Introduction
Male infertility is a complex reproductive disorder
caused by hereditary, physiological, pathological and environmental
factors. According to a 2018 estimate, male infertility affects
8-12% of couples globally, with the male factor dominating in ~50%
of the cases (1). Semen analysis
is the key method to diagnose male infertility. The 6th edition of
the World Health Organization (WHO) laboratory manual for examining
and processing human semen has recently updated the sperm vitality
criterion during semen analysis. Sperm vitality is estimated by
assessing the membrane integrity of the cells, and it should be
tested examined sperm motility is <40%. The reasons for
decreased sperm vitality include structural defects in the sperm
flagellum, epididymal defects and immunological reactions due to
infection (2).
Asthenozoospermia is one of the most common types of
male infertility (1), and it is
characterized by low sperm motility. A retrospective study
conducted in 2018 on 117,979 male semen samples collected from 1989
to 1993 revealed that the incidence of male infertility was 45%,
which included asthenozoospermia (11%), oligozoospermia (22%) and
azoospermia (12%) as the main causes (3). Asthenozoospermia can also coexist
with oligozoospermia and teratozoospermia. In a multicenter study
in 2001(4), the incidence of
asthenozoospermia, oligozoospermia or asthenoteratozoospermia, and
oligoasthenoteratozoospermia was 2- to 3-fold, 5- to 7-fold, and
16-fold higher, respectively, in the infertile group compared with
the normal group.
Several pathogenic factors can influence the
development of asthenozoospermia, such as increased scrotal
temperature due to varicocele and renal and adrenal metabolite
reflux, which can effectively induce apoptosis of genital cells
through enhanced oxidative stress. An imbalance in the levels of
reactive oxygen species (ROS) and antioxidants can induce anomalous
changes in sperm morphology and vitality through fatty acid
oxidation of the sperm cell membrane (5). The presence of cysts in the
reproductive tract (for example, in the ejaculatory duct and
seminal vesicles) can lead to low semen volume and
asthenozoospermia due to compression or blockage of the ejaculatory
duct (6).
Numerous factors, including immunological, genetic,
microbial, physiological and environmental factors, affect sperm
health. An imbalance or damage to the immune system in humans can
cause sperm antigens to stimulate the production of anti-sperm
antibodies, resulting in sperm agglutination and asthenozoospermia
(7). Genetic mutations can cause
structural and functional changes in microtubules of the sperm
flagella, resulting in multiple morphological abnormalities of the
sperm flagella and cilia and leading to primary ciliary dyskinesia
(8). Escherichia coli,
Chlamydia trachomatis and Ureaplasma urealyticum can
infect the reproductive system, particularly the accessory gonads,
causing inflammation and leukocytosis in the reproductive system.
Furthermore, the abundant content of peroxidase in the cytoplasm of
leukocytes accelerates ROS production and enhances oxidative stress
reactions. These microbial and biochemical effects can damage sperm
quality, sperm integrity, and the secretory function of accessory
gonadal structures, resulting in a decline in sperm viability
(9). The decline in semen volume,
sperm concentration and viability, and normal sperm rate is also
associated with long-term exposure to polluted air (10). Environmental exposure to heavy
metals (such as Pb and Cd) and plasticizers (such as phthalate) can
induce DNA damage. Moreover, excessive use of synthetic rubber or
polyester can cause a high environmental concentration of bisphenol
A, which damages the integrity of sperm DNA. Tobacco metabolites
(including nicotine, Cd and benzopyrene) can damage sperm DNA,
while alcohol can accelerate the apoptosis of sperm cells (11-13).
Although the effects of physiological and
pathological changes, environmental conditions, lifestyle and other
exposure factors on male fertility have been progressively
confirmed by numerous studies, asthenozoospermia remains a poorly
understood disorder because of its complex pathogenetic mechanism
and ambiguous treatment effects. Hence, the investigation of sperm
quality is critical to improve male fertility. Bioinformatics
combines molecular biology techniques and information technology to
study the molecular mechanisms of diseases. In the present study,
bioinformatics techniques were utilized to elucidate the underlying
genetic markers and pathogenetic mechanisms associated with
asthenozoospermia. Differentially expressed genes (DEGs) were
obtained using the GSE160749 dataset, a newly updated sperm
transcriptome profile of infertile men, from the Gene Expression
Omnibus (GEO) database (last updated: November 05, 2021).
Furthermore, epithelial-mesenchymal transition (EMT) is known to
influence embryo and organ development, inflammatory injury and
organ repair. Therefore, the obtained DEGs were intersected with
EMT datasets to filter candidate DEGs of asthenozoospermia.
Subsequently, bioinformatics techniques were used to analyze the
key genes, pathways, transcription regulation, immune regulation
and other factors involved in asthenozoospermia. These results
could enable providing novel clues to treat asthenozoospermia.
Materials and methods
Dataset selection and analysis of
DEGs
The GSE160749 dataset (GPL17692, [HuGene-2_1-st]
Affymetrix Human Gene 2.1 ST Array [transcript (gene) version]) was
downloaded from the GEO database. A total of six fertile control
sample files and five asthenozoospermia sample files were selected
from this dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE160749).
The EMT datasets (1184 EMT-related genes) were downloaded from the
dbEMT2 database (http://www.dbemt.bioinfo-minzhao.org/dbemt2.txt). DEGs
from the GSE160749 dataset were filtered using the limma package
(v3.42.2) (14) in R software
(v3.6; https://cran.r-project.org/bin/windows/base/old/3.6.0/)
based on the following preset thresholds: |log Fold Change| >1.0
and P<0.05. Venn analysis was used to filter the candidate DEGs
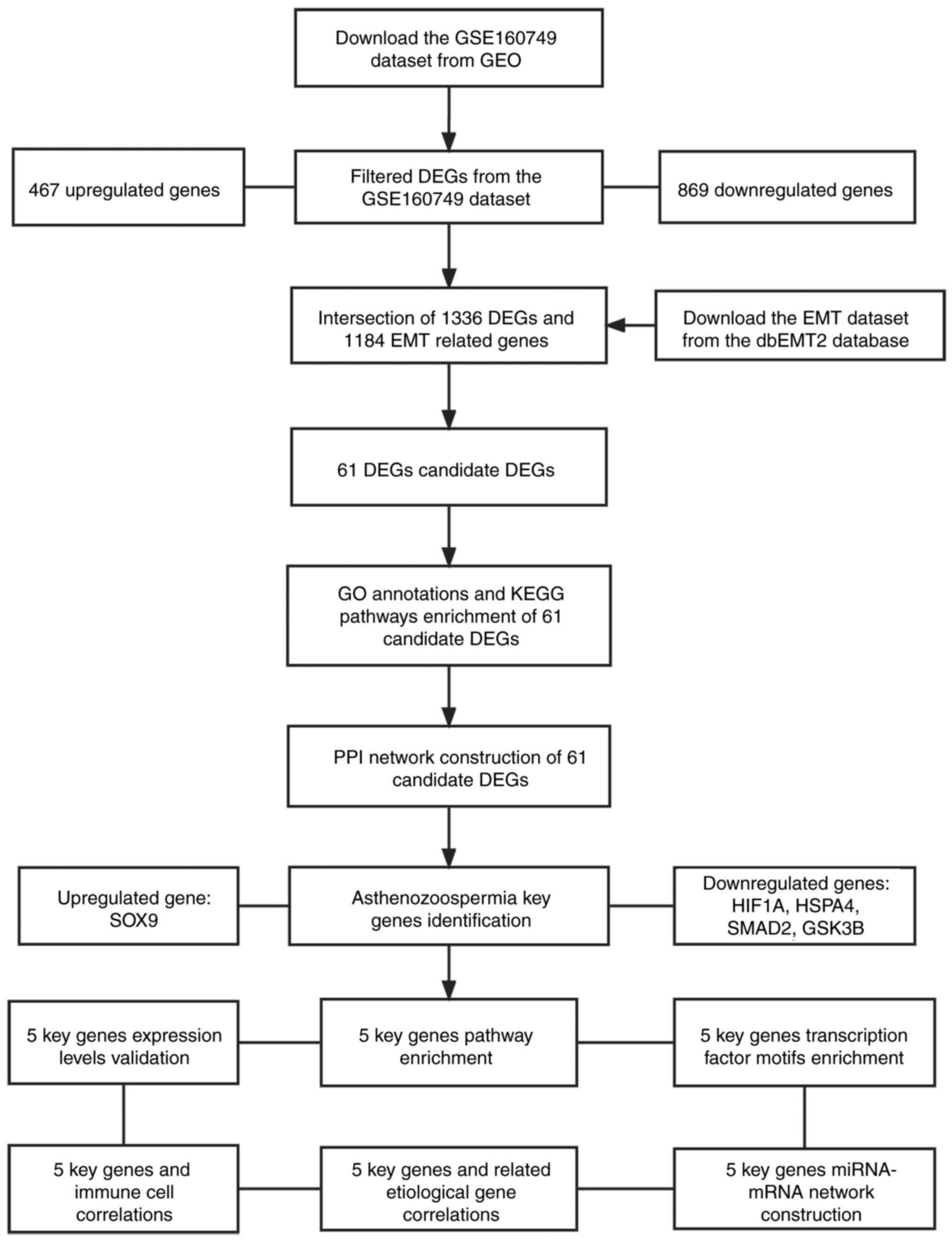
that intersected with the EMT datasets. The study design and data
processing are demonstrated in Fig.
1.
Functional enrichment analyses of the
candidate DEGs
To investigate the biological functions and pathways
of the candidate DEGs, Gene Ontology (GO) and Kyoto Encyclopedia of
Genes and Genomes (KEGG) pathway enrichment analyses were performed
using the clusterProfiler package (v4.1.4) in R software (15) (P<0.05). The Metascape database
(www.metascape.org) was also used to analyze the
candidate DEGs. The preset thresholds of functional enrichment were
min overlap ≥3 and P≤0.01.
Construction of the protein-protein
interaction (PPI) network
A PPI network was constructed by using the STRING
database (https://www.string-db.org/) to
determine the correlations among the candidate DEGs. To identify
the key genes in asthenozoospermia, the candidate DEGs were
filtered based on the constructed PPI network by using the Mcode
function of Cytoscape software (v3.7; https://cytoscape.org/). The confidence level was set
at 0.5.
Key gene pathway enrichment
analysis
By using predefined gene sets, gene set enrichment
analysis (GSEA) ranks genes according to their expression patterns
in two groups and then checks whether the predefined gene set is
overrepresented at the top or bottom of the gene rankings. In the
present study, the GSEA database (https://www.gsea-msigdb.org/) was used to find the
enriched key gene pathways in asthenozoospermia, and the molecular
mechanisms of the key genes were determined. To compare the
variations in the enriched KEGG pathways between the control and
asthenozoospermia groups, the number of permutations was preset to
1,000, and type was the inputted phenotype.
