Introduction
Neutrophils constitute 50-70% of white blood cells
in humans (1), and are short-lived
effector cells of the innate immune system that play an important
role in the response to extracellular pathogens (2). These pathogens can be recognized by
pattern recognition receptors on neutrophils, which subsequently
activate anti-pathogen responses, including the production of
reactive oxidative species (ROS) and inflammatory mediators, the
release of lytic enzymes from granules and the formation of
neutrophil extracellular traps (NETs) (3-5).
The toll-like receptor (TLR) family is a class of pattern
recognition receptors (6). With
the exception of TLR3, all TLRs are expressed in neutrophils
(7). The TLR family plays a
critical role in innate bacterial recognition (8). For example, TLR2 recognizes the cell
wall components of Gram-positive bacteria including lipopeptides,
peptidoglycan and lipoteichoic acids (LTAs) (9-11).
By contrast, TLR4 senses lipopolysaccharide, a component of the
outer membrane of Gram-negative bacteria (12-14).
Thus, TLR-mediated signaling pathways are important for regulating
antibacterial immune responses in neutrophils.
LTAs are found in the cell walls of many
gram-positive bacteria, such as Staphylococci,
Streptococci, Bacilli and Listeria (15). Different types of LTAs found in
different bacterial species can be grouped according to their
chemical structure (16). For
example, type I LTAs are found in Bacillus subtilis,
Staphylococcus aureus and Listeria monocytogenes,
whereas type IV LTAs are found in Streptococcus pneumoniae
(16). Exposure to S.
aureus-derived LTA activates the TLR2 and NF-κB signaling
pathways, increases production of ROS and induces the secretion of
inflammatory molecules such as tumor necrosis factor-α (TNF-α),
interleukin (IL)-1β and chemokine (C-X-C motif) ligand 8 (CXCL8, or
IL-8) in human neutrophils (17-20).
Experimental studies on the molecular mechanisms and
cell behaviors of primary human neutrophils are typically limited
due to the short lifespan of human neutrophils (21,22).
Therefore, a surrogate neutrophil-like cell line, such as the human
promyelocytic leukemia cell line, HL-60, has been developed
(23). HL-60 cells differentiate
into a neutrophil-like phenotype in vitro (23). Differentiated HL-60 (dHL-60) cells
serve as a good model for studying the phenotypes of human
neutrophils, including chemotaxis, phagocytosis and the responses
of TLR signaling pathways (24-27).
Several protocols have been established for differentiating HL-60
cells into a neutrophil-like state using dimethyl sulfoxide (DMSO),
N,N-dimethylformamide and all-trans retinoic acid (23,28,29).
Reports have shown that different reagents induce the
differentiation of HL-60 cells via different mechanisms (30,31),
and that the characteristics of HL-60 cells differentiated by
different methods are not completely identical to those of primary
human neutrophils.
Although LTA-induced inflammatory responses have
been studied in human neutrophils, comprehensive transcriptional
regulation in LTA-treated neutrophil-like cells is not currently
well understood. Specifically, there is a lack of relevant studies
comparing LTA treatment at different time points and
concentrations. Since DMSO-differentiated HL-60 cells can respond
to TLR2 and TLR4 ligands (26),
DMSO-induced neutrophil-like cells served as the experimental model
in the present study. The present study investigated the
transcriptional profiles of LTA-treated dHL-60 and LTA-treated
undifferentiated HL-60 (uHL-60) cells, and further evaluated
whether dHL-60 cells could be an alternative cell model for TLR
studies in human neutrophils.
Materials and methods
Cell culture and differentiation
HL-60 cells were obtained from the Bioresource
Collection and Research Center. The HL-60 cells were maintained in
RPMI 1640 medium with L-glutamine (GeneDireX, Inc.) supplemented
with 20% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.)
and antibiotics, including 100 U/ml penicillin, 100 µg/ml
streptomycin and 0.25 µg/ml amphotericin B, at 37˚C in a humidified
atmosphere with 5% CO2. The cell density was maintained
between 1x105 and 1x106 cells/ml. To
differentiate the HL-60 cells into a neutrophil-like phenotype, the
cells were cultured at a density of 1x106 cells/ml in
RPMI 1640 medium containing L-glutamine supplemented with 10% fetal
bovine serum, 10 mM HEPES (Gibco; Thermo Fisher Scientific, Inc.),
the aforementioned antibiotics and 1.25% DMSO for 6 days.
Evaluating NET formation, NETosis and
ROS production in dHL-60 cells
To observe the induction of NETs in dHL-60 cells,
dHL-60 cells were collected 6 days after DMSO stimulation.
Extracellular DNA of NET were then visualized using SYTOX Green
staining. Briefly, dHL-60 cells were seeded into 24-well plates
(2x105 cells per well) in serum-free RPMI 1640 medium
containing 1% bovine serum albumin and 1 mM calcium chloride, then
treated with vehicle (DMSO) or 20 nM phorbol myristate acetate
(PMA; MedChemExpress) at 37˚C. After 4 h, dHL-60 cells were stained
with 5 µM SYTOX Green (Invitrogen; Thermo Fisher Scientific, Inc.)
for 10 min in the dark at room temperature. Images were then
obtained using a Nikon Eclipse TE2000-S inverted fluorescence
microscope equipped with x10 magnification objectives. The
experiment was repeated 3 times.
NET formation in dHL-60 cells was evaluated using a
NETosis Assay Kit (Cayman Chemical Company). Briefly,
2x105 dHL-60 cells were treated with either vehicle or
PMA and then incubated at 37˚C for 4 h to induce NET formation,
according to the manufacturer's instructions. After 4 h, the
culture supernatant was collected and NET-associated neutrophil
elastase activity was detected at 405 nm. The experiment was
repeated 3 times.
ROS production in dHL-60 cells was detected by
staining with a DCFDA/H2DCFDA-Cellular ROS Assay Kit (Abcam). After
a 4-h stimulation with vehicle, 2 or 20 nM PMA, the dHL-60 cells
were stained with 10 µM DCFDA for 30 min at 37˚C and then analyzed
on a BD Accuri C6 Flow Cytometer (BD Biosciences), and the data
were analyzed using FCSalyzer ver. 0.9.22-alpha (https://sourceforge.net/projects/fcsalyzer/). The
experiment was repeated 2 times.
RNA sequencing
For RNA sequencing, 2x106 uHL-60 cells
were treated with 1 µg/ml S. aureus LTA (cat.no. L2515;
Sigma-Aldrich; Merck KGaA) or vehicle (ddH2O) for 4 and 24 h (n=1),
and 2x106 dHL-60 cells were treated with 1 µg/ml or 10
µg/ml S. aureus LTA or vehicle (ddH2O) for 4 and 24 h (n=2)
in 37˚C incubator with 5% CO2. Total RNA from
2x106 uHL-60 and dHL-60 cells was extracted using a
Total RNA Purification Kit (Norgen Biotek Corp.). The purified RNA
was used for the preparation of the sequencing library by TruSeq
Stranded mRNA Library Prep Kit (Illumina, Inc.) following the
manufacturer's recommendations. After the generation of
double-strand cDNA and adenylation on 3' ends of DNA fragments, the
adaptors were ligated and purified with AMPure XP system (Beckman
Coulter, Inc.). The quality of the libraries was assessed on the
Agilent Bioanalyzer 2100 system and a Real-Time PCR system. RNA
sequencing was performed using the Illumina NovaSeq 6000 platform
with 150 bp paired end read lengths and 20 million clean reads per
sample by a commercial vendor (Genomics BioSci & Tech Co.
Ltd.). The bases with low quality and sequences from adapters in
raw data were removed using program fastp (version 0.20.0;
https://github.com/OpenGene/fastp). The
filtered reads were aligned to the reference genomes using HISAT2
(version 2.1.0; https://daehwankimlab.github.io/hisat2/). The software
FeatureCounts (v2.0.1; https://subread.sourceforge.net) in Subread package
was applied for the quantification of the gene abundance. For
identification of differentially expressed genes (DEGs), DESeq2
(version 1.28.0; https://bioconductor.org/packages/release/bioc/html/DESeq2.html)
and EdgeR (version 3.36.0; https://bioconductor.org/packages/release/bioc/html/edgeR.html)
were used to analyze samples with and without biological replicates
respectively. The criteria for DEGs were set at fold change ≥2.0
and P<0.05 in the dHL-60 cell group. Compared with the dHL-60
cells, the gene expression of uHL-60 was less affected by LTA
treatment. Therefore, the criterion of DEGs for the uHL-60 cells
was set at P<0.05, owing to the small number of DEGs. Raw and
processed RNA sequencing data were uploaded to the Gene Expression
Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE239859).
Bioinformatic analysis
The processed RNA sequencing data deposited as
GSE239859 in GEO database was further analyzed through the
following methods. Principal Component Analysis (PCA), heatmaps and
bubble plots were constructed using the online web tool, Srplot
(http://www.bioinformatics.com.cn/srplot), and Venn
diagrams were compiled using the website http://bioinformatics.psb.ugent.be/webtools/Venn/. To
perform Gene Ontology (GO) analysis (biological processes) and
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, the DEGs
were analyzed using the Database for Annotation, Visualization, and
Integrated Discovery (DAVID; v6.8; https://david.ncifcrf.gov/) (32). The identified DEGs of uHL-60 and
dHL-60 were further processed as input data for the DAVID analyses.
