Introduction
Inflammatory bowel diseases (IBDs) are recognized as
chronic inflammatory disorders of the gastrointestinal tract, and
include ulcerative colitis (UC) and Crohn's disease (CD) (1). Notably, the global incidence of CD
has been on the rise in recent years. Over the past six decades in
Western countries, the incidence of CD has gradually risen to
50-200/100,000 individuals. In some other countries with fewer
reported cases of CD, such as Japan and South Korea, there has also
been a trend of increasing incidence (2,3). CD
typically manifests in the terminal ileum, with invasion into the
surrounding intestinal tissues (4). In addition, CD carries a
predisposition for dysplasia and colorectal cancer (CRC)
development (5), with a ~22% risk
of cancer development in patients with long-standing CD (6). Furthermore, the manifestations of CD
commonly emerge in the advanced stages, frequently with patients
already presenting severe intestinal lesions when symptoms become
evident (2). Current therapeutic
approaches include surgical resection, administration of
anti-inflammatory medications or immunosuppressants (2). However, the outcomes have been
somewhat unsatisfactory, as a majority of patients encounter
short-term recurrence (7).
Ferroptosis, a newly discovered form of programmed
cell death, has been implicated in various benign and malignant
diseases of the digestive system (8); however, the mechanisms underlying CD
associated with ferroptosis-related genes remain incompletely
understood. Ferroptosis is a form of cell death that differs from
apoptosis and pyroptosis. The main causes are lipid peroxidation,
iron accumulation and cell membrane breakdown (9). Furthermore, it has been reported that
ferroptosis can participate in the regulation of IBD (10).
Although ferroptosis is strongly associated with
certain digestive tract-related diseases, the mechanisms involved
in ferroptosis associated with CD are not understood. Therefore,
ferroptosis-related genes in CD were searched for in order to
determine the association between CD and ferroptosis. GSE102133 and
GSE75214 were used as target datasets to identify differentially
expressed genes (DEGs) in CD and these genes were cross-compared
with ferroptosis-related genes to determine the object of study.
The present study provided new insights into the relationship
between CD and ferroptosis, and may provide novel biomarkers for
further study in CD research to validate their potential effect on
clinical diagnosis and treatment.
Materials and methods
Source of data acquisition
CD-related microarray sequencing datasets were
searched in the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) for analysis
in the present study; the key word was ‘crohn disease’. The
following screening criteria were used: i) samples from human
tissues; ii) the dataset contained microarray expression data; iii)
the dataset was acquired from CD ileal mucosa and healthy ileal
mucosa; iv) the total number of samples was >10; and v) the
number of DEGs was >100. In this manner, two GEO datasets,
GSE75214 and GSE102133 (11,12),
were enrolled in the present study. These two datasets were based
on the GPL6244 platform (Affymetrix Human Gene 1.0 ST Array;
Affymetrix; Thermo Fisher Scientific, Inc.); GSE102133 included 65
patients with CD and 12 controls, and GSE75214 contained 67
patients with CD and 11 controls. Specific information regarding
the two GSE datasets is shown in Table
I. Ferroptosis-related genes were obtained from the FerrDb
website (http://www.zhounan.org/ferrdb/index.html), and 259
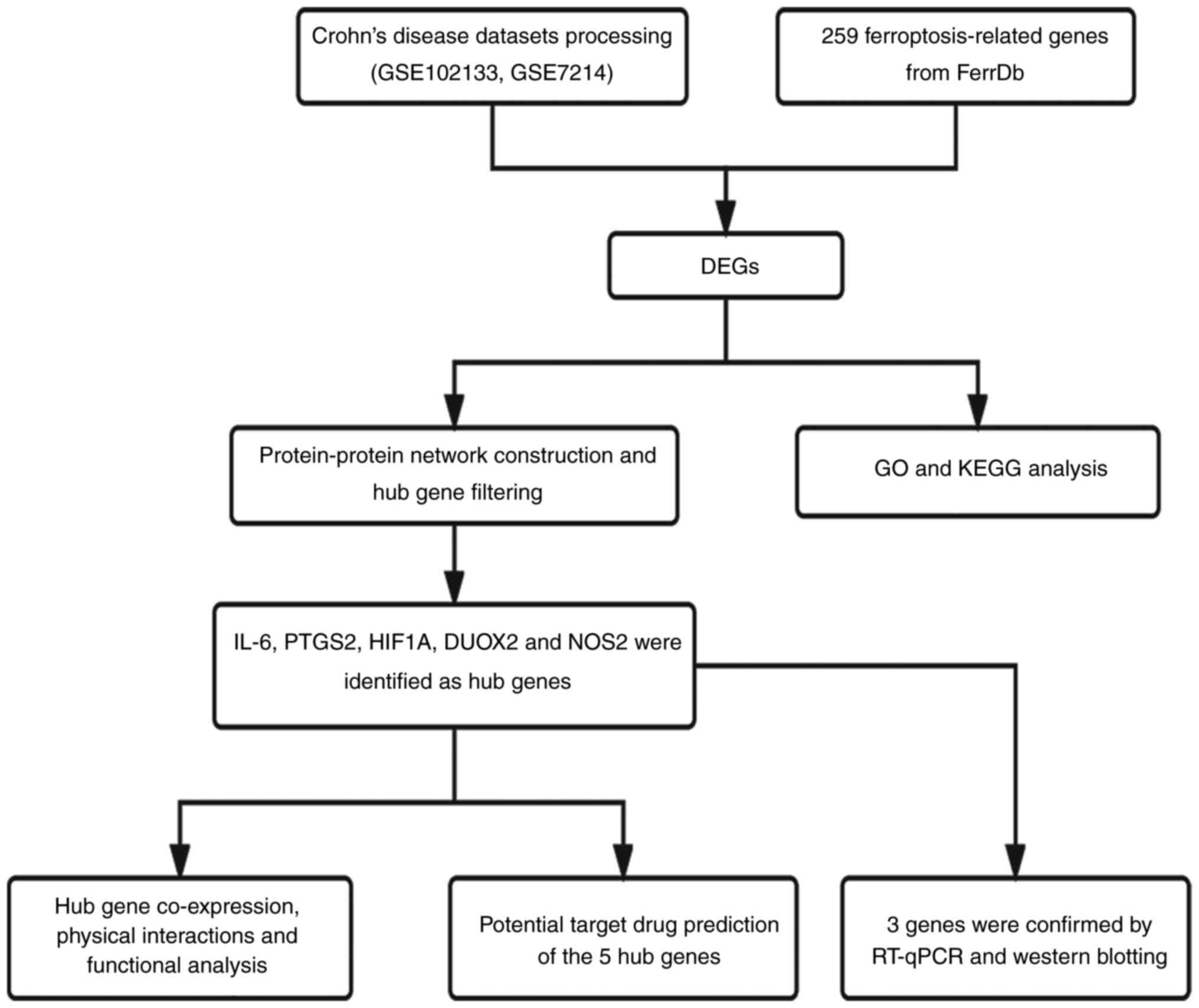
genes were found. The overall research process of the present study
is shown in Fig. 1.
 | Table IGEO dataset information. |
Table I
GEO dataset information.
| | GEO dataset |
|---|
| Variable | GSE75214 | GSE102133 |
|---|
| Platform | GPL6244 | GPL6244 |
| Organism | Homo
sapiens | Homo
sapiens |
| Contributor(s) | Vancamelbeke et
al, 2017(11) | Verstockt et
al, 2019(12) |
| Organization
name | TARGID-IBD Leuven,
Clinical and Experimental Medicine of KU Leuven | KU Leuven |
| Sample site | Terminal ileum | Ileal mucosal
biopsies |
| Sample number
(Crohn's disease and control) | 78 (67 CD and 11
Control) | 77 (65 CD and12
Control) |
| Last update | July 26, 2018 | July 25, 2021 |
Identification of ferroptosis-related
DEGs in CD
All microarray data were obtained from the GEO
database and the extracted data were normalized by log2
transformation. Probes were converted to gene symbols according to
the annotation information for the normalized data in the platform.
A principal component analysis (PCA) plot was generated between the
two datasets. Data were standardized using the Limma toolkit
(https://bioconductor.org/packages/release/bioc/html/limma.html)
in R software (version 4.1.2; http://www.R-project.org/). |Log2 fold change
(FC)|>1 and P<0.05 were used as the criteria for differential
gene expression. The heatmap and volcano plot were drawn using the
‘ComplexHeatmap’ (https://bioconductor.org/packages/release/bioc/html/ComplexHeatmap.html)
and ‘ggplot2’ (https://ggplot2.tidyverse.org/) packages in R
software. The DEGs were intersected with ferroptosis-related genes
from FerrDb, and a Venn diagram was generated using the online
Bioinformatics tool (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Gene Ontology (GO) functional
enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway analyses
To further investigate the biological function of
the genes involved, GO enrichment analysis was performed, which
included biological process (BP), molecular function (MF) and
cellular component (CC), and KEGG pathway enrichment analysis
(13-15)
was conducted to evaluate related functions. The R package
‘clusterProfiler’(https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html)
toolkit in R software was used for these calculations. P<0.05
was considered to indicate a significant difference. The chord
diagram of GO enrichment terms was generated using the circlize R
package (version 0.4.1; https://cran.r-project.org/web/packages/circlize/index.html).