Transcription factor motif enrichment
based on key genes
RcisTarget package (v1.6.0) in R software was
utilized (16), to analyze
transcription factors (TFs). DNA motifs with significant
over-representation in the transcription start site of the genes in
the gene set were selected using a database containing genome-wide
cross-species rankings for each motif. The selected motifs were
then annotated to TFs, and those with a high normalized enrichment
score (NES) were retained. Briefly, the area under the curve (AUC)
of the motif-motif set pair was computed to estimate the
overexpression of each motif in the gene set. This was calculated
from the recovery curves of the gene sets used for the ordering of
motifs. The NES for each motif was estimated from the AUC
distribution of all motifs in the gene set. In the present study,
the 4.6-kb motif (rcistarget.hg19.motifdb.cisbpont.500bp) was used,
which is applied to human TFs in the gene-motif ranking
database.
miRNA-mRNA network construction based
on key genes
To analyze the asthenozoospermia regulatory
mechanisms, the miRWalk database (http://129.206.7.150/) was advised to extract
miRNA-mRNA pairs. The obtained results were then entered in the
TargetScan (https://www.targetscan.org/) or miRDB database
(http://www.mirdb.org/). The miRNA-mRNA network
was constructed using Cytoscape software (v3.7).
Correlations between key genes and
related etiological genes
Related etiological genes (relevance score: top 20)
of asthenozoospermia were obtained from the GeneCards database
(https://www.genecards.org/). Pearson's
correlation analysis was performed to determine the correlations
between the key genes and the related etiological genes.
Correlations between key genes and
immune cells
The ssGSEA package in R software was used to analyze
the proportion and type of immune cells in the semen based on six
fertile control sample files and five asthenozoospermia sample
files in the GSE160749 dataset. Pearson's correlation analysis was
used to analyze the correlations among the key genes and multiple
immune cells.
Clinical semen sample collection and
validation by reverse-transcription quantitative PCR (RT-qPCR)
The present study was approved (approval no.
chrec-2018-6) by the Ethics Committee of the Chinese Naval Medical
University (Shanghai, China) and was performed in accordance with
the principles of the Declaration of Helsinki as revised in 2013.
Informed consent was obtained from all participants before sample
collection. All patients were Han Chinese with no genetic
association. Patients without diseases of the reproductive system
(e.g., tumor, inflammation, and varicocele) were included in the
healthy control group. Semen samples were collected from 8 healthy
individuals and 8 asthenozoospermia patients diagnosed in The First
Affiliated Hospital of Naval Medical University (Shanghai, China)
in April 2023. Clinicopathological parameters including age
distribution are presented in Table
II. The samples were obtained through masturbation after 2 to 7
days of abstinence. The samples were processed according to the
guidelines of the WHO Laboratory Manual on Human Semen Examination
and Processing (6th Edition 2021), and the 5th percentile of the
semen parameter values was chosen as the lower reference limit
(2). The following parameters were
considered for asthenozoospermia semen samples: Samples collected
at ≥2 different time points and sperm progressive motility (PR)
<30%. The following parameters were considered for normal semen
samples: Semen volume ≥1.4 ml, sperm concentration
≥16x106/ml, pH≥7.2, PR≥0%, and PR + non-progressive
motility (NP) ≥42%.
 | Table IIAge and semen parameters in normal
and asthenozoospermia groups. |
Table II
Age and semen parameters in normal
and asthenozoospermia groups.
| No. | Parameters | Normal (n=8) | Asthenozoospermia
(n=8) | P-value |
|---|
| 1 | Age | 31.75±3.45 | 33.88±3.83 | 0.390 |
| 2 | Semen volume
(ml) | 4.39±1.52 | 3.31±1.68 | 0.220 |
| 3 | pH | 7.20 | 7.20 | NA |
| 4 | Sperm concentration
(x106/ml) | 78.24±35.42 | 19.32±16.12 | 0.005 |
| 5 | Progressive
motility (%) | 70.38±5.93 | 4.75±4.56 | <0.001 |
| 6 | Motility (%) | 75.88±4.52 | 11.00±9.35 | <0.001 |
In the validation test, by using phosphate buffered
saline and somatic cell lysis buffer successively, seminal plasma
was removed and somatic cells from the semen and extracted pure
sperm cells from the samples (17). Subsequently, TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) was selected
to extract RNA from sperm cells. A commercial kit (cat. no.
AG11706; Hunan Accurate Bio-Medical Co., Ltd.) was used for the
reverse transcription of RNA into cDNA according to the
manufacturer's instructions. The SYBR qPCR mix kit (cat. no.
AG11702; Hunan Accurate Bio-Medical Co., Ltd.) was used for RT-qPCR
with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a
reference gene. The primers (Sangon Biotech Co., Ltd.) for five
target genes are listed in Table
I. A total of 20 µl reaction mixture volume was used for
RT-qPCR performed with the ABI PRISM 7500 Real-Time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The qPCR
conditions were as follows: Stage 1, initial denaturation at 95˚C
for 2 min; Stage 2, 40 cycles of denaturation at 95˚C for 30 sec,
annealing at 60˚C for 30 sec, and elongation at 72˚C for 30 sec.
All samples were normalized according to GAPDH expression,
and the relative expression of these genes was quantified using the
2-IICq method (18).
 | Table IPrimers used in reverse
transcription-quantitative PCR. |
Table I
Primers used in reverse
transcription-quantitative PCR.
| No. | Gene name | Primer sequence
(5'→3') |
|---|
| 1 | SOX9 | F:
AAGCTCTGGAGACTTCTGAACG |
| | | R:
CTGCCCGTTCTTCACCGACT |
| 2 | HSPA4 | F:
GTGGACCTGCCAATCGAGAA |
| | | R:
CCTCCACTGCGTTCTTAGCA |
| 3 | SMAD2 | F:
TGGGGACTGAGTACACCAAA |
| | | R:
GGGATACCTGGAGACGACCA |
| 4 | HIF1A | F:
TTTGGCAGCAACGACACAGA |
| | | R:
TTTCAGCGGTGGGTAATGGA |
| 5 | GSK3B | F:
TGTGTGTTGGCTGAGCTGTT |
| | | R:
TCCCTTGTTGGAGTTCCCAG |
Statistical analysis
All the experimental data were recorded in Excel
2007 (Microsoft Corp.). SPSS version 28.0 (IBM Corp.) was used to
compare the data between the semen parameters of the normal and
asthenozoospermia groups. All measurement data are presented as the
mean ± standard deviation which represent the average level and
dispersion tendency. The unpaired t-test was performed for
comparison between measurement data from clinical semen sample
parameter detection and RT-qPCR validation in the normal and
asthenozoospermia groups. The DEGs were determined using the
Benjamini-Hochberg method. The Pearson's method was performed to
analyze the correlation between the expression levels of five key
genes and the top 20 related etiological genes in
asthenozoospermia. The Wilcoxon rank-sum test and Pearson's method
were used to analyze the differential expression levels and
correlations of immune cells in six fertile control sample files
and five asthenozoospermia sample files from the GSE160749 dataset.
For all results, P<0.05 was considered to indicate a
statistically significant difference.
Results
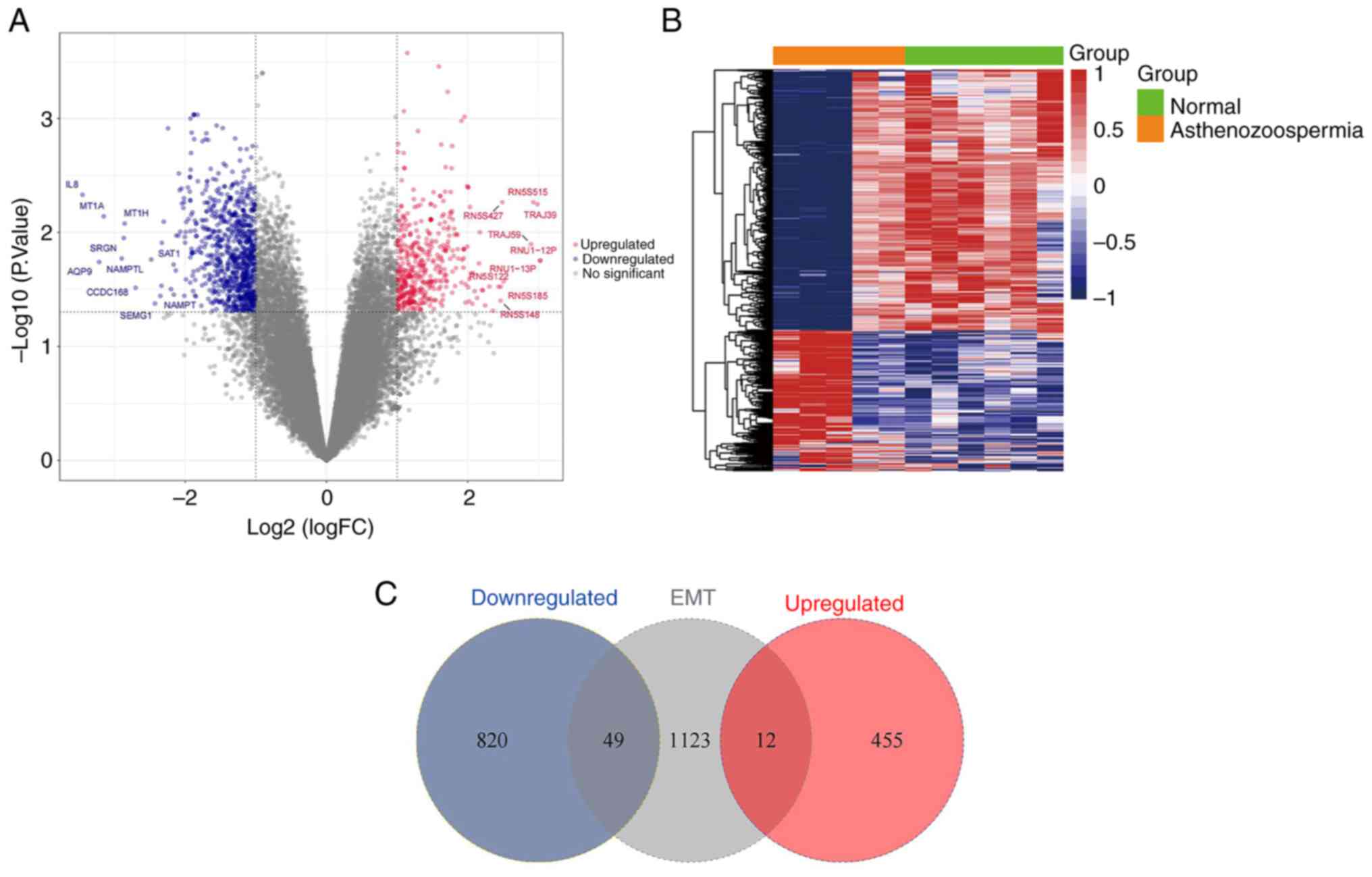
Filtered DEGs
A total of 1,336 DEGs were successfully filtered
from the GSE160749 dataset (Fig.
