In spite of extensive efforts and progress in the
field of cancer research, the mortality rate due to tumors remains
high (1). According to the
recently published official data from the National Bureau of
Statistics of China, in 2018-2020, malignant tumors ranked first
for the constituted ratio of deaths from diseases in cities and
towns in China, and the death rate due to malignant tumors
consistently ranked second or third in rural areas of China
(2). Therefore, in-depth study of
the mechanisms of the occurrence and development of tumors and the
search for novel therapeutic targets and prognostic biomarkers are
of great significance to reduce the mortality rate and improve the
survival rate of patients with tumors. Long non-coding RNAs
(lncRNAs) are a crucial class of RNA molecules and an increasing
number of lncRNAs have been demonstrated to have important roles in
the development and progression of tumors. They may serve as
diagnostic or prognostic biomarkers and therapeutic targets for
cancer (3). Dysregulated lipid
metabolism is a prominent metabolic alteration in cancer, and
increased synthesis or uptake and storage of lipids occur in
tumors, promoting rapid cancer cell growth and tumor formation
(4,5). lncRNAs can reprogram cancer lipid
metabolism pathways and are important factors regulating abnormal
lipid metabolism in cancer (6). In
the present review, the progress in understanding the mechanisms of
lncRNA action and lipid metabolism in cancer were summarized.
lncRNAs are RNA molecules transcribed from the
mammalian genome with a length of >200 nucleotides (7). Although they are void of any
protein-coding function and the roles of most lncRNAs have remained
to be elucidated, they are newly discovered functional ncRNAs that
may exert a wide range of regulatory effects through several
mechanisms of action and are important regulators in gene
expression networks (8-10).
lncRNAs are widely known for their key role in human cancer
progression (11). With the
development of molecular biology and genomics, lncRNAs have emerged
as promising biomarkers for cancer and individual lncRNAs have
different characteristics in different cancers (12). Certain lncRNAs are involved in
important biological processes in cancer development through the
regulation of target genes, including DNA methylation, histone
modification and chromatin remodeling (13), and have considerable roles in the
processes of proliferation, migration and invasion in a variety of
tumors, which are closely related to treatment strategies and
prognoses of these patients. For instance, lncRNA hypoxia-inducible
factor 1α-antisense 2 (AS2) is an oncogenic KRAS-inducible lncRNA
and its high expression may promote lung cancer cell proliferation
and tumor metastasis (14).
Therefore, lncRNAs may be potential therapeutic and prognostic
targets for tumors (15-18).
The pathogenesis of cancer involves precancerous
lesions, gene mutations, oncogenic activation and tumor-suppressor
gene inactivation, which is a complex process. Aberrant expression
of multiple lncRNAs has been implicated in the development of
cancer and is associated with various biological behaviors of
cancer cells, including enhanced metastasis, upregulation of
proliferation, inhibition of apoptosis and altered cell cycle
(19). Both overexpression of
certain lncRNAs with oncogenic functions and inactivation of
lncRNAs with tumor-suppressor properties may promote tumor
development. lncRNAs are able to regulate the activity and
expression of oncogenes by targeting tumor-related genes or
signaling pathways, thereby affecting downstream processes of
autophagy, proliferation and migration of tumor cells (20). lncRNAs may be utilized as
biomarkers for the effectiveness of therapies for the treatment of
patients with tumors, such as immunotherapy, chemotherapy and
surgery (21). For instance,
lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) was
observed to have an important role in patients with tumors treated
with immune checkpoint inhibitors, and those with high NEAT1
expression had good treatment outcomes (22).
LOC100133669 is an lncRNA with a length of 831 nt.
An experimental study indicated that LOC100133669 is mainly
localized in the cytoplasm and upregulated in esophageal cancer
(EC) tissues, as evidenced by an RNA in situ hybridization
assay (23). High LOC100133669
expression promotes the progression of EC, indicating poor
prognosis of patients with EC. The LOC100133669 gene promotes EC
cell proliferation and cell cycle progression, whereas knocking
down LOC100133669 had the opposite effect. LOC100133669, as a novel
oncogene of EC, is a potential diagnostic marker and therapeutic
target for EC.
CASC15 is highly expressed and has an oncogenic role
in lung cancer, breast cancer, GC, CRC, hepatocellular carcinoma
(HCC) and cervical cancer. Furthermore, aberrant expression of
CASC15 is associated with tumorigenesis, progression and patient
prognosis through the regulation of several target genes and
signaling pathways (27). Wu et
al (28) found significantly
elevated CASC15 expression in GC tissues, and high expression of
CASC15 promoted the proliferation, migration and
epithelial-mesenchymal transition (EMT) of GC cells. lncRNAs
contribute to EMT and promote GC progression by regulating cell
cycle progression and affecting the downstream signaling pathways
to promote GC progression through mechanisms involving competing
endogenous RNA (ceRNA). High expression of CASC15 is associated
with poor prognosis (poor survival) of patients with GC and by
analyzing The Cancer Genome Atlas data, expression of CASC15 was
found to be negatively correlated with the overall survival of
patients with GC and positively correlated with tumor size and TNM
stage (29). The findings suggest
that CASC15 has a relatively powerful prognostic value for patients
with GC, a molecular marker with potential oncogenic effects in GC,
and a promising therapeutic target.