A significant difference was indicated by a P<0.001. The
predicted gene-targeted microRNAs (miRNAs) were analyzed using the
miRNet 2.0 website (https://www.mirnet.ca) (33). The interaction network was
constructed using Cytoscape software 3.9.1 (https://cytoscape.org). Gene Set Enrichment Analysis
(GSEA) was performed using the WeB-based GEne SeT Analysis Toolkit
(http://www.webgestalt.org) (34). The gene set for the KEGG pathway
analysis was obtained from the Molecular Signatures Database
[curated gene sets, canonical pathways, KEGG analysis for human
gene symbols; v2022.1; https://www.gsea-msigdb.org/gsea/index.jsp] (35), while a false discovery rate of
<0.05 was set as the significance level. For GSEA analysis, all
genes identified by RNA sequencing were as input data.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA of dHL-60 was extracted using a Total RNA
Purification Kit (Norgen Biotek Corp.) and then reverse-transcribed
to cDNA using an iScript™ cDNA Synthesis Kit (Bio-Rad
Laboratories, Inc.) according to the manufacturer's instructions.
All qPCR experiments were run on a QuantStudio 5 Real-Time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.) using
the SYBR Green I-based TB Green Premix Ex Taq II (Takara Bio, Inc.)
and the program, 95˚C for 30 sec, 40 cycles of 95˚C for 3 sec, and
60˚C for 30 sec. The relative mRNA expression was normalized to the
expression of the internal control, b-actin, using by
2-∆∆Cq method (36).
The nucleotide sequences for the primers used are listed in
Table I.
 | Table IPrimer sequences used in reverse
transcription-quantitative polymerase chain reaction. |
Table I
Primer sequences used in reverse
transcription-quantitative polymerase chain reaction.
| Primer name | Sequence
(5'-3') |
|---|
| CCL2 forward |
TCTGTGCCTGCTGCTCATAG |
| CCL2 reverse |
TGGAATCCTGAACCCACTTC |
| CCL5 forward |
CGTGCCCACATCAAGGAGTAT |
| CCL5 reverse |
CGGTTCTTTCGGGTGACAAA |
| CXCL8 forward |
TGTGTGTAAACATGACTTCCAAGCT |
| CXCL8 reverse |
GCAAAACTGCACCTTCACACAG |
| IL-1β forward |
TGAAAGATGATAAGCCCACTCTACA |
| IL-1β reverse |
AGACTCAAATTCCAGCTTGTTATTG |
| TNF forward |
CCCAGGCAGTCAGATCATCTTC |
| TNF reverse |
GCTTGAGGGTTTGCTACAACATG |
| β-actin
forward |
TTAGTTGCGTTACACCCTTTCTTG |
| β-actin
reverse |
TCACCTTCACCGTTCCAGTTT |
Statistical analysis
Bar plots and statistical analyses were performed
using GraphPad Prism 8.4.3 software (Dotmatics). A two-tailed
unpaired t-test was used to analyze the differences between two
groups. One-way ANOVA and Tukey's multiple-comparison test were
used to compare three groups. P<0.05 was considered to indicate
a statistically significant difference.
Results
Evaluation of the neutrophil-like
phenotypes of dHL-60 and the transcriptomes of differentiated and
undifferentiated HL-60 cells
A flowchart of the present study is presented in
Fig. 1A. To obtain neutrophil-like
phenotypes, uHL-60 cells were differentiated by DMSO treatment. The
neutrophil-like phenotype of dHL-60 cells was then further
evaluated. PMA is a known inducer of NET formation and ROS
production (37,38). Fluorescence microscopy revealed the
formation of NETs in PMA-stimulated dHL-60 cells (Fig. 1B). The NETosis assay also
demonstrated that PMA stimulation significantly increased
NET-associated elastase activity (Fig.
1C). In addition, ROS production was increased following PMA
stimulation (Fig. 1D). These
results indicated that DMSO-differentiated HL-60 cells had
neutrophil-like phenotypes.
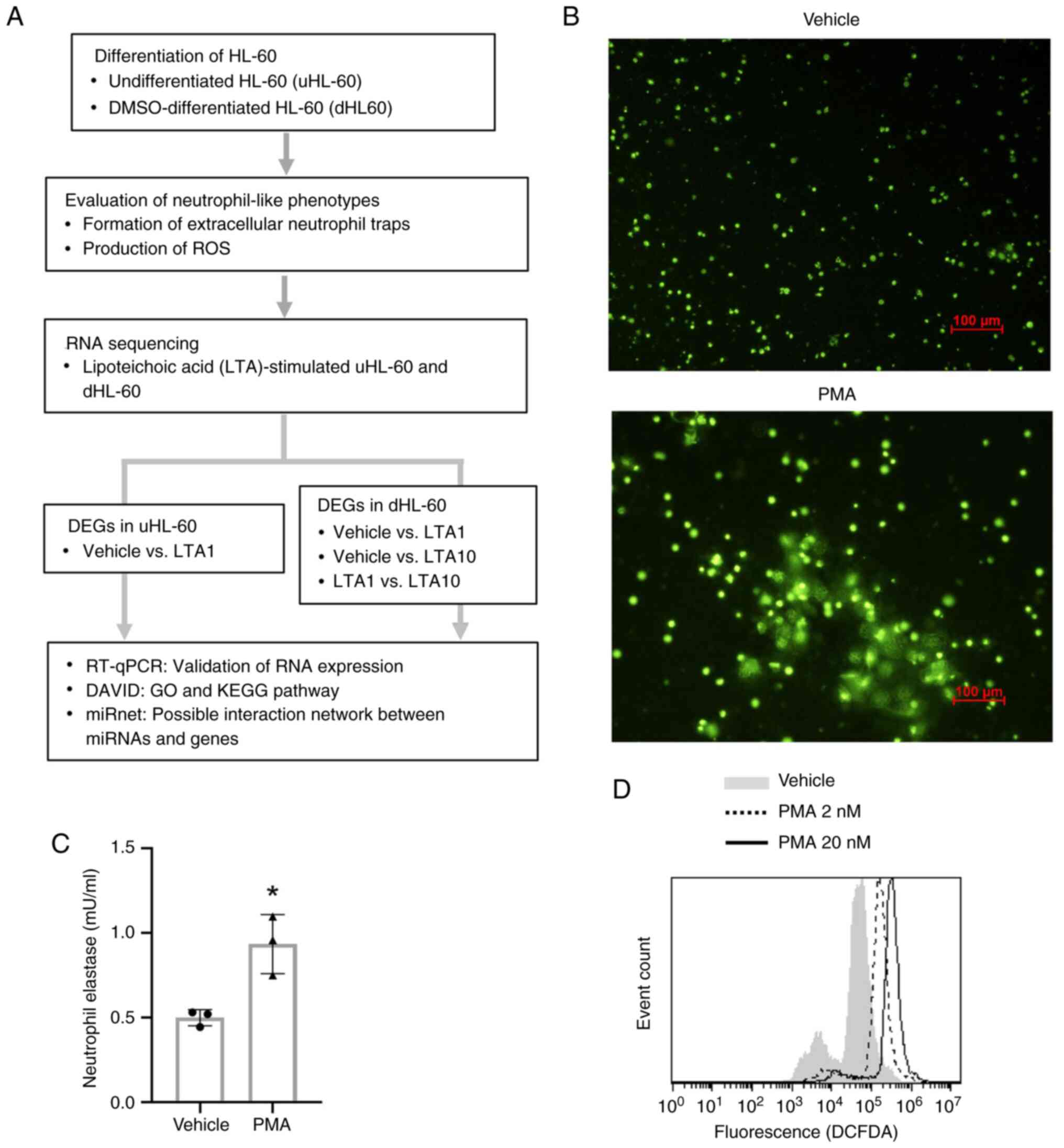 | Figure 1Overall study design and the
neutrophil-like phenotypes of dHL-60 cells. (A) Flowchart of the
present study. Differentiation of HL-60 cells was induced by DMSO
treatment, and then the neutrophil-like phenotypes of dHL-60 cells
were confirmed. To investigate the gene expression profiles of
LTA-treated uHL-60 and dHL-60 cells, uHL-60 cells were treated with
vehicle (ddH2O) or 1 µg/ml S. aureus LTA for 4
and 24 h (n=1), and dHL-60 cells were treated with vehicle
(ddH2O), 1 µg or 10 µg of S. aureus LTA (n=2).