2A and B); these DEGs
comprised 467 upregulated genes (Table SI) and 869 downregulated genes
(Table SII). In total, 61
candidate DEGs that intersected with the EMT datasets were filtered
for analysis, which included 12 upregulated genes and 49
downregulated genes (Fig. 2C).
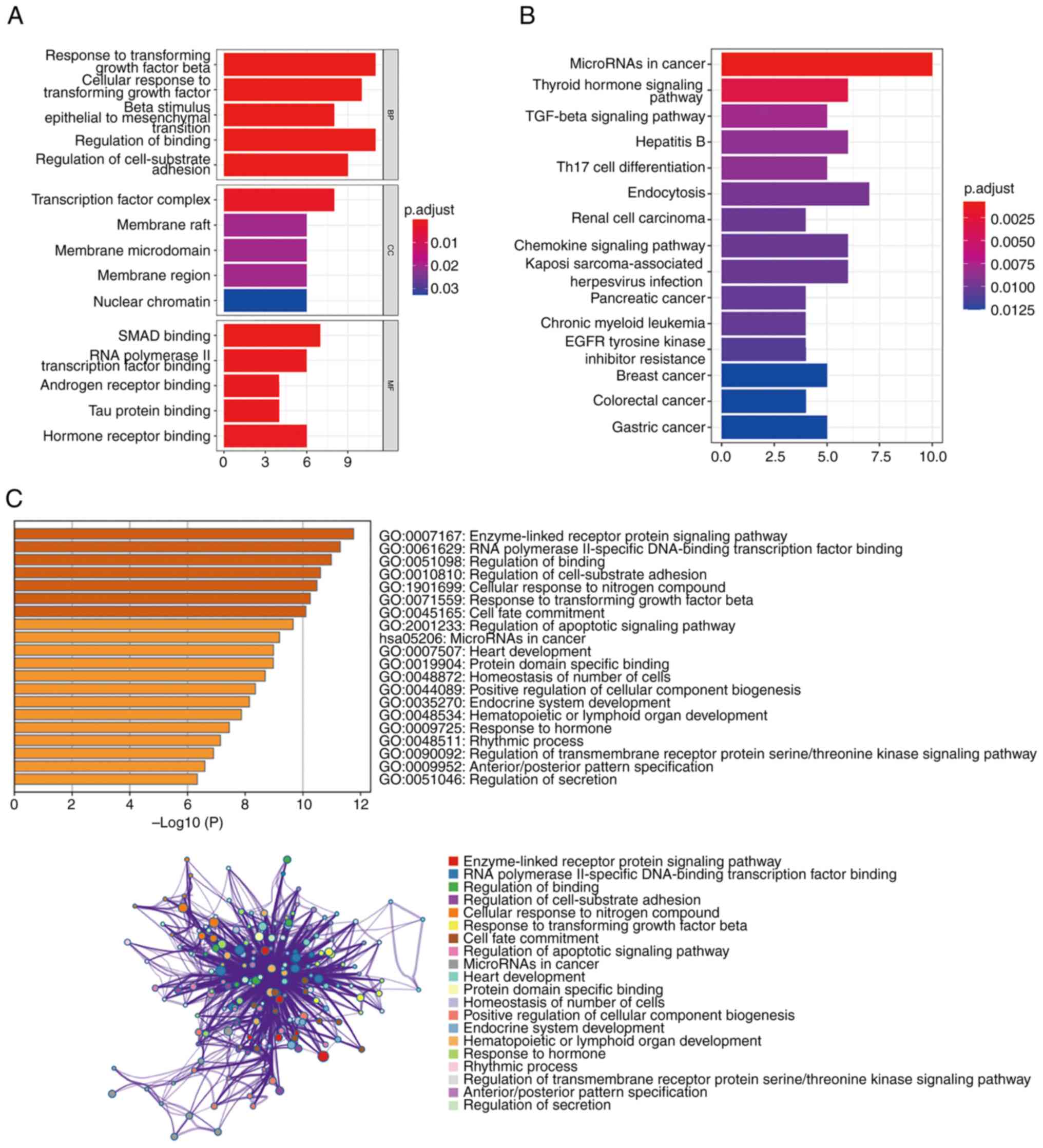
Functional enrichment of the candidate
DEGs
GO and KEGG pathway enrichment analyses were
conducted to determine functional enrichment of the 61 candidate
DEGs (Fig. 3A and B). In the biological processes' category,
the 61 DEGs were significantly enriched in the regulation of
binding. In the cellular components' category, the DEGs were
significantly enriched in the TF complex. In the molecular
functions' category, the DEGs were significantly enriched in SMAD
binding and hormone receptor binding. Additionally, the pathways
that were significantly enriched in the 61 DEGs were the thyroid
hormone signaling pathway, TGF-β signaling pathway and Th17 cell
differentiation pathway. The Metascape database was also used to
determine pathway enrichment of the 61 DEGs. The results
demonstrated that the pathways were mainly enriched in the
enzyme-linked receptor protein signaling pathway and RNA polymerase
II-specific DNA binding TF binding (Fig. 3C).
Construction of the PPI network
To conduct in-depth analysis of the relationship
among the 61 candidate DEGs, a PPI network was constructed that
included 40 nodes and 80 edges (Fig.
4). It is necessary to explain that each node represents a
protein and each edge represents a link with two proteins,
indicating functional association between proteins. The gene
cluster with the highest score was identified from the PPI network
by using the MCODE function. The gene cluster included
HSPA4, SOX9, SMAD2, HIF1A and
GSK3B (score=5.00), there was five nodes and 10 edges.
SOX9 was upregulated, while HSPA4, SMAD2,
HIF1A and GSK3B were downregulated in
asthenozoospermia.
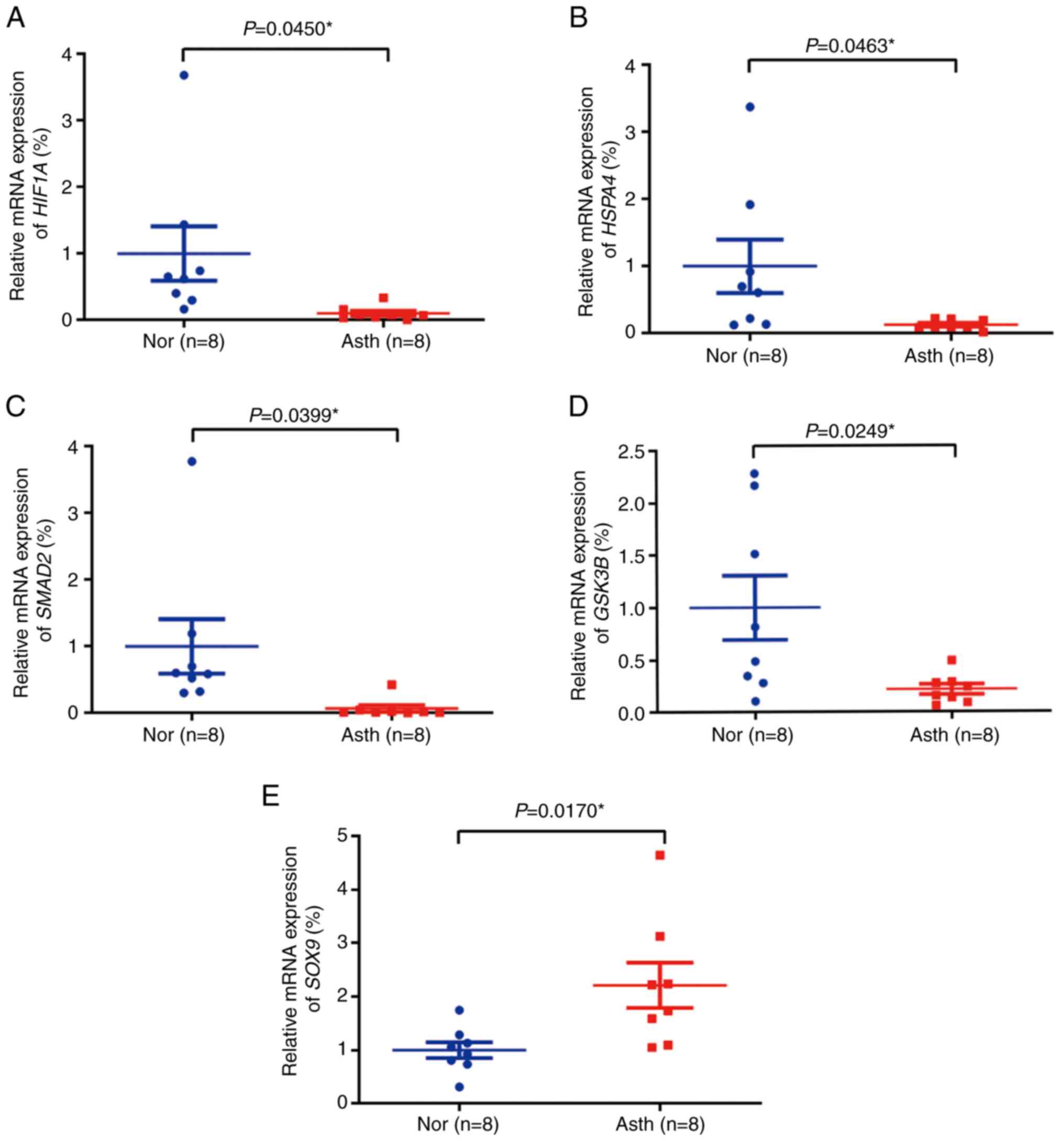
Validation of clinical semen samples
by RT-qPCR
The age and semen parameters in the normal and
asthenozoospermia groups are listed in Table II. The RT-qPCR results revealed
that the expression levels of HIF1A, HSPA4,
SMAD2 and GSK3B (Fig.
5A-D, respectively) were lower in the asthenozoospermia group
than in the normal group; however, SOX9 (Fig. 5E) expression was significantly
higher in the asthenozoospermia group. The validation results of
the 16 clinical semen samples were consistent with those of the
bioinformatics analysis for downregulated and upregulated genes.
The Cq values for RT-qPCR validation for clinical semen samples are
included in Table SIII.