Gene correlation analysis was performed using the Pearson
correlation test to assess reciprocal relationships at the mRNA
level in CD.
Constructing a protein-protein
interaction (PPI) network and hub gene selection
To further investigate the interactions between DEG
proteins, PPI network analysis was performed using the Search Tool
for the Retrieval of Interacting Genes (STRING) database
(https://string-db.org/). The PPI network analysis
map was visualized by Cytoscape software (version 3.9.1; https://cytoscape.org/). First, the STRING database
was used to establish the PPI network according to the DEGs, after
which the analysis results were exported to Cytoscape for mapping.
Nodes represent proteins, lines represent interactions between
proteins and different colors represent different functions. Hub
genes were obtained from the cytoHubba plugin (https://apps.cytoscape.org/apps/cytohubba) in
Cytoscape (version 3.9.1) through maximal clique centrality
methods. The GeneMANIA website (https://genemania.org/) was used for coexpression,
physical interaction, colocalization, pathway and other analyses of
the five hub genes.
Potential target drug prediction
To predict potential targeted drugs for the five hub
genes of CD, predictive analysis was conducted using the DSigDB
database. DSigDBv1.0 (http://dsigdb.tanlab.org/DSigDBv1.0/) was used to
predict the potential targeted drugs for the five hub genes, and
the top 10 scores were selected as candidate drugs.
CD and control samples from
patients
For the present study, a total of six tissue samples
were chosen from the biobank of Qingdao Municipal Hospital
(Qingdao, China). Samples were collected from patients between
October 2021 and September 2022 and stored in the biobank, and
subsequently retrieved for experiments performed in the present
study. This comprised three specimens of CD tissue and three
specimens of normal intestinal tissue. The use of samples was
granted permission by the biobank. Written informed consent was
obtained from the patients for inclusion of their samples in the
biobank. The research protocol and utilization of samples were both
approved and formally authorized by the Ethics Committee of Qingdao
Municipal Hospital (Qingdao, China) in October 2022. RT-qPCR was
completed in October 2022, and western blotting was completed in
November 2023 (approval no. 2022-066).
Hematoxylin and eosin (H&E)
staining
All of the tissues were fixed in 10% buffered
formalin for 48 h at 4˚C, embedded in paraffin and sectioned at a
thickness of 4 µm. The slides were then dewaxed twice in 100%
xylene for 30 min at 56˚C and rehydrated through graded alcohol
solutions (100, 90, 80, 70 and 50%). The slides were then rinsed in
H2O for 5 min, stained with H&E (cat. no. C0105S;
Beyotime Institute of Biotechnology) for 10 min at room
temperature, serially dehydrated in ethanol, cleared in 100% xylene
and mounted with a coverslip using Permount (Wuhan Servicebio
Technology Co., Ltd.) at room temperature. Finally, the sections
were imaged under a TE2000 microscope under white light (Nikon
Corporation).
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
A Kz-111-fp high-speed low-temperature grinding
instrument (Wuhan Servicebio Technology Co., Ltd.) was used to
grind the tissues, and VeZol Reagent (R411-01; Vazyme Biotech Co.,
Ltd.) was used to extract total RNA. RT to generate cDNA was
performed with HiScript II Q Select RT SuperMix (Vazyme Biotech
Co., Ltd.) according to the manufacturer's instructions. AceQ
Universal SYBR qPCR Master Mix (cat. no. Q511-02; Vazyme Biotech
Co., Ltd.) was used for the qPCR analysis process and the reaction
was run at 94˚C for 3 min, followed by 40 cycles of 94˚C for 10 sec
and 60˚C for 30 sec. All experimental data are presented as the
mean values obtained from three independent experiments and were
analyzed statistically using the 2-ΔΔCq cycle threshold
method (16). Endogenous GAPDH
(17) was used as an internal
control. All primer sequences are listed in Table II.
 | Table IIPrimer sequences. |
Table II
Primer sequences.
| Gene | Sequence,
5'-3' | RefSeq | Amplicon size,
bp |
|---|
| IL-6-F |
ACTCACCTCTTCAGAACGAATTG | NG_011640.1 | 149 |
| IL-6-R |
CCATCTTTGGAAGGTTCAGGTTG | | |
| PTGS2-F |
CTGGCGCTCAGCCATACAG | NG_028206.2 | 94 |
| PTGS2-R |
CGCACTTATACTGGTCAAATCCC | | |
| DUOX2-F |
CTGGGTCCATCGGGCAATC | NG_009447.1 | 144 |
| DUOX2-R |
GTCGGCGTAATTGGCTGGTA | | |
| HIF1A-F |
GAACGTCGAAAAGAAAAGTCTCG | NG_029606.1 | 124 |
| HIF1A-R |
CCTTATCAAGATGCGAACTCACA | | |
| NOS2-F |
TTCAGTATCACAACCTCAGCAAG | NG_011470.1 | 207 |
| NOS2-R |
TGGACCTGCAAGTTAAAATCCC | | |
| GAPDH-F |
GGAGCGAGATCCCTCCAAAAT | NG_007073.2 | 197 |
| GAPDH-R |
GGCTGTTGTCATACTTCTCATGG | | |
Western blotting and antibodies
The tissue grinding method was the same as described
in the qPCR section. The ground tissue was lysed using RIPA Lysis
Buffer (cat. no. R0010; Beijing Solarbio Science & Technology
Co., Ltd.), which contained PMSF (cat. no. P0100; Beijing Solarbio
Science & Technology Co., Ltd.), an inhibitor of serine
proteases and acetylcholinesterase, on ice. Protein concentration
was measured using the BCA Protein Assay kit (cat. no. MA0082;
Dalian Meilun Biology Technology Co., Ltd.). The aliquots of
lysates (20 µg protein) were boiled with sample loading buffer
(cat. no. LT101S; Epizyme Biotech) for 5 min. Equal amounts of
total protein (20 µg/lane) from different samples were separated by
10% SDS-PAGE at 120 V for 1.5 h and transferred onto 0.22-µm
polyvinylidene difluoride membranes (cat. no. ISEQ00010;
MilliporeSigma) at 280 mA for 1.5 h. Then, the membranes were
blocked with 5% skimmed milk powder in TBS with 0.05% Tween-20
(TBST) for 1 h at room temperature and treated with specific
primary antibodies overnight at 4˚C. The next day, the membranes
were washed with TBST and incubated with an HRP-conjugated
secondary antibody [1:5,000; cat. nos. SA00001-1 (mouse) and
SA00001-2 (rabbit); Proteintech Group, Inc.]. Each band was
detected using an enhanced chemiluminescence kit (BeyoECL Star;
cat. no. P0018AS; Beyotime Institute of Biotechnology).
Anti-β-actin (1:20,000; cat. no. 66009-1-Ig),
anti-prostaglandin-endoperoxide synthase 2 (PTGS2)/COX2 (1:1,000;
cat. no. 27308-1-AP) and anti-IL-6 (1:1,000; cat. no. 21865-1-AP)
were purchased from Proteintech Group, Inc. Anti-dual oxidase 2
(DUOX2; 1:500; cat. no. ab97266) was purchased from Abcam. Antibody
Dilution Buffer was purchased from Epizyme Biotech (cat. no.
PS119). ImageJ software (version 2.14.0; National Institutes of
Health) was used to conduct semi-quantification analysis of
blots.
Statistical analysis
R software (version 4.1.2), SPSS 26.0 (IBM Corp.)
and Prism 8 (Dotmatics) statistical software were used to perform
statistical analysis. In the qPCR experiment, the results were
analyzed by unpaired Student's t-test. For the western blot
experiment, the results were analyzed by unpaired Student's t-test.
Each experiment was repeated three times. P<0.05 was considered
to indicate a statistically significant difference.
Results
Analysis of genes associated with
ferroptosis in CD
The microarray expression profiling datasets
GSE102133 and GSE75214 were downloaded from the GEO database. The
present study included a total of 155 samples, consisting of 132 CD
samples and 23 control samples. PCA was conducted on the samples
from the two GEO datasets. Batch effects identified in the PCA were
subsequently removed to ensure data could be further analyzed. The
processed PCA plot is presented in Fig. S1A.