The cell samples were then analyzed via RNA sequencing. DEGs in
each group were merged using Venn diagrams, and then the expression
levels of genes were further validated by RT-qPCR. The GO and KEGG
pathway analysis was performed using DAVID, and the predicted
miRNA-gene interaction was analyzed by miRnet. (B) Formation of
NETs were detected by fluorescent microscopy. Following treatment
with 20 nM PMA or vehicle (DMSO) for 4 h, the samples were stained
with SYTOX Green (magnification, x10; scale bar, 100 µm). Because
SYTOX Green stain does not penetrate living dHL-60, untreated live
dHL-60 cells showed little or no fluorescence. By contrast,
extracellular DNA of NET induced by PMA were stained with SYTOX
Green fluorescence. (C) The activity of neutrophil elastase was
measured following treatment with 20 nM PMA or vehicle (DMSO) for 4
h. Data presented as the mean ± standard deviation.
*P<0.05, determined using an unpaired t-test. (D)
Flow cytometry of H2DCFDA staining for ROS in dHL-60 cells
following treatment with 2 or 20 nM PMA or vehicle (DMSO) for 4 h.
DAVID, Database for Annotation, Visualization; DEGs, differentially
expressed genes; DMSO, dimethyl sulfoxide; GO, Gene Ontology;
dHL-60, differentiated HL-60; LTA, lipoteichoic acid; KEGG, Kyoto
Encyclopedia of Genes and Genomes; LTA1, cells treated with 1 µg/ml
LTA; LTA10, cells treated with 10 µg/ml LTA; miRNAs, microRNAs;
PMA, phorbol myristate acetate; ROS, reactive oxygen species;
RT-qPCR, reverse transcription-quantitative PCR; uHL-60,
undifferentiated HL-60. |
The doses of 0.1-10 µg/ml S. aureus LTA are
used in several published studies (18,20),
and treatment with LTA >1 µg/ml induces relatively significant
immune responses in immune cells. However, the present pretest
experiments showed that treatments with 1 or 10 µg/ml S.
aureus did not induce the expression of inflammatory molecules
in uHL-60 as in human primary neutrophils (data not shown).
Therefore, the current study focused on LTA-treated dHL-60, while
udHL-60 was tested only at a single dose of LTA using a single
sample. For RNA sequencing analysis, uHL-60 cells were treated with
1 µg/ml S. aureus LTA or vehicle (ddH2O) for 4
and 24 h (n=1), and dHL-60 cells were treated with 1 µg/ml or 10
µg/ml S. aureus LTA or vehicle (ddH2O) for 4 and
24 h (n=2). Subsequently, the gene expression profiles of the
harvested cells were analyzed by RNA sequencing, and PCA was
performed to examine the distribution of DEGs in each group.
Fig. 2A and B show the differences in gene expression
profiles between the uHL-60 and dHL-60 cells. The PCA plot
demonstrated a clear separation between the uHL-60 and dHL-60
cells, and LTA-treated dHL-60 and vehicle-treated dHL-60 were also
distributed in two clusters. By contrast, there was no clear
separation of LTA-treated uHL-60 and vehicle-treated uHL-60 cells
at either time point. A heatmap plot revealed similar clusters
among the uHL-60 and dHL-60 cell samples (Fig. S1).
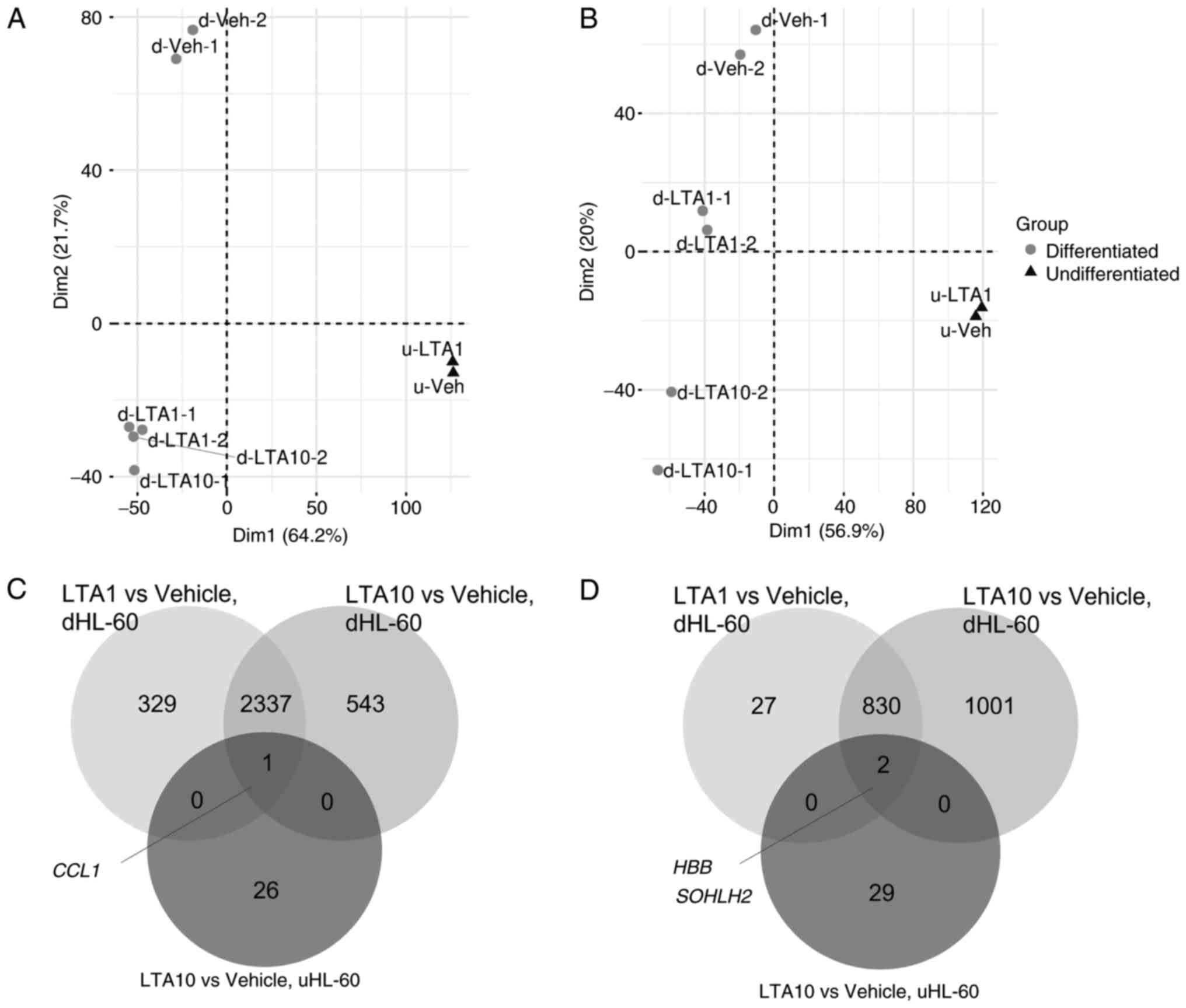 | Figure 2Principal component analysis of
uHL-60 and dHL-60 cells treated with LTA and vehicle (DMSO) for (A)
4 and (B) 24 h. Differentially expressed genes from ‘LTA1 vs.
Vehicle’ in uHL-60 cells, ‘LTA1 vs. Vehicle’ in dHL-60 cells and
‘LTA10 vs. Vehicle’ in dHL-60 cells were merged in a the Venn
diagram to identify the shared genes among these three groups
following (C) 4 and (D) 24 h of LTA treatment. CCL1, C-C motif
chemokine ligand 1; DMSO, dimethyl sulfoxide; dHL-60,
differentiated HL-60; HBB, hemoglobin subunit b; LTA, lipoteichoic
acid; LTA1, cells treated with 1 µg/ml LTA; LTA10, cells treated
with 10 µg/ml LTA; SOHLH2, spermatogenesis and oogenesis specific
basic helix-loop-helix 2; uHL-60, undifferentiated HL-60. |
The Venn diagram demonstrated that various shared
DEGs were identified in the ‘LTA1 vs. Vehicle’ and ‘LTA10 vs.
Vehicle’ groups for the dHL-60 cells at both time points (Fig. 2C and D). However, only 1 shared DEG [C-C motif
chemokine ligand 1 (CCL1)] and 2 shared DEGs (hemoglobin
subunit β and spermatogenesis and oogenesis specific basic
helix-loop-helix 2 genes) were identified in the three groups
comparison at 4 and 24 h, respectively. The results showed that the
transcript levels of uHL-60 cells were completely different from
those of dHL-60 cells after LTA treatment. The results suggested
that uHL60 and dHL60 indeed had different characteristics, and
their responses to LTA treatment were almost completely different.
Therefore, the enriched pathways in uHL-60 and dHL-60 cells were
investigated separately.
LTA-affected pathways in uHL-60
cells
The functions of the identified DEGs in uHL-60 cells
after 4 and 24 h of LTA treatment were annotated by GO analysis
using DAVID. No significant pathways were enriched after 4 h of LTA
treatment. The enriched pathways in uHL-60 cells after 24 h of LTA
treatment are shown in Table II.
