Introduction
Diabetes mellitus (DM) can result in cardiomyopathy,
kidney failure, retinopathy and other complications of chronic
hyperglycemia, severely affecting life quality and expectancy in
this patient population (1,2).
Hyperglycemia-induced endothelial dysfunction causes
diabetes-related micro- and macrovascular complications in patients
with long-term DM (3). However,
endothelial dysfunction persists if hyperglycemia is not controlled
in a timely manner; this phenomenon is known as metabolic memory
(MM) (4). MM is an obstacle in the
treatment of diabetic complications. Oxidative stress,
non-enzymatic glycation of proteins, epigenetic changes and chronic
inflammation are the four basic mechanisms playing vital roles in
MM (5-9).
Epigenetic mechanisms, including those involving non-coding RNAs,
histone modifications and DNA methylation, are crucial components
in the pathology of diabetic complications (7). The roles of long non-coding RNAs
(lncRNAs) in diabetes-associated endothelial dysfunction and the
effects of lncRNAs on MM-related mechanisms are currently being
investigated (10,11).
lncRNAs, which are long transcripts containing
>200 nucleotides, are structurally similar to, but functionally
different from mRNAs. By acting as molecular sponges or host genes
for microRNAs (miRNAs/miRs), serving as scaffolds for specific
protein complexes and guiding histone-modifying complexes, lncRNAs
serve key roles in the epigenetic mechanisms mediating numerous
physiological and pathological processes (12-14).
Several previous studies have indicated that lncRNAs are involved
in diabetes-related nephropathy, retinopathy and cardiovascular
diseases, and numerous lncRNAs, such as ENST00000600527,
NONHSAT037576.2 and NONHSAT135706.2, have been reported to
demonstrate abnormal expression in vascular endothelial and smooth
muscle cells placed under high glucose (HG) culture conditions
(14-17).
The functions of lncRNAs in diabetic endothelial dysfunction and MM
have been gathering increasing attention. In human retinal
endothelial cells, lncRNA antisense non-coding RNA in the INK4
locus is highly expressed under HG conditions, causing the
overexpression of vascular endothelial growth factors (VEGFs),
which induces angiogenesis (18).
HG-treated retinal endothelial cells express high levels of the
lncRNA myocardial infarction associated transcript (MIAT), which
functions as a competing endogenous RNA (ceRNA) by absorbing
miR-150-5p, thereby weakening the miRNA-mediated inhibition of VEGF
expression. Accordingly, knockdown of lncRNA MIAT alleviates
microvascular dysfunction in vivo (19). HG levels can also upregulate the
expression of lncRNA metastasis-associated lung adenocarcinoma
transcript 1 (MALAT1) in the retinas of rats with diabetes.
Silencing MALAT1 expression ameliorates diabetic-induced retinal
vasculopathy and inflammation in vivo and endothelial
dysfunction in vitro (20).
Furthermore, lncRNA maternally expressed gene 3 (MEG3) levels are
significantly reduced in the retinas of mice with diabetes and in
endothelial cells subjected to HG concentrations or oxidative
stress. Knockdown of lncRNA MEG3 enhances inflammation and retinal
vascular dysfunction, and promotes the proliferation and migration
of retinal endothelial cells and endothelial tube formation
(21). Overexpression of MEG3
alleviates diabetic retinopathy by reducing the expression of
TGF-β1 and VEGFs (22).
Determining which lncRNAs are expressed in MM can
help uncover their roles in the molecular mechanisms driving MM and
may facilitate the therapeutic use of these lncRNAs in the
treatment of patients with MM. Therefore, in the present study, RNA
sequencing and bioinformatics analyses were used to identify
lncRNAs with upregulated [(up)-MM-involved differentially expressed
lncRNAs (MMDELs)] and downregulated expression (down-MMDELs) in MM
induced by HG conditions.
Materials and methods
Cell groups and establishment of the
MM model
Primary human umbilical vein endothelial cells
(HUVECs) were purchased from the China Center for Type Culture
Collection and were maintained in Modified Eagle's Medium (Hyclone;
Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin
(Gibco; Thermo Fisher Scientific, Inc.) and 100 µg/ml streptomycin
(Gibco; Thermo Fisher Scientific, Inc.) at 37˚C in a humidified, 5%
CO2 chamber. Cell culture and establishment of the MM
model were performed as described previously (16,23).
Briefly, HUVEC cells in the low glucose (LG) group were cultured
with 5 mM glucose and 20 mM mannitol for 6 days. The cells in the
HG group were cultured with 25 mM glucose for 6 days. The cells in
the MM group were treated for 3 days using 25 mM glucose, followed
by another 3 days of treatment using 5 mM glucose and 20 mM
mannitol, to induce the state of MM in these cells.
RNA preparation and sequencing
Total RNA was isolated from HUVECs and purified
using TRIzol® (cat. no. 15596026, Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions, the quantity and quality of the RNA were determined
using an Agilent 4200 Bioanalyzer (Thermo, Fisher Scientific,
Inc.). The RNA integrity was assessed by electrophoresis with
denaturing agarose gels. Libraries were constructed using a VAHTS
Total RNA-Seq(H/M/R) Library PrepKit for Illumina (cat. no.
NR603-02; Vazyme Biotech Co., Ltd.) according to the manufacturer's
protocol, and libraries were identified using a Qubit™ dsDNA HS
Assay Kit (cat. no. Q32854; Invitrogen; Thermo Fisher Scientific,
Inc.) and Agilent High Sensitivity DNA Kit (cat. no. 5067-4626;
Agilent Technologies, Inc.). The qualities of the libraries were
assessed using a Qubit® 2.0 Fluorometer (Thermo Fisher
Scientific, Inc.) and the concentration of DNA in the libraries was
analyzed using an Agilent 2100 bioanalyzer (Agilent Technologies,
Inc.). Clusters were generated bycBot (Illumina, Inc.) and the
libraries were diluted to 10 pm, then sequencing was performed on
the Illumina HiSeq 2500 according to the manufacturer's protocol at
Shanghai Ao-Ji Biotechnology Co., Ltd (2x150 bp, paired-end).
Details of the experimental procedures and instruments used are
described in our previous study (24).
Identification and analysis of the
parental genes of MMDELs
FastQC (version 0.11.3; https://www.bioinformatics.babraham.ac.uk/projects/fastqc/)
was used to ensure the quality of RNA-sequencing (RNA-seq) reads.
The reads were trimmed using seqtk (https://github.com/lh3/seqtk), and then Illumina
TruSeq adapter sequences, poor reads and ribosome RNA reads were
removed. The trimmed reads were mapped onto the Homo sapiens
genome (hg38) using Hisat2 (version 2.0.4) (25). StringTie (version 1.3.0) and
gffcompare (version 0.9.8) were used to assemble and compile
transcripts from the trimmed reads (26,27).
These transcripts were then compared with the reference annotation
databases NONCODE (version 5, http://v5.noncode.org/introduce.php) and Ensembl
(http://grch37.ensembl.org/index.html). EdgeR and Venny
were used to screen out the differentially expressed lncRNAs in the
HG vs. LG and MM vs. LG groups (P<0.05 and fold change >2)
(28,29). Non-significant differentially
expressed lncRNAs were then filtered through the comparison of HG
vs. MM group (P>0.05), and the intersection of these
differentially expressed lncRNAs was illustrated using a Venn
diagram. Gene Ontology (GO) and KEGG (https://www.kegg.jp/) analysis were used to examine
the enrichment of parental genes from which the lncRNAs are
transcribed and the function of MMDELs using clusterProfiler
(version 3.16, https://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html)
(30,31).