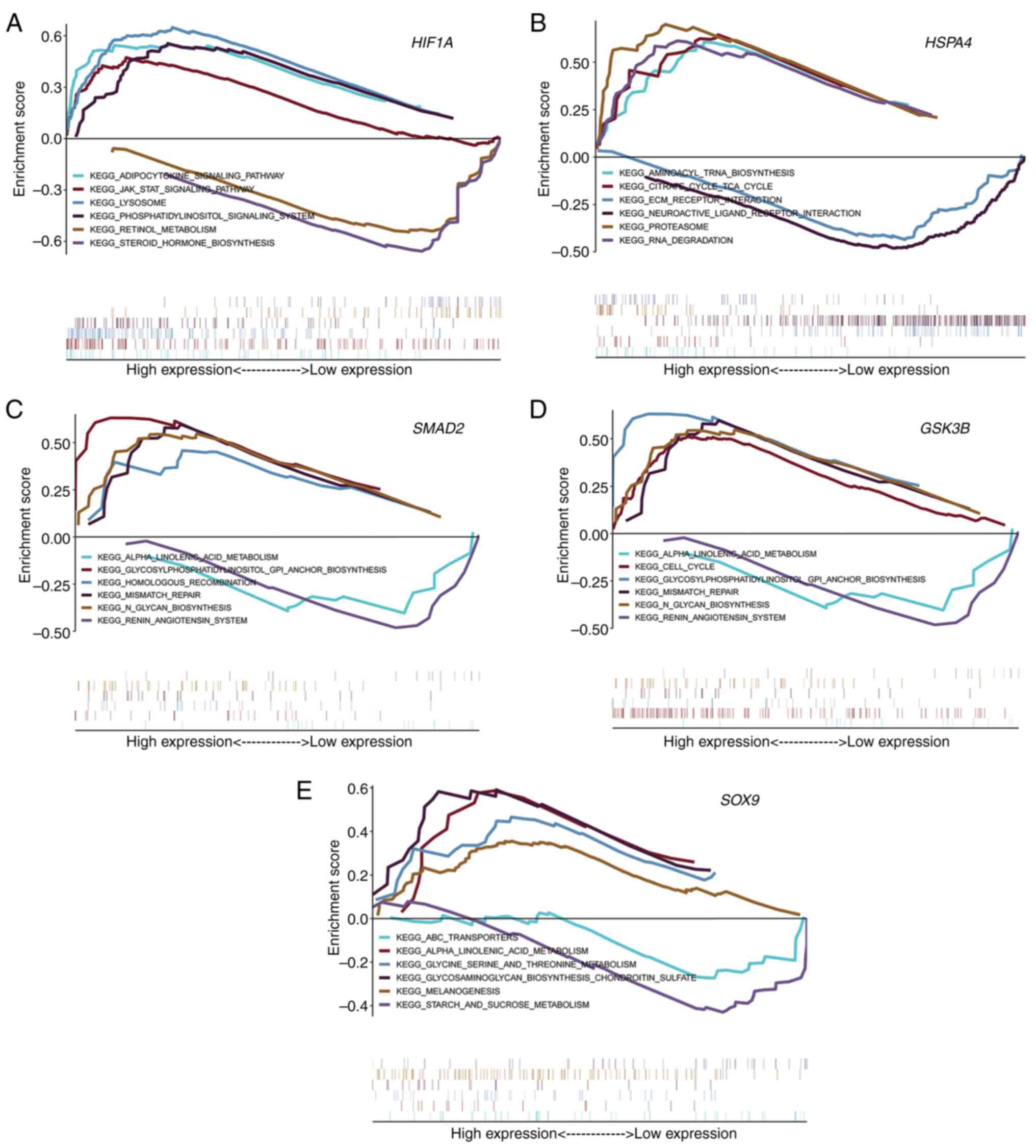
Key gene pathway enrichment
Pathway enrichment analysis was performed using GSEA
on five key genes of asthenozoospermia. HIF1A was enriched
mainly in the adipocytokine cytokine signaling pathway, JAK-STAT
signaling pathway and retinol metabolism (Fig. 6A). HSPA4 was enriched mainly
in aminoacyl tRNA biosynthesis and the tricarboxylic acid (TCA)
cycle (Fig. 6B). SMAD2 was
enriched mainly in α-linolenic acid metabolism,
glycosylphosphatidylinositol GPI anchor biosynthesis and the
renin-angiotensin system (Fig.
6C). GSK3B was enriched mainly in α-linolenic acid
metabolism, the cell cycle and the renin-angiotensin system
(Fig. 6D). SOX9 was
enriched mainly in ABC transporters, α-linolenic acid metabolism,
and glycine serine and threonine metabolism (Fig. 6E).
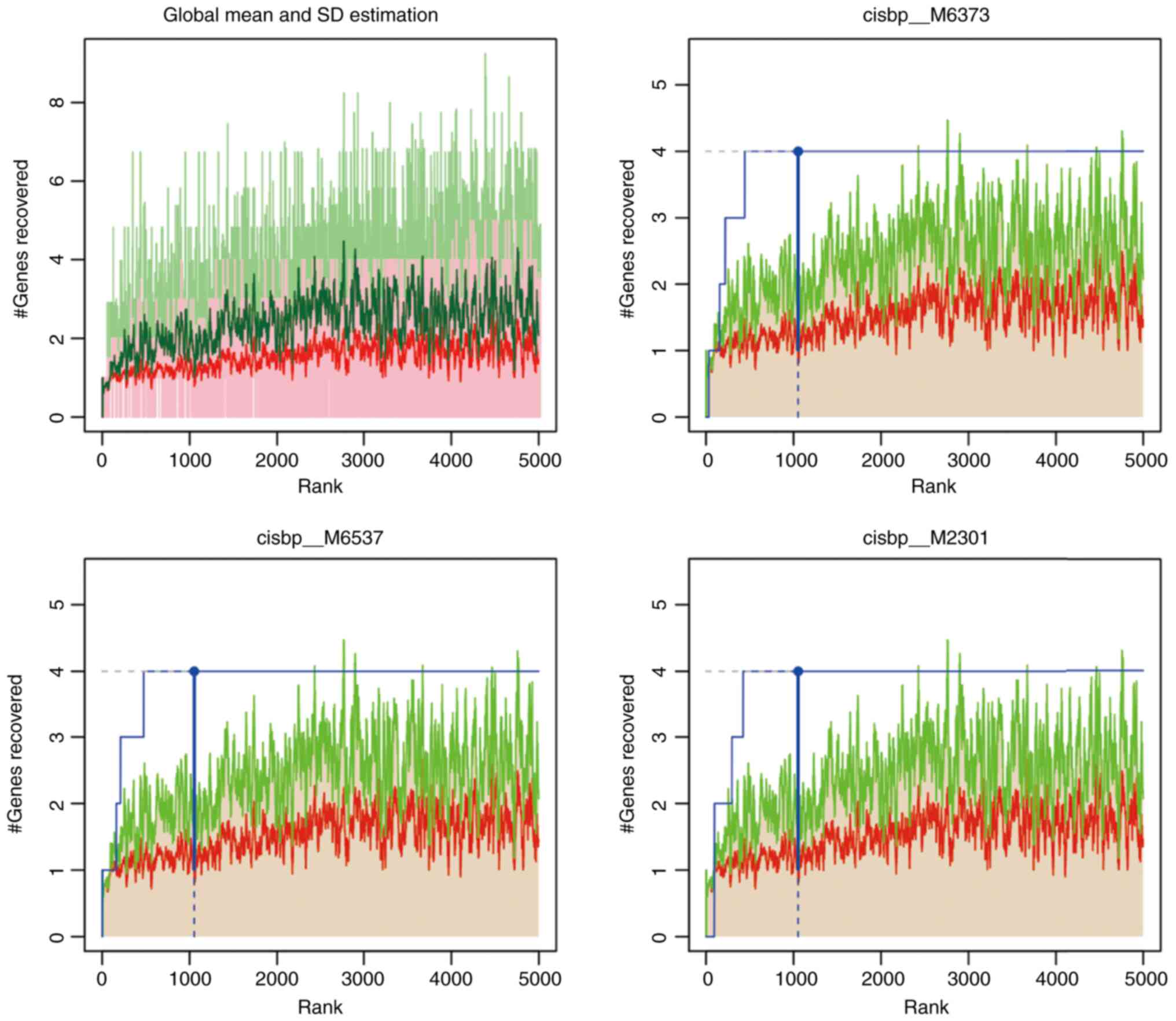
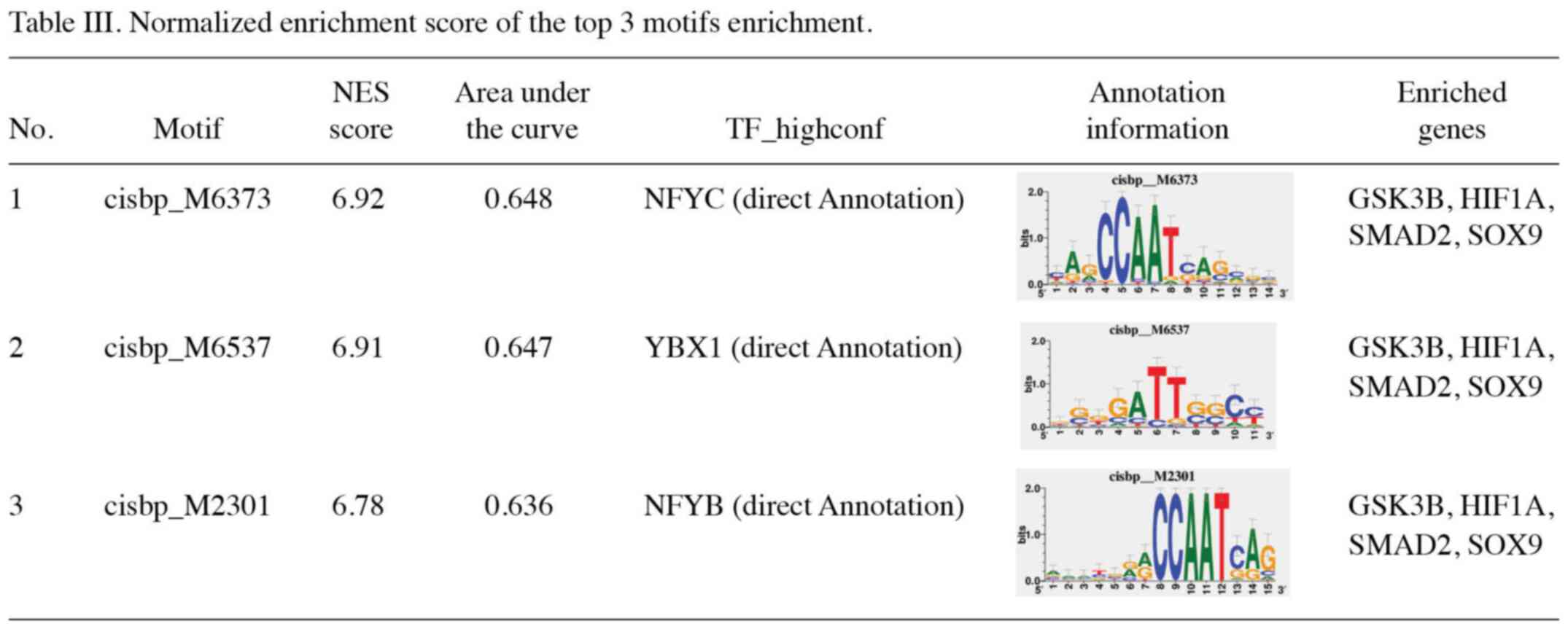
TF motif enrichment for key genes
A total of five key genes were selected from the
gene set to analyze their regulation mechanisms. These genes were
regulated by multiple TFs. Therefore, cumulative recovery curves
were used to enrich these TFs (Fig.