Most of the identified pathways were associated with survival of
motor neuron 1, telomeric (SMN1) and survival of motor
neuron 2, centromeric (SMN2) genes. The results therefore
suggested that treatment with 1 µg/ml LTA affected only a small
proportion of genes in uHL-60 cells.
 | Table IIGO enriched pathways in
undifferentiated HL-60 cells following 24 h of lipoteichoic acid
treatment. |
Table II
GO enriched pathways in
undifferentiated HL-60 cells following 24 h of lipoteichoic acid
treatment.
| GO term | Count | P-value | Genes |
|---|
| GO:0006353,
DNA-templated transcription, termination (BP) | 2 | 0.0075 | SMN2, SMN1 |
| GO:0000245,
spliceosomal complex assembly (BP) | 2 | 0.0161 | SMN2, SMN1 |
| GO:0000387,
spliceosomal snRNP assembly (BP) | 2 | 0.0173 | SMN2, SMN1 |
| GO:0032797, SMN
complex (CC) | 2 | 0.0092 | SMN2, SMN1 |
| GO:0097504, Gemini
of coiled bodies (CC) | 2 | 0.0111 | SMN2, SMN1 |
| GO:0031083, BLOC-1
complex (CC) | 2 | 0.0120 | BLOC1S5,
BLOC1S5-TXNDC5 |
| GO:0034719, SMN-Sm
protein complex (CC) | 2 | 0.0166 | SMN2, SMN1 |
LTA-affected pathways in dHL-60
cells
A large number of common DEGs were identified in the
1 µg/ml LTA (LTA1) and 10 µg/ml LTA (LTA10) treated dHL-60 cells at
both time points (Fig. 2C and
D). Compared with the vehicle
group, increased expression levels of known LTA-induced
inflammatory molecules were observed in the LTA1 and LTA10 groups,
according to the RNA sequencing data (Fig. 3A and B). To further confirm the observed
expression profiles, the mRNA expression levels of CCL2, CCL5,
CXCL8, IL-1β and TNF were quantified using RT-qPCR. In general, the
expression patterns of these molecules in the RT-qPCR analysis were
similar to those observed in the RNA sequencing analysis (Fig. 3C-G).
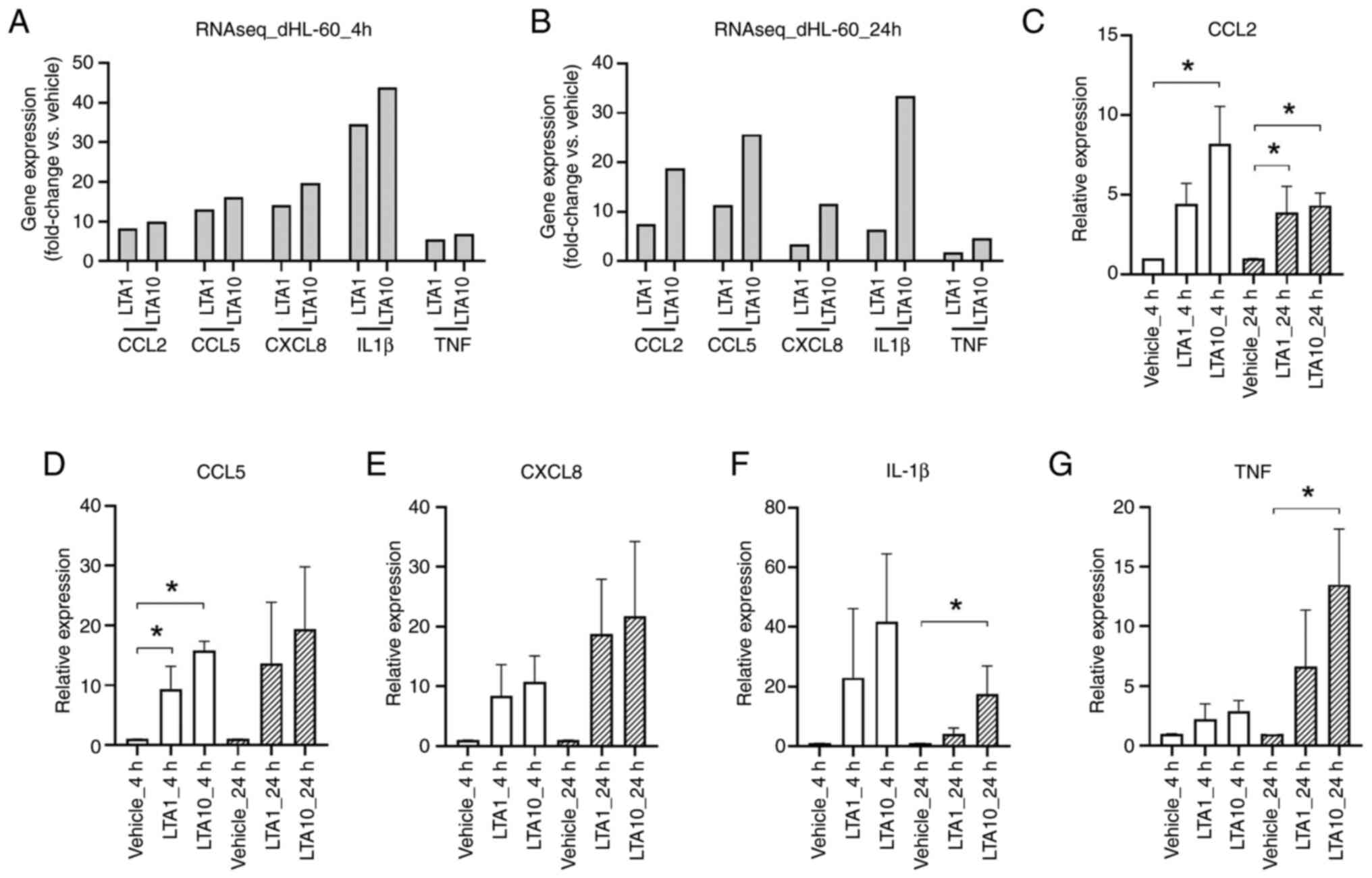 | Figure 3Expression of LTA-induced
inflammatory molecules in dHL-60 cells. After RNAseq analysis, the
fold change (LTA-treated group/vehicle group) of each gene was
determined following (A) 4 and (B) 24 h of LTA treatment. The
relative expression levels of (C) CCL2, (D) CCL5, (E) CXCL8, (F)
IL-1β and (G) TNF were determined using reverse
transcription-quantitative polymerase chain reaction. Data are
presented as the mean ± standard deviation. *P<0.05
vs. Vehicle, determined using one-way ANOVA and Tukey's
multiple-comparison test. CCL, C-C motif chemokine ligand; CXCL8,
C-X-C motif ligand 8; dHL-60, differentiated HL-60; IL-1β,
interleukin-1β; LTA1, cells treated with 1 µg/ml LTA; LTA10, cells
treated with 10 µg/ml LTA; TNF, tumor necrosis factor; RNAseq, RNA
sequencing. |
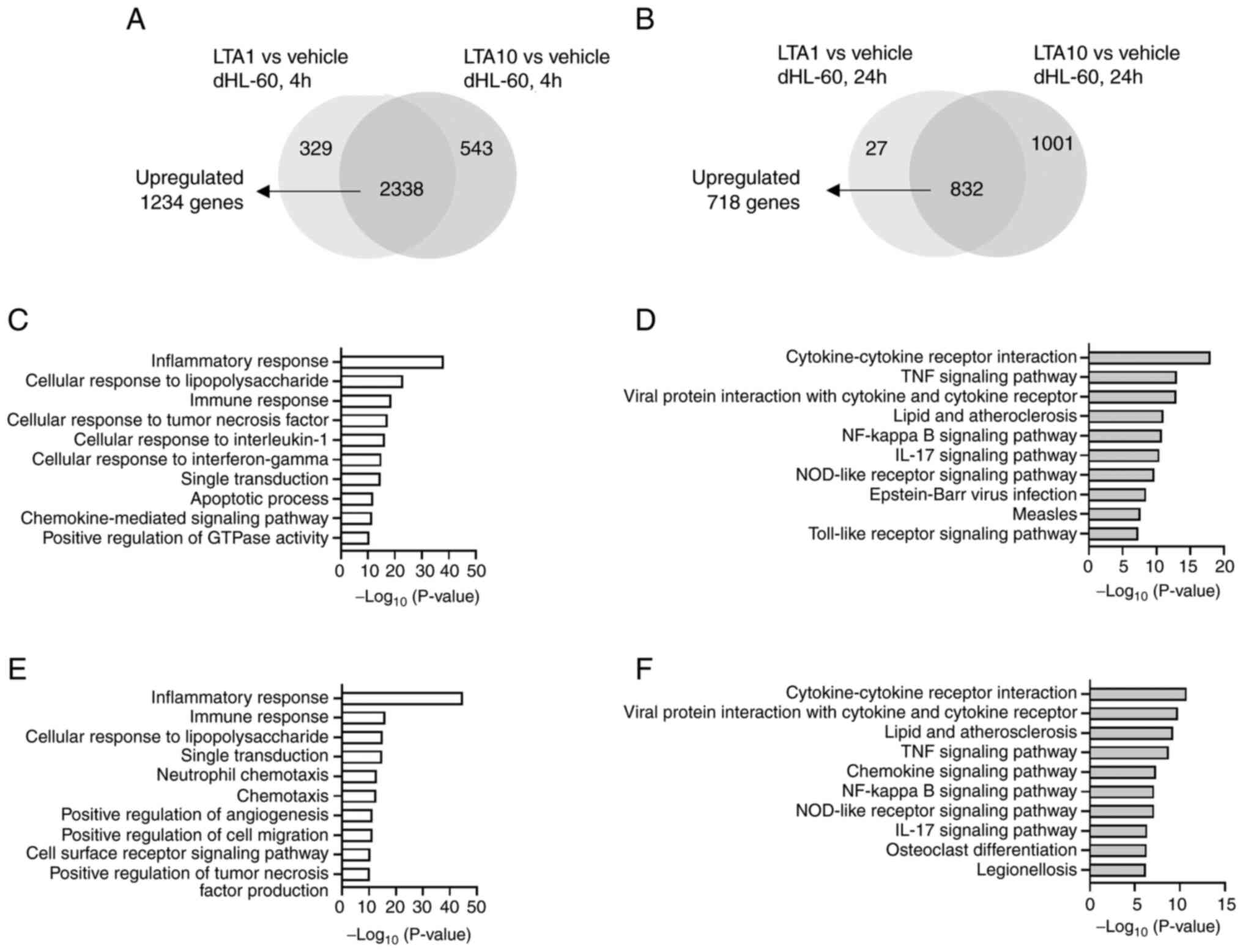
To further investigate the enriched pathways in
LTA-treated dHL-60 cells, GO (biological processes) and KEGG
pathway analyses were performed using DAVID, according to the
common upregulated 1,234 and 718 DEGs in the 4 and 24 h LTA-treated
groups, respectively (Fig. 4A and
B). Fig. 4C-F show the identified top 10
significantly enriched biological processes and KEGG pathways. The
top 20 biological processes, KEGG pathways and detailed gene lists
are presented in Table SI,
Table SII, Table SIII and Table SIV. Numerous identical biological