Construction of
lncRNA-mRNA-co-expression network
The Pearson correlation coefficient (PCC) was
calculated for the analysis of the correlation between the
expression of lncRNAs and mRNAs. LncRNA-mRNA pairs with PCC >0.9
and P<0.01 were selected for the construction of the
co-expression network, which was visualized using Cytoscape
(version 2.8.3) (32). The node
degree indicates the number of directly linked neighbors for each
node. GO and pathway analyses were also used to estimate the highly
correlated candidate coding genes using clusterProfiler (version
3.16).
Reverse transcription-quantitative PCR
(RT-qPCR) validation
To minimize selection bias, 3 upregulated and 3
downregulated MMDELs were randomly selected. The expression level
of these six MMDELs was quantified using qPCR on a Roche
LightCycler 480 (Roche Applied Science). The sequences of the
specific primers used and the lengths of the products are indicated
in Table I. After the total RNA
was isolated from HUVECs according to the aforementioned method.
RT-qPCR was performed according to the manufacturer's protocol
using a ABScript II cDNA First-Strand Synthesis Kit (cat. no.
RK20400; Abclonal Biotech Co., Ltd.) and qPCR was performed using a
ABScript II One Step SYBR Green RT-qPCR Kit (cat. no. RK20404;
Abclonal Biotech Co., Ltd.). PCR reactions were implemented using
the following temperature protocol: 95˚C for 10 min, followed by 40
cycles of 95˚C for 15 sec and 60˚C for 20 sec. Expression levels of
target genes were normalized to that of GAPDH, which used as an
internal reference gene. The relative expression of each lncRNA was
calculated using the 2-ΔΔCq method (33).
 | Table ISequences of PCR primers used in the
present study. |
Table I
Sequences of PCR primers used in the
present study.
| Gene | Primer sequences
(5' to 3') | PCR product length
(bp) |
|---|
| GAPDH | F:
CCTGGTATGACAACGAATTTG | 131 |
| | R:
CAGTGAGGGTCTCTCTCTTCC | |
|
NONHSAT180590.1 | F:
TCCATTCAGAGAACAGGCCC | 189 |
| | R:
TGTGTTGAGTGATCTCCCCG | |
|
NONHSAT175141.1 | F:
AAACAGGGGTGTCAGGGTTG | 154 |
| | R:
CAAGCCCTGTAGGAAGACGG | |
|
ENST00000603538 | F:
GGCTTCCTTCTATCCCGCTC | 92 |
| | R:
CCGGAGTGCAACAAAATCCG | |
|
ENST00000530490 | F:
CGGAAACGCCAGAAAAGTCG | 103 |
| | R:
GTCAACTCGGGCCACATGAT | |
|
ENST00000621248 | F:
ATGCTCGGAAAAGCCTCTGG | 92 |
| | R:
AGACAGGCCAAAACCCACAA | |
|
ENST00000537869 | F:
CTGTTCCCGTCATGAGCCTT | 83 |
| | R:
GCAAGGCCCTGAATGAGCTA | |
Statistical analysis
SPSS software (version 24.0; IBM Corp.) was used for
data analysis, and the results are presented as the mean ± SEM.
There were three repeats per group. To analyze the RT-qPCR
validation data, sample distribution was examined using
Shapiro-Wilk test, and Levene's test was used to analyze the
homogeneity of variance. One-way ANOVA analysis was used to test
overall statistical significance, followed by the Least
Significance Difference test for pairwise comparisons. P<0.05
was considered to indicate a statistically significant
difference.
Results
Identification of HG-induced
MMDELs
Three samples each of normal HUVECs (LG group),
HG-induced HUVECs (HG group) and MM-induced HUVECs (MM group) were
analyzed using RNA-seq to characterize the differences in the
lncRNA expression levels. In a total of nine samples, 41,484
lncRNAs were identified and of these, 36,387 lncRNAs were shared
between the three groups examined in the present study (Fig. 1A). These lncRNAs were widely
distributed on all chromosomes, with higher numbers of lncRNAs
revealed on chromosomes 1 and 2 (Fig.
1B). LncRNAs are classified into six types: Intergenic,
exonic-antisense, exonic-sense, bidirectional, intronic-antisense
and intronic-sense (34). The most
numerous species of the identified lncRNAs were intergenic,
followed by the exonic-sense and exonic-antisense types (Fig. 1C). These findings suggested the
potential diversity and complexity of regulatory mechanisms.
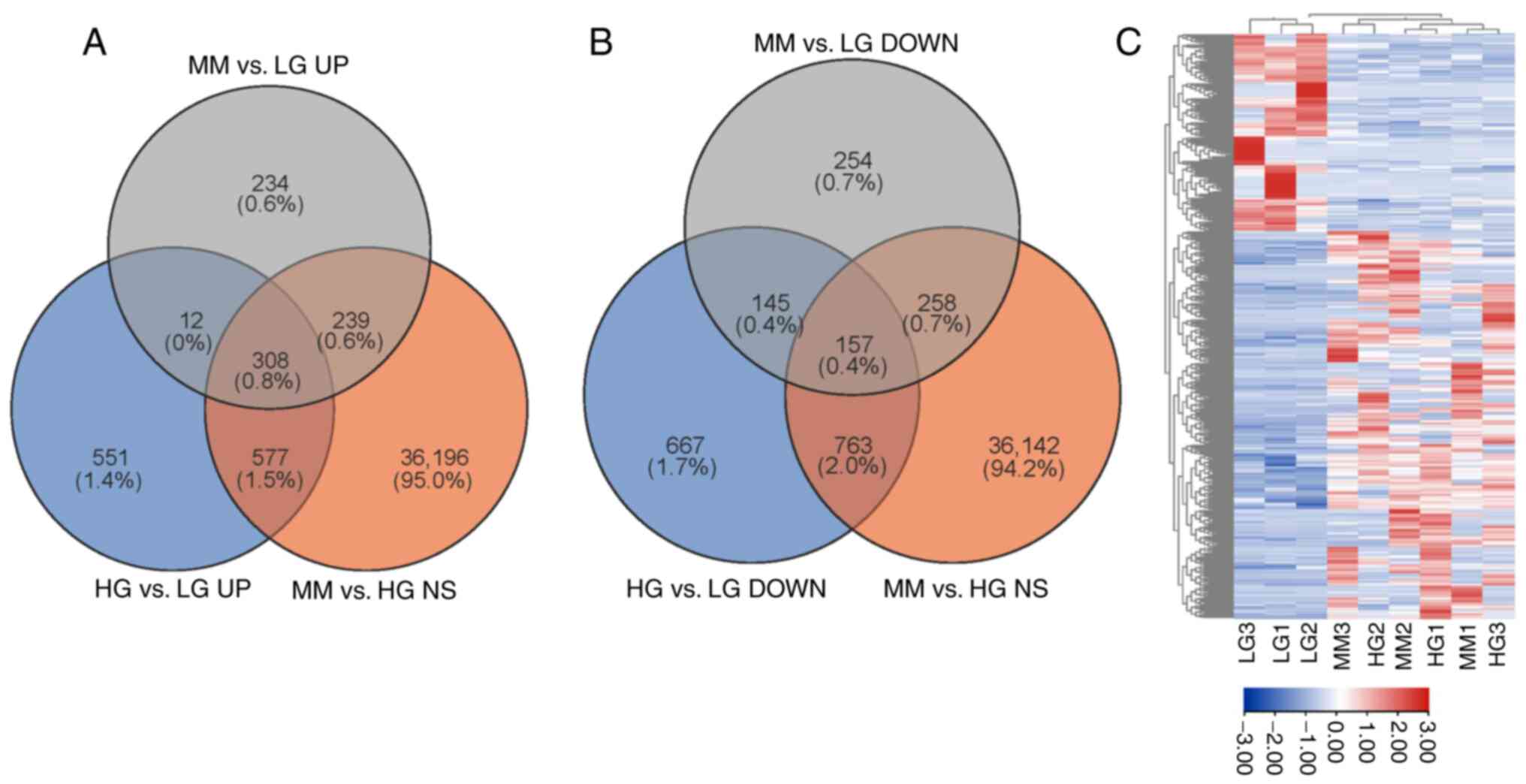
Using edgeR analysis and comprehensive filtration,
308 up- and 157 downregulated MMDELs were identified. As presented
in Fig. 2A, 308 upregulated MMDELs
were screened from the intersection of upregulated lncRNAs in all
groups; among them, 1,448 lncRNAs were upregulated in the HG vs. LG
group (fold-change >2, P<0.05), 793 lncRNAs were upregulated
in the MM vs. LG group (fold-change >2, P<0.05) and 37,320
lncRNAs were non-significantly differentially expressed (MM vs. HG;
P>0.05). Similarly, 157 downregulated MMDELs were identified
from the intersection of downregulated lncRNAs; among them, 1,732
lncRNAs were downregulated in the HG vs. LG group (fold-change
<-2, P<0.05), 814 lncRNAs were downregulated in the MM vs. LG
group (fold-change <-2, P<0.05) and 37,320 lncRNAs were
non-significantly differentially expressed (MM vs. HG; P>0.05)
(Fig. 2B). The MMDEL expression
profile was presented in a heatmap (Fig. 2C).