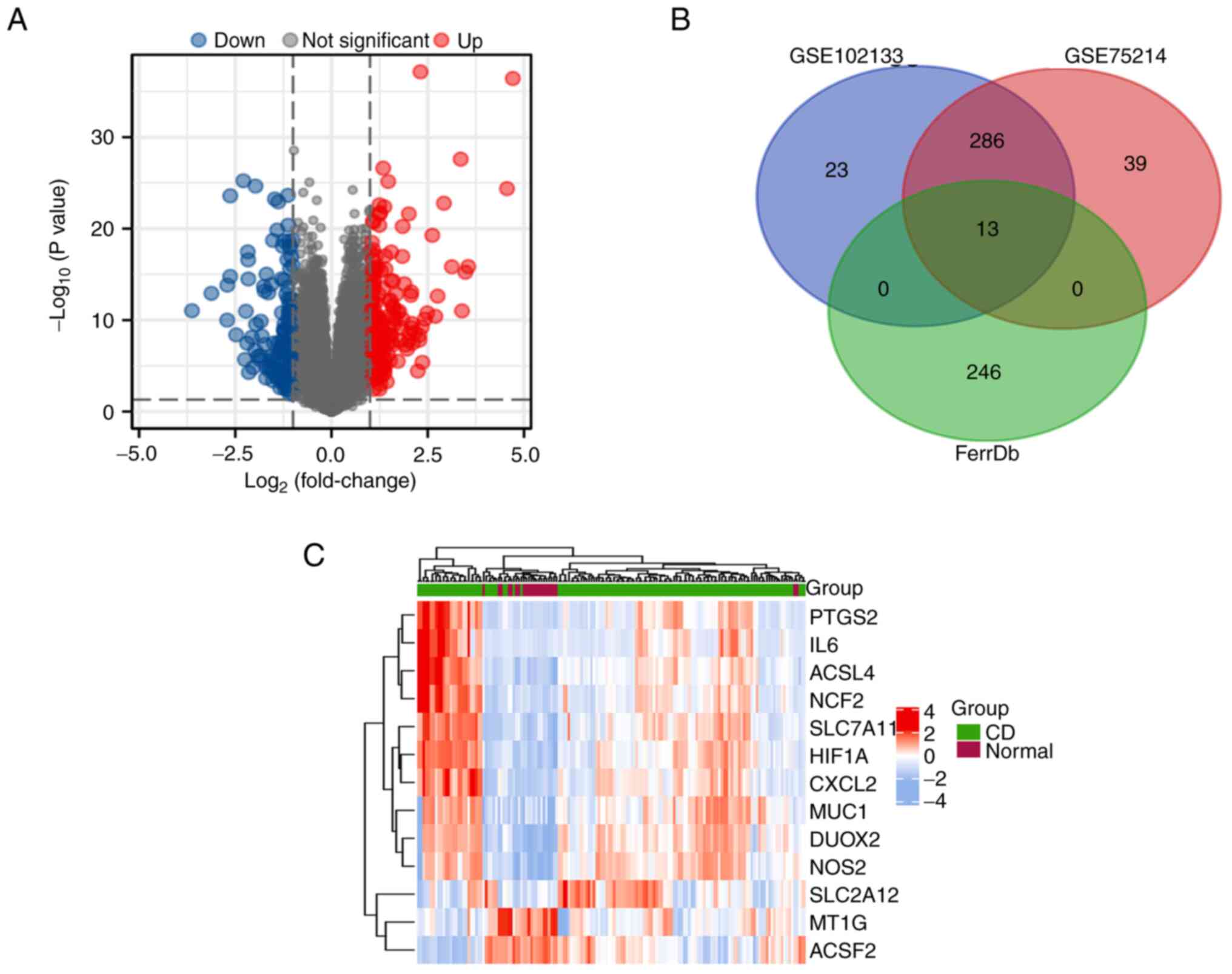
Subsequently, |log2 FC|>1 and adjusted P<0.05
were used as the criteria to select the DEGs in these two datasets.
A volcano plot (Fig. 2A) was
generated to display the DEGs between the two groups of samples.
After performing a crossover analysis with 259 ferroptosis-related
genes, a total of 13 DEGs were identified and are shown in a Venn
diagram (Fig. 2B). Among these
genes, 11 were found to be upregulated, while two were
downregulated. A heatmap was generated to illustrate the expression
patterns of these genes between the CD group and the normal group
(Fig. 2C). The expression levels
of the 13 DEGs in the CD samples and normal samples are depicted in
box plots (Fig. 3A and B). Significant changes were observed in
upregulated genes, including DUOX2 and MUC1, while downregulated
genes, such as MT1G and ACSF2, also exhibited significant changes.
Detailed information regarding these genes can be found in Table III.
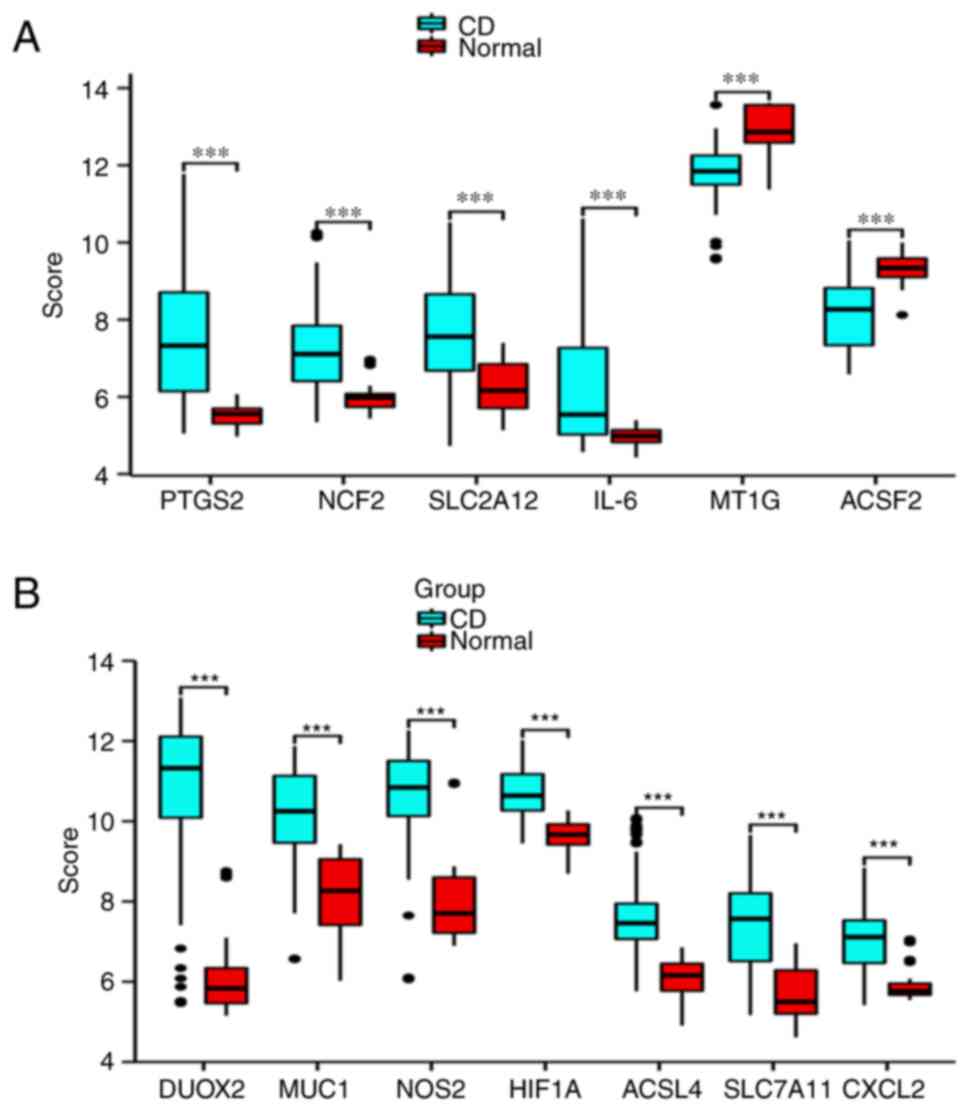 | Figure 3Box plots of the expression of 13
ferroptosis-related genes in CD and normal samples. (A) Expression
of PTGS2, NCF2, SLC2A12, IL-6, MTIG and ACSF2. (B) Expression of
DUOX2, MUC1, NOS2, HIF1A, ACSL4, SLC7A11 and CXCL2. Blue represents
CD group, red represents normal group and the score on the Y-axis
represents the relative expression level of genes
(***P<0.001). CD, Crohn's disease. |
 | Table IIIComparison of 13 genes associated
with ferroptosis between Crohn's disease samples and normal
samples. |
Table III
Comparison of 13 genes associated
with ferroptosis between Crohn's disease samples and normal
samples.