processes were observed including ‘inflammatory response’,
‘cellular response to lipopolysaccharide’, ‘immune response’,
‘cellular response to tumor necrosis factor’, ‘signal
transduction’, ‘apoptotic process’, ‘chemokine-mediated signaling
pathway’, ‘positive regulation of inflammatory response’,
‘neutrophil chemotaxis’, ‘positive regulation of cell migration’,
‘chemotaxis’ and ‘positive regulation of ERK1 and ERK2 cascade’ in
the 4 or 24 h LTA-treated groups. Furthermore, identical KEGG
pathways including ‘Cytokine-cytokine receptor interaction’, ‘TNF
signaling pathway’, ‘Viral protein interaction with cytokine and
cytokine receptor’, ‘Lipid and atherosclerosis’, ‘NF-kappa B
signaling pathway’, ‘IL-17 signaling pathway’, ‘NOD-like receptor
signaling pathway’, ‘Toll-like receptor signaling pathway’,
‘Chemokine signaling pathway’ and ‘Rheumatoid arthritis’ were
observed in the top 20 enriched KEGG pathways. Therefore, the
results demonstrated that the enriched pathways following 4 and 24
h LTA treatment of dHL-60 cells were similar.
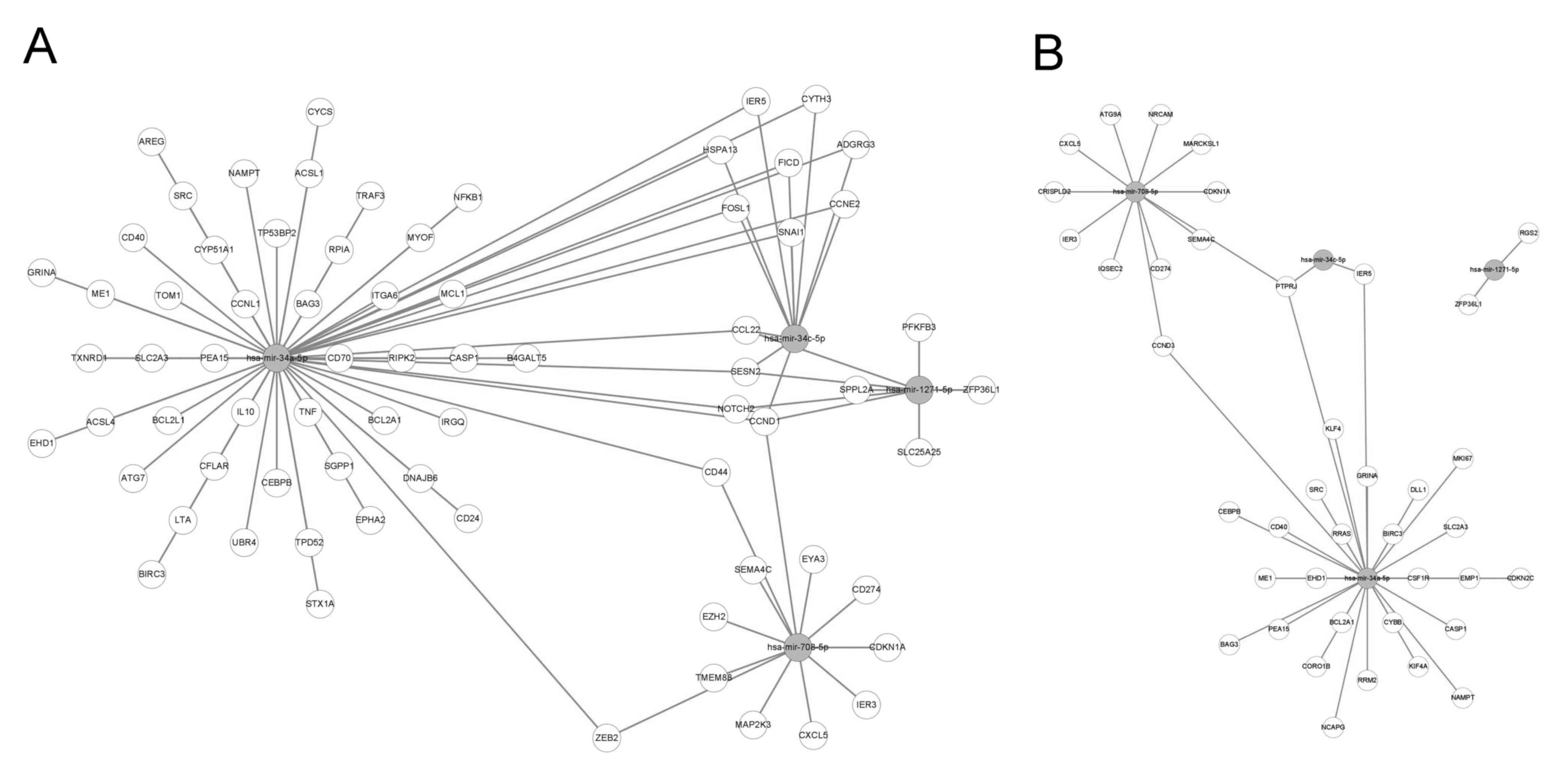
Our previous study demonstrated that the expression
levels of four miRNAs, including hsa-miR-34a-5p, hsa-miR-34c-5p,
hsa-miR-708-5p and hsa-miR-1271-5p, are affected by LTA treatment
in human primary neutrophils (39). Although the present study aimed to
determine whether the same miRNA is regulated in LTA-treated dHL-60
cells, microRNA sequencing was not performed in this study.
Therefore, the miRNAs that were predicted to interact with the
commonly upregulated DEGs in the 4 or 24 h LTA-treated groups were
analyzed using the miRnet website. The above four miRNAs were also
included in the list of miRNAs predicted to interact with the
upregulated DEGs. These results implied that similar miRNA-gene
interactions can be found in both human primary neutrophils and
dHL-60 cells treated with LTA. A possible interaction network
between the shared upregulated DEGs in LTA-treated dHL-60 and the
aforementioned four miRNAs is shown in Fig. 5.
Effect of low and high dose LTA
treatment in dHL-60 cells
The impact of high and low concentrations of LTA on
dHL-60 cells was further investigated by comparing the enriched
pathways between the two groups. A volcano plot presenting the DEGs
between the LTA1 and LTA10-treated dHL-60 cells is shown in
Fig. S2. The results revealed
that less DEGs were found in the samples treated for 4 h compared
with the samples treated for 24 h. GSEA was performed to further
investigate the enriched pathways in both groups. The ‘Complement
and coagulation cascades’ was the only pathway that reached
statistical significance (false discovery rate <0.05) in the
LTA1 cells treated for 4 h (Fig.
6A). By contrast, multiple significantly enriched KEGG pathways
were observed in the LTA10 cells treated for 24 h (Fig. 6B-L). Several immune
response-related pathways were enriched, including
‘Cytokine-cytokine receptors interaction,’ ‘Chemokine signaling
pathways,’ ‘Toll-like receptor signaling pathway,’ ‘NOD like
receptor signaling pathway’ and ‘Cytosolic DNA sensing pathway.’
These enriched pathways were associated with innate immune
responses.