GO and KEGG analyses of parental genes
of MMDELs
To understand the characteristics and functions of
lncRNAs, it is important to investigate their parental genes. In
the present study, 465 MMDELs were identified; the parental genes
of these 465 MMDELs were annotated using GO and pathway enrichment
analysis.
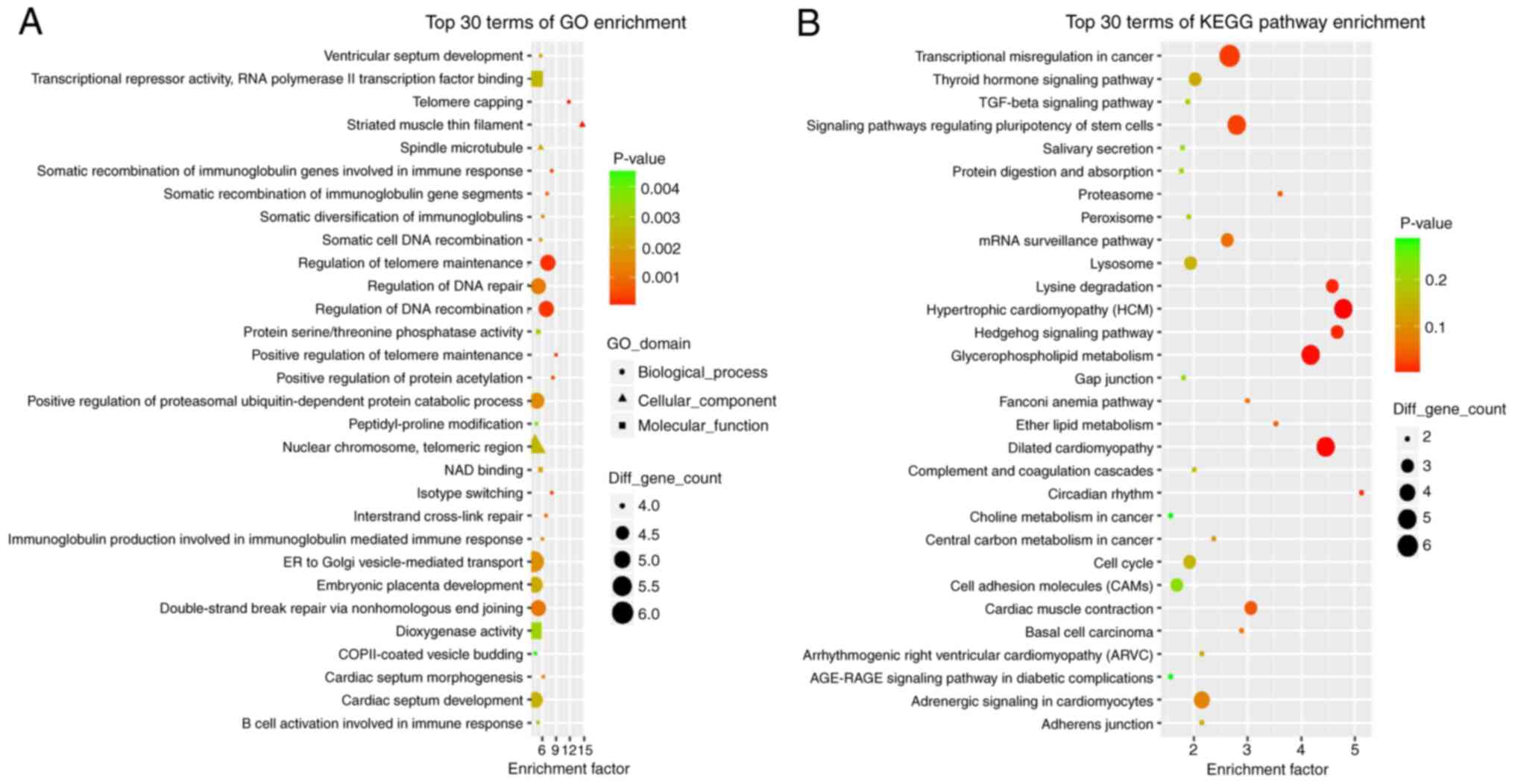
The top 30 GO terms with the highest enrichment
factors identified using GO analysis included ‘positive regulation
of telomere maintenance’, ‘positive regulation of protein
acetylation’, ‘regulation of DNA recombination’ and ‘interstrand
cross-link repair’ in biological process (BP) terms, ‘striated
muscle thin filament’, ‘spindle microtubule’ and ‘nuclear
chromosome, telomeric region’ in cellular component (CC) terms,
‘NAD binding’, ‘protein serine/threonine phosphatase activity’,
‘transcriptional repressor activity, RNA polymerase II
transcription factor binding’ and ‘dioxygenase activity’ in
molecular function (MF) terms (Fig.
3A). These results indicated disturbances in cell cycle and
proliferation.
KEGG pathway enrichment analysis indicated that a
total of 126 pathway terms were enriched with MMDELs. The top 30
pathways with the highest enrichment factors are demonstrated in
Fig. 3B; significantly enriched
terms included ‘glycerophospholipid metabolism’, ‘proteasome’,
‘cardiac muscle contraction’, ‘signaling pathways regulating
pluripotency of stem cells’, ‘mRNA surveillance pathway’, ‘cell
cycle’, ‘peroxisome’, ‘TGF-β signaling pathway’ and ‘AGE-RAGE
signaling pathway in diabetic complications’.