| Gene symbol | Description | Log fold
change | Up/Down
regulated | P-value |
|---|
| DUOX2 | Dual oxidase 2 | 4.643206134 | Up |
4.23x10-25 |
| MUC1 | Mucin 1, cell
surface associated | 2.986183308 | Up |
1.65188x10-23 |
| NOS2 | Nitric oxide
synthase 2 | 2.605279822 | Up |
5.37157x10-20 |
| HIF1A | Hypoxia inducible
factor 1 subunit α | 1.055152853 | Up |
8.37052x10-15 |
| ACSL4 | Acyl-CoA synthetase
long chain family member 4 | 1.553297113 | Up |
6.5226x10-13 |
| SLC7A11 | Solute carrier
family 7 member 11 | 1.72699847 | Up |
1.37298x10-11 |
| CXCL2 | C-X-C motif
chemokine ligand 2 | 1.109787323 | Up |
1.3473x10-10 |
| PTGS2 |
Prostaglandin-endoperoxide synthase 2 | 2.034361857 | Up |
1.76391x10-8 |
| NCF2 | Neutrophil
cytosolic factor 2 | 1.213650296 | Up |
2.63299x10-7 |
| SLC2A12 | Solute carrier
family 7 member 12 | 1.23521218 | Up |
3.29996x10-5 |
| IL-6 | Interleukin 6 | 1.432941749 | Up |
7.12667x10-5 |
| MT1G | Metallothionein
1G | -1.077752314 | Down |
1.2887x10-11 |
| ACSF2 | Acyl-CoA synthetase
family member 2 | -1.171871909 | Down |
9.68187x10-9 |
GO enrichment, KEGG pathway and gene
interrelationship analyses of DEGs related to ferroptosis in
CD
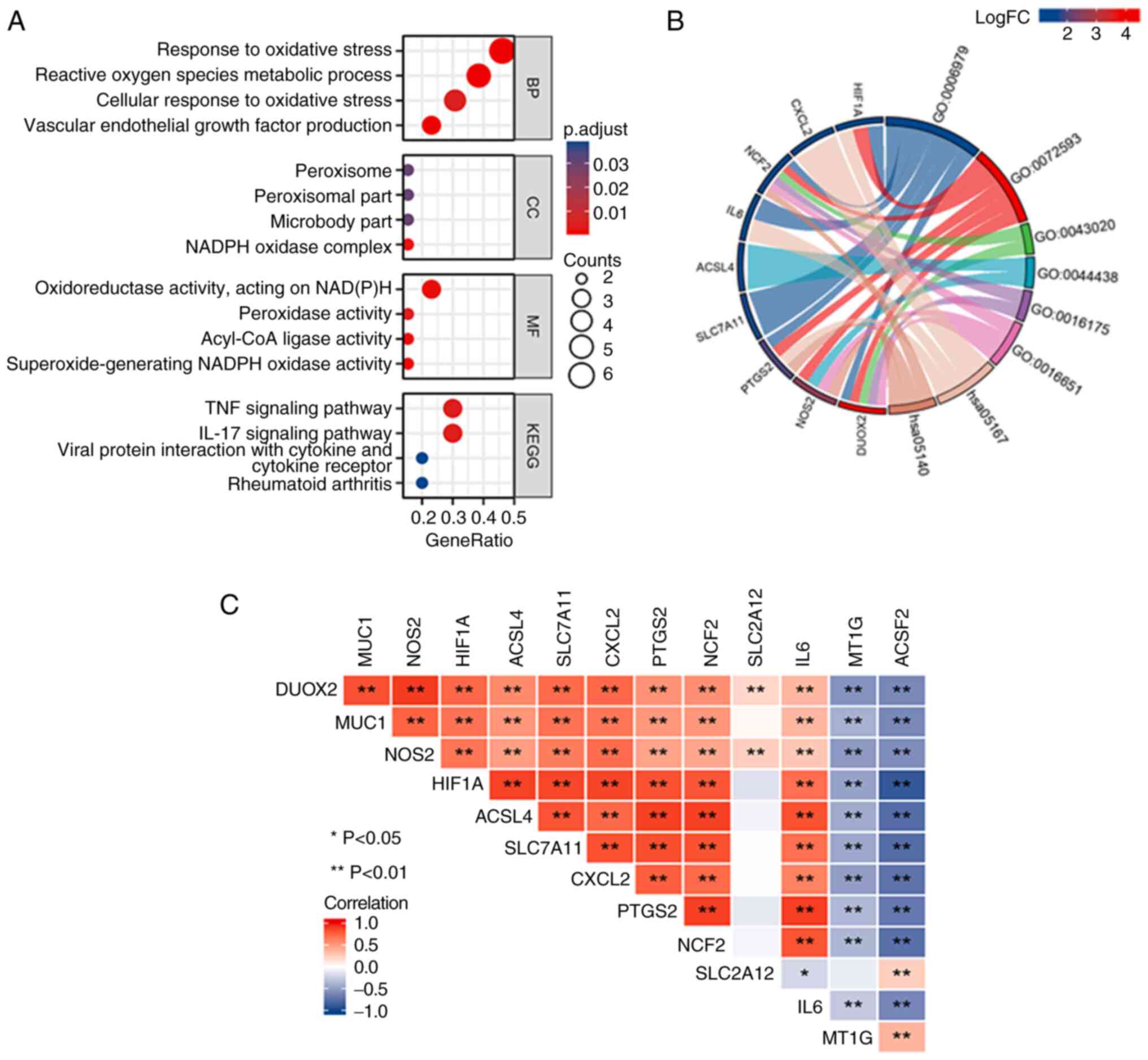
According to the GO enrichment analysis results
(Fig. 4A and B), the BP terms that were primarily
enriched in the DEGs were ‘response to oxidative stress’, ‘reactive
oxygen metabolic process’, ‘cellular response to oxidative stress’
and ‘vascular endothelial growth factor production’. The MF and CC
terms that were primarily enriched in the DEGs were ‘oxidoreductase
activity, acting on NAD(P)H’ and the ‘NADPH oxidase complex’. The
KEGG pathway analysis (Fig. 4A)
revealed that the enriched genes were predominantly associated with
the ‘TNF signaling pathway’ and the ‘IL-17 signaling pathway’. As
depicted in the Chord diagram generated using the circlize R
package (version 0.4.1) of GO enrichment terms (Fig. 4B), the relationships between genes
and biological functions within GO are illustrated through
connecting lines, with hypoxia inducible factor 1 subunit α
(HIF1A), NCF2, PTGS2, nitric oxide synthase 2 (NOS2) and DUOX2
being enriched in multiple GO biological functions.
The Pearson correlation analysis indicated that
several of the 13 ferroptosis-related genes exhibited significant
positive and negative correlations in CD (Fig. 4C). Red and blue colors represent
positive and negative correlations, respectively.
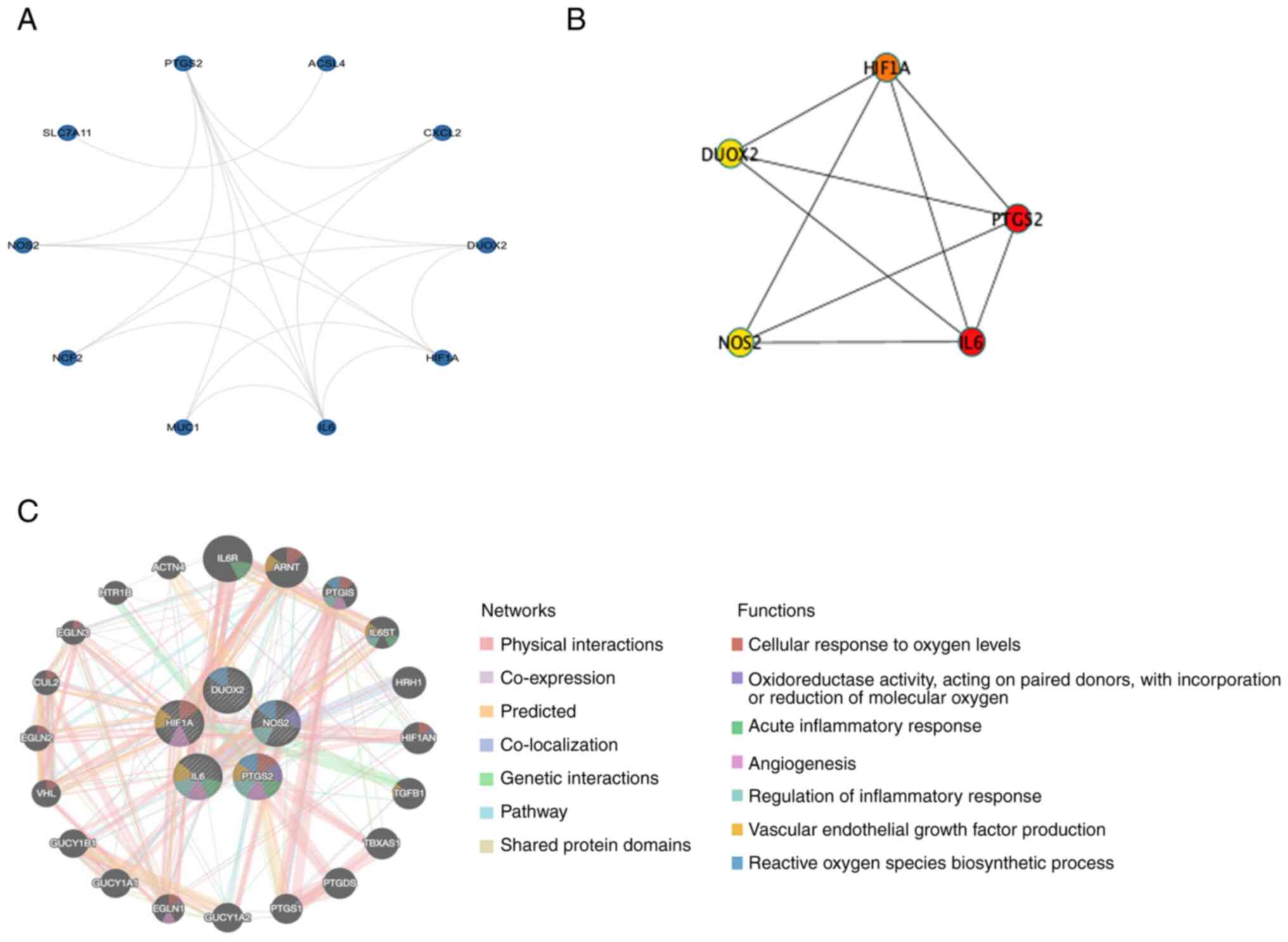
PPI network generation and filtering
of hub genes associated with ferroptosis
PPI analysis was conducted to demonstrate the
relationships among the genes of interest (Fig. S1B). Out of the 13 genes, three
were excluded from the analysis because they did not exhibit any
interactions with each other. The remaining 10 genes were used to
construct the PPI network. Genes are represented by nodes, and the
interactions among the genes are indicated by lines (Fig. 5A). Next, cytoHubba plugin in
Cytoscape software identified the following five hub genes: IL-6,
PTGS2, HIF1A, DUOX2 and NOS2 (Fig.