FENDRR is a novel lncRNA that is significantly
downregulated in EC and CRC cells, inhibits their proliferation,
migration and invasion, and serves as a prognostic marker for
patients with CRC (33). Luo et
al (34) performed
high-throughput sequencing of tumor tissues obtained from six
patients with diffuse GC, analyzed the expression patterns of
transcriptomes from matched adjacent tissues and identified a group
of representative lncRNAs that were potentially associated with
gastric carcinogenesis. Furthermore, a co-expression network for
dysregulated lncRNAs and mRNAs was constructed and FENDRR was found
to be significantly downregulated in GC, thus inhibiting its
development.
Lipids are a group of highly complex biomolecules
that not only constitute the structural basis of biological
membranes but also function as signaling molecules and energy
sources (38). Lipids or lipid
metabolites promote the proliferation and invasion of cancer cells
by assisting in the synthesis of biofilms and the production of
cholesterol lipids (39). Lipid
metabolism has been a hot spot of research in recent years and
reprogramming lipid metabolism has an important role in the
proliferation and migration of cancer cells. Lipid metabolites also
alter the tumor microenvironment (TME). For instance, the
progression of CRC is accompanied by substantial changes in the
cellular lipidome, including changes in the composition of fatty
acids (FAs), phospholipids and sphingolipids in CRC tissues
(40). Lipid metabolism is a
complex process. Altered lipid metabolism in cancer cells affects
numerous cellular functions, such as autophagy, apoptosis,
necrosis, growth, proliferation, differentiation and chemotherapy
drug resistance (41). Therefore,
elucidating the molecular mechanism by which lipid metabolism
regulates tumor immunity is of great value for understanding tumor
cell immune escape and targeting lipid metabolism, along with
developing novel and promising anticancer treatment regimens
(42-44).
A long-term follow-up of patients with EC who had
undergone radical esophagectomy was performed to investigate the
association between recurrence and lipid metabolism after curative
esophagectomy. The results indicated that an earlier time of
postoperative recurrence was associated with higher serum total
cholesterol levels in patients, suggesting that hyperlipidemia is a
high-risk factor for postoperative recurrence of EC. It was
demonstrated that high and low serum total cholesterol levels have
a certain prognostic value for patients with EC (45).
Adipose tissue triglyceride (TG) lipase (ATGL) is a
novel CRC oncogene and upregulation of ATGL expression was
indicated to promote the proliferation of CRC cells, and
conversely, its knockdown inhibited the proliferation and promote
apoptosis of CRC cells (52). ATGL
is a key lipase for TG hydrolysis and CRC cells with high
expression of ATGL have a greater lipolysis ability. ATGL may also
enhance the proliferation of CRC cells by promoting lipolytic
pathways [degradation of genes related to sphingolipid metabolism
and coenzyme A (CoA) biosynthetic pathway]. ATGL levels are
significantly inversely associated with overall survival of
patients with CRC, and may thus represent a novel and valuable
target for the treatment and prognostication of patients with CRC.
Wen et al (53) found that
increased lipogenesis mediated by SREBP may promote the rapid
proliferation of colon cancer cells, along with the occurrence and
development of colon cancer. Knockdown of SREBP-1 or SREBP-2
inhibited lipid synthesis in colon cancer cells, and the FA and
cholesterol indices were consequently decreased, leading to
inhibition of colon cancer cell proliferation and tumorigenesis.
Therefore, targeting lipid synthesis by SREBP may provide a novel
and promising therapeutic strategy for the treatment of colon
cancer and prognosis of patients.
Numerous studies have indicated that lncRNAs, in
addition to regulating key lipid transcription factors, also alter
lipid storage and FA oxidation by modulating de novo
lipogenesis. lncRNAs reprogram a large portion of cancer lipid
metabolism pathways, regulate lipid metabolism and are key
regulators of abnormal lipid metabolism in cancer. lncRNAs and
lipid metabolism are also involved in the expression of multiple
signaling pathways in the process of tumor occurrence and
development, leading to the activation of oncogenes or the
inactivation of tumor suppressor genes, and promoting rapid growth
of tumors (60,61).
