Introduction
In total, ~80% of malignant brain tumors are gliomas
(1). The histological subtypes of
glioma include astrocytoma, oligodendroglioma, ependymoma and mixed
oligoastrocytoma. The World Health Organization (WHO) has also
classified glioma into four grades according to the degree of
malignancy (2). Glioblastoma
(GBM), which has the highest frequency of occurrence, is the most
aggressive cerebral malignancy (3). The prognosis of patients with GBM is
highly unfavorable, with a median survival time of 1 year (4). Based on specific genetic mutation
profiles, GBM can be further subdivided into four molecular
subtypes: Proneural, mesenchymal, neural and classical (5). Furthermore, other molecular
subpopulations of GBM have been proposed by Lee et al
(6). Numerous studies have
indicated that various signaling pathways act in the progression of
glioma, including purinergic, receptor tyrosine kinase, TGF-β and
STAT signaling pathways (7).
Ubiquitin-specific protease 18 (USP18) (enzyme
commission: 3.4.19.12), originally termed UBP43, was primitively
discovered as a differentially expressed gene in mice with acute
myeloid leukemia (AML) carrying the AML t(8;21) gene translocation
(8). Subsequently, Zhang et
al (9) and Kang et al
(10) verified that USP18 exists
in virus-infected porcine alveolar macrophages and human melanoma
cells, respectively. USP18 is expressed in multiple tissues; for
example, USP18 is upregulated in the spleen and liver, while it is
downregulated in the lungs and bone marrow (8). At present, the known primary function
of USP18 is the role it has in the de-ISGylation mediated by
interferon (IFN)-stimulated gene 15 (ISG15) (11). Furthermore, USP18 inhibits
ISGylation-independent IFN-I signaling through the IFNAR2/JAK/STAT
pathway and it mediates the inflammatory response accordingly
(12,13). The mechanism by which USP18
inhibits the IFN-I response to promote cancer progression has been
elucidated in detail (14).
Furthermore, USP18 is involved in several mechanisms involved in
carcinogenesis, such as in promoting breast cancer growth and
epithelial-mesenchymal transition (EMT) in glioma (15).
In the present study, a detailed analysis of the
role of USP18 in glioma was performed. USP18 was upregulated in
glioma and was associated with clinical traits such as tumor stage
and a worse prognosis. USP18 was also associated with the IFN-I
pathway in glioma T cells. In glioma, USP18 expression was also
associated with the proportion of a number of immune cells. Genes
co-expressed with USP18 were mainly enriched in the immune pathways
such as IFN-I and complement activation. Cell Counting Kit-8
(CCK-8) experiments demonstrated that USP18 knockdown reduced
glioma cell viability. Furthermore, colony formation assays
demonstrated that knockdown of USP18 significantly reduced the
proliferation of glioma cells. Specifically, the results of the
present study suggested that USP18 may influence glioma initiation
and progression, with potential regulatory effects on IFN-I
signaling pathways.
Materials and methods
Data preparation
Transcripts for 33 types of cancer were obtained
from the University of California Santa Cruz (UCSC) database
(https://xenabrowser.net/datapages/),
all of which were uniformly processed using the TOIL workflow
(16). All transcriptome profiles
[in high throughput sequencing (seq) transcripts per million (TPM)
formats] and clinical information were obtained from The Cancer
Genome Atlas (TCGA; https://portal.gdc.cancer.gov/) lower grade glioma
(LGG)/GBM projects. The immunophenoscore (IPS) of TCGA glioma
samples was obtained from The Cancer Immunome Atlas (https://tcia.at/) (17). The transcript and clinical data of
glioma samples from TCGA with complete survival information and
clinical features were extracted using R software (https://www.R-project.org/) (v.4.2.1). The RNAseq and
clinical information of 693 and 325 glioma samples were acquired
from the Chinese Glioma Genome Atlas [CGGA; Dataset IDs,
mRNAseq_693 (CGGA_693) and mRNAseq_325 (CGGA_325); http://www.cgga.org.cn/download.jsp] as
additional test sets. Visualization of USP18 expression in the
pan-cancer analysis was conducted using the R package, ‘ggplot2’
(https://ggplot2.tidyverse.org/).
Differential expression and clinical
correlation analyses
A pan-cancer analysis of USP18 expression in 33
types of cancer and the corresponding normal tissues was conducted
using the R package, ‘ggplot2’. To perform clinical correlation
analyses, all patients with glioma were separated into subgroups
based on clinical features and were analyzed using R software
(18).
Single-cell sequencing analysis
The single cell RNAseq glioma tissue dataset,
GSE131928_10X and the glioma T cell dataset, GSE163108_10X, were
obtained from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo) (19). The single-cell expression matrix,
Uniform Manifold Approximation and Projection (UMAP) dimensionality
reduction and cell type metadata were downloaded from the Tumor
Immune Single-cell Hub 2 (TISCH2) database (http://tisch.comp-genomics.org/) (20). The dimensionality reduction and
cell annotation of single-cell matrices were conducted using the R
package, ‘Seurat’ (21). The UMAP
visualization, violin plots, bubble plots and differential
expression analyses were also conducted using the R package,
‘Seurat’. Differential expression analyses were performed in cell
groups with specific USP18 expression patterns. The threshold for
differential genes was set to |log fold change (FC)|>0.5 and
adjusted P<0.05. Kyoto Encyclopedia of Genes and Genomes (KEGG;
https://www.genome.jp/kegg/) analysis
was conducted using the R package, ‘clusterProfiler’ (22). Histogram visualization