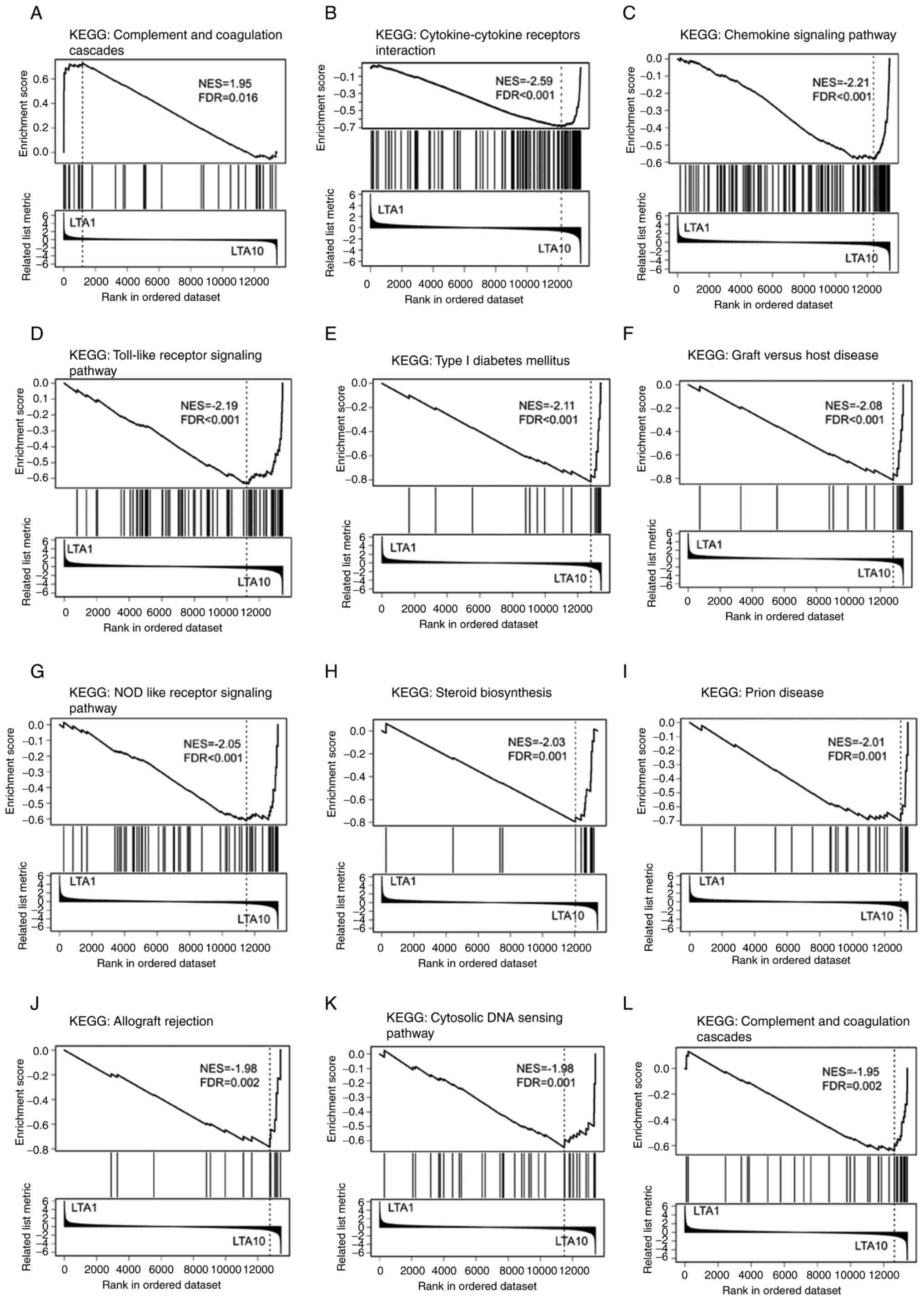 | Figure 6Analytical results of the Gene Set
Enrichment Analysis of DEGs between the LTA1 and LTA10 samples. The
KEGG pathway gene set was used to analyze the DEGs. Enrichment
plots showing the significantly enriched KEGG terms (FDR<0.05),
including (A) ‘Complement and coagulation cascades’ pathway
enriched in LTA1, and (B) ‘Cytokine-cytokine receptors
interaction’, (C) ‘Chemokine signaling pathway’, (D) ‘Toll-like
receptor signaling pathway’, (E) ‘Type I diabetes mellitus’, (F)
‘Graft vs. host disease’, (G) ‘NOD like receptor signaling
pathway’, (H) ‘Steroid biosynthesis’, (I) ‘Prion disease’, (J)
‘Allograft rejection’, (K) ‘Cytosolic DNA sensing pathway’ and (L)
‘Complement and coagulation cascades’ pathways enriched in LTA10.
DEGs, differentially expressed genes; FDR, false discovery rate;
LTA, lipoteichoic acid; KEGG, Kyoto Encyclopedia of Genes and
Genomes; LTA1, cells treated with 1 µg/ml LTA; LTA10, cells treated
with 10 µg/ml LTA; NES, normalized enrichment score. |
Discussion
In the present study, uHL-60 cells were only treated
with a single concentration of LTA (1 µg/ml) for 4 and 24 h. RNA
sequencing revealed that only a few genes were affected by this LTA
treatment. No significantly enriched pathways were found following
GO and KEGG analyses in the 4-h LTA treatment group. By contrast,
some biological processes reached statistical significance in the
24-h LTA treatment group. In total, 4 genes, including the
protein-coding genes SMN1, SMN2 and biogenesis of
lysosomal organelles complex 1 subunit 5 (BLOC1S5), and the
non-protein-coding gene BLOC1S5-TXNDC5 readthrough (NMD candidate)
(BLOC1S5-TXNDC5), are involved in these biological
processes. SMN1 and SMN2 play important roles in small nuclear
ribonucleoproteins (40), and the
functions of BLOC1S5 and BLOC1S5-TXNDC5 are associated with the
BLOC-1 complex (41). However, the
mechanism by which SMN1, SMN2 and BLOC-1 complexes are regulated by
LTA treatment remains unknown. Overall, uHL-60 cells treated with
LTA showed a transcriptional profile completely different from that
of LTA-treated dHL-60.
In previous studies, the S. aureus
LTA-induced immune responses in human neutrophils have been
investigated over <6 h or following 16 and 24 h of treatment at
concentrations of LTA ranging 1-10 µg/ml (17,18,39).
A DMSO-differentiated HL-60 cell model has also been used
previously to investigate the functions of neutrophils, including
cell polarization, ROS production, chemotaxis, NETosis and
phagocytosis, which the present study aimed to further confirm
(42). The present study
investigated the effects of S. aureus LTA on DMSO-treated
dHL-60 cells. The experimental conditions used in the present
study, including the LTA concentrations and time points, were set
according to previous reports.
LTA stimulation activates TLR2 signaling and the
pro-inflammatory cytokine response, including TNF-α, IL-1β and
CXCL8, in human neutrophils (17-20).
In addition, CXCL8 recruits more neutrophils and immune cells
(43), and prolongs the lifespan
of neutrophils (17). In addition,
S. aureus infection can be recognized by TLR, nucleotide
binding oligomerization domain (NOD) and C-type lectin (CLR)
receptors (44). Th17 signaling is
also activated following LTA stimulation (45). In the present study, the RNA
sequencing results demonstrated that hundreds of identical DEGs
were found in samples treated with 1 and 10 µg/ml LTA for 4 and 24
h. According to the GO and KEGG pathway analyses of DEGs with
upregulated expression, biological processes and KEGG pathways,
such as ‘immune response’, ‘inflammatory response’,
‘Cytokine-cytokine receptor interaction’, ‘TNF signaling pathway’,
‘Toll-like receptor pathway’, ‘IL-17 signaling pathway’, ‘NOD-like
receptor signaling pathway’ and ‘NF-kappa B signaling pathway’ were
significantly enriched in LTA-treated dHL-60 cells at both time
points. Interferon γ (IFN-γ) is a cytokine that promotes Th1 cell
development (46), and the
presence of IFN-γ could enhance CCL2 production in LTA-treated
neutrophils (47). In the present
study, significantly high CCL2 expression was observed by RNA
sequencing and RT-qPCR analysis, which may suggest that the
enriched IFN-γ signaling pathways also enhanced CCL2 and chemokine
signaling pathways in LTA-treated dHL-60 cells. In summary, the
known LTA-induced signaling pathways in human neutrophils were also
observed in 1 and 10 µg/ml LTA-treated dHL-60 cells following 4 and
24 h of treatment. This suggested that dHL-60 cells could serve as
an in vitro model for investigating TLR2 signaling pathways
in human neutrophils.
The effects of different LTA concentrations on
dHL-60 cells were also determined in the present study. Due to the
relatively small number of DEGs observed between the 1 and 10 µg/ml
treated groups, GSEA analysis was performed instead of
over-representation analysis. Only the ‘Complement and coagulation
cascades’ was a significantly enriched KEGG pathway in the 1 µg/ml
4-h LTA-treated dHL-60 cells, while no KEGG pathways were
significantly enriched in the 1 µg/ml 4-h LTA-treated uHL-60 cells.