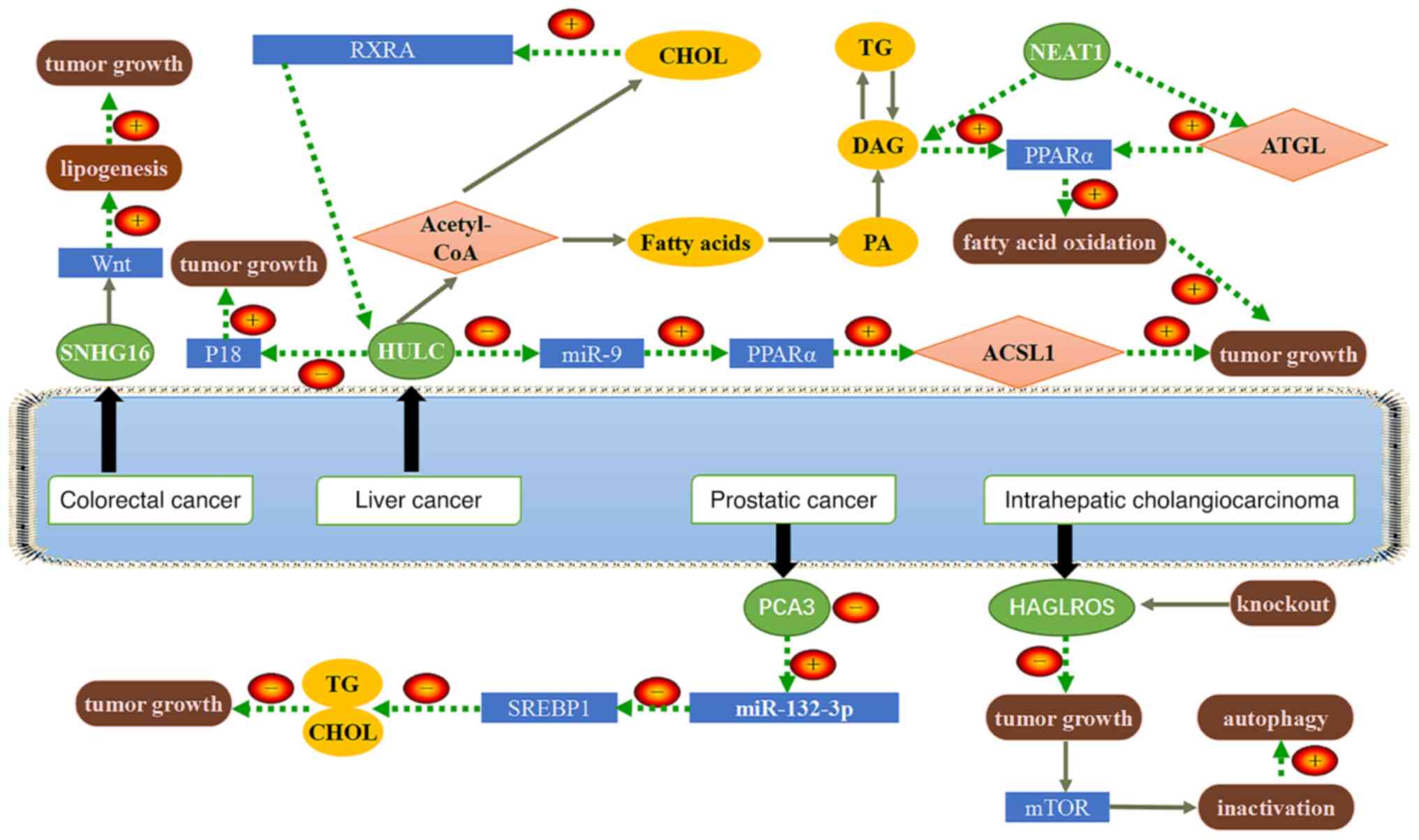
Small nucleolar RNA host gene 16 (SNHG16) is located
at 17q25.1 and is an oncogenic lncRNA. It has been indicated that
SNHG16 is aberrantly expressed in gastrointestinal tumors and
associated with poor prognosis (64). An experimental study demonstrated
that SNHG16 and SREBP-2 promote the proliferation, migration,
invasion and lipogenesis of pancreatic cancer cells, while miR-195
inhibits the above processes (65). SREBP-2 is a potential downstream
target of miR-195 and SNHG16 regulates SREBP-2 expression by
directly targeting miR-195, thereby inducing SREBP-2 to promote
pancreatic cancer growth and lipogenesis in pancreatic cancer
cells.
It has been indicated that SNHG16 may have an
oncogenic role in CRC, targeting the Wnt signaling pathway to
promote lipogenesis (70).
Therefore, SNHG16 is a promising tumor biomarker and therapeutic
target. Experimental results have demonstrated that miR-132-3p can
directly interact with SREBP-1; high expression of the lncRNA
prostate cancer antigen 3 promoted the growth of prostate cancer
and inhibited TG and cholesterol levels in prostate cancer cells
through the miR-132-3p/SREBP-1 axis (71). HAGLR opposite strand long
non-coding RNA (lncRNA HAGLROS) is highly expressed in intrahepatic
cholangiocarcinoma (ICC) and has an oncogenic role. Knockdown of
HAGLROS inhibited ICC proliferation, inactivated the mTOR axis and
promoted autophagy. Lipid metabolic reprogramming of ICC was
indicated to be promoted by inactivation of the mTOR pathway.
Patients with ICC having high expression of HAGLROS had poor
prognosis and HAGLROS may be used as a biomarker to predict the
treatment efficacy of ICC (72).
NEAT1 is able to increase FA oxidation through the direct
activation of ATGL or indirect activation of the PPARα signaling
pathway by FFA and DAG, whereas miR-124-3p, a gene downstream of
NEAT1, inhibits ATGL, thereby attenuating FA oxidation, so the
tumor grows (60). The above
findings indicated that lncRNAs and lipid metabolism are associated
with multiple signal transduction pathways and the downstream
specific signal transduction mechanisms require further
investigation.