LncRNA-mRNA co-expression network and
functional analysis of target mRNAs
To further explore the regulatory role of these
lncRNAs, the differentially expressed genes in the same HUVEC
samples were evaluated and Cytoscape was used to select the
lncRNA-mRNA pairs with PPC (Pearson correlation coefficient)
>0.9 and P<0.01 to construct and visualize the co-expression
network. Consequently, 397 lncRNA nodes, 708 mRNA nodes and 8,303
edges were used to build the network (Fig. S1). The top 10 up- and
downregulated MMDELs with the highest node degree are presented in
Table II.
 | Table IITop 10 up-MMDELs and top 10
down-MMDELs with the highest node degree score. |
Table II
Top 10 up-MMDELs and top 10
down-MMDELs with the highest node degree score.
| A, HG vs. LG |
|---|
| lncRNA ID | Degree | Type | Locus | Fold change | P-value | Up- or
downregulated |
|---|
|
NONHSAT107810.2 | 210 | Bidirectional | chr6 | 2.56318202 | 0.003 | UP |
|
NONHSAT180590.1 | 186 | Exonic_sense | chr19 | 3.56619419 | ≤0.001 | UP |
|
ENST00000537869 | 180 | Exonic_sense | chr11 | 3.33917148 | ≤0.001 | UP |
|
NONHSAT156713.1 | 148 | Exonic_sense | chr10 | 3.18029669 | ≤0.001 | UP |
|
NONHSAT118785.2 | 125 | Exonic_sense | chr7 | 6.98152837 | ≤0.001 | UP |
|
NONHSAT028252.2 | 96 |
Exonic_antisense | chr12 | 5.99732944 | ≤0.001 | UP |
|
NONHSAT135407.2 | 83 | Intronic_sense | chr9 | NA | 0.006 | UP |
|
NONHSAT135581.2 | 80 | Exonic_sense | chr9 | 2.69833263 | 0.001 | UP |
|
NONHSAT183181.1 | 75 | Exonic_sense | chr2 | 3.28481839 | 0.008 | UP |
|
NONHSAT188701.1 | 74 | Exonic_sense | chr20 | 13.06448443 | ≤0.001 | UP |
|
NONHSAT163874.1 | 240 | Intronic_sense | chr12 | 0.17285088 | ≤0.001 | DOWN |
|
NONHSAT182446.1 | 209 | Intergenic | chr2 | 0.14367777 | 0.016 | DOWN |
|
ENST00000623851 | 205 | Intergenic | chr3 | 0.23109513 | ≤0.001 | DOWN |
|
NONHSAT194515.1 | 198 | Intergenic | ch3 | 0.09350689 | 0.002 | DOWN |
|
NONHSAT215705.1 | 175 | Exonic_sense | chr8 | 0.07170152 | ≤0.001 | DOWN |
|
NONHSAT155504.1 | 169 | Intergenic | chr10 | 0.1565883 | ≤0.001 | DOWN |
|
NONHSAT175141.1 | 164 | Intergenic | chr17 | 0.31294430 | 0.005 | DOWN |
|
NONHSAT200243.1 | 155 | Exonic_sense | chr4 | 0.02487727 | ≤0.001 | DOWN |
|
NONHSAT156901.1 | 154 | Intergenic | chr10 | 0.41563310 | 0.028 | DOWN |
|
ENST00000621248 | 151 | Intergenic | chr12 | 0.21394491 | ≤0.001 | DOWN |
| B, HG vs. MM |
| lncRNA ID | Node Degree | Type | Locus | Fold change | P-value | Up- or
downregulated |
|
NONHSAT107810.2 | 210 | Bidirectional | chr6 | 1.25578160 | 0.453 | NS |
|
NONHSAT180590.1 | 186 | Exonic_sense | chr19 | 1.40275545 | 0.292 | NS |
|
ENST00000537869 | 180 | Exonic_sense | chr11 | 1.41459655 | 0.252 | NS |
|
NONHSAT156713.1 | 148 | Exonic_sense | chr10 | 1.50549064 | 0.230 | NS |
|
NONHSAT118785.2 | 125 | Exonic_sense | chr7 | 1.87617204 | 0.115 | NS |
|
NONHSAT028252.2 | 96 |
Exonic_antisense | chr12 | 1.62237440 | 0.136 | NS |
|
NONHSAT135407.2 | 83 | Intronic_sense | chr9 | 1.06453167 | 1.000 | NS |
|
NONHSAT135581.2 | 80 | Exonic_sense | chr9 | 1.22268316 | 0.544 | NS |
|
NONHSAT183181.1 | 75 | Exonic_sense | chr2 | 1.36461675 | 0.351 | NS |
|
NONHSAT188701.1 | 74 | Exonic_sense | chr20 | 0.82783165 | 0.674 | NS |
|
NONHSAT163874.1 | 240 | Intronic_sense | chr12 | 0.47487294 | 0.156 | NS |
|
NONHSAT182446.1 | 209 | Intergenic | chr2 | 0.65481627 | 1.000 | NS |
|
ENST00000623851 | 205 | Intergenic | chr3 | 0.60246918 | 0.203 | NS |
|
NONHSAT194515.1 | 198 | Intergenic | ch3 | 0.49615362 | 0.760 | NS |
|
NONHSAT215705.1 | 175 | Exonic_sense | chr8 | 1.02467761 | 0.853 | NS |
|
NONHSAT155504.1 | 169 | Intergenic | chr10 | 0.52492177 | 0.274 | NS |
|
NONHSAT175141.1 | 164 | Intergenic | chr17 | 0.71489774 | 0.440 | NS |
|
NONHSAT200243.1 | 155 | Exonic_sense | chr4 | 0.94930333 | 1.000 | NS |
|
NONHSAT156901.1 | 154 | Intergenic | chr10 | 0.94316562 | 0.950 | NS |
|
ENST00000621248 | 151 | Intergenic | chr12 | 0.51438924 | 0.064 | NS |
| C, LG vs. MM |
| lncRNA ID | Node Degree | Type | Locus | Fold change | P-value | Up- or
downregulated |
|
NONHSAT107810.2 | 210 | Bidirectional | chr6 | 2.04110494 | 0.023 | UP |
|
NONHSAT180590.1 | 186 | Exonic_sense | chr19 | 2.54227790 | 0.006 | UP |
|
ENST00000537869 | 180 | Exonic_sense | chr11 | 2.36051154 | 0.008 | UP |
|
NONHSAT156713.1 | 148 | Exonic_sense | chr10 | 2.11246527 | 0.021 | UP |
|
NONHSAT118785.2 | 125 | Exonic_sense | chr7 | 3.72115576 | ≤0.001 | UP |
|
NONHSAT028252.2 | 96 |
Exonic_antisense | chr12 | 3.69663713 | 0.002 | UP |
|
NONHSAT135407.2 | 83 | Intronic_sense | chr9 | NA | 0.009 | UP |
|
NONHSAT135581.2 | 80 | Exonic_sense | chr9 | 2.20689441 | 0.018 | UP |
|
NONHSAT183181.1 | 75 | Exonic_sense | chr2 | 2.40713620 | 0.046 | UP |
|
NONHSAT188701.1 | 74 | Exonic_sense | chr20 | 15.78157158 | ≤0.001 | UP |
|
NONHSAT163874.1 | 240 | Intronic_sense | chr12 | 0.36399395 | 0.022 | DOWN |
|
NONHSAT182446.1 | 209 | Intergenic | chr2 | 0.21941693 | 0.042 | DOWN |
|
ENST00000623851 | 205 | Intergenic | chr3 | 0.38358000 | 0.015 | DOWN |
|
NONHSAT194515.1 | 198 | Intergenic | ch3 | 0.18846359 | 0.025 | DOWN |
|
NONHSAT215705.1 | 175 | Exonic_sense | chr8 | 0.06997471 | ≤0.001 | DOWN |
|
NONHSAT155504.1 | 169 | Intergenic | chr10 | 0.29830789 | 0.006 | DOWN |
|
NONHSAT175141.1 | 164 | Intergenic | chr17 | 0.43774693 | 0.045 | DOWN |
|
NONHSAT200243.1 | 155 | Exonic_sense | chr4 | 0.02620582 | ≤0.001 | DOWN |
|
NONHSAT156901.1 | 154 | Intergenic | chr10 | 0.440678802 | 0.033 | DOWN |
|
ENST00000621248 | 151 | Intergenic | chr12 | 0.41592027 | 0.022 | DOWN |
The top 30 GO terms revealed using GO analysis of
target mRNAs included ‘negative regulation of telomerase activity’,
‘regulation of double-strand break repair via homologous
recombination’, ‘mitotic cytokinesis’, ‘negative regulation of DNA
biosynthetic process’ and ‘exit from mitosis’ in BP terms,
‘condensed nuclear chromosome’, ‘centromeric region,’ ‘nuclear
nucleosome’ and ‘mitotic spindle’ in CC terms and ‘14-3-3 protein
binding’ in MF terms, which were also demonstrated in our previous
study (24). This may be
attributed to the similar screening criteria used in the two
bioinformatics analyses. In this study, we focused on the mRNA
whose expression was positively correlated with MMDELs, KEGG
pathway enrichment terms included ‘cell cycle’, ‘p53 signaling
pathway’, ‘Notch signaling pathway’, ‘β-alanine metabolism’,
‘glutathione metabolism’, ‘oxidative phosphorylation’, ‘arginine
and proline metabolism’, ‘glyoxylate and dicarboxylate metabolism’,
‘insulin secretion’ and ‘homologous recombination’ (Fig. 4).
