CJ, Hendrich BD, Rupert JL,
Lafrenière RG, Xing Y, Lawrence J and Willard HF: The human XIST
gene: Analysis of a 17 kb inactive X-specific RNA that contains
conserved repeats and is highly localized within the nucleus. Cell.
71:527–542. 1992.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Yang Z, Jiang X, Jiang X and Zhao H:
X-inactive-specific transcript: A long noncoding RNA with complex
roles in human cancers. Gene. 679:28–35. 2018.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Mutzel V and Schulz EG: Dosage sensing,
threshold responses, and epigenetic memory: A systems biology
perspective on random X-Chromosome inactivation. Bioessays.
42(e1900163)2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Creytens D: NKX2.2 immunohistochemistry in
the distinction of Ewing sarcoma from cytomorphologic mimics:
Diagnostic utility and pitfalls-Comment on Russell-Goldman et
al. Cancer Cytopathol. 127(202)2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4(e05005)2015.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Wang X: Improving microRNA target
prediction by modeling with unambiguously identified
microRNA-target pairs from CLIP-ligation studies. Bioinformatics.
32:1316–1322. 2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Krek A, Grun D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005.PubMed/NCBI View
Article : Google Scholar
|
|
47
|
Zhang R, Xia LQ, Lu WW, Zhang J and Zhu
JS: LncRNAs and cancer. Oncol Lett. 12:1233–1239. 2016.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Jali I, Vanamamalai VK, Garg P, Navarrete
P, Gutierrez-Adan A and Sharma S: Identification and differential
expression of long non-coding RNAs and their association with XIST
gene during early embryonic developmental stages of Bos taurus. Int
J Biol Macromol. 229:896–908. 2023.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Raglan O, Kalliala I, Markozannes G,
Cividini S, Gunter MJ, Nautiyal J, Gabra H, Paraskevaidis E,
Martin-Hirsch P, Tsilidis KK and Kyrgiou M: Risk factors for
endometrial cancer: An umbrella review of the literature. Int J
Cancer. 145:1719–1730. 2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Banno K, Kisu I, Yanokura M, Tsuji K,
Masuda K, Ueki A, Kobayashi Y, Yamagami W, Nomura H, Tominaga E, et
al: Biomarkers in endometrial cancer: Possible clinical
applications (Review). Oncol Lett. 3:1175–1180. 2012.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Schmidt TI, Kleylein-Sohn J, Westendorf J,
Clech ML, Lavoie SB, Stierhof YD and Nigg EA: Control of centriole
length by CPAP and CP110. Curr Biol. 19:1005–1011. 2009.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Spektor A, Tsang WY, Khoo D and Dynlacht
BD: Cep97 and CP110 suppress a cilia assembly program. Cell.
130:678–690. 2007.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Plotnikova OV, Golemis EA and Pugacheva
EN: Cell cycle-dependent ciliogenesis and cancer. Cancer Res.
68:2058–2061. 2008.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Michaud EJ and Yoder BK: The primary
cilium in cell signaling and cancer. Cancer Res. 66:6463–6467.
2006.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Fesler A, Zhai H and Ju J: miR-129 as a
novel therapeutic target and biomarker in gastrointestinal cancer.
Onco Targets Ther. 7:1481–1485. 2014.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Tsai CH, Liu SC, Wang YH, Su CM, Huang CC,
Hsu CJ and Tang CH: Osteopontin inhibition of miR-129-3p enhances
IL-17 expression and monocyte migration in rheumatoid arthritis.
Biochim Biophys Acta Gen Subj. 1861:15–22. 2017.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Chen R, Ye B, Xie H, Huang Y, Wu Z, Wu H,
Wang X, Miao H and Liang W: miR-129-3p alleviates chondrocyte
apoptosis in knee joint fracture-induced osteoarthritis through
CPEB1. J Orthop Surg Res. 15(552)2020.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Zou Y and Kong M: Tetrahydroxy stilbene
glucoside alleviates palmitic acid-induced inflammation and
apoptosis in cardiomyocytes by regulating miR-129-3p/Smad3
signaling. Cell Mol Biol Lett. 24(5)2019.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Wang B, Li Y and You C: MiR-129-3p
targeting of MCU protects against glucose fluctuation-mediated
neuronal damage via a mitochondrial-dependent intrinsic apoptotic
pathway. Diabetes Metab Syndr Obes. 14:153–163. 2021.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Jia Y, Gao Y and Dou J: Effects of
miR-129-3p on biological functions of prostate cancer cells through
targeted regulation of Smad3. Oncol Lett. 19:1195–1202.
2020.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Zhang M, Jiang D, Xie X, He Y, Lv M and
Jiang X: miR-129-3p inhibits NHEJ pathway by targeting SAE1 and
represses gastric cancer progression. Int J Clin Exp Pathol.