5B). The deeper color of the dot indicates that the rank order
of the hub gene is more advanced. The GeneMANIA website was used to
predict the gene coexpression, functional analysis and physical
binding properties of these five hub genes, as illustrated in
Fig. 5C. Notably, the coexpressed
genes for the five genes were functionally enriched in cellular
response to oxygen levels, oxidoreductase activity, acute
inflammatory response and other related functions.
Targeted drug prediction for related
genes
The DSigDB database was used to predict target drugs
associated with the five hub genes, which have the potential to
control the occurrence of ferroptosis and treat CD. A total of
1,378 potential drugs were predicted, and among them, zinc
protoporphyrin, chitosamine, benzoyl peroxide, ferric citrate and
haem were identified as the top-rated drugs. The specific ratings
and statistical values are shown in Table IV.
 | Table IVTop 10 predicted target drugs based
on five hub genes. |
Table IV
Top 10 predicted target drugs based
on five hub genes.
| Drug name | P-value | Odds ratio | Combined score |
|---|
| Zinc protoporphyrin
CTD 00000915 |
2.74x10-8 | 1,152.057692 | 20,062.49288 |
| Chitosamine CTD
00006030 |
1.66x10-9 | 959.6144578 | 19,397.65623 |
| Benzoyl peroxide
CTD 00005495 |
2.75x10-6 | 1,480.444444 | 18,956.85145 |
| Ferric citrate CTD
00001186 |
3.30x10-6 | 1,332.333333 | 16,817.52991 |
| Heme TTD
00008407 |
3.30x10-6 | 1,332.333333 | 16,817.52991 |
| Flufenamic acid CTD
00005976 |
4.48x10-8 | 966 | 16,346.25293 |
| Roxithromycin CTD
00007073 |
3.90x10-6 | 1,211.151515 | 15,085.68949 |
| Kaempferol CTD
00000297 |
3.85x10-9 | 772.5048544 | 14,966.43303 |
| NSC267099 CTD
00001568 |
4.54x10-6 | 1,110.166667 | 1,3656.83282 |
| Deferoxamine CTD
00005759 |
7.00x10-9 | 641 | 1,1953.71327 |
Validation of the differential
expression of the five hub genes in CD clinical samples by
RT-qPCR
All collected clinical tissue samples of CD were
definitively diagnosed by two pathologists through observation
under a colonoscope, tissue sectioning and H&E staining. The
clinicopathological features of the patients are shown in Table V. The H&E staining and
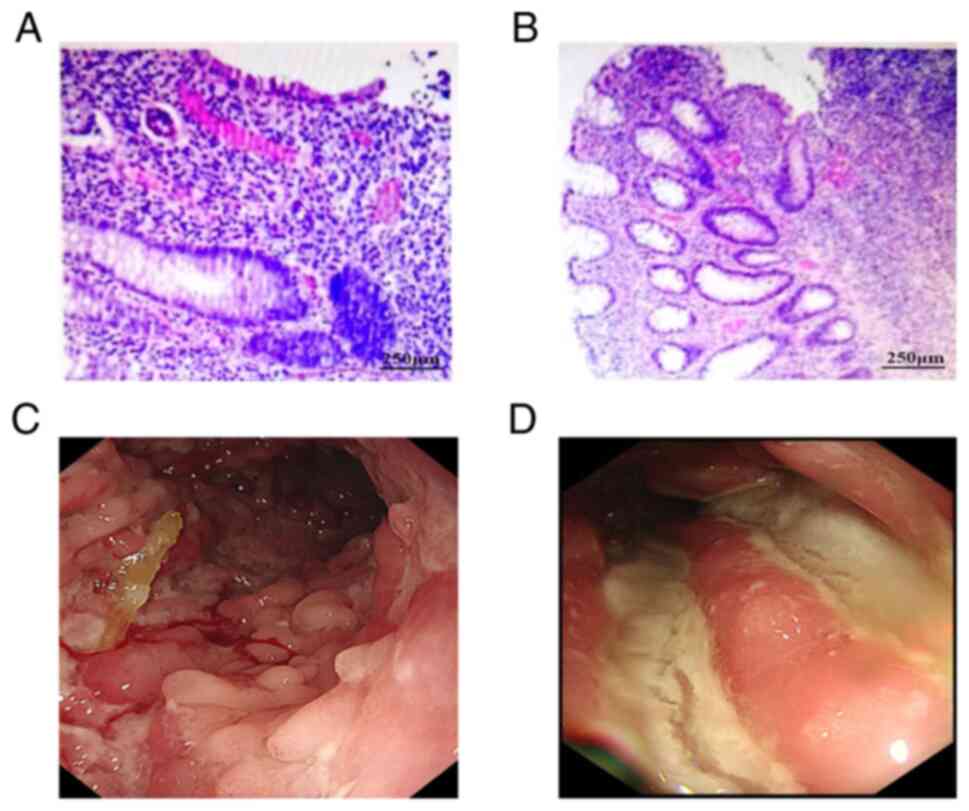
colonoscopy findings are shown in Fig.
6. Under the microscope, visible infiltration of inflammatory
cells and lymphocytes was observed. Colonoscopy indicated that the
intestinal lumen exhibited a coarse and congested surface,
accompanied by focal mucosal ulceration and numerous irregular
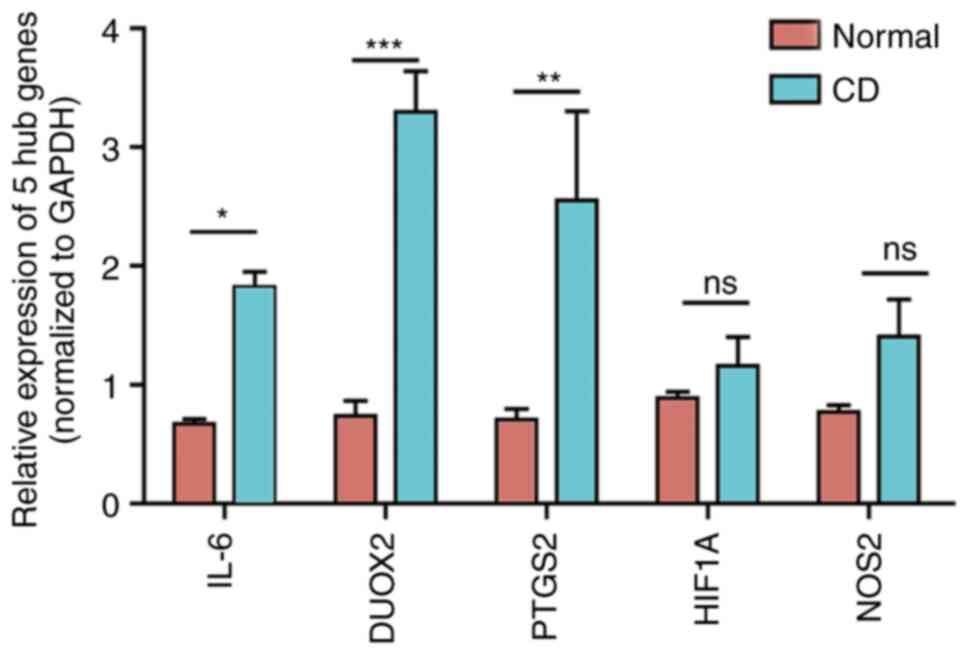
protuberant lesions. RT-qPCR analysis was performed to detect the
expression levels of the five hub genes in three pairs of CD
tissues and healthy control tissues. The statistical analysis
revealed significant changes in the expression levels of IL-6,
PTGS2 and DUOX2 between the samples. Although the expression
patterns of HIF1A and NOS2 were consistent with the bioinformatics
analysis results, the difference in their expression between groups
was not significant (Fig. 7).
 | Table VClinicopathological characteristics
of patients with Crohn's disease. |
Table V
Clinicopathological characteristics
of patients with Crohn's disease.
| Characteristic | CD Patient 1 | CD Patient 2 | CD Patient 3 | Control 1 | Control 2 | Control 3 |
|---|
| Age, years | 32 | 53 | 41 | 42 | 38 | 44 |
| Sex | Female | Male | Female | Male | Male | Female |
| Sample
location | Ileocecal
region | Ascending
colon | Terminal ileum | Ileocecal
region | Terminal ileum | Terminal ileum |
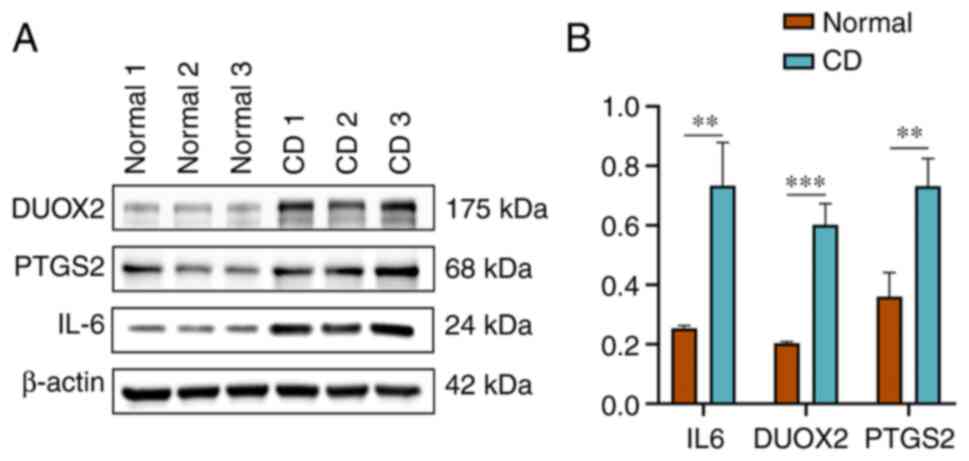
Validation of the protein expression
levels of the three genes in CD clinical samples
Protein expression level validation of the three
target genes identified through RT-qPCR experiments was conducted
in normal and CD tissues using western blotting (Fig. 8A and B). The protein expression level results
were in accordance with the qPCR findings.