Long intergenic non-protein coding RNA 00514
(LINC00514) is a novel lncRNA. Overexpression of LINC00514 promotes
the proliferation and lipogenesis of EC cells. Conversely, knocking
down LINC00514 inhibits EC cell proliferation and reduces
lipogenesis. These results suggest that LINC00514 may participate
in EC lipogenesis and regulate alterations in EC lipid metabolism,
and thus, targeting LINC00514 may provide a new strategy for the
treatment of patients with EC (73). The lncRNA RP11-386G11.10 is a ceRNA
for miR-345-3p and high expression of RP11-386G11.10 results in
elevated expression of both heterogeneous nuclear ribonucleoprotein
U (HNRNPU), a multifunctional protein that regulates precursor
messenger ribonucleic acid, and its downstream lipogenic enzymes,
further causing lipid accumulation in HCC cells. HNRNPU also
promotes the expression of the transcription factor of
RP11-386G11.10, as well as zinc finger and BTB domain containing 7A
(ZBTB7A), in HCC cells. The ZBTB7A/RP11-386G11.10/HNRNPU positive
feedback loop promotes HCC cell growth and metastasis by regulating
lipid anabolism (74). Certain
lncRNAs promote cancer development by regulating sphingolipid
metabolism-related enzymes, such as ceramide synthase (CERS) and
targeting these lncRNAs may provide new avenues for treating
cancer. For instance, CERS6-AS1, an antisense lncRNA for CERS6, is
significantly upregulated in breast cancer cells and binds to
insulin-like growth factor 2 mRNA-binding protein 3 to maintain
CERS6 mRNA stability and promote cancer progression (75). In Table I (76-87),
certain lncRNAs that regulate tumor lipid metabolism are presented,
aiming to more comprehensively elaborate on the current research
progress in this field. For instance, lncROPM is highly expressed
in breast cancer cells, and high expression of ROPM activates the
phosphatidylinositol 3-kinase/AKT, Wnt/β-catenin and Hippo/YAP
signaling pathways, thereby promoting adipogenesis and the
development of breast cancer (78). Zuo et al (82) found that the expression of
LINC00958, an lncRNA associated with adipogenesis, was upregulated
in HCC cells. Dual luciferase reporter, RNA immunoprecipitation,
biotin-labelled miRNA pull-down and fluorescence in situ
hybridisation showed that LINC00958 sponged miR-3619-5p to promote
HCC progression. Bioinformatics and RNA sequencing results
identified hepatocellular carcinoma-derived growth factor as a
downstream signalling molecule of the LINC00958/miR-3619-5p axis.
In vitro cellular experiments demonstrated that the knocked
down expression of miR-3619-5p resulted in increased expression
levels of hepatocellular carcinoma-derived growth factor. In
summary, overexpression of LINC00958 inhibited miR-3619-5p
expression, which upregulated hepatocellular carcinoma-derived
growth factor expression and promoted HCC adipogenesis and
progression. Lu et al (86)
found that increased HAGLR expression may activate the expression
of FASN, MMP-9 and p21 in non-small cell lung cancer (NSCLC) cells,
which in turn promotes NSCLC progression.
In summary, lncRNAs are involved in the
proliferation, migration and invasion of cancer cells.
Simultaneously, our understanding of the changes in lipid
metabolism during the occurrence and development of tumors has
markedly improved and lipid metabolism is one of the most
significant metabolic alterations in tumors. Existing studies have
indicated that lipid metabolic reprogramming and lncRNAs have a key
role in the occurrence and development of tumors; however, the
underlying mechanism has remained to be fully elucidated and
requires further research. Targeting lncRNAs and reprogramming
lipid metabolism may provide potential therapeutic targets and
prognostic monitoring indicators for cancer treatment. Therefore,
in-depth research on the role of lncRNAs and lipid metabolism in
tumor development is expected to facilitate the development of
clinical applications of lncRNAs, thereby providing a reference for
early diagnosis and prognostic evaluation of patients with cancer
and the development of novel anti-tumor drugs.
Not applicable.
Funding: The present study was supported by grants from the
Natural Science Foundation Project of Xizang Autonomous Region
(grant no. XZ202101ZR0074G), the major cultivation project of
Xizang Minzu University (grant no. 20MDT02), the National
Innovation and Entrepreneurship Training Program for College
Students in 2022 (grant no. 202210695033) and the Natural Science
Basic Research Plan in Shaanxi Province of China (grant nos.
2020JM-590 and 2022JM-465).
Not applicable.
ZZ, XH, XC and XW retrieved the relevant literature
and drafted the manuscript. ZZ and XW participated in the design of
the review and drafted the manuscript. ZZ critically revised the
manuscript for important intellectual content. All authors have
read and approved the final manuscript. Data authentication is not
applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Xia C, Dong X, Li H, Cao M, Sun D, He S,
Yang F, Yan X, Zhang S, Li N and Chen W: Cancer statistics in China
and United States, 2022: Profiles, trends, and determinants. Chin
Med J (Engl). 135:584–590. 2022.PubMed/NCBI View Article : Google Scholar
|
|
2
|
National Bureau of Statistics of China.
[Cited 2023 June 30]. China Statistical Yearbook, 2023. Available
from: http://www.stats.gov.cn/tjsj/ndsj/.
|
|
3
|
McCabe EM and Rasmussen TP: lncRNA
involvement in cancer stem cell function and epithelial-mesenchymal
transitions. Semin Cancer Biol. 75:38–48. 2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Cheng C, Geng F, Cheng X and Guo D: Lipid
metabolism reprogramming and its potential targets in cancer.
Cancer Commun (Lond). 38(27)2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Bian X, Liu R, Meng Y, Xing D, Xu D and Lu
Z: Lipid metabolism and cancer. J Exp Med.
218(e20201606)2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Liu C, Li H, Chu F, Zhou X, Xie R, Wei Q,
Yang S, Li T, Liang S and Lü M: Long non-coding RNAs: Key
regulators involved in metabolic reprogramming in cancer (review).