12:1539–1547. 2019.PubMed/NCBI
|
|
62
|
Shaker OG, Abdelwahed MY, Ahmed NA, Hassan
EA, Ahmed TI, Abousarie MA and Ayoub SE: Evaluation of serum long
noncoding RNA NEAT and MiR-129-5p in hepatocellular carcinoma.
IUBMB Life. 71:1571–1578. 2019.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Zhai J, Qu S, Li X, Zhong J, Chen X, Qu Z
and Wu D: miR-129 suppresses tumor cell growth and invasion by
targeting PAK5 in hepatocellular carcinoma. Biochem Biophys Res
Commun. 464:161–167. 2015.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Zhang D, Cao J, Zhong Q, Zeng L, Cai C,
Lei L, Zhang W and Liu F: Long noncoding RNA PCAT-1 promotes
invasion and metastasis via the miR-129-5p-HMGB1 signaling pathway
in hepatocellular carcinoma. Biomed Pharmacother. 95:1187–1193.
2017.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Chen C, Jiang J, Fang M, Zhou L, Chen Y,
Zhou J, Song Y, Kong G, Zhang B, Jiang B, et al: MicroRNA-129-2-3p
directly targets Wip1 to suppress the proliferation and invasion of
intrahepatic cholangiocarcinoma. J Cancer. 11:3216–3224.
2020.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Chen X, Xiong D, Ye L, Wang K, Huang L,
Mei S, Wu J, Chen S, Lai X, Zheng L and Wang M: Up-regulated lncRNA
XIST contributes to progression of cervical cancer via regulating
miR-140-5p and ORC1. Cancer Cell Int. 19(45)2019.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Xu Z, Xu J, Lu H, Lin B, Cai S, Guo J,
Zang F and Chen R: LARP1 is regulated by the XIST/miR-374a axis and
functions as an oncogene in non-small cell lung carcinoma. Oncol
Rep. 38:3659–3667. 2017.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Wang H, Shen Q, Zhang X, Yang C, Cui S,
Sun Y, Wang L, Fan X and Xu S: The long non-coding RNA XIST
controls non-small cell lung cancer proliferation and invasion by
modulating miR-186-5p. Cell Physiol Biochem. 41:2221–2229.
2017.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Zhang YL, Li XB, Hou YX, Fang NZ, You JC
and Zhou QH: The lncRNA XIST exhibits oncogenic properties via
regulation of miR-449a and Bcl-2 in human non-small cell lung
cancer. Acta Pharmacol Sin. 38:371–381. 2017.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Fang J, Sun CC and Gong C: Long noncoding
RNA XIST acts as an oncogene in non-small cell lung cancer by
epigenetically repressing KLF2 expression. Biochem Biophys Res
Commun. 478:811–817. 2016.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Chen Z, Hu X, Wu Y, Cong L, He X, Lu J,
Feng J and Liu D: Long non-coding RNA XIST promotes the development
of esophageal cancer by sponging miR-494 to regulate CDK6
expression. Biomed Pharmacother. 109:2228–2236. 2019.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Wang H, Li H, Yu Y, Jiang Q, Zhang R, Sun
H, Xing W and Li Y: Long non-coding RNA XIST promotes the
progression of esophageal squamous cell carcinoma through sponging
miR-129-5p and upregulating CCND1 expression. Cell Cycle. 20:39–53.
2021.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Tang Y, He R, An J, Deng P, Huang L and
Yang W: lncRNA XIST interacts with miR-140 to modulate lung cancer
growth by targeting iASPP. Oncol Rep. 38:941–948. 2017.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Wang W, Shen H, Cao G and Huang J: Long
non-coding RNA XIST predicts poor prognosis and promotes malignant
phenotypes in osteosarcoma. Oncol Lett. 17:256–262. 2019.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Han J and Shen X: Long noncoding RNAs in
osteosarcoma via various signaling pathways. J Clin Lab Anal.
34(e23317)2020.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Wu D, Nie X, Ma C, Liu X, Liang X, An Y,
Zhao B and Wu X: RSF1 functions as an oncogene in osteosarcoma and
is regulated by XIST/miR-193a-3p axis. Biomed Pharmacother.
95:207–214. 2017.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Xu T, Jiang W, Fan L, Gao Q and Li G:
Upregulation of long noncoding RNA Xist promotes proliferation of
osteosarcoma by epigenetic silencing of P21. Oncotarget.
8:101406–101417. 2017.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Yang C, Wu K, Wang S and Wei G: Long
non-coding RNA XIST promotes osteosarcoma progression by targeting
YAP via miR-195-5p. J Cell Biochem. 119:5646–5656. 2018.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Zhang R and Xia T: Long non-coding RNA
XIST regulates PDCD4 expression by interacting with miR-21-5p and
inhibits osteosarcoma cell growth and metastasis. Int J Oncol.
51:1460–1470. 2017.PubMed/NCBI View Article : Google Scholar
|