Discussion
CD is a chronic, long-term and recurring intestinal
disease that belongs to a group of conditions collectively known as
IBD (18). Prior to the 20th
century, the incidence of CD was more prevalent in the Americas and
Europe; however, in recent years, there has been a steady rise in
the prevalence of CD in Asian countries (19). CD primarily develops in the
terminal ileum and can lead to lesions that extend from the mouth
to the anus, as well as potential extraintestinal complications. CD
often presents subtle or atypical symptoms in a clinical setting,
which can pose challenges in its detection and diagnosis.
Consequently, patients are frequently overlooked or missed during
colonoscopy procedures (20).
Patients commonly experience symptoms, such as diarrhea, abdominal
pain, fever and anemia. Additionally, CD can present with
extraintestinal manifestations, including inflammatory arthropathy,
osteoporosis, scleritis and IgA nephropathy (21,22).
A review of the PubMed database revealed that the current findings
on risk factors for CD include environmental factors, genetic
factors, nutritional status, intestinal barrier disorders, immune
response, microbial dysbiosis, gut viruses and fungi (3,23).
However, the specific mechanisms or pathways involved in these risk
factors remain to be elucidated.
Ferroptosis is a recently discovered form of
iron-dependent, nonapoptotic cell death that is primarily triggered
by intracellular lipid peroxidation. Ferroptosis is regulated by
various oxidative and antioxidant systems in vivo, including
redox homeostasis, mitochondrial activation, amino acid metabolism,
lipid metabolism and sugar metabolism (7). Ferroptosis has been observed to
regulate various benign or malignant diseases through multiple
pathways, such as disrupting cellular metabolic homeostasis,
causing mitochondrial damage or activating other cell death
pathways. These diseases include but are not limited to tumors,
neurological disorders, kidney damage and cardiovascular diseases
(24-26).
Huang et al (27) indicated that the
ferroptosis-related hub gene STAT3 can mediate the process of
ferroptosis and promote UC. Ferroptosis also plays a significant
role in the regulation of CRC. Glutathione peroxidase 4, an
inhibitor of ferroptosis, is an important central regulatory
molecule involved in the progression of CRC (28).
Regarding the intestinal barrier and immune
response, which are key mechanisms contributing to CD, ferroptosis
also plays an important role. Liu et al (29) discovered that ferroptosis can
reverse the maintenance of intestinal mucosal barrier integrity
mediated by Sestrin2. Ma et al (30) reported that CD36-directed
CD8+ T-cell ferroptosis suppresses antitumor effects
in vivo. Xu et al (31) suggested that ferroptosis could
exert antitumor effects by reversing the immune microenvironment.
For example, CD8+ T cells activated during the immune
regulation process can induce cellular ferroptosis by promoting
lipid peroxidation in cells and finally enhancing its antitumor
effect. The present study conducted an analysis to investigate the
potential interaction between CD and ferroptosis.
Based on the GEO datasets and the FerrDb, a total of
13 DEGs that are associated with ferroptosis were identified. The
present study further investigated the functional implications of
these genes through GO and KEGG enrichment analyses. The GO
analysis indicated that the BPs associated with ferroptosis were
related to oxidative stress. These processes include the ‘response
to oxidative stress’, ‘reactive oxygen species metabolic process’
and ‘cellular response to oxidative stress’. This finding suggested
that ferroptosis may be implicated in the onset of Crohn's disease,
as oxidative stress processes play a crucial role in the occurrence
of ferroptosis. The accumulation of iron during cellular metabolism
can lead to impaired redox homeostasis, thereby inducing
ferroptosis in cells (32,33). Oxidative stress is also an
important factor in inflammation, which activates a variety of
proinflammatory factors, such as NF-κB and p53(34). The increased expression of p53 in
CD has been shown to promote apoptosis of intestinal epithelial
cells and aggravate inflammation (35,36).
Therefore, it was hypothesized that the oxidative stress process
associated with the ferroptosis pathway might be implicated in the
occurrence and progression of CD.
Another noteworthy result obtained in terms of BP
enrichment specifically related to ‘vascular endothelial growth
factor production’. Vascular endothelial growth factor (VEGF) is a
biomolecule that is crucial for mitosis and preventing apoptosis in
vascular endothelial cells (37)
and includes several members, including VEGF-A, VEGF-B and VEGF-C.
Inflammation and tumors typically promote pathological vascular
proliferation, leading to an increased oxygen demand in the local
microenvironment (38). The
concentration of reactive oxygen species (ROS) that depends on the
production of NADPH oxidase during pathological vascular
proliferation also increases, which appears to establish a
reciprocal relationship with ferroptosis (39). High production of ROS increases the
likelihood of ferroptosis in cells (40). This is also consistent with the
enrichment results obtained for MF) and CC.
In other studies, the biomodulatory role of VEGF in
IBD has been reported. Scaldaferri et al (41) reported that overexpression of
VEGF-A in mice with DSS-induced IBD inflammation can exacerbate
disease severity. According to Eder et al (42), the degree of VEGF expression can
affect the severity of CD. However, the existing mechanism between
VEGF and ferroptosis remains to be elucidated. It was hypothesized
that VEGF may lead to ferroptosis of intestinal epithelial cells
and promote the occurrence of CD.
KEGG pathway analysis further revealed that the
relevant genes were predominantly abundant in the ‘TNF pathway’,
‘IL-17 pathway’ and ‘rheumatoid arthritis’. TNF is an important
cell regulatory molecule involved in the regulation of cell
survival, apoptosis, inflammation and the immune response (43). IL-7 is an inflammatory factor
secreted by T helper 17 cells (44). Bishu et al (45) detected elevated expression levels
of both TNF and IL-17 in CD, which may be associated with a type of
CD 4+ T cells referred to as CD4+
tissue-resident memory T cells (TRM cells). CD4+ TRM
cells proliferate in CD and serve as the primary sources of TNF and
IL-17. However, the regulatory relationship between ferroptosis and
TNF and IL-17 in the currently available studies of CD remains to
be elucidated.
A PPI network map was generated in Cytoscape and
five hub genes, IL-6, PTGS2, HIF1A, DUOX2 and NOS2, were identified
using cytoHubba software. Subsequently, RT-qPCR and western
blotting experiments were conducted on three pairs of patient
samples to analyze the expression of these genes. The results
demonstrated significant differences in the expression levels of
IL-6, DUOX2 and PTGS2.
IL-6 is an inflammatory cytokine belonging to the IL
family. It serves a crucial role in stimulating anti-inflammatory
processes and immune responses in the human body (46). Shi et al (47) suggested that Toll-like receptor 4
aggravates intestinal injury by regulating the expression of IL-6.
Gross et al (48)
identified significantly elevated IL-6 in CD in both the active and
inactive phases. IL-6 plays a role in other processes besides
inflammation. Han et al (49) revealed that, in asthma, IL-6
disrupts iron homeostasis in BEAS-2B cells by promoting lipid
peroxidation, which in turn promotes ferroptosis in bronchial
epithelial cells. Li et al (50) recently demonstrated that the
expression of an amino acid counter transporter (xCT) in head and
neck squamous cell carcinoma was associated with the levels of IL-6
and that xCT can induce cellular ferroptosis. These findings
indirectly confirm that IL-6 can regulate ferroptosis by promoting
lipid peroxidation, thereby exacerbating the severity of
inflammation. However, there is no conclusive evidence for an
association between IL-6 and ferroptosis in CD, and further study
is required to determine the specific mechanism between IL-6 and
ferroptosis in CD.