Oncol Rep. 45(54)2021.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Ghafouri-Fard S and Taheri M: Long
non-coding RNA signature in gastric cancer. Exp Mol Pathol.
113(104365)2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Taniue K and Akimitsu N: The functions and
unique features of LncRNAs in cancer development and tumorigenesis.
Int J Mol Sci. 22(632)2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Statello L, Guo CJ, Chen LL and Huarte M:
Gene regulation by long non-coding RNAs and its biological
functions. Nat Rev Mol Cell Bio. 22:96–118. 2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Park EG, Pyo SJ, Cui Y, Yoon SH and Nam
JW: Tumor immune microenvironment lncRNAs. Brief Bioinform.
23(bbab504)2022.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Shi L, Duan R, Sun Z, Jia Q, Wu W, Wang F,
Liu J, Zhang H and Xue X: LncRNA GLTC targets LDHA for
succinylation and enzymatic activity to promote progression and
radioiodine resistance in papillary thyroid cancer. Cell Death
Differ. 30:1517–1532. 2023.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Mas AM and Huarte M: Long Noncoding RNA
signatures as cancer biomarkers. J Clin Oncol. 41:3059–3062.
2023.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Huang Z, Zhang M, Li J and Lou C: Long
non-coding RNA MAFG-AS1: A promising therapeutic target for human
cancers. Biomed Pharmacother. 163(114756)2023.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Yang K, Zhang W, Zhong L, Xiao Y, Sahoo S,
Fassan M, Zeng K, Magee P, Garofalo M and Shi L: Long non-coding
RNA HIF1A-As2 and MYC form a double-positive feedback loop to
promote cell proliferation and metastasis in KRAS-driven non-small
cell lung cancer. Cell Death Differ. 30:1533–1549. 2023.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Abdi E, Latifi-Navid S, Abdi F and
Taherian-Esfahani Z: Emerging circulating MiRNAs and LncRNAs in
upper gastrointestinal cancers. Expert Rev Mol Diagn. 20:1121–1138.
2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Qi FF, Yang Y, Zhang H and Chen H: Long
non-coding RNAs: Key regulators in oxaliplatin resistance of
colorectal cancer. Biomed Pharmacother. 128(110329)2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Chen M, Zhang C, Liu W, Du X, Liu X and
Xing B: Long noncoding RNA LINC01234 promotes hepatocellular
carcinoma progression through orchestrating aspartate metabolic
reprogramming. Mol Ther. 30:2354–2369. 2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhang Y, Liu Q and Liao Q: Long noncoding
RNA: A dazzling dancer in tumor immune microenvironment. J Exp Clin
Canc Res. 39(231)2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Xue W, Zheng Y, Shen Z, Li L, Fan Z, Wang
W, Zhu Z, Zhai Y, Zhao J and Kan Q: Involvement of long non-coding
RNAs in the progression of esophageal cancer. Cancer Commun (Lond).
41:371–388. 2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhang H, Wang J, Wang Y, Li J, Zhao L,
Zhang T and Liao X: Long non-coding LEF1-AS1 sponge miR-5100
regulates apoptosis and autophagy in gastric cancer cells via the
miR-5100/DEK/AMPK-mTOR axis. Int J Mol Sci. 23(4787)2022.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Verma S, Sahu BD and Mugale MN: Role of
lncRNAs in hepatocellular carcinoma. Life Sci.
325(121751)2023.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Toker J, Iorgulescu JB, Ling AL, Villa GR,
Gadet JAMA, Parida L, Getz G, Wu CJ, Reardon DA, Chiocca EA and
Mineo M: Clinical importance of the lncRNA NEAT1 in cancer patients
treated with immune checkpoint inhibitors. Clin Cancer Res.
29:2226–2238. 2023.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Guan Z, Wang Y, Wang Y, Liu X, Wang Y,
Zhang W, Chi X, Dong Y, Liu X, Shao S and Zhan Q: Long non-coding
RNA LOC100133669 promotes cell proliferation in oesophageal
squamous cell carcinoma. Cell Prolif. 53(e12750)2020.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Xing C, Sun SG, Yue ZQ and Bai F: Role of
lncRNA LUCAT1 in cancer. Biomed Pharmacother.
134(111158)2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Chi J, Liu T, Shi C, Luo H, Wu Z, Xiong B,
Liu S and Zeng Y: Long non-coding RNA LUCAT1 promotes proliferation
and invasion in gastric cancer by regulating miR-134-5p/YWHAZ axis.
Biomed Pharmacother. 118(109201)2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhou Q, Hou Z, Zuo S, Zhou X, Feng Y, Sun
Y and Yuan X: LUCAT1 promotes colorectal cancer tumorigenesis by
targeting the ribosomal protein L40-MDM2-p53 pathway through
binding with UBA52. Cancer Sci. 110:1194–1207. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Gu X, Chu Q, Zheng Q, Wang J and Zhu H:
The dual functions of the long noncoding RNA CASC15 in malignancy.
Biomed Pharmacother. 135(111212)2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Wu Q, Xiang S, Ma J, Hui P, Wang T, Meng
W, Shi M and Wang Y: Long non-coding RNA CASC15 regulates gastric
cancer cell proliferation, migration and epithelial mesenchymal
transition by targeting CDKN1A and ZEB1. Mol Oncol. 12:799–813.