By contrast, several significantly enriched pathways were observed
in the 10 µg/ml 24-h LTA-treated dHL-60 cells. These enriched
pathways, including ‘Cytokine-cytokine receptor interaction’,
‘Chemokine signaling pathway’, ‘Toll-like receptor signaling
pathway’, ‘NOD-like receptor signaling pathway’ and ‘Cytosolic DNA
sensing pathway’ are associated with or are downstream of TLR
signaling pathways (6).
Furthermore, other identified pathways, such as ‘Type I diabetes
mellitus’, ‘Graft vs. host disease’, ‘Prion disease’, ‘Steroid
biosynthesis’ and ‘Allograft rejection’ are associated with innate
immune responses (48-52).
The ‘Complement and coagulation cascade’ was also enriched in both
the 1 µg/ml 4-h LTA-treated dHL-60 cells and the 10 µg/ml 24-h
LTA-treated dHL-60 cells. The complement and coagulation pathways
are important for the host defense functions of neutrophils
(53). However, this is related to
the mechanism by which S. aureus evades phagocytosis
(54). Altogether, it was
discovered that all enriched pathways were associated with TLR
signaling pathways in neutrophils, although there were some
differences in gene expression between the high and low
concentrations of LTA treatment for 4 and 24 h.
Bacterial infection alters the expression of
inflammation-related miRNAs in vivo (55). For example, miR-142 is essential
for the clearance of S. aureus infections at skin wound
sites via neutrophil regulation (56). In addition, targeting miR-223 and
miR-139-5p may serve as a therapeutic strategy to enhance the
clearance of S. aureus infections in skin wounds (57,58).
Our previous study identified several differentially expressed
miRNAs using small RNA sequencing in LTA-stimulated primary human
neutrophils (39). Although small
RNA sequencing was not performed in the present study, predictive
tools were used to determine whether dHL-60 cells have a regulatory
network similar to that of human neutrophils. In total, hundreds of
miRNAs were identified in the prediction results, which also
included the same miRNAs as previously identified in primary human
neutrophils (39). However, these
interactions should be experimentally verified in future
studies.
The present study had some limitations. For example,
there is a gap between transcription, translation and
post-translational processes. Therefore, the change in
transcriptional levels may not necessarily be reflected in the
protein levels. As such, the protein levels of LTA-treated dHL-60
cells need to be validated. Furthermore, the effect of LTA on
primary human neutrophils under similar experimental conditions was
not investigated. These issues require further investigation.
In summary, RNA sequencing of LTA-treated dHL-60
cells confirmed that the enriched pathways following treatment were
associated with TLR signaling pathways at the two tested time
points. A comparison of different LTA concentrations also revealed
that TLR and TLR-related signaling pathways were enriched following
treatment. This further suggested that DMSO-differentiated HL-60
cells may be a suitable alternative model for studying human
neutrophils.
Supplementary Material
Transcriptome profiles of uHL-60 and
dHL-60 following vehicle control or LTA treatment. Heat map
representing transcripts per kilobase million expression values of
genes in dHL-60 and uHL-60 cells after (A) 4 and (B) 24 h of LTA
treatment. dHL-60, differentiated HL-60; LTA, lipoteichoic acid;
LTA1, cells treated with 1 μg/ml LTA; LTA10, cells treated
with 10 μg/ml LTA; uHL-60, undifferentiated HL-60; Veh,
vehicle.
Volcano plots of upregulated (red) and
downregulated (blue) genes showing the differential gene
distribution following (A) 4 and (B) 24 h of LTA treatment. The
numbers in brackets represent the number of genes. LTA,
lipoteichoic acid; LTA1, cells treated with 1 μg/ml LTA;
LTA10, cells treated with 10 μg/ml LTA.
Top 20 enriched biological processes
following GO analysis of the shared upregulated 1,234 genes in
differentiated HL-60 cells after 4-h treatment with 1 and 10
μg lipoteichoic acid.
Top 20 enriched biological processes
following GO analysis of the shared upregulated 718 genes in
differentiated HL-60 cells after 24-h treatment with 1 and 10
μg lipoteichoic acid.
Top 20 enriched KEGG pathways of the
shared upregulated 1,234 genes in differentiated HL-60 cells after
4-h treatment with 1 and 10 μg lipoteichoic acid.
Top 20 enriched KEGG pathways of the
shared upregulated 718 genes in differentiated HL-60 cells after
24-h treatment with 1 and 10 μg lipoteichoic acid.
Acknowledgements
Not applicable.
Funding
Funding: This research was supported by grants from the Ministry
of Science and Technology (MOST) of Taiwan (grant no. MOST
107-2320-B-037-011-MY3) and Kaohsiung Medical University Hospital
(grant nos. KMUH107-7M36, KMUH109-9R82, KMUH110-0M75 and
KMUH111-1M61).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
KL and MY conceived and designed the study. KL, IY,
YH and MY acquired, analyzed and interpreted the data. KL and MY
drafted the manuscript. KL and MY confirm the authenticity of all
the raw data. All the authors have read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that that they have no competing
interests.
References
|
1
|
Sagiv JY, Michaeli J, Assi S, Mishalian I,
Kisos H, Levy L, Damti P, Lumbroso D, Polyansky L, Sionov RV, et
al: Phenotypic diversity and plasticity in circulating neutrophil
subpopulations in cancer. Cell Rep. 10:562–573. 2015.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Mantovani A, Cassatella MA, Costantini C
and Jaillon S: Neutrophils in the activation and regulation of
innate and adaptive immunity. Nat Rev Immunol. 11:519–531.
2011.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Brubaker SW, Bonham KS, Zanoni I and Kagan
JC: Innate immune pattern recognition: A cell biological
perspective. Annu Rev Immunol. 33:257–290. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Mogensen TH: Pathogen recognition and
inflammatory signaling in innate immune defenses. Clin Microbiol
Rev. 22:240–273, table of Contents. 2009.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Borregaard N: Neutrophils, from marrow to
microbes. Immunity. 33:657–670. 2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kawasaki T and Kawai T: Toll-like receptor
signaling pathways. Front Immunol. 5(461)2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Hayashi F, Means TK and Luster AD:
Toll-like receptors stimulate human neutrophil function. Blood.
102:2660–2669. 2003.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Cook DN, Pisetsky DS and Schwartz DA:
Toll-like receptors in the pathogenesis of human disease. Nat
Immunol. 5:975–979. 2004.PubMed/NCBI View
Article : Google Scholar
|
|
9
|
Seo HS, Michalek SM and Nahm MH:
Lipoteichoic acid is important in innate immune responses to
gram-positive bacteria. Infect Immun. 76:206–213. 2008.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Schwandner R, Dziarski R, Wesche H, Rothe
M and Kirschning CJ: Peptidoglycan- and lipoteichoic acid-induced
cell activation is mediated by toll-like receptor 2. J Biol Chem.
274:17406–17409. 1999.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Oliveira-Nascimento L, Massari P and
Wetzler LM: The role of TLR2 in infection and immunity. Front
Immunol. 3(79)2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Lu YC, Yeh WC and Ohashi PS: LPS/TLR4
signal transduction pathway. Cytokine. 42:145–151. 2008.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Park BS and Lee JO: Recognition of
lipopolysaccharide pattern by TLR4 complexes. Exp Mol Med.
45(e66)2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Miller SI, Ernst RK and Bader MW: LPS,
TLR4 and infectious disease diversity. Nat Rev Microbiol. 3:36–46.
2005.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Weidenmaier C and Peschel A: Teichoic
acids and related cell-wall glycopolymers in gram-positive
physiology and host interactions. Nat Rev Microbiol. 6:276–287.
2008.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Percy MG and Gründling A: Lipoteichoic
acid synthesis and function in gram-positive bacteria. Annu Rev
Microbiol. 68:81–100. 2014.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lotz S, Aga E, Wilde I, van Zandbergen G,
Hartung T, Solbach W and Laskay T: Highly purified lipoteichoic
acid activates neutrophil granulocytes and delays their spontaneous
apoptosis via CD14 and TLR2. J Leukoc Biol. 75:467–477.
2004.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Hattar K, Grandel U, Moeller A, Fink L,
Iglhaut J, Hartung T, Morath S, Seeger W, Grimminger F and Sibelius
U: Lipoteichoic acid (LTA) from Staphylococcus aureus stimulates
human neutrophil cytokine release by a CD14-dependent,
Toll-like-receptor-independent mechanism: Autocrine role of tumor
necrosis factor-[alpha] in mediating LTA-induced interleukin-8
generation. Crit Care Med. 34:835–841. 2006.PubMed/NCBI View Article : Google Scholar
|
|
19
|
van Kessel KP, Bestebroer J and van Strijp
JA: Neutrophil-mediated phagocytosis of Staphylococcus aureus.