PTGS2, also called COX-2, is an enzyme that can be
suppressed by nonsteroidal anti-inflammatory drugs, such as aspirin
and ibuprofen, and serves a regulatory role in a number of
inflammatory or neoplastic diseases (51). Roberts et al (52) revealed through densitometric
analysis that the expression of COX-2 was increased by 6-8 times in
IBD tissues compared with normal tissues. Singer et al
(53) demonstrated that COX-2 was
predominantly present in apical epithelial cells and in lamina
propria mononuclear cells in CD. Notably, neither study detected
COX-2 expression in normal tissues. Furthermore, it has been
confirmed in the literature that COX-2-induced ferroptosis may be
associated with heme oxygenase (HO)-1, which has been identified as
a crucial mediator of ferroptosis (54). Chen et al (55) discovered that the use of astragalus
polysaccharides, by inhibiting HO-1 expression, can treat
ferroptosis and reduce COX-2 expression. It was hypothesized that
there may be a certain interaction between COX-2 and HO-1, thereby
regulating the relationship between CD and ferroptosis. These
results suggested that PTGS2/COX-2 may have an important regulatory
role in CD. However, to the best of our knowledge, no related
research has determined whether the upregulation of COX-2 in CD is
associated with ferroptosis. This notion provides a foundation for
future mechanistic studies on the occurrence of CD.
DUOX2 is an enzyme that was initially discovered in
the mammalian thyroid and belongs to the NADPH oxidase family,
responsible for the production of ROS. ROS are an important
component of the immune system in eukaryotic organisms. However,
excessive levels of ROS in cells may induce and promote
ferroptosis. Lipinski et al (56) demonstrated that DUOX2 is an
important factor involved in NOD2-dependent ROS production. A
separate study on pediatric CD conducted by Haberman et al
(57) observed strong and robust
expression of DUOX2 in the villous enterocytes of the ileum
affected by CD. This increased expression of DUOX2 could
potentially contribute to the worsening of intestinal mucosal
damage. This finding seems to further support the hypothesis of the
present study that the upregulation of DUOX2 in intestinal mucosal
cells may have a role in the pathogenesis of CD. The increased
expression of DUOX2 in the villous enterocytes of the small
intestine could lead to the accumulation of ROS within the cells,
consequently promoting iron-mediated cell death and resulting in
the loss of intestinal mucosal barrier function.
Current treatment options for CD are typically
limited to anti-inflammatory medications, immunosuppressive agents
and surgical interventions. However, the treatment outcomes are
often unsatisfactory, with high rates of recurrence and severe side
effects (58,59). Based on the results of the present
study, drug targets for the five hub genes were inferred using the
DSigDB database. Based on existing research on the target drugs,
kaempferol was found to inhibit the initiation of cellular
ferroptosis. Additionally, other studies have shown that kaempferol
can be used to improve intestinal inflammation and intestinal
barrier disorders (60,61).
The present study has several limitations. First,
the datasets used in the present study were obtained from the GEO
database and both datasets were generated using the same microarray
platform. In addition, there was a lack of additional validation
through cell or animal experiments, as well as clinical validation.
Moreover, the number of tissue samples obtained for clinical
experiments and testing was relatively small. Therefore, further
investigations are needed in future studies to explore the
relationship between ferroptosis and CD.
In conclusion, 13 ferroptosis-associated genes were
identified in CD in the present bioinformatics analysis. Among
them, IL-6, DUOX2 and PTGS2 may be most likely to regulate
ferroptosis in CD, and to be involved in CD development and
progression. However, specific mechanistic studies need to be
further explored. These results provide an experimental foundation
for targeted therapy in CD, introducing novel ideas, insights and
methods for understanding the development and occurrence of CD.
Supplementary Material
PCA image and PPI network based on
STRING. (A) PCA result. (B) PPI network between the 13
ferroptosis-related genes based on STRING. PCA, principal component
analysis; PPI, protein-protein interaction; STRING, Search Tool for
the Retrieval of Interacting Genes.
Acknowledgements
Not applicable.
Funding
Funding: The present study was financially supported by the
National Natural Science Foundation of China (grant nos. 81270448
and 81470890).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the GEO repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE102133
and https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE75214.
Other datasets used and/or analyzed during the current study are
available from the corresponding authors on reasonable request.
Authors' contributions
WZ, ZL, HL and DZ contributed to the study
conception and design. Material preparation, data collection and
analysis were performed by WZ and ZL. HL and ZL confirm the
authenticity of all the raw data. The first draft of the manuscript
was written by WZ and all authors commented on previous versions of
the manuscript. HL conceived the concept, designed the manuscript,
coordinated and critically revised the manuscript. DZ provided
valuable suggestions and assistance during the revision processes
of the manuscript, offered financial support for the experiments
and collaborated with all authors to finalize the content of the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was performed in line with the
principles of The Declaration of Helsinki. Approval was granted by
the Ethics Committee of Qingdao Municipal Hospital (approval no.
2022-066). Written informed consent was obtained from the patients
for inclusion of their samples in the biobank.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Geremia A, Biancheri P, Allan P, Corazza
GR and Di Sabatino A: Innate and adaptive immunity in inflammatory
bowel disease. Autoimmun Rev. 13:3–10. 2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Cosnes J, Gower-Rousseau C, Seksik P and
Cortot A: Epidemiology and natural history of inflammatory bowel
diseases. Gastroenterology. 140:1785–1794. 2011.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Roda G, Chien Ng S, Kotze PG, Argollo M,
Panaccione R, Spinelli A, Kaser A, Peyrin-Biroulet L and Danese S:
Crohn's disease. Nat Rev Dis Primer. 6(22)2020.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Dulai PS, Singh S, Vande Casteele N,
Boland BS, Rivera-Nieves J, Ernst PB, Eckmann L, Barrett KE, Chang
JT and Sandborn WJ: Should we divide Crohn's disease into
ileum-dominant and isolated colonic diseases? Clin Gastroenterol
Hepatol. 17:2634–2643. 2019.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Friedberg S and Rubin DT: Intestinal
cancer and dysplasia in Crohn's disease. Gastroenterol Clin North
Am. 51:369–379. 2022.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Friedman S: Cancer in Crohn's disease.
Gastroenterol Clin North Am. 35:621–639. 2006.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Baumgart DC and Sandborn WJ: Crohn's
disease. Lancet. 380:1590–1605. 2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Jiang X, Stockwell BR and Conrad M:
Ferroptosis: Mechanisms, biology and role in disease. Nat Rev Mol
Cell Biol. 22:266–282. 2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Li J, Cao F, Yin HL, Huang ZJ, Lin ZT, Mao
N, Sun B and Wang G: Ferroptosis: Past, present and future. Cell
Death Dis. 11(88)2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Xu C, Liu Z and Xiao J: Ferroptosis: A
double-edged sword in gastrointestinal disease. Int J Mol Sci.
22(12403)2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Vancamelbeke M, Vanuytsel T, Farré R,
Verstockt S, Ferrante M, Van Assche G, Rutgeerts P, Schuit F,
Vermeire S, Arijs I and Cleynen I: Genetic and transcriptomic bases
of intestinal epithelial barrier dysfunction in inflammatory bowel
disease. Inflamm Bowel Dis. 23:1718–1729. 2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Verstockt S, De Hertogh G, Van der Goten
J, Verstockt B, Vancamelbeke M, Machiels K, Van Lommel L, Schuit F,
Van Assche G, Rutgeerts P, et al: Gene and mirna regulatory
networks during different stages of Crohn's disease. J Crohns
Colitis. 13:916–930. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kanehisa M, Furumichi M, Sato Y, Kawashima
M and Ishiguro-Watanabe M: KEGG for taxonomy-based analysis of
pathways and genomes. Nucleic Acids Res. 51:D587–D592.
2023.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kanehisa M: Toward understanding the
origin and evolution of cellular organisms. Protein. 28:1947–1951.
2019.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Yao B, Zhang Q, Yang Z, An F, Nie H, Wang
H, Yang C, Sun J, Chen K, Zhou J, et al: CircEZH2/miR-133b/IGF2BP2
aggravates colorectal cancer progression via enhancing the
stability of m6A-modified CREB1 mRNA. Mol Cancer.
21(140)2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Veauthier B and Hornecker JR: Crohn's
disease: Diagnosis and management. Am Fam Physician. 98:661–669.
2018.PubMed/NCBI
|
|
19
|
Chiba M, Morita N, Nakamura A, Tsuji K and
Harashima E: Increased incidence of inflammatory bowel disease in
association with dietary transition (westernization) in Japan. JMA
J. 4:347–357. 2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Chetcuti Zammit S, Ellul P and Sidhu R:
The role of small bowel endoscopy for Crohn's disease. Curr Opin
Gastroenterol. 35:223–234. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wilkins T, Jarvis K and Patel J: Diagnosis
and management of Crohn's disease. Am Fam Physician. 84:1365–1375.
2011.PubMed/NCBI
|
|
22
|
Ambruzs JM and Larsen CP: Renal
manifestations of inflammatory bowel disease. Rheum Dis Clin North
Am. 44:699–714. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Prieto JMI, Andrade AR, Magro DO, Imbrizi
M, Nishitokukado I, Ortiz-Agostinho CL, Dos Santos FM, Luzia LA,
Rondo PHC, Leite AZA, et al: Nutritional global status and its
impact in Crohn's disease. J Can Assoc Gastroenterol. 4:290–295.
2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wu X, Li Y, Zhang S and Zhou X:
Ferroptosis as a novel therapeutic target for cardiovascular
disease. Theranostics. 11:3052–3059. 2021.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Mahoney-Sánchez L, Bouchaoui H, Ayton S,
Devos D, Duce JA and Devedjian JC: Ferroptosis and its potential
role in the physiopathology of Parkinson's disease. Prog Neurobiol.