7). The analysis demonstrated that the motif with the highest
NES value (6.92) was cisbp_M6373. In total, four genes, namely
GSK3B, HIF1A, SMAD2 and SOX9, were
enriched in this motif. TFs of the key genes were identified in all
enriched motifs (Tables III and
SIV).
miRNA-mRNA network construction for
key genes
Several miRNA-mRNA pairs were associated with the
mRNAs of the five key genes. A total of 384 miRNA-mRNA pairs
(Table SV) were retained from the
TargetScan and miRDB databases, including five mRNAs and 361
miRNAs. In this network, 21 miRNAs acted as coregulators among the
five key genes (Table IV).
 | Table IVCo-regulators of the five key genes
in the miRNA-mRNA network. |
Table IV
Co-regulators of the five key genes
in the miRNA-mRNA network.
| No. | Genes | miRNAs |
|---|
| 1 | HSPA4, SMAD2 and
GSK3B | hsa-miR-8085,
hsa-miR-6788-5p |
| 2 | SOX9, GSK3B | hsa-miR-4306 |
| 3 | SOX9, SMAD2 | hsa-miR-8485,
hsa-miR-5195-3p |
| 4 | HSPA4, SMAD2 |
hsa-miR-30c-2-3p |
| 5 | HIF1A, SMAD2 | hsa-miR-3121-3p,
hsa-miR-6807-5p |
| 6 | GSK3B, SMAD2 | hsa-miR-3190-3p,
hsa-miR-4728-5p, hsa-miR-30c-1-3p, hsa-miR-6881-3p,
hsa-miR-5002-5p, hsa-miR-4768-5p, hsa-miR-6731-5p, hsa-miR-6733-5p,
hsa-miR-6833-3p, hsa-miR-641, hsa-miR-5584-5p, hsa-miR-6513-3p,
hsa-miR-6766-3p |
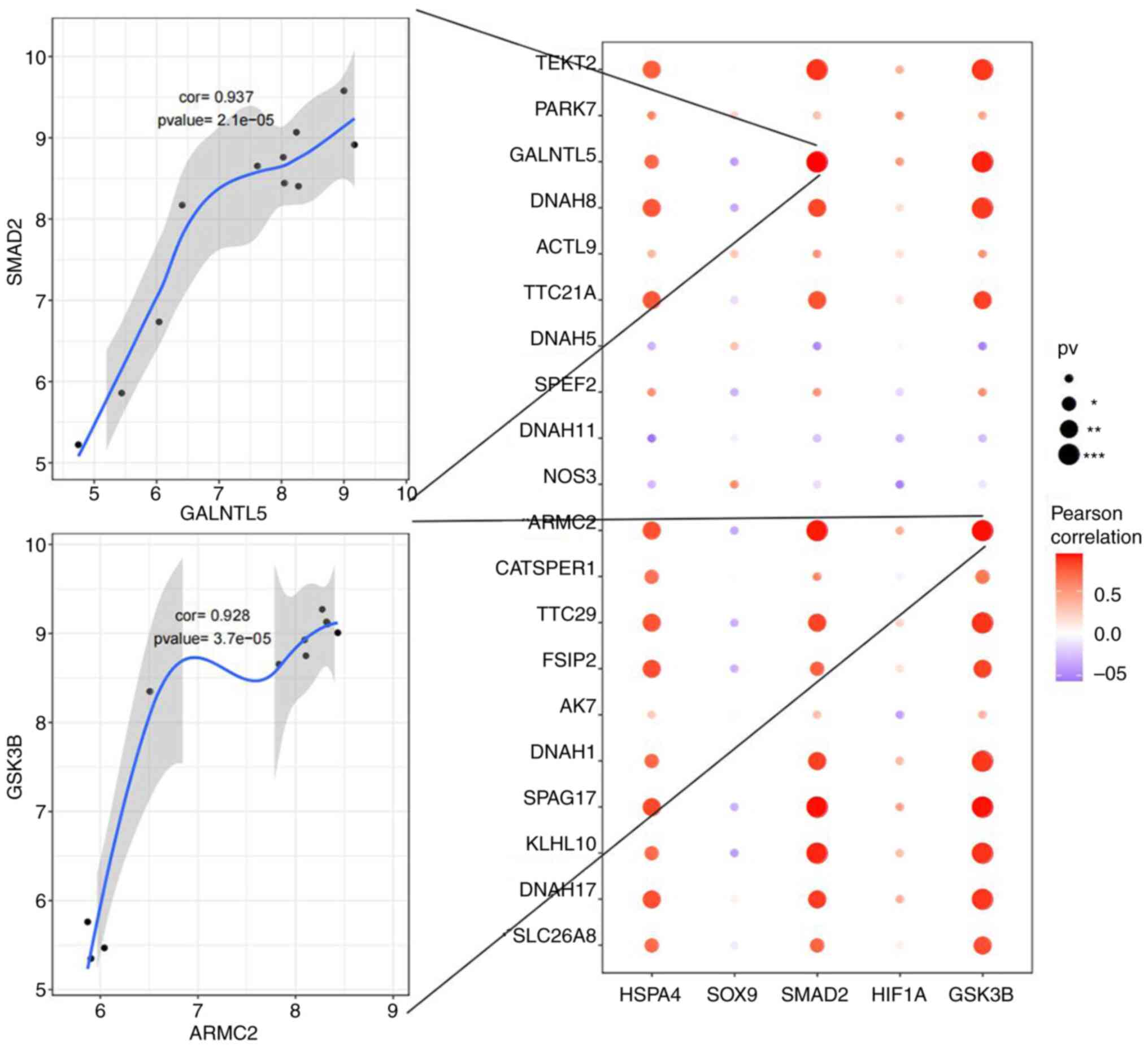
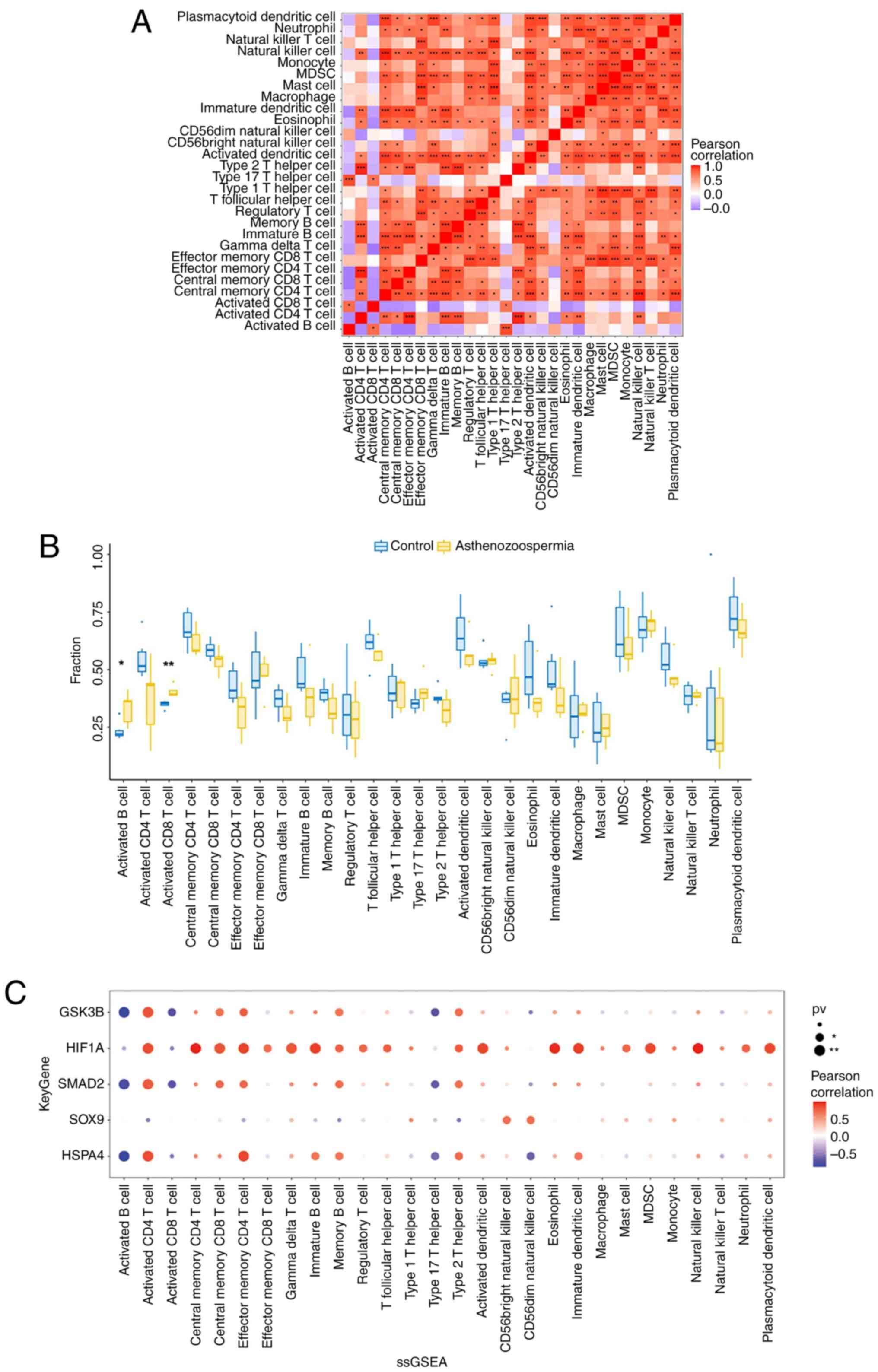
Correlations between key genes and
related etiological genes
A correlation analysis was performed between the
expression levels of five key genes and the top 20 related
etiological genes in asthenozoospermia. HSPA4, SMAD2
and GSK3B revealed a positive correlation with multiple
related etiological genes, including SMAD2 and
GALNTL5 (r=0.937, P<0.001) and GSK3B and
ARMC2 (r=0.928, P<0.001) (Fig. 8 and Table SVI).
Correlations between key genes and
immune cells
The correlations of immune cells in six fertile
control sample files and five asthenozoospermia sample files from
the GSE160749 dataset were analyzed (Fig. 9A). Higher levels of activated B
cells and activated CD8 T cells were observed in the five
asthenozoospermia samples than in the six control samples (Fig. 9B). Correlations among the
expression levels of the five key genes and multiple immune cells
were also analyzed in 11 semen samples from the GSE160749 dataset.
The expression levels of HSPA4, SMAD2 and
GSK3B were positively correlated with activated CD4 T cells
and effector memory CD4 T cells but negatively correlated with
activated B cells and type 17 T helper cells. The HIF1A
expression level was positively correlated with numerous immune
cells, such as activated CD4 T cells, central memory CD4 T cells,
central memory CD8 T cells and natural killer (NK) cells. The
expression level of SOX9 was positively correlated with CD56
(bright or dim) NK cells (Fig.
9C).
Discussion
Asthenozoospermia is mainly characterized by poor
sperm motility in clinical examination, and it is generally caused
by defects in the function or structure of sperms and abnormal
spermatoplasm (19). Studies
investigating the molecular mechanisms of asthenozoospermia may
help discover more novel effective treatments for male infertility.