2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Yao XM, Tang JH, Zhu H and Jing Y: High
expression of LncRNA CASC15 is a risk factor for gastric cancer
prognosis and promote the proliferation of gastric cancer. Eur Rev
Med Pharmaco. 21:5661–5667. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ghafouri-Fard S, Esmaeili M and Taheri M:
H19 lncRNA: Roles in tumorigenesis. Biomed Pharmacother.
123(109774)2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Hashemi M, Moosavi MS, Abed HM, Dehghani
M, Aalipour M, Heydari EA, Behroozaghdam M, Entezari M,
Salimimoghadam S, Gunduz ES, et al: Long non-coding RNA (lncRNA)
H19 in human cancer: From proliferation and metastasis to therapy.
Pharmacol Res. 184(106418)2022.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ding D, Li C, Zhao T, Li D, Yang L and
Zhang B: LncRNA H19/miR-29b-3p/PGRN axis promoted
epithelial-mesenchymal transition of colorectal cancer cells by
acting on Wnt signaling. Mol Cells. 41:423–435. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zheng Q, Zhang Q, Yu X, He Y and Guo W:
FENDRR: A pivotal, cancer-related, long non-coding RNA. Biomed
Pharmacother. 137(111390)2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Luo T, Zhao J, Lu Z, Bi J, Pang T, Cui H,
Yang B, Li W, Wang Y, Wu S and Xue X: Characterization of long
non-coding RNAs and MEF2C-AS1 identified as a novel biomarker in
diffuse gastric cancer. Transl Oncol. 11:1080–1089. 2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Ghafouri-Fard S, Shoorei H, Dashti S,
Branicki W and Taheri M: Expression profile of lncRNAs and miRNAs
in esophageal cancer: Implications in diagnosis, prognosis, and
therapeutic response. J Cell Physiol. 235:9269–9290.
2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Yang Y, Jin L, He J, Wang R, Wang Y, Bai
J, Chen Y and Luo Z: Upregulation LncRNA MEG3 expression suppresses
proliferation and metastasis in melanoma via miR-208/SOX4. Mol Cell
Biochem. 478:407–414. 2023.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Soghala S, Harsiny K, Momeni P, Hatami M,
Kholghi Oskooei V, Hussen BM, Taheri M and Ghafouri-Fard S:
Down-regulation of LINC-ROR, HOXA-AS2 and MEG3 in gastric cancer.
Heliyon. 8(e11155)2022.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Butler LM, Perone Y, Dehairs J, Lupien LE,
de Laat V, Talebi A, Loda M, Kinlaw WB and Swinnen JV: Lipids and
cancer: Emerging roles in pathogenesis, diagnosis and therapeutic
intervention. Adv Drug Deliver Rev. 159:245–293. 2020.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Liu X, Hu J and Liu B: Characteristics and
clinical significance of lipid metabolism in patients with
gastrointestinal stromal tumor. Lipids Health Dis.
21(1)2022.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Machala M, Procházková J, Hofmanová J,
Králiková L, Slavík J, Tylichová Z, Ovesná P, Kozubík A and
Vondráček J: Colon cancer and perturbations of the sphingolipid
metabolism. Int J Mol Sci. 20(6051)2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Pakiet A, Kobiela J, Stepnowski P,
Sledzinski T and Mika A: Changes in lipids composition and
metabolism in colorectal cancer: A review. Lipids Health Dis.
18(29)2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Zheng M, Wang W, Liu J, Zhang X and Zhang
R: Lipid metabolism in cancer cells. Adv Exp Med Biol. 1316:49–69.
2021.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Broadfield LA, Pane AA, Talebi A, Swinnen
JV and Fendt SM: Lipid metabolism in cancer: New perspectives and
emerging mechanisms. Dev Cell. 56:1363–1393. 2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Bacci M, Lorito N, Smiriglia A and Morandi
A: Fat and furious: Lipid metabolism in antitumoral therapy
response and resistance. Trends Cancer. 7:198–213. 2021.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Zheng L, Jiang J, Liu Y, Zheng X and Wu C:
Correlations of recurrence after radical surgery for esophageal
cancer with glucose-lipid metabolism, insulin resistance,
inflammation, stress and serum p53 expression. J BUON.
24:1666–1672. 2019.PubMed/NCBI
|
|
46
|
Luo Q, Zheng N, Jiang L, Wang T, Zhang P,
Liu Y, Zheng P, Wang W, Xie G, Chen L, et al: Lipid accumulation in
macrophages confers protumorigenic polarization and immunity in
gastric cancer. Cancer Sci. 111:4000–4011. 2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Mejia JC and Pasko J: Primary liver
cancers: intrahepatic cholangiocarcinoma and hepatocellular
carcinoma. Surg Clin North Am. 100:535–549. 2020.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Tiong TY, Weng PW, Wang CH, Setiawan SA,
Yadav VK, Pikatan NW, Fong IH, Yeh CT, Hsu CH and Kuo KT: Targeting
the SREBP-1/Hsa-Mir-497/SCAP/FASN oncometabolic axis inhibits the
cancer stem-like and chemoresistant phenotype of non-small cell
lung carcinoma cells. Int J Mol Sci. 23(7283)2022.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Sun Q, Yu X, Peng C, Liu N, Chen W, Xu H,
Wei H, Fang K, Dong Z, Fu C, et al: Activation of SREBP-1c alters
lipogenesis and promotes tumor growth and metastasis in gastric
cancer. Biomed Pharmacother. 128(110274)2020.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Pascual G, Domínguez D, Elosúa-Bayes M,
Beckedorff F, Laudanna C, Bigas C, Douillet D, Greco C, Symeonidi
A, Hernández I, et al: Dietary palmitic acid promotes a
prometastatic memory via Schwann cells. Nature. 599:485–490.