Verification of the expression levels
of MMDELs using RT-qPCR
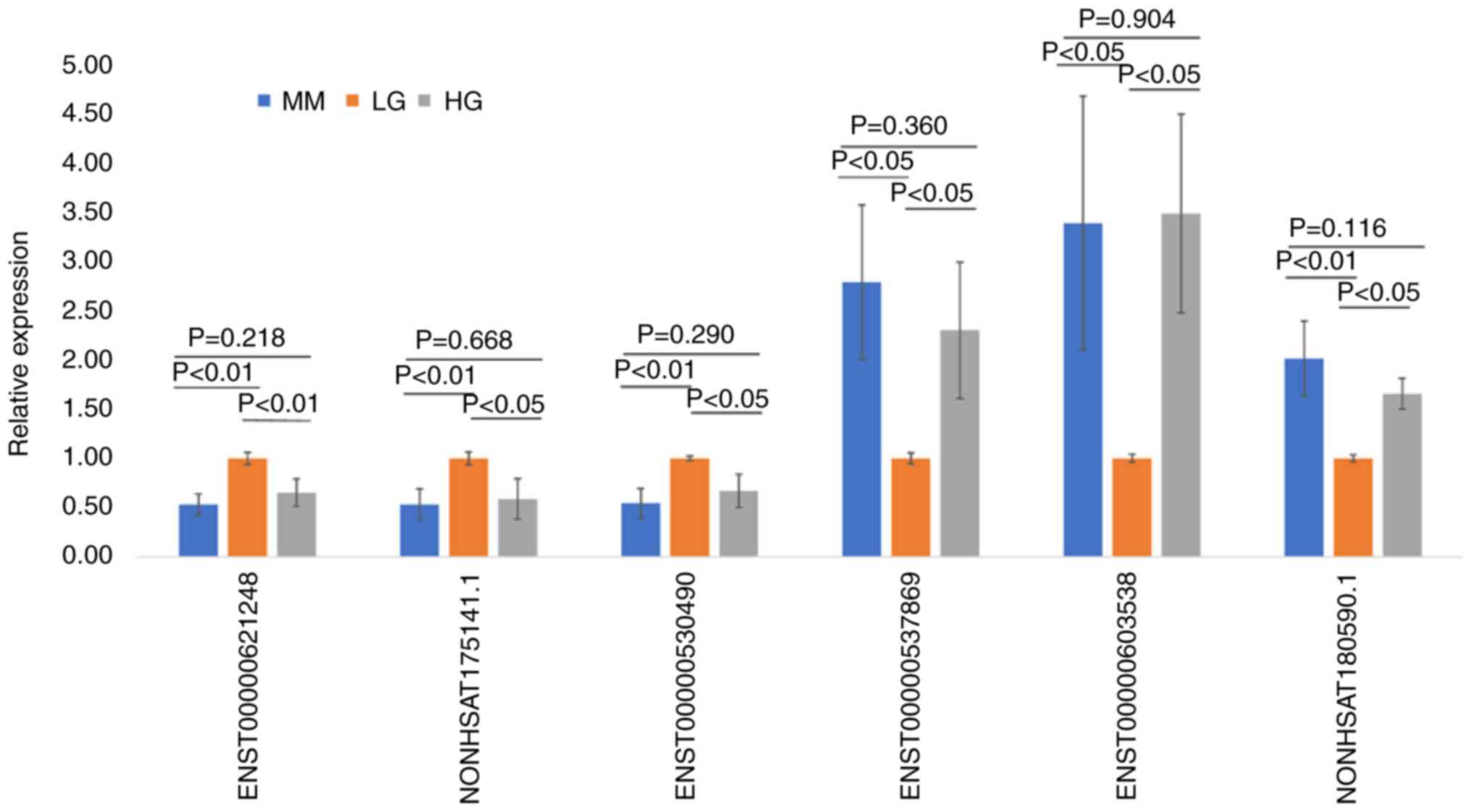
To minimize selection bias, the expression levels of
the six randomly selected MMDELs were verified using RT-qPCR. As
presented in Fig. 5, the
expression levels of these six MMDELs were in accordance with the
results obtained using high-throughput RNA-seq. The expression
levels of ENST00000621248, NONHSAT175141.1 and ENST00000530490 were
significantly downregulated in the MM and HG groups compared with
those of the LG group (P<0.05). However, the expression levels
of ENST00000537869, ENST00000603538 and NONHSAT180590.1 were
significantly increased in the MM and HG groups compared with those
of the LG group (P<0.05). The expression levels of the six
selected lncRNAs did not differ significantly between the MM and HG
groups (P>0.05), indicating that the results of RNA-seq analysis
were reliable.
Discussion
Various vascular complications, such as nephropathy,
retinopathy and atherosclerosis, are responsible for the decreased
quality of life and increased mortality in patients with diabetes.
Increased levels of inflammation, non-enzymatic glycation of
proteins and oxidative stress are common characteristics of the
majority of complications observed in patients with diabetes. At
present, MM is considered a major obstacle to implementing
effective control of diabetes-related complications (35,36).
Therefore, it is important to delineate the mechanisms underlying
vascular complications and MM. In the present study, the expression
profiles of lncRNAs were comprehensively analyzed in HUVECs from
the LG, HG and MM groups, identifying a total of 308 up- and 157
downregulated MMDELs. GO analysis of the parental genes of these
MMDELs suggested that the regulation of proteasomal
ubiquitin-dependent catabolic process and DNA repair may
participate in stress-caused vascular damage induced by exposure to
HG in HUVECs. KEGG analysis indicated that in diabetic
complications, the TGF-β and advanced glycation end products
(AGE)-receptor for AGE (RAGE) signaling pathways may participate in
MM-mediated pathogenic mechanisms induced by HG environments.
Previous studies have demonstrated that AGE-modified proteins
remain in the vessels, kidneys and hearts of patients with diabetes
for extended periods of time, even after control of hyperglycemia
is achieved in these patients. In addition, AGEs can induce
oxidative stress and inflammation by interacting with RAGE
receptors on the cellular surface. Therefore, the AGE-RAGE
signaling pathway provides a clinical link between MM and diabetic
complications (37-39).