Front Immunol. 5(467)2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Schröder NW, Morath S, Alexander C, Hamann
L, Hartung T, Zähringer U, Göbel UB, Weber JR and Schumann RR:
Lipoteichoic acid (LTA) of Streptococcus pneumoniae and
Staphylococcus aureus activates immune cells via Toll-like receptor
(TLR)-2, lipopolysaccharide-binding protein (LBP), and CD14,
whereas TLR-4 and MD-2 are not involved. J Biol Chem.
278:15587–15594. 2003.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Summers C, Rankin SM, Condliffe AM, Singh
N, Peters AM and Chilvers ER: Neutrophil kinetics in health and
disease. Trends Immunol. 31:318–324. 2010.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kolaczkowska E and Kubes P: Neutrophil
recruitment and function in health and inflammation. Nat Rev
Immunol. 13:159–175. 2013.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Collins SJ, Ruscetti FW, Gallagher RE and
Gallo RC: Terminal differentiation of human promyelocytic leukemia
cells induced by dimethyl sulfoxide and other polar compounds. Proc
Natl Acad Sci USA. 75:2458–2462. 1978.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wang D, Sennari Y, Shen M, Morita K,
Kanazawa T and Yoshida Y: ERK is involved in the differentiation
and function of dimethyl sulfoxide-induced HL-60 neutrophil-like
cells, which mimic inflammatory neutrophils. Int Immunopharmacol.
84(106510)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Poplutz MK, Wessels I, Rink L and
Uciechowski P: Regulation of the interleukin-6 gene expression
during monocytic differentiation of HL-60 cells by chromatin
remodeling and methylation. Immunobiology. 219:619–626.
2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Shuto T, Furuta T, Cheung J, Gruenert DC,
Ohira Y, Shimasaki S, Suico MA, Sato K and Kai H: Increased
responsiveness to TLR2 and TLR4 ligands during
dimethylsulfoxide-induced neutrophil-like differentiation of HL-60
myeloid leukemia cells. Leuk Res. 31:1721–1728. 2007.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wen SH, Hong ZW, Chen CC, Chang HW and Fu
HW: Helicobacter pylori neutrophil-activating protein directly
interacts with and activates Toll-like receptor 2 to induce the
secretion of interleukin-8 from neutrophils and ATRA-induced
differentiated HL-60 cells. Int J Mol Sci. 22(11560)2021.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Graziano RF, Ball ED and Fanger MW: The
expression and modulation of human myeloid-specific antigens during
differentiation of the HL-60 cell line. Blood. 61:1215–1221.
1983.PubMed/NCBI
|
|
29
|
Atkinson JP and Jones EA: Biosynthesis of
the human C3b/C4b receptor during differentiation of the HL-60 cell
line. Identification and characterization of a precursor molecule.
J Clin Invest. 74:1649–1657. 1984.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Manda-Handzlik A, Bystrzycka W, Wachowska
M, Sieczkowska S, Stelmaszczyk-Emmel A, Demkow U and Ciepiela O:
The influence of agents differentiating HL-60 cells toward
granulocyte-like cells on their ability to release neutrophil
extracellular traps. Immunol Cell Biol. 96:413–425. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Babatunde KA, Wang X, Hopke A, Lannes N,
Mantel PY and Irimia D: Chemotaxis and swarming in differentiated
HL-60 neutrophil-like cells. Sci Rep. 11(778)2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Chang L, Zhou G, Soufan O and Xia J:
miRNet 2.0: Network-based visual analytics for miRNA functional
analysis and systems biology. Nucleic Acids Res. 48 (W1):W244–W251.
2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Liao Y, Wang J, Jaehnig EJ, Shi Z and
Zhang B: WebGestalt 2019: Gene set analysis toolkit with revamped
UIs and APIs. Nucleic Acids Res. 47 (W1):W199–W205. 2019.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Kuwabara WMT, Zhang L, Schuiki I, Curi R,
Volchuk A and Alba-Loureiro TC: NADPH oxidase-dependent production
of reactive oxygen species induces endoplasmatic reticulum stress
in neutrophil-like HL60 cells. PLoS One.
10(e0116410)2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Guo Y, Gao F, Wang Q, Wang K, Pan S, Pan
Z, Xu S, Li L and Zhao D: Differentiation of HL-60 cells in
serum-free hematopoietic cell media enhances the production of
neutrophil extracellular traps. Exp Ther Med.
21(353)2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Yen MC, Yeh IJ, Liu KT, Jian SF, Lin CJ,
Tsai MJ and Kuo PL: Next-generation sequencing predicts interaction
network between miRNA and target genes in lipoteichoic
acid-stimulated human neutrophils. Int J Mol Med. 44:1436–1446.
2019.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Coady TH and Lorson CL: SMN in spinal
muscular atrophy and snRNP biogenesis. Wiley Interdiscip Rev RNA.
2:546–564. 2011.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Pennamen P, Le L, Tingaud-Sequeira A,
Fiore M, Bauters A, Van Duong Béatrice N, Coste V, Bordet JC,
Plaisant C, Diallo M, et al: BLOC1S5 pathogenic variants cause a
new type of Hermansky-Pudlak syndrome. Genet Med. 22:1613–1622.
2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Blanter M, Gouwy M and Struyf S: Studying
neutrophil function in vitro: Cell models and environmental
factors. J Inflamm Res. 14:141–162. 2021.PubMed/NCBI View Article : Google Scholar
|
|
43
|
von Aulock S, Morath S, Hareng L, Knapp S,
van Kessel KP, van Strijp JA and Hartung T: Lipoteichoic acid from
Staphylococcus aureus is a potent stimulus for neutrophil
recruitment. Immunobiology. 208:413–422. 2003.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Askarian F, Wagner T, Johannessen M and
Nizet V: Staphylococcus aureus modulation of innate immune
responses through Toll-like (TLR), (NOD)-like (NLR) and C-type
lectin (CLR) receptors. FEMS Microbiol Rev. 42:656–671.
2018.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Damsker JM, Hansen AM and Caspi RR: Th1
and Th17 cells: Adversaries and collaborators. Ann N Y Acad Sci.
1183:211–221. 2010.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Bradley LM, Dalton DK and Croft M: A
direct role for IFN-gamma in regulation of Th1 cell development. J
Immunol. 157:1350–1358. 1996.PubMed/NCBI
|
|
47
|
Yoshimura T and Takahashi M:
IFN-gamma-mediated survival enables human neutrophils to produce
MCP-1/CCL2 in response to activation by TLR ligands. J Immunol.
179:1942–1949. 2007.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Donath MY, Dinarello CA and
Mandrup-Poulsen T: Targeting innate immune mediators in type 1 and
type 2 diabetes. Nat Rev Immunol. 19:734–746. 2019.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Penack O, Holler E and van den Brink MRM:
Graft-versus-host disease: Regulation by microbe-associated
molecules and innate immune receptors. Blood. 115:1865–1872.
2010.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Aguzzi A, Nuvolone M and Zhu C: The
immunobiology of prion diseases. Nat Rev Immunol. 13:888–902.
2013.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Murphy SP, Porrett PM and Turka LA: Innate
immunity in transplant tolerance and rejection. Immunol Rev.
241:39–48. 2011.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Ito Y and Amagai M: Controlling skin
microbiome as a new bacteriotherapy for inflammatory skin diseases.
Inflamm Regen. 42(26)2022.PubMed/NCBI View Article : Google Scholar
|
|
53
|
de Bont CM, Boelens WC and Pruijn GJM:
NETosis, complement, and coagulation: A triangular relationship.
Cell Mol Immunol. 16:19–27. 2019.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Ko YP, Kuipers A, Freitag CM, Jongerius I,
Medina E, van Rooijen WJ, Spaan AN, van Kessel KP, Höök M and
Rooijakkers SH: Phagocytosis escape by a Staphylococcus aureus
protein that connects complement and coagulation proteins at the
bacterial surface. PLoS Pathog. 9(e1003816)2013.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Mori R, Tanaka K and Shimokawa I:
Identification and functional analysis of inflammation-related
miRNAs in skin wound repair. Dev Growth Differ. 60:306–315.
2018.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Tanaka K, Kim SE, Yano H, Matsumoto G,
Ohuchida R, Ishikura Y, Araki M, Araki K, Park S, Komatsu T, et al:
MiR-142 is required for Staphylococcus aureus clearance at skin
wound sites via small GTPase-mediated regulation of the neutrophil
actin cytoskeleton. J Invest Dermatol. 137:931–940. 2017.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Zhang W, Qu X, Zhu Z, Wang L, Qi Q, Zhou
P, Wang X and Li W: Inhibition of miR-139-5p by topical JTXK gel
promotes healing of Staphylococcus aureus-infected skin wounds.
Cells Dev. 166(203658)2021.PubMed/NCBI View Article : Google Scholar
|
|
58
|
de Kerckhove M, Tanaka K, Umehara T,
Okamoto M, Kanematsu S, Hayashi H, Yano H, Nishiura S, Tooyama S,
Matsubayashi Y, et al: Targeting miR-223 in neutrophils enhances
the clearance of Staphylococcus aureus in infected wounds. EMBO Mol
Med. 10(e9024)2018.PubMed/NCBI View Article : Google Scholar
|