2021.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Zhang X, Li X, Xiong G, Yun F, Feng Y, Ni
Q, Wu N, Yang L, Yi Z, Zhang Q, et al: Palmitic acid promotes lung
metastasis of melanomas via the TLR4/TRIF-Peli1-pNF-κB pathway.
Metabolites. 12(1132)2022.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Yin H, Li W, Mo L, Deng S, Lin W, Ma C,
Luo Z, Luo C and Hong H: Adipose triglyceride lipase promotes the
proliferation of colorectal cancer cells via enhancing the
lipolytic pathway. J Cell Mol Med. 25:3963–3975. 2021.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Wen YA, Xiong X, Zaytseva YY, Napier DL,
Vallee E, Li AT, Wang C, Weiss HL, Evers BM and Gao T:
Downregulation of SREBP inhibits tumor growth and initiation by
altering cellular metabolism in colon cancer. Cell Death Dis.
9(265)2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Hall Z, Chiarugi D, Charidemou E, Leslie
J, Scott E, Pellegrinet L, Allison M, Mocciaro G, Anstee QM, Evan
GI, et al: Lipid remodeling in hepatocyte proliferation and
hepatocellular carcinoma. Hepatology. 73:1028–1044. 2021.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Wang Z, Qin H, Liu S, Sheng J and Zhang X:
Precision diagnosis of hepatocellular carcinoma. Chin Med J (Engl).
136:1155–1165. 2023.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Pope ER III, Kimbrough EO, Vemireddy LP,
Surapaneni PK, Copland JR III and Mody K: Aberrant lipid metabolism
as a therapeutic target in liver cancer. Expert Opin Ther Tar.
23:473–483. 2019.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Li D and Li Y: The interaction between
ferroptosis and lipid metabolism in cancer. Signal Transduct Target
Ther. 5(108)2020.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Chen J, Ding C, Chen Y, Hu W, Yu C, Peng
C, Feng X, Cheng Q, Wu W, Lu Y, et al: ACSL4 reprograms fatty acid
metabolism in hepatocellular carcinoma via c-Myc/SREBP1 pathway.
Cancer Lett. 502:154–165. 2021.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Buechler C and Aslanidis C: Role of lipids
in pathophysiology, diagnosis and therapy of hepatocellular
carcinoma. Biochim Biophys Acta Mol Cell Biol Lipids.
1865(158658)2020.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Lin W, Zhou Q, Wang CQ, Zhu L, Bi C, Zhang
S, Wang X and Jin H: LncRNAs regulate metabolism in cancer. Int J
Biol Sci. 16:1194–1206. 2020.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Xu Y, Qiu M, Shen M, Dong S, Ye G, Shi X
and Sun M: The emerging regulatory roles of long non-coding RNAs
implicated in cancer metabolism. Mol Ther. 29:2209–2218.
2021.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Li D, Cheng M, Niu Y, Chi X, Liu X, Fan J,
Fan H, Chang Y and Yang W: Identification of a novel human long
non-coding RNA that regulates hepatic lipid metabolism by
inhibiting SREBP-1c. Int J Biol Sci. 13:349–357. 2017.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Li D, Guo L, Deng B, Li M, Yang T, Yang F
and Yang Z: Long non-coding RNA HR1 participates in the expression
of SREBP-1c through phosphorylation of the PDK1/AKT/FoxO1 pathway.
Mol Med Rep. 18:2850–2856. 2018.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Gong CY, Tang R, Nan W, Zhou KS and Zhang
HH: Role of SNHG16 in human cancer. Clin Chim Acta. 503:175–180.
2020.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Yu Y, Dong JT, He B, Zou YF, Li XS, Xi CH
and Yu Y: LncRNA SNHG16 induces the SREBP2 to promote lipogenesis
and enhance the progression of pancreatic cancer. Future Oncol.
15:3831–3844. 2019.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Panzitt K, Tschernatsch MM, Guelly C,
Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder
R, Trauner M and Zatloukal K: Characterization of HULC, a novel
gene with striking up-regulation in hepatocellular carcinoma, as
noncoding RNA. Gastroenterology. 132:330–342. 2007.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Du Y, Kong G, You X, Zhang S, Zhang T, Gao
Y, Ye L and Zhang X: Elevation of highly up-regulated in liver
cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell
proliferation via down-regulating p18. J Biol Chem.
287:26302–26311. 2012.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Cui M, Xiao Z, Wang Y, Zheng M, Song T,
Cai X, Sun B, Ye L and Zhang X: Long noncoding RNA HULC modulates
abnormal lipid metabolism in hepatoma cells through an
miR-9-mediated RXRA signaling pathway. Cancer Res. 75:846–857.
2015.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Hu B, Lin JZ, Yang XB and Sang XT:
Aberrant lipid metabolism in hepatocellular carcinoma cells as well
as immune microenvironment: A review. Cell Prolif.
53(e12772)2020.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Christensen LL, True K, Hamilton MP,
Nielsen MM, Damas ND, Damgaard CK, Ongen H, Dermitzakis E, Bramsen
JB, Pedersen JS, et al: SNHG16 is regulated by the Wnt pathway in
colorectal cancer and affects genes involved in lipid metabolism.