Additionally, the findings of the present study indicated that
these pathways were enriched on the parental genes of MMDELs, which
suggested that these lncRNAs may affect vascular endothelial cell
function, cellular proliferation and apoptosis by altering the
expression of parental genes.
In the present study, a bioinformatics analysis of
target mRNAs was performed, which indicated that several
DM-associated pathways, including cell cycle, p53 signaling and
oxidative phosphorylation, may play a role in MM.
The present study indicated that 15 target genes
were enriched in the cell cycle pathway, 14 of which had
upregulated expression levels in MM compared with LG, including
cyclin dependent kinase 1 (CDK1), Cyclin B1 (CCNB1) and CCNB2
(Table SI). This finding
suggested that abnormal proliferation of vascular endothelial cells
may lead to pathological angiogenesis, which is a crucial component
of the pathological alterations observed in diabetic retinopathy
(22). Furthermore, high levels of
the CCNB1/CDK1 complex contribute to apoptosis by promoting cell
cycle arrest at the mitotic prometaphase and induce the
phosphorylation of antiapoptotic proteins, such as Mcl-1 and
Bcl-xl, which can activate the subsequent intrinsic cell death
pathway (40-42).
CDK1 is an essential regulator of mitosis; therefore, CDK1
expression was predicted to be associated with 24 of the MMDELs
identified in the present study. The present study indicated that
20 of these MMDELs had downregulated expression and were negatively
correlated with CDK1 levels in MM compared with LG. The other four
MMDELs, including lncRNA SNHG1 (ENST00000537869; node degree=180;
up-MMDELs), had upregulated expression levels in MM compared with
LG, a positive correlation with CDK1 levels and a strong positive
correlation with the levels of CCNB1 and CCNB2.
The lncRNA SNHG1 is implicated in the progression of
various cancers and is associated with a poor prognosis in patients
with cancer. SNHG1 acts as a ceRNA to inhibit the expression of
miR-140, thereby upregulating the expression of its downstream
target, A disintegrin and metalloproteinase domain-containing
protein 10, and promoting the proliferation and invasion of gastric
cancer cells (43). Toll-like
receptor 4 (TLR4) is a target of miR-140(44). TLR4 expression can also be
increased by SNHG1, which activates the NF-κB signaling pathway to
regulate growth and tumorigenesis in cholangiocarcinoma tissues
(44). SNHG1 promotes the
expression of NUAK family SNF1-like kinase 1 by downregulating
miR-145-5p expression, thereby promoting the invasion of
nasopharyngeal carcinoma cells via the AKT signaling pathway
(45). SNHG1 knockdown inhibits
cancer cell migration and proliferation in vitro and in
vivo (45,46). These findings suggest that
upregulation of SNHG1 expression in HG environments may lead to
proliferation abnormalities in vascular endothelial cells via
mechanisms similar to those found in cancers. Specifically,
interference with the expression of cell cycle-related regulatory
molecules may occur via adsorption of specific miRNAs and the
resultant upregulated expression of their target molecules.
The p53 signaling pathway, which mediates cell cycle
arrest, cellular senescence and apoptosis, can be activated by
various stressors including oxidative stress and DNA damage
(47). The analysis of the present
study indicated that eight target genes were enriched in this
pathway and seven of these genes (Table SI), including CDK1, G2 and S
phase-expressed-1 (GTSE1) and DNA damage-binding protein 2 (DDB2),
had upregulated expression in MM compared with the LG group. GTSE1
leads to cell cycle arrest by inhibiting the expression of
CDK1/CCNB. However, several previous studies have demonstrated that
GTSE1 is overexpressed in several malignant tumors and is closely
associated with tumor cell migration and invasiveness (48,49).
Knockdown of GTSE1 expression suppresses the proliferation,
migration and invasiveness of tumor cells (48). In the present study, 28 MMDELs were
predicted to be correlated with GTSE1 expression; however, only
four MMDELs, including lnc-FCN2-4:1 (NONHSAT135407.2; node
degree=83; up-MMDELs), were upregulated and positively correlated
with GTSE1 expression (Table SI
and SII). Furthermore,
lnc-FCN2-4:1 was also predicted to be correlated with CCNB1, CCNB2
and cell division cycle 20 expression. The expression of DDB2,
which serves a key role in the repair of DNA (50), was associated with 15 of the MMDELs
examined in the present study. Among these 15 MMDELs, eight had
upregulated expression and were positively correlated with related
lncRNAs, such as lncRNA SNHG1. The mechanism underlying the
dysregulated expression of these lncRNAs, which were observed in
HUVECs of the MM group in the present study, needs to be further
explored in future studies.
Compared with the LG group, a total of 11 target
genes were enriched in oxidative phosphorylation (OXPHOS) and five
of these genes had upregulated expression in the MM group.
Ubiquinol-cytochrome c reductase core protein 1 (UQCRC1),
one of the five genes with upregulated expression, is a subunit of
complex III of the respiratory chain in the mitochondria (51). UQCRC1 serves a fundamental role in
normal mitochondrial function and cellular metabolism. Mutations
and abnormal expression of UQCRC1 are associated with Parkinson's
disease and multiple malignancies (52-54).
In pancreatic ductal adenocarcinoma, increased expression of UQCRC1
mRNA promotes cellular proliferation by generating excessive levels
of ATP and OXPHOS via the ATP/RTK/AKT pathway (53). In the present study, UQCRC1
expression was predicted to be positively associated with four
MMDELs including lncRNA SNHG7 (NONHSAT135581.2; node degree=80;
up-MMDELs). SNHG7, which is highly expressed in certain neoplastic
diseases, drives the occurrence and development of tumors by
sponging miRNAs, such as miR-34a, miR-2682-5p and miR-449a, thereby
promoting proliferation and suppressing apoptosis in cancer cells
(55-57).
SNHG7 is overexpressed in the areas surrounding the site of
myocardial infarction. By acting as a ceRNA and targeting miR-34-5p
expression, SNHG7 can promote cardiac fibrosis. Therefore,
silencing SNHG7 expression can improve cardiac function (58). The present study indicated that the
expression levels of MT-ND2, MT-ND3, MT-ND4L, MT-ATP8, MT-CYB and
ATP6V1E1 were downregulated among 11 target genes in MM than LG
group that were enriched in Oxidative phosphorylation (Table SI). The expression of MT-ND2,
MT-ND3, MT-CYB and MT-ATP8 was positively associated with that of
lncRNA XIST (NONHSAT137546.2; node degree=11; down-MMDELs)
(Table SII). A previous study
reported that knockdown of lncRNA XIST expression can inhibit
proliferation, promote apoptosis, and even increase the production
of reactive oxygen species (ROS) in non-small-cell lung cancer
cells (59). It has been also
demonstrated that mitochondrial dysfunction and fission can
increase the levels of mitochondrial ROS (60,61).
Excess accumulation of mitochondrial ROS in endothelial cells from
diabetic patients results in cell death by damaging DNA, lipids and
proteins, and leads to vasoconstriction caused by the decreased
bioavailability of nitric oxide (62). These findings may partially explain
the impairment of endothelial function in DM. Furthermore, based on
the aforementioned findings and the results of the present study,
it is proposed that the expression of OXPHOS-related mRNAs may be
regulated by certain lncRNAs. These currently unknown and complex
regulatory networks may be retained for extended periods of time in
MM.
In summary, the present study analyzed the
expression of MMDELs and the co-expression of mRNA networks in
HUVECs subjected to conditions of LG, HG and induction of MM. The
present study indicated that several MMDELs were abnormally
expressed in experimentally-induced diabetic MM and may be involved
in the regulation of mRNA expression through various mechanisms in
MM-related pathways, which may play roles in the damage associated
with long-term exposure to HG environments. The findings obtained
in the present study contribute to improving the understanding of
the molecular mechanisms underlying the pathology of MM. Further
investigation into the functions of these lncRNAs may result in
novel insights and treatments, which could help control MM in
patients with diabetes.