196(101890)2021.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Martin-Sanchez D, Fontecha-Barriuso M,
Martinez-Moreno JM, Ramos AM, Sanchez-Niño MD, Guerrero-Hue M,
Moreno JA, Ortiz A and Sanz AB: Ferroptosis and kidney disease.
Nefrologia (Engl Ed). 40:384–394. 2020.PubMed/NCBI View Article : Google Scholar : (In English,
Spanish).
|
|
27
|
Huang F, Zhang S, Li X, Huang Y, He S and
Luo L: STAT3-mediated ferroptosis is involved in ulcerative
colitis. Free Radic Biol Med. 188:375–385. 2022.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Sui X, Zhang R, Liu S, Duan T, Zhai L,
Zhang M, Han X, Xiang Y, Huang X, Lin H and Xie T: RSL3 drives
ferroptosis through GPX4 inactivation and ROS production in
colorectal cancer. Front Pharmacol. 9(1371)2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Liu W, Xu C, Zou Z, Weng Q and Xiao Y:
Sestrin2 suppresses ferroptosis to alleviate septic intestinal
inflammation and barrier dysfunction. Immunopharmacol
Immunotoxicol. 45:123–132. 2023.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ma X, Xiao L, Liu L, Ye L, Su P, Bi E,
Wang Q, Yang M, Qian J and Yi Q: CD36-mediated ferroptosis dampens
intratumoral CD8+ T cell effector function and impairs
their antitumor ability. Cell Metab. 33:1001–1012.e5.
2021.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Xu H, Ye D, Ren M, Zhang H and Bi F:
Ferroptosis in the tumor microenvironment: Perspectives for
immunotherapy. Trends Mol Med. 27:856–867. 2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yu Y, Yan Y, Niu F, Wang Y, Chen X, Su G,
Liu Y, Zhao X, Qian L, Liu P and Xiong Y: Ferroptosis: A cell death
connecting oxidative stress, inflammation and cardiovascular
diseases. Cell Death Discov. 7(193)2021.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Mancardi D, Mezzanotte M, Arrigo E,
Barinotti A and Roetto A: Iron overload, oxidative stress, and
ferroptosis in the failing heart and liver. Antioxidants (Basel).
10(1864)2021.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Reuter S, Gupta SC, Chaturvedi MM and
Aggarwal BB: Oxidative stress, inflammation, and cancer: How are
they linked? Free Radic Biol Med. 49:1603–1616. 2010.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Gu L, Ge Z, Wang Y, Shen M and Zhao P:
Activating transcription factor 3 promotes intestinal epithelial
cell apoptosis in Crohn's disease. Pathol Res Pract. 214:862–870.
2018.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhang J, Xu M, Zhou W, Li D, Zhang H, Chen
Y, Ning L, Zhang Y, Li S, Yu M, et al: Deficiency in the
anti-apoptotic protein DJ-1 promotes intestinal epithelial cell
apoptosis and aggravates inflammatory bowel disease via p53. J Biol
Chem. 295:4237–4251. 2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Melincovici CS, Boşca AB, Şuşman S,
Mărginean M, Mihu C, Istrate M, Moldovan IM, Roman AL and Mihu CM:
Vascular endothelial growth factor (VEGF)-key factor in normal and
pathological angiogenesis. Rom J Morphol Embryol. 59:455–467.
2018.PubMed/NCBI
|
|
38
|
Kvietys PR and Granger DN: Role of
reactive oxygen and nitrogen species in the vascular responses to
inflammation. Free Radic Biol Med. 52:556–592. 2012.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Fukai T and Ushio-Fukai M: Cross-talk
between nadph oxidase and mitochondria: Role in ros signaling and
angiogenesis. Cells. 9(1849)2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Su LJ, Zhang JH, Gomez H, Murugan R, Hong
X, Xu D, Jiang F and Peng ZY: Reactive oxygen species-induced lipid
peroxidation in apoptosis, autophagy, and ferroptosis. Oxid Med
Cell Longev. 2019(5080843)2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Scaldaferri F, Vetrano S, Sans M, Arena V,
Straface G, Stigliano E, Repici A, Sturm A, Malesci A, Panes J, et
al: VEGF-A links angiogenesis and inflammation in inflammatory
bowel disease pathogenesis. Gastroenterology. 136:585–95.e5.
2009.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Eder P, Korybalska K, Lykowska-Szuber L,
Krela-Kazmierczak I, Stawczyk-Eder K, Klimczak K, Szymczak A, Linke
K and Witowski J: Association of serum VEGF with clinical response
to anti-TNFα therapy for Crohn's disease. Cytokine. 76:288–293.
2015.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Bradley JR: TNF-mediated inflammatory
disease. J Pathol. 214:149–160. 2008.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Iwakura Y, Ishigame H, Saijo S and Nakae
S: Functional specialization of interleukin-17 family members.
Immunity. 34:149–162. 2011.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Bishu S, El Zaatari M, Hayashi A, Hou G,
Bowers N, Kinnucan J, Manoogian B, Muza-Moons M, Zhang M and
Grasberger H: , et al: CD4+ tissue-resident memory T cells
expand and are a major source of mucosal tumour necrosis factor α
in active Crohn's disease. J Crohns Colitis. 13:905–915.
2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Tanaka T, Narazaki M and Kishimoto T: IL-6
in inflammation, immunity, and disease. Cold Spring Harb Perspect
Biol. 6(a016295)2014.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Shi YJ, Hu SJ, Zhao QQ, Liu XS, Liu C and
Wang H: Toll-like receptor 4 (TLR4) deficiency aggravates dextran
sulfate sodium (DSS)-induced intestinal injury by down-regulating
IL6, CCL2 and CSF3. Ann Transl Med. 7(713)2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Gross V, Andus T, Caesar I, Roth M and
Schölmerich J: Evidence for continuous stimulation of interleukin-6
production in Crohn's disease. Gastroenterology. 102:514–519.
1992.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Han F, Li S, Yang Y and Bai Z:
Interleukin-6 promotes ferroptosis in bronchial epithelial cells by
inducing reactive oxygen species-dependent lipid peroxidation and
disrupting iron homeostasis. Bioengineered. 12:5279–5288.
2021.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Li M, Jin S, Zhang Z, Ma H and Yang X:
Interleukin-6 facilitates tumor progression by inducing ferroptosis
resistance in head and neck squamous cell carcinoma. Cancer Lett.
527:28–40. 2022.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Kunzmann AT, Murray LJ, Cardwell CR,
McShane CM, McMenamin UC and Cantwell MM: PTGS2 (Cyclooxygenase-2)
expression and survival among colorectal cancer patients: A
systematic review. Cancer Epidemiol Biomarkers Prev. 22:1490–1497.
2013.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Roberts PJ, Morgan K, Miller R, Hunter JO
and Middleton SJ: Neuronal COX-2 expression in human myenteric
plexus in active inflammatory bowel disease. Gut. 48:468–472.
2001.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Singer II, Kawka DW, Schloemann S, Tessner
T, Riehl T and Stenson WF: Cyclooxygenase 2 is induced in colonic
epithelial cells in inflammatory bowel disease. Gastroenterology.
115:297–306. 1998.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Chiang SK, Chen SE and Chang LC: A dual
role of heme oxygenase-1 in cancer cells. Int J Mol Sci.
20(39)2018.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Chen Y, Wang J, Li J, Zhu J, Wang R, Xi Q,
Wu H, Shi T and Chen W: Astragalus polysaccharide prevents
ferroptosis in a murine model of experimental colitis and human
Caco-2 cells via inhibiting NRF2/HO-1 pathway. Eur J Pharmacol.
911(174518)2021.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Lipinski S, Till A, Sina C, Arlt A,
Grasberger H, Schreiber S and Rosenstiel P: DUOX2-derived reactive
oxygen species are effectors of NOD2-mediated antibacterial
responses. J Cell Sci. 122:3522–3530. 2009.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Haberman Y, Tickle TL, Dexheimer PJ, Kim
MO, Tang D, Karns R, Baldassano RN, Noe JD, Rosh J, Markowitz J, et
al: Pediatric Crohn disease patients exhibit specific ileal
transcriptome and microbiome signature. J Clin Invest.
124:3617–3633. 2014.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Gajendran M, Loganathan P, Catinella AP
and Hashash JG: A comprehensive review and update on Crohn's
disease. Dis Mon. 64:20–57. 2018.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Rolak S and Kane SV: Conventional
therapies for Crohn's disease. Gastroenterol Clin North Am.
51:271–282. 2022.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Bian Y, Dong Y, Sun J, Sun M, Hou Q, Lai Y
and Zhang B: Protective effect of kaempferol on LPS-induced
inflammation and barrier dysfunction in a coculture model of
intestinal epithelial cells and intestinal microvascular
endothelial cells. J Agric Food Chem. 68:160–167. 2020.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Yuan Y, Zhai Y, Chen J, Xu X and Wang H:
Kaempferol ameliorates oxygen-glucose
deprivation/reoxygenation-induced neuronal ferroptosis by
activating Nrf2/SLC7A11/GPX4 axis. Biomolecules.
11(923)2021.PubMed/NCBI View Article : Google Scholar
|