Mol Oncol. 10:1266–1282. 2016.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Guo S, Zhang Y, Wang S, Yang T, Ma B, Li
X, Zhang Y and Jiang X: LncRNA PCA3 promotes antimony-induced lipid
metabolic disorder in prostate cancer by targeting MIR-132-3
P/SREBP1 signaling. Toxicol Lett. 348:50–58. 2021.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Ma J, Feng J and Zhou X: Long non-coding
RNA HAGLROS regulates lipid metabolism reprogramming in
intrahepatic cholangiocarcinoma via the mTOR signaling pathway. Exp
Mol Pathol. 115(104466)2020.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Wang X, Liu H, Zhang Q, Zhang X, Qin Y,
Zhu G, Dang J, Wang F, Yang X and Fan R: LINC00514 promotes
lipogenesis and tumor progression in esophageal squamous cell
carcinoma by sponging miR-378a-5p to enhance SPHK1 expression. Int
J Oncol. 59(86)2021.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Xu K, Xia P, Gongye X, Zhang X, Ma S, Chen
Z, Zhang H, Liu J, Liu Y, Guo Y, et al: A novel lncRNA
RP11-386G11.10 reprograms lipid metabolism to promote
hepatocellular carcinoma progression. Mol Metab.
63(101540)2022.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Duan J, Huang Z, Nice EC, Xie N, Chen M
and Huang C: Current advancements and future perspectives of long
noncoding RNAs in lipid metabolism and signaling. J Adv Res.
48:105–123. 2023.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Chen J, Alduais Y, Zhang K, Zhu X and Chen
B: CCAT1/FABP5 promotes tumour progression through mediating fatty
acid metabolism and stabilizing PI3K/AKT/mTOR signalling in lung
adenocarcinoma. J Cell Mol Med. 25:9199–9213. 2021.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Wang H, Zhang Y, Guan X, Li X, Zhao Z, Gao
Y, Zhang X and Chen R: An integrated transcriptomics and proteomics
analysis implicates lncRNA MALAT1 in the regulation of lipid
metabolism. Mol Cell Proteomics. 20(100141)2021.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Liu S, Sun Y, Hou Y, Yang L, Wan X, Qin Y,
Liu Y, Wang R, Zhu P, Teng Y and Liu M: A novel lncRNA
ROPM-mediated lipid metabolism governs breast cancer stem cell
properties. J Hematol Oncol. 14(178)2021.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Liu Y, Li C, Fang L, Wang L, Liu H, Tian
H, Zheng Y, Fan T and He J: Lipid metabolism-related lncRNA
SLC25A21-AS1 promotes the progression of oesophageal squamous cell
carcinoma by regulating the NPM1/c-Myc axis and SLC25A21
expression. Clin Transl Med. 12(e944)2022.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Tito C, Ganci F, Sacconi A, Masciarelli S,
Fontemaggi G, Pulito C, Gallo E, Laquintana V, Iaiza A, De Angelis
L, et al: LINC00174 is a novel prognostic factor in thymic
epithelial tumors involved in cell migration and lipid metabolism.
Cell Death Dis. 11(959)2020.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Bo H, Zhang W, Zhong X, Chen J, Liu Y,
Cheong KL, Fan P and Tang S: LINC00467, driven by copy number
amplification and DNA demethylation, is associated with oxidative
lipid metabolism and immune infiltration in breast cancer. Oxid Med
Cell Longev. 2021(4586319)2021.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Zuo X, Chen Z, Gao W, Zhang Y, Wang J,
Wang J, Cao M, Cai J, Wu J and Wang X: M6A-mediated upregulation of
LINC00958 increases lipogenesis and acts as a nanotherapeutic
target in hepatocellular carcinoma. J Hematol Oncol.
13(5)2020.PubMed/NCBI View Article : Google Scholar
|
|
83
|
He W, Liang B, Wang C, Li S, Zhao Y, Huang
Q, Liu Z, Yao Z, Wu Q, Liao W, et al: MSC-regulated lncRNA
MACC1-AS1 promotes stemness and chemoresistance through fatty acid
oxidation in gastric cancer. Oncogene. 38:4637–4654.
2019.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Lin Y, Xiao Y, Liu S, Hong L, Shao L and
Wu J: Role of a lipid metabolism-related lncRNA signature in risk
stratification and immune microenvironment for colon cancer. Bmc
Med Genomics. 15(221)2022.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Liu X, Liang Y, Song R, Yang G, Han J, Lan
Y, Pan S, Zhu M, Liu Y, Wang Y, et al: Long non-coding RNA
NEAT1-modulated abnormal lipolysis via ATGL drives hepatocellular
carcinoma proliferation. Mol Cancer. 17(90)2018.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Lu C, Ma J and Cai D: Increased HAGLR
expression promotes non-small cell lung cancer proliferation and
invasion via enhanced de novo lipogenesis. Tumour Biol.
39(1010428317697574)2017.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Mazar J, Zhao W, Khalil AM, Lee B, Shelley
J, Govindarajan SS, Yamamoto F, Ratnam M, Aftab MN, Collins S, et
al: The functional characterization of long noncoding RNA SPRY4-IT1
in human melanoma cells. Oncotarget. 5:8959–8969. 2014.PubMed/NCBI View Article : Google Scholar
|