Supplementary Material
lncRNA-mRNA co-expression network of
the MMDELs analyzed in the present study. Circles represent mRNAs
and squares represent lncRNAs. Red color indicates RNAs with
upregulated expression and green color indicates RNAs with
downregulated expression. The size of the nodes is positively
associated with the node degree. lncRNA, long non-coding RNA;
MMDEL, metabolic memory-involved differentially expressed
lncRNA.
KEGG analysis of target mRNAs of
MM-involved differentially expressed lncRNAs
Analysis of correlated expression of
mRNAs and LncRNAs
Acknowledgements
The authors would like to acknowledge Mr. Qiang Fan
(Shanghai Ao-Ji Biotechnology Co., Ltd., China) for technical
assistance with RNA-seq experiments.
Funding
Funding: The present study was funded by the Key Program of
Nature Science Foundation of Anhui Education Committee (grant no.
KJ2019A0353), Key Project of Translational Medicine of Bengbu
Medical College (grant no. BYTM2019039) and Key Project of Natural
Science Foundation of Bengbu Medical College (grant no.
2020BYZD020).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the National Center for
Biotechnology Information, BioProject repository (accession no.
PRJNA534362; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA534362).
Authors' contributions
JinC and JiC contributed to acquisition of data for
the work, wrote the draft of the paper and contributed to the
literature review of the study. JiC performed the bioinformatics
analysis. AH performed formal analysis and validation of sequencing
data. LY and EX collected and analyzed RT-qPCR data. XP and AH
contributed to formal bioinformatics analysis and the literature
review. GJ conceived and designed the study. GJ and XP confirm the
authenticity of all the raw data. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
Ethics approval for the use of commercially
purchased primary human cells was waived by the Ethics Committee of
Bengbu Medical College (Bengbu, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jia G, Hill MA and Sowers JR: Diabetic
cardiomyopathy: An update of mechanisms contributing to this
clinical entity. Circ Res. 122:624–638. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Wong TY, Cheung CM, Larsen M, Sharma S and
Simo R: Diabetic retinopathy. Nat Rev Dis Primers.
2(16012)2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Shi Y and Vanhoutte PM: Macro- and
microvascular endothelial dysfunction in diabetes. J Diabetes.
9:434–449. 2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Testa R, Bonfigli AR, Prattichizzo F, La
Sala L, De Nigris V and Ceriello A: The ‘Metabolic Memory’ theory
and the early treatment of hyperglycemia in prevention of diabetic
complications. Nutrients. 9(437)2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Giacco F and Brownlee M: Oxidative stress
and diabetic complications. Circ Res. 107:1058–1070.
2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Chilelli NC, Burlina S and Lapolla A:
AGEs, rather than hyperglycemia, are responsible for microvascular
complications in diabetes: A ‘glycoxidation-centric’ point of view.
Nutr Metab Cardiovasc Dis. 23:913–919. 2013.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Reddy MA, Zhang E and Natarajan R:
Epigenetic mechanisms in diabetic complications and metabolic
memory. Diabetologia. 58:443–455. 2015.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Reddy MA and Natarajan R: Epigenetic
mechanisms in diabetic vascular complications. Cardiovasc Res.
90:421–429. 2011.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Thompson JA and Webb RC: Potential role of
Toll-like receptors in programming of vascular dysfunction. Clin
Sci (Lond). 125:19–25. 2013.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhang HN, Xu QQ, Thakur A, Alfred MO,
Chakraborty M, Ghosh A and Yu XB: Endothelial dysfunction in
diabetes and hypertension: Role of microRNAs and long non-coding
RNAs. Life Sci. 213:258–268. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Leung A, Amaram V and Natarajan R: Linking
diabetic vascular complications with LncRNAs. Vascul Pharmacol.
114:139–144. 2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Biswas S, Thomas AA and Chakrabarti S:
LncRNAs: Proverbial genomic ‘Junk’ or key epigenetic regulators
during cardiac fibrosis in diabetes? Front Cardiovasc Med.
5(28)2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: Past, present, and future. Genetics. 193:651–669.
2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhang X, Hong R, Chen W, Xu M and Wang L:
The role of long noncoding RNA in major human disease. Bioorg Chem.
92(103214)2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Singh KK, Mantella LE, Pan Y, Quan A,
Sabongui S, Sandhu P, Teoh H, Al-Omran M and Verma S: A global
profile of glucose-sensitive endothelial-expressed long non-coding
RNAs. Can J Physiol Pharmacol. 94:1007–1014. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Xu E, Hu X, Li X, Jin G, Zhuang L, Wang Q
and Pei X: Analysis of long non-coding RNA expression profiles in
high-glucose treated vascular endothelial cells. BMC Endocr Disord.
20(107)2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Leung A and Natarajan R: Long noncoding
RNAs in diabetes and diabetic complications. Antioxid Redox Signal.
29:1064–1073. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Thomas AA, Feng B and Chakrabarti S:
ANRIL: A regulator of VEGF in diabetic retinopathy. Invest
Ophthalmol Vis Sci. 58:470–480. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li
YJ, Tao ZF, Song YC, Chen Q and Jiang Q: lncRNA-MIAT regulates
microvascular dysfunction by functioning as a competing endogenous
RNA. Circ Res. 116:1143–1156. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li
YJ, Yan B and Jiang Q: Pathogenic role of lncRNA-MALAT1 in
endothelial cell dysfunction in diabetes mellitus. Cell Death Dis.
5(e1506)2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Qiu GZ, Tian W, Fu HT, Li CP and Liu B:
Long noncoding RNA-MEG3 is involved in diabetes mellitus-related
microvascular dysfunction. Biochem Biophys Res Commun. 471:135–141.
2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhang D, Qin H, Leng Y, Li X, Zhang L, Bai
D, Meng Y and Wang J: LncRNA MEG3 overexpression inhibits the
development of diabetic retinopathy by regulating TGF-β1 and VEGF.
Exp Ther Med. 16:2337–2342. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Zhang E, Guo Q, Gao H, Xu R, Teng S and Wu
Y: Metformin and resveratrol inhibited high glucose-induced
metabolic memory of endothelial senescence through
SIRT1/p300/p53/p21 pathway. PLoS One. 10(e0143814)2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Jin G, Wang Q, Pei X, Li X, Hu X, Xu E and
Li M: mRNAs expression profiles of high glucose-induced memory in
human umbilical vein endothelial cells. Diabetes Metab Syndr Obes.
12:1249–1261. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Pertea M, Kim D, Pertea GM, Leek JT and
Salzberg SL: Transcript-level expression analysis of RNA-seq
experiments with HISAT, StringTie and Ballgown. Nat Protoc.
11:1650–1667. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Pertea M, Pertea GM, Antonescu CM, Chang
TC, Mendell JT and Salzberg SL: StringTie enables improved
reconstruction of a transcriptome from RNA-seq reads. Nat
Biotechnol. 33:290–295. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
28
|
Nikolayeva O and Robinson M: edgeR for
differential RNA-seq and ChIP-seq analysis: An application to stem
cell biology. Methods Mol Biol. 1150:45–79. 2014.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wang C and Li Q, Yang H, Gao C, Du Q,
Zhang C, Zhu L and Li Q: MMP9, CXCR1, TLR6, and MPO participant in
the progression of coronary artery disease. J Cell Physiol.