Abnormal spermatogenesis and reproductive tract infections are the
main causative factors of asthenozoospermia. The critical roles of
EMT in embryo and organ development, inflammatory injury, and organ
repair have received widespread attention. Cunha et al
(20) reported the presence of an
androgen-receptor (AR)-positive testicular medulla in the
developing human testis; this medulla acted as a zone of
mesenchymal to epithelial transition and a zone from which
AR-positive cells appeared to migrate into the human testicular
cortex. The human testicular medulla is a known source of Sertoli
cells in seminiferous cords. Klein et al (21) noted that dexamethasone therapy
improved the outcomes of mice with preclinical uropathogenic E.
coli-induced epididymitis and found that EMT was involved in the
interstitial fibrosis transformation process of this epididymitis.
Therefore, in the present study, the dbEMT2 database was selected
to explore the pathogenic genes related to asthenozoospermia. The
GSE160749 dataset from the GEO database is a recently updated sperm
transcriptome profile of samples from infertile men, and it
includes five asthenozoospermia files, seven
asthenoteratozoospermia files, six infertile files and six fertile
control sample files. A PubMed search revealed that a limited
number of relevant studies have contributed to this dataset
(22). Therefore, in the present
study, five asthenozoospermia files and six fertile control sample
files were selected as a dataset to analyze novel key genes in
asthenozoospermia. Subsequently, by using bioinformatics
techniques, 1,336 DEGs associated with asthenozoospermia were
identified, including 467 upregulated genes and 869 downregulated
genes. Furthermore, 61 EMT-related genes were identified among
these 1,336 DEGs, which included 12 upregulated genes and 49
downregulated genes. The STRING database was utilized to construct
a PPI network for the 61 candidate DEGs. Five genes were most
closely associated with asthenozoospermia after filtering; this
included one upregulated gene (SOX9) and four downregulated
genes (HSPA4, SMAD2, HIF1A and GSK3B).
Moreover, the bioinformatics analysis results for these five genes
were consistent with the RT-qPCR results for the 16 clinical semen
samples used for validation. Therefore, these results may provide
novel clues regarding the molecular mechanisms of
asthenozoospermia.
Among the five key genes, SOX9, an autosomal
gene and a primary downstream target of SRY, plays a pivotal role
in male sexual development. SOX9 is involved in the development or
maintenance of Sertoli cells and in gonadal dysgenesis and XY and
XX sex reversal (23,24). HSPA4 plays an important role in
spermatogenesis and is also involved in the development of
oligozoospermia and varicocele (25). Compared with wild-type littermates,
Hspa4-deficient mice showed a drastic reduction in the total number
of spermatozoa and their motility. The majority of pachytene
spermatocytes in juvenile Hspa4(-/-) mice failed to
complete the first meiotic prophase and underwent apoptosis
(26). SMAD2 is mainly localized
in the cytoplasm of meiotic genital, Sertoli and Leydig cells. It
plays an important role in testicular development and
spermatogenesis (27). HIF1A was
robustly expressed in spermatogonial cells of the testis in both
juvenile (6 weeks old) and adult (3 months old) male mice (28). Gorga et al (29) suggested that HIFs may be involved
in regulating the proliferation of Sertoli cells by
follicle-stimulating hormone (FSH). However, Ghandehari-Alavijeh
et al (30) reported a
different result for the association of HIF1A expression with
asthenozoospermia; these authors found a significant negative
correlation between HIF1A expression (r=-0.403, P=0.046) and sperm
motility. Therefore, the function of HIF1A in asthenozoospermia
will require further investigations. GSK3A and GSK3B are two
isomers of GSK3. Both these isomers are related to spermatogenesis
and sperm motility, and the effect of GSK3A was found to be more
significant than that of GSK3B. Following GSK3A knockout in the
testis, testicular weight and sperm counts were normal in
GSK3A(-/-) mice; however, the number of infertile male
mice increased because of decreased sperm motility. Compared with
wild-type mice, GSK3A-/- mice exhibited lower sperm ATP
levels and flagellar beat amplitude (31,32).
As one of the key downregulated genes in asthenozoospermia, the
role of GSK3B in spermatogenesis requires confirmation in future
research.
In the present study, several pathways that may
provide clues to the molecular mechanisms of asthenozoospermia were
identified. The thyroid hormone signaling pathway affects the
testis in various ways, including its effects on Leydig cells,
Sertoli cells and spermatogenic cells. A surplus or deficiency of
thyroid hormone can lead to abnormal testicular function.
Hyperthyroidism is associated with decreased semen volume, sperm
density, motility and abnormal sperm morphology (33). In vitro experiments have
revealed that in male patients with idiopathic infertility, 0.9
pmol/l of levothyroxine (LT4) significantly increased the
percentage of spermatozoa with high mitochondrial membrane
potential and improved sperm motility. LT4 also reduced sperm
necrosis and lipid peroxidation, thereby ameliorating chromatin
compactness. LT4 exerted these effects at 2.9 pmol/l concentration,
which is close to the physiological concentration of free thyroxine
(FT4) in the seminal fluid of euthyroid subjects. Thyroid hormones
play a beneficial role in sperm mitochondrial function, oxidative
stress and DNA integrity (34). In
a cross-sectional study of 5401 infertile men, subclinical
hypothyroidism was significantly associated with an increased risk
of an abnormal DNA fragmentation index (DFI) (35). The enzyme-linked receptor signaling
pathway is related to cell reproduction, growth and differentiation
processes (36). The enzyme-linked
receptor is a transmembrane protein, and the intracellular domain
usually exhibits some enzyme activity. Enzyme-linked receptors
respond slowly to extracellular signaling (measured in h) and
require coordination among numerous intracellular transduction
steps. A number of experiments have confirmed that receptor
tyrosine kinase (RTK) activity may affect primordial germ cell
migration through the RTK-Ras signaling pathway. Mitogen-activated
protein kinase (MAPK) can promote genital cell proliferation,
meiosis and Sertoli cell proliferation. Differential miRNA
expression is associated with the PI3K-AKT and MAPK signaling
pathways in asthenozoospermia, and sperm motility is regulated
through the concerted efforts of these signaling pathways (37-39).
Transforming growth factor-beta (TGF-β) family members and their
receptors are expressed in the testis and play important paracrine
and autocrine roles in testicular development and spermatogenesis.
SMAD is a downstream protein of the TGF-β family, and the
TGF-β-SMAD pathway plays an important role in testicular
development and spermatogenesis (27). Inhibin B, a member of the TGF-β
family, is involved in the regulation of spermatogenesis. A
previous study revealed a high expression of inhibin B in Sertoli
cells, Leydig cells, and the cytoplasm of spermatogonia in patients
with focal spermatogenesis disorders (40). The JAK-STAT signaling pathway is
stimulated by cytokines and plays a role in cell proliferation,
differentiation, apoptosis and immune regulation. The JAK-STAT
pathway may also be involved in the pathogenic mechanism of
asthenozoospermia. The protein and mRNA levels of both Janus kinase
(JAK) and signal transducer and activator of transcription (STAT)
were significantly reduced in asthenozoospermia (41).
The five identified key genes were also found to be
enriched in several other signaling pathways. HSPA4 was
enriched in the TCA cycle. As the common pathway for the catabolism
of sugars, lipids and proteins in humans, the TCA cycle is the main
pathway of energy production. Recent studies have demonstrated that
this pathway is involved in spermatogenesis in asthenozoospermia
patients. The semen of asthenozoospermia patients showed decreased
expression of four enzymes (fructokinase, citrate synthase,
succinate dehydrogenase and spermine synthase) related to energy
metabolism (42). The levels of
citrate, malate, succinate and pyruvate were substantially
decreased; however, lactate levels were increased. These results
suggested that asthenozoospermic patients had reduced energy
produced by aerobic metabolism through the TCA cycle, which was
compensated by the anaerobic glycolytic pathway, resulting in
reduced sperm capacity. Isocitrate dehydrogenase 3 is a key enzyme
in the mitochondrial TCA cycle, and its reduced levels can cause
sperm energy deficits and disrupt acrosome and flagellogenesis,
resulting in spermatogenesis arrest (43). Furthermore, extramitochondrial
citrate synthase is abundantly found in the sperm head. A
metabolomics analysis of aged sperms revealed that the loss of
extramitochondrial citrate synthase enhances the TCA cycle in the
mitochondria with age, presumably leading to the depletion of
extramitochondrial citrate; this might lead to age-dependent male
infertility (44).
SOX9, SMAD2 and GSK3B were
enriched in the α-linoleic acid metabolic signaling pathway, which
can be regarded as the focus of the molecular mechanism in the
pathogenesis of asthenozoospermia. The lipid composition of the
sperm membrane significantly affects sperm quality and function.
Sperm samples from asthenozoospermic patients revealed a lower
level of polyunsaturated fatty acids (such as docosahexaenoic acid)
and a higher level of saturated fatty acids (such as palmitic acid)
than those from normal individuals (45). A randomized controlled study that
evaluated the effect of long-term nut consumption on changes in
semen parameters reported that nuts were rich in unsaturated fatty
acids such as α-linoleic acid, which increased sperm count,
vitality, motility and morphology, and the DFI level of sperm was
significantly reduced (46).
Similar results were obtained in long-term observations of the
effects of animal diets on sperm parameters (47). SMAD2 and GSK3B were
simultaneously enriched in the renin-angiotensin system (RAS)
signaling pathway in the male reproductive system, which regulates
male fertility through paracrine and autocrine mechanisms. RAS is
present in Leydig, Sertoli and spermatogenic cells in the testis
and regulates testosterone and spermatogenesis.
Angiotensin-converting enzyme, angiotensin II type 2 receptor and
aminopeptidase N in the testicular RAS can be used as potential
biological markers of high-quality embryos to assess and diagnose
male fertility (48). SOX9
was enriched in the glycine-serine-threonine metabolism signaling
pathway (49). This pathway is
activated and affects sperm motility in asthenozoospermia,
oligozoospermia, teratozoospermia and azoospermia.
Leptin is a member of the adipocytokine signaling
pathway enriched by HIF1A. Human and animal studies have
identified that leptin correlates with male infertility and
obesity. Increased leptin levels are associated with low sperm
count, high abnormal sperm count, enhanced sperm oxidative stress
effects and obesity (50). Daily
intraperitoneal administration of 5-30 mg/kg body weight leptin for
42 days in adult rats decreased the sperm count and increased the
fraction of abnormal sperms (51).
Obesity-related diseases are associated with dysregulated adipocyte
function and microenvironmental inflammatory processes.