235:8283–8292. 2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ashburner M, Ball C, Blake J, Botstein D,
Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et
al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000.PubMed/NCBI View
Article : Google Scholar
|
|
31
|
Yu G, Wang L, Han Y and He Q:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Livak K and Schmittgen T: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Knauss JL and Sun T: Regulatory mechanisms
of long noncoding RNAs in vertebrate central nervous system
development and function. Neuroscience. 235:200–214.
2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Singh R, Chandel S, Dey D, Ghosh A, Roy S,
Ravichandiran V and Ghosh D: Epigenetic modification and
therapeutic targets of diabetes mellitus. Biosci Rep.
40(BSR20202160)2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Berezin A: Metabolic memory phenomenon in
diabetes mellitus: Achieving and perspectives. Diabetes Metab
Syndr. 10 (2 Suppl 1):S176–S183. 2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Yamagishi S, Fukami K and Matsui T:
Crosstalk between advanced glycation end products (AGEs)-receptor
RAGE axis and dipeptidyl peptidase-4-incretin system in diabetic
vascular complications. Cardiovasc Diabetol. 14(2)2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Yamagishi S and Matsui T: Role of receptor
for advanced glycation end products (RAGE) in liver disease. Eur J
Med Res. 20(15)2015.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Koulis C, Watson AMD, Gray SP and
Jandeleit-Dahm KA: Linking RAGE and Nox in diabetic micro- and
macrovascular complications. Diabetes Metab. 41:272–281.
2015.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Choi HJ and Zhu BT: Upregulated cyclin
B1/CDK1 mediates apoptosis following 2-methoxyestradiol-induced
mitotic catastrophe: Role of Bcl-XL phosphorylation.
Steroids. 150(108381)2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Choi HJ and Zhu BT: Role of cyclin B1/Cdc2
in mediating Bcl-XL phosphorylation and apoptotic cell death
following nocodazole-induced mitotic arrest. Mol Carcinog.
53:125–137. 2014.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Harley ME, Allan LA, Sanderson HS and
Clarke PR: Phosphorylation of Mcl-1 by CDK1-cyclin B1 initiates its
Cdc20-dependent destruction during mitotic arrest. EMBO J.
29:2407–2420. 2010.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Guo W, Huang J, Lei P, Guo L and Li X:
LncRNA SNHG1 promoted HGC-27 cell growth and migration via the
miR-140/ADAM10 axis. Int J Biol Macromol. 122:817–823.
2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Li Z, Li X, Du X, Zhang H, Wu Z, Ren K and
Han X: The Interaction Between lncRNA SNHG1 and miR-140 in
Regulating Growth and Tumorigenesis via the TLR4/NF-κB pathway in
Cholangiocarcinoma. Oncol Res. 27:663–672. 2019.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Lan X and Liu X: LncRNA SNHG1 functions as
a ceRNA to antagonize the effect of miR-145a-5p on the
down-regulation of NUAK1 in nasopharyngeal carcinoma cell. J Cell
Mol Med. 23:2351–2361. 2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Yu Y, Zhang M, Wang N, Li Q, Yang J, Yan
S, He X, Ji G and Miao L: Epigenetic silencing of tumor suppressor
gene CDKN1A by oncogenic long non-coding RNA SNHG1 in
cholangiocarcinoma. Cell Death Dis. 9(746)2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Liu J, Zhang C, Wang J, Hu W and Feng Z:
The regulation of ferroptosis by tumor suppressor p53 and its
pathway. Int J Mol Sci. 21(8387)2020.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Lai W, Zhu W, Li X, Han Y, Wang Y, Leng Q,
Li M and Wen X: GTSE1 promotes prostate cancer cell proliferation
via the SP1/FOXM1 signaling pathway. Lab Invest. 101:554–563.
2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Chen W, Wang H, Lu Y, Huang Y, Xuan Y, Li
X, Guo T, Wang C, Lai D, Wu S, et al: GTSE1 promotes tumor growth
and metastasis by attenuating of KLF4 expression in clear cell
renal cell carcinoma. Lab Invest. 102:1011–1022. 2022.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Stoyanova T, Roy N, Kopanja D,
Raychaudhuri P and Bagchi S: DDB2 (damaged-DNA binding protein 2)
in nucleotide excision repair and DNA damage response. Cell Cycle.
8:4067–4071. 2009.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Hung Y, Huang K, Chen P, Li JL, Lu SH,
Chang JC, Lin HY, Lo WC, Huang SY, Lee TT, et al: UQCRC1 engages
cytochrome c for neuronal apoptotic cell death. Cell Rep.
36(109729)2021.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Lin CH, Tsai PI, Lin HY, Hattori N,
Funayama M, Jeon B, Sato K, Abe K, Mukai Y, Takahashi Y, et al:
Mitochondrial UQCRC1 mutations cause autosomal dominant
parkinsonism with polyneuropathy. Brain. 143:3352–3373.
2020.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Wang Q, Li M, Gan Y, Jiang S, Qiao J,
Zhang W, Fan Y, Shen Y, Song Y, Meng Z, et al: Mitochondrial
Protein UQCRC1 is Oncogenic and a potential therapeutic target for
pancreatic cancer. Theranostics. 10:2141–2157. 2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Torricelli F, Saxena A, Nuamah R, Neat M,
Harling L, Ng W, Spicer J, Ciarrocchi A and Bille A: Genomic
analysis in short- and long-term patients with malignant pleura
mesothelioma treated with palliative chemotherapy. Eur J Cancer.
132:104–111. 2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Sun X, Huang T, Liu Z, Sun M and Luo S:
LncRNA SNHG7 contributes to tumorigenesis and progression in breast
cancer by interacting with miR-34a through EMT initiation and the
Notch-1 pathway. Eur J Pharmacol. 856(172407)2019.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Wang W, Chen S, Song X, Gui J, Li Y and Li
M: ELK1/lncRNA-SNHG7/miR-2682-5p feedback loop enhances bladder
cancer cell growth. Life Sci. 262(118386)2020.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Guo L, Lu J, Gao J, Li M, Wang H and Zhan
X: The function of SNHG7/miR-449a/ACSL1 axis in thyroid cancer. J
Cell Biochem. 121:4034–4042. 2020.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Wang J, Zhang S, Li X and Gong M: LncRNA
SNHG7 promotes cardiac remodeling by upregulating ROCK1 via
sponging miR-34-5p. Aging (Albany NY). 12:10441–10456.
2020.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Liu J, Yao L, Zhang M, Jiang J, Yang M and
Wang Y: Downregulation of LncRNA-XIST inhibited development of
non-small cell lung cancer by activating miR-335/SOD2/ROS signal
pathway mediated pyroptotic cell death. Aging (Albany NY).
11:7830–7846. 2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Shenouda SM, Widlansky ME, Chen K, Xu G,
Holbrook M, Tabit CE, Hamburg NM, Frame AA, Caiano TL, Kluge MA, et
al: Altered mitochondrial dynamics contributes to endothelial
dysfunction in diabetes mellitus. Circulation. 124:444–453.
2011.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Rovira-Llopis S, Banuls C, Diaz-Morales N,
Hernandez-Mijares A, Rocha M and Victor VM: Mitochondrial dynamics
in type 2 diabetes: Pathophysiological implications. Redox Biol.
11:637–645. 2017.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Pinti MV, Fink GK, Hathaway QA, Durr AJ,
Kunovac A and Hollander JM: Mitochondrial dysfunction in type 2
diabetes mellitus: An organ-based analysis. Am J Physiol Endocrinol
Metab. 316:E268–E285. 2019.PubMed/NCBI View Article : Google Scholar
|