Dysregulated adipocytokines notably influence the insulin signaling
pathway and may induce adverse effects on testicular function
(52). Although obesity factors
negatively affect sperm quality and function, they can still be
transferred as epigenetic factors to the offspring (53). In a previous study, patients with
varicocele and leukocytospermia revealed significantly higher
seminal plasma leptin levels and sperm apoptosis rates than the
control group (54). Seminal
plasma leptin levels were significantly associated with the sperm
apoptotic rate, and leptin may promote sperm cell apoptosis. It was
also discovered that leptin induces sperm cell apoptosis through
TNF-α in leukozoospermic patients (54). However, adiponectin and its
receptors, expressed in male genital cells, can promote
spermatogenesis and maturation (55). Treatment of elderly mice with
exogenous adiponectin significantly improved testicular mass,
genital cell proliferation, insulin receptor expression, testicular
glucose uptake, antioxidant enzyme activity and testosterone
synthesis. Thus, adiponectin therapy could serve as an effective
therapeutic strategy to improve sperm and testosterone levels in
the testis (56). HIF1A was
also enriched in the retinol signaling pathway. Retinol is also
known as vitamin A, and its metabolite retinoic acid (RA) occurs in
the seminiferous tubules. RA is stimulated by hormones such as FSH
and testosterone and can regulate the processes of proliferation
and differentiation of spermatogonia, meiosis, spermiogenesis and
sperm release. RA regulates the expression of Stra8, Kit, GDNF and
BMP4 to promote or inhibit spermatogenesis through various
pathways. RA also inhibits spermatogonial renewal by directly or
indirectly inhibiting the expression of DMRT, GDNF and cyclin. RA
controls spermatogonial stem cell differentiation through Kit
induction and Nanos2 inhibition and regulates spermatogonia meiosis
through Stra8 upregulation. At the spermatogenesis stage, RARα
binds to RA as a key regulator and upregulates Stra8 to control
spermatogenesis. Although RA plays an important role in all stages
of spermatogenesis, it has more critical involvement in
spermatogonia differentiation and early meiosis of spermatocytes
(57). Although the role of
adiponectin and retinol in supporting male reproductive function
has been observed in some clinical treatments or in animal studies,
the appropriateness of these drugs remains to be validated by more
clinical trials and results.
Hernández-Silva et al (58) found that human spermatozoa RNA
includes both non-coding and protein-coding RNAs that play a
potential regulatory function in male fertility. They identified
100 transcripts with consistent differential expression as
candidates for the molecular source of asthenozoospermia. As
underlying biomarkers, miRNAs may provide evidence of the
pathophysiological changes occurring during the spermatogenesis
process. Corral-Vazquez et al (59) analyzed 48 clinical semen samples to
search new molecular biomarkers for diagnosing male infertility,
and they finally detected 2 pairs of miRNAs
(hsa-miR-942-5p/hsa-miR-1208 and hsa-miR-34b-3p/hsa-miR-93-3p). In
the present study, a regulatory network of TFs and miRNAs was
analyzed and constructed, and it was proposed that the synergistic
effect of these TFs and miRNAs could serve as a novel approach to
investigate the underlying mechanisms and molecular targets of
asthenozoospermia.
To date, very few studies have investigated the
effects of the testis immune microenvironment on asthenozoospermia.
As a complete and stable immune microenvironment, the human
blood-testis barrier is jointly regulated by humoral and cellular
immunity, and immune cells, immunoglobulins and immunoregulatory
factors play a coordinated role in maintaining this immune
microenvironment. Inflammatory factors and oxidative stress play an
etiological role in asthenozoospermia and can stimulate B cell and
T cell activation (60-62).
This finding is consistent with the results of the present study,
where high levels of activated B cells and CD8 T cells were
detected in the semen samples of the asthenozoospermia group.
Activated B cells can synthesize and secrete immunoglobulins, which
are involved in the humoral immune response. As important
components in cellular immunity, CD8 T cells mediate cytotoxic
effects and play a critical role in infection and inflammation.
Based on the GSE160749 dataset, correlations were found among the
five identified key genes and different immune cells, thus
providing new clues regarding the role of cellular immunity in the
mechanism of asthenozoospermia and clinical therapeutic
sensitivity. An analysis of seminal leukocyte subsets of 70
sub-fertile men identified that the levels of T (CD3 and CD69), B
(CD20 and CD69), NK (CD56) and CD8 T cells were significantly
elevated in the oligoasthenospermia group (63). Leukocytospermia may impair sperm
function through enhanced helper T-cell regulation. An increase in
B cells may induce the secretion of more anti-sperm antibodies,
thereby leading to low fertility. NK cells may mediate the entire
process of sperm damage. This is consistent with the results
obtained for CD4 T, CD8 T and CD56 (bright or dim) NK cells in the
present study. These observations suggested that the five
identified key genes in asthenozoospermia are strongly associated
with the expression of immune cells and play an important
regulatory role in the testis immune microenvironment. Furthermore,
in the study of the correlations among multiple etiological genes,
HSPA4, SMAD2 and GSK3B were significantly
positively associated with five spermatogenesis-related genes
(SLC26A8, GALNTL5, ARMC2, FSIP2 and
KLHL10) and eight sperm flagellar function-related genes
(TEKT2, DNAH8, TTC21A, CATSPER1,
TTC29, DNAH1, SPAG17 and DNAH17). The
knowledge of gene interactions in asthenozoospermia is currently
limited, and the findings of the present study may provide a new
avenue to study the interactions of etiological genes of
asthenozoospermia.
Previous studies have suggested that the ordered
expression of key genes is crucial during spermatogenesis and that
abnormal gene expression may lead to poor sperm quality or
functional defects (64-66).
The present study has certain limitations. First,
the GSE160749 dataset does not contain abundant sample profiles,
which may limit the accuracy of our results to some extent. Second,
although bioinformatics evidence suggests that the disturbed
expression of the five key genes affects male spermatogenesis and
sperm function, additional in vivo experiments are required
to confirm these results.
In conclusion, in the present study, bioinformatics
techniques were used to identify five key genes and signaling
pathways. The underlying molecular mechanisms of asthenozoospermia
were explored, and correlations were found between the expression
of these key genes with multiple related etiological genes and
immune cells in patients with asthenozoospermia. These findings
could provide clues to identify novel diagnostic genetic biomarkers
and treatment strategies for asthenozoospermia.
Supplementary Material
467 upregulated genes.
869 downregulated genes.
Cq values of clinical semen samples
obtained by reverse transcription-quantitative PCR.
Motifs.
MRNA-miRNA.
Genes correlation analysis.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported (grant. no. 23JSZ03) by
the Family Planning Program of Military Medical Innovation Project
of Chinese People's Liberation Army.
Availability of data and materials
The data generated in the present study may be found
in the Gene Expression Omnibus databased under accession number
GSE160749 or at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE160749
and in the epithelial-mesenchymal transition gene database at the
following URL: http://www.dbemt.bioinfo-minzhao.org/dbemt2.txt. The
other data generated in the present study may be requested from the
corresponding author.
Authors' contributions
YZ and HY designed the present study. JX and YW
contributed to the detection of clinical semen sample parameters,
the extraction of pure sperm cells and the collection of clinical
data. YZ and YP participated in RT-qPCR analysis. YZ, YP, YW and JX
drafted the manuscript and prepared figures and tables. YZ, YP, YW,
JX and HY revised the paper. YZ and HY confirm the authenticity of
all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved (approval no.
chrec-2018-6) by the Ethics Committee of the Chinese Naval Medical
University and was performed in accordance with the principles of
the Declaration of Helsinki. Informed consent was obtained from all
participants before sample collection.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Agarwal A, Baskaran S, Parekh N, Cho CL,
Henkel R, Vij S, Arafa M, Panner Selvam MK and Shah R: Male
infertility. Lancet. 397:319–333. 2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
WHO-World Health Organization. WHO
Laboratory Manual for the Examination and Processing of Human Semen
(6th ed). In: Examination and post-examination procedures. WHO.
2021. https://apps.who.int/iris/rest/bitstreams/1358672/retrieve.
Accessed 27 Jul, 2021.
|
|
3
|
Mehra BL, Skandhan KP, Prasad BS,
Pawankumar G, Singh G and Jaya V: Male infertility rate: A
retrospective study. Urologia. 85:22–24. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Guzick DS, Overstreet JW, Factor-Litvak P,
Brazil CK, Nakajima ST, Coutifaris C, Carson SA, Cisneros P,
Steinkampf MP, Hill JA, et al: Sperm morphology, motility, and
concentration in fertile and infertile men. N Engl J Med.
345:1388–1393. 2001.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Cho CL, Esteves SC and Agarwal A: Novel
insights into the pathophysiology of varicocele and its association
with reactive oxygen species and sperm DNA fragmentation. Asian J
Androl. 18:186–193. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Belet U, Danaci M, Sarikaya S, Odabaş F,
Utaş C, Tokgöz B, Sezer T, Turgut T, Erdoğan N and Akpolat T:
Prevalence of epididymal, seminal vesicle, prostate, and testicular
cysts in autosomal dominant polycystic kidney disease. Urology.
60:138–141. 2002.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Li N, Wang T and Han D: Structural,
cellular and molecular aspects of immune privilege in the testis.
Front Immunol. 3(152)2012.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Touré A, Martinez G, Kherraf ZE, Cazin C,
Beurois J, Arnoult C, Ray PF and Coutton C: The genetic
architecture of morphological abnormalities of the sperm tail. Hum
Genet. 140:21–42. 2021.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Condorelli RA, Russo GI, Calogero AE,
Morgia G and La Vignera S: Chronic prostatitis and its detrimental
impact on sperm parameters: A systematic review and meta-analysis.
J Endocrinol Invest. 40:1209–1218. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhang J, Cai Z, Ma C, Xiong J and Li H:
Impacts of outdoor air pollution on human semen quality: A
meta-analysis and systematic review. Biomed Res Int.
2020(7528901)2020.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Rahman MS, Kwon WS, Lee JS, Yoon SJ, Ryu
BY and Pang MG: Bisphenol-A affects male fertility via
fertility-related proteins in spermatozoa. Sci Rep.
5(9169)2015.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Pant N, Kumar G, Upadhyay AD, Patel DK,
Gupta YK and Chaturvedi PK: Reproductive toxicity of lead, cadmium,
and phthalate exposure in men. Environ Sci Pollut Res Int.
21:11066–11074. 2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Aboulmaouahib S, Madkour A, Kaarouch I,
Sefrioui O, Saadani B, Copin H, Benkhalifa M, Louanjli N and Cadi
R: Impact of alcohol and cigarette smoking consumption in male
fertility potential: Looks at lipid peroxidation, enzymatic
antioxidant activities and sperm DNA damage. Andrologia: Nov 21,
2018 (Epub ahead of print).
|
|
14
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43(e47)2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z,
Feng T, Zhou L, Tang W, Zhan L, et al: clusterProfiler 4.0: A
universal enrichment tool for interpreting omics data. Innovation
(Camb). 2(100141)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Aibar S, Hulselmans G and Aerts S:
RcisTarget: Identify transcription factor binding motifs enriched
on a gene list. Laboratory of Computational Biology; VIB-KU Leuven
Center for Brain & Disease Research. 2016. Available from:
https://bioconductor.org/packages/3.12/bioc/html/RcisTarget.html.
|
|
17
|
Goodrich R, Johnson G and Krawetz SA: The
preparation of human spermatozoal RNA for clinical analysis. Arch
Androl. 53:161–167. 2007.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Krausz C and Riera-Escamilla A: Genetics
of male infertility. Nat Rev Urol. 15:369–384. 2018.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Cunha GR, Cao M, Aksel S, Derpinghaus A
and Baskin LS: Mouse-human species differences in early testicular
development and its implications. Differentiation. 129:79–95.
2023.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Klein B, Pant S, Bhushan S, Kautz J, Rudat
C, Kispert A, Pilatz A, Wijayarathna R, Middendorff R, Loveland KL,
et al: Dexamethasone improves therapeutic outcomes in a preclinical
bacterial epididymitis mouse model. Hum Reprod. 34:1195–1205.
2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Chen Y, Sun T, Liu K, Yuan P and Liu C:
Exploration of the common genetic landscape of COVID-19 and male
infertility. Front Immunol. 14(1123913)2023.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Major AT, Estermann MA and Smith CA:
Anatomy, endocrine regulation, and embryonic development of the
rete testis. Endocrinology. 162(bqab046)2021.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Jedidi I, Ouchari M and Yin Q: Autosomal
single-gene disorders involved in human infertility. Saudi J Biol
Sci. 25:881–887. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ferlin A, Speltra E, Patassini C, Pati MA,
Garolla A, Caretta N and Foresta C: Heat shock protein and heat
shock factor expression in sperm: relation to oligozoospermia and
varicocele. J Urol. 183:1248–1252. 2010.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Held T, Barakat AZ, Mohamed BA, Paprotta
I, Meinhardt A, Engel W and Adham IM: Heat-shock protein HSPA4 is
required for progression of spermatogenesis. Reproduction.
142:133–144. 2011.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Xu J, Beyer AR, Walker WH and McGee EA:
Developmental and stage-specific expression of Smad2 and Smad3 in
rat testis. J Androl. 24:192–200. 2003.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Takahashi N, Davy PM, Gardner LH, Mathews
J, Yamazaki Y and Allsopp RC: Hypoxia inducible factor 1 alpha is
expressed in germ cells throughout the murine life cycle. PLoS One.
11(e0154309)2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Gorga A, Rindone G, Regueira M, Riera MF,
Pellizzari EH, Cigorraga SB, Meroni SB and Galardo MN: HIF
involvement in the regulation of rat Sertoli cell proliferation by
FSH. Biochem Biophys Res Commun. 502:508–514. 2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Ghandehari-Alavijeh R, Zohrabi D, Tavalaee
M and Nasr-Esfahani MH: Association between expression of TNF-α,
P53 and HIF1α with asthenozoospermia. Hum Fertil (Camb).
22:145–151. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Freitas MJ, Silva JV, Brothag C,
Regadas-Correia B, Fardilha M and Vijayaraghavan S:
Isoform-specific GSK3A activity is negatively correlated with human
sperm motility. Mol Hum Reprod. 25:171–183. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Bhattacharjee R, Goswami S, Dudiki T,
Popkie AP, Phiel CJ, Kline D and Vijayaraghavan S: Targeted
disruption of glycogen synthase kinase 3A (GSK3A) in mice affects
sperm motility resulting in male infertility. Biol Reprod.
92(65)2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
La Vignera S and Vita R: Thyroid
dysfunction and semen quality. Int J Immunopathol Pharmacol.
32(2058738418775241)2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Condorelli RA, La Vignera S, Mongioì LM,
Alamo A, Giacone F, Cannarella R and Calogero AE: Thyroid hormones
and spermatozoa: In VitroEffects on Sperm mitochondria, viability
and DNA Integrity. J Clin Med. 8(756)2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhao S, Tang L, Fu J, Yang Z, Su C and Rao
M: Subclinical Hypothyroidism and Sperm DNA Fragmentation: A
Cross-sectional Study of 5401 men seeking infertility care. J Clin
Endocrinol Metab. 107:e4027–e4036. 2022.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Silver-Morse L and Li WX: The role of
receptor tyrosine kinases in primordial germ cell migration.
Methods Mol Biol. 750:291–306. 2011.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Ni FD, Hao SL and Yang WX: Multiple
signaling pathways in Sertoli cells: Recent findings in
spermatogenesis. Cell Death Dis. 10(541)2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Liang G and Wang Q, Zhang G, Li Z and Wang
Q: Differentially expressed miRNAs and potential therapeutic
targets for asthenospermia. Andrologia. 54(e14265)2022.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Parte PP, Rao P, Redij S, Lobo V, D'Souza
SJ, Gajbhiye R and Kulkarni V: Sperm phosphoproteome profiling by
ultra performance liquid chromatography followed by data
independent analysis (LC-MS(E)) reveals altered proteomic
signatures in asthenozoospermia. J Proteomics. 75:5861–5871.
2012.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Demyashkin GA: Inhibin B in seminiferous
tubules of human testes in normal spermatogenesis and in idiopathic
infertility. Syst Biol Reprod Med. 65:20–28. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Li J, Zhang L and Li B: Correlative study
on the JAK-STAT/PSMβ3 signal transduction pathway in
asthenozoospermia. Exp Ther Med. 13:127–130. 2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Chen L, Wen CW, Deng MJ, Ping-Li Zhang ZD,
Zhou ZH and Wang X: Metabolic and transcriptional changes in
seminal plasma of asthenozoospermia patients. Biomed Chromatogr.
34(e4769)2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhu S, Huang J, Xu R, Wang Y, Wan Y,
McNeel R, Parker E, Kolson D, Yam M, Webb B, et al: Isocitrate
dehydrogenase 3b is required for spermiogenesis but dispensable for
retinal viability. J Biol Chem. 298(102387)2022.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Kang W, Harada Y, Yamatoya K, Kawano N,
Kanai S, Miyamoto Y, Nakamura A, Miyado M, Hayashi Y, Kuroki Y, et
al: Extra-mitochondrial citrate synthase initiates calcium
oscillation and suppresses age-dependent sperm dysfunction. Lab
Invest. 100:583–595. 2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Eslamian G, Amirjannati N, Rashidkhani B,
Sadeghi MR, Baghestani AR and Hekmatdoost A: Dietary fatty acid
intakes and asthenozoospermia: A case-control study. Fertil Steril.
103:190–198. 2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Salas-Huetos A, Moraleda R, Giardina S,
Anton E, Blanco J, Salas-Salvadó J and Bulló M: Effect of nut
consumption on semen quality and functionality in healthy men
consuming a Western-style diet: A randomized controlled trial. Am J
Clin Nutr. 108:953–962. 2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Qi X, Shang M, Chen C, Chen Y, Hua J,
Sheng X, Wang X, Xing K, Ni H and Guo Y: Dietary supplementation
with linseed oil improves semen quality, reproductive hormone, gene
and protein expression related to testosterone synthesis in aging
layer breeder roosters. Theriogenology. 131:9–15. 2019.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Gianzo M and Subirán N: Regulation of male
fertility by the renin-angiotensin system. Int J Mol Sci.
21(7943)2020.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Ma P, Zhang Z, Zhou X, Luo J, Lu H and
Wang Y: Characterizing semen abnormality male infertility using
non-targeted blood plasma metabolomics. PLoS One.
14(e0219179)2019.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Malik IA, Durairajanayagam D and Singh HJ:
Leptin and its actions on reproduction in males. Asian J Androl.
21:296–299. 2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Haron MN, D'Souza UJ, Jaafar H, Zakaria R
and Singh HJ: Exogenous leptin administration decreases sperm count
and increases the fraction of abnormal sperm in adult rats. Fertil
Steril. 93:322–324. 2010.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Barbagallo F, Condorelli RA, Mongioì LM,
Cannarella R, Cimino L, Magagnini MC, Crafa A, La Vignera S and
Calogero AE: Molecular mechanisms underlying the relationship
between obesity and male infertility. Metabolites.
11(840)2021.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Leisegang K, Sengupta P, Agarwal A and
Henkel R: Obesity and male infertility: Mechanisms and management.
Andrologia. 53(e13617)2021.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Wang H, Lv Y, Hu K, Feng T, Jin Y, Wang Y,
Huang Y and Chen B: Seminal plasma leptin and spermatozoon
apoptosis in patients with varicocele and leucocytospermia.
Andrologia. 47:655–661. 2015.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Martin LJ: Implications of adiponectin in
linking metabolism to testicular function. Endocrine. 46:16–28.
2014.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Choubey M, Ranjan A, Bora PS, Baltazar F,
Martin LJ and Krishna A: Role of adiponectin as a modulator of
testicular function during aging in mice. Biochim Biophys Acta Mol
Basis Dis. 1865:413–427. 2019.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Zhang HZ, Hao SL and Yang WX: How does
retinoic acid (RA) signaling pathway regulate spermatogenesis?
Histol Histopathol. 37:1053–1064. 2022.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Hernández-Silva G, Caballero-Campo P and
Chirinos M: Sperm mRNAs as potential markers of male fertility.
Reprod Biol. 22(100636)2022.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Corral-Vazquez C, Salas-Huetos A, Blanco
J, Vidal F, Sarrate Z and Anton E: Sperm microRNA pairs: New
perspectives in the search for male fertility biomarkers. Fertil
Steril. 112:831–841. 2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Courey-Ghaouzi AD, Kleberg L and Sundling
C: Alternative B cell differentiation during infection and
inflammation. Front Immunol. 13(908034)2022.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Tohyama Y, Takano T and Yamamura H: B cell
responses to oxidative stress. Curr Pharm Des. 10:835–839.
2004.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Yu W, Li C, Zhang D, Li Z, Xia P, Liu X,
Cai X, Yang P, Ling J, Zhang J, et al: Advances in T cells based on
inflammation in metabolic diseases. Cells. 11(3554)2022.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Seshadri S, Flanagan B, Vince G and
Lewis-Jones DJ: Detection of subpopulations of leucocytes in
different subgroups of semen sample qualities. Andrologia. 44
(Suppl 1):S354–S361. 2012.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Chu DS and Shakes DC: Spermatogenesis. Adv
Exp Med Biol. 757:171–203. 2013.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Coutton C, Vargas AS, Amiri-Yekta A,
Kherraf ZE, Ben Mustapha SF, Le Tanno P, Wambergue-Legrand C,
Karaouzène T, Martinez G, Crouzy S, et al: Mutations in CFAP43 and
CFAP44 cause male infertility and flagellum defects in Trypanosoma
and human. Nat Commun. 9(686)2018.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Ben Khelifa M, Coutton C, Zouari R,
Karaouzène T, Rendu J, Bidart M, Yassine S, Pierre V, Delaroche J,
Hennebicq S, et al: Mutations in DNAH1, which encodes an inner arm
heavy chain dynein, lead to male infertility from multiple
morphological abnormalities of the sperm flagella. Am J Hum Genet.
94:95–104. 2014.PubMed/NCBI View Article : Google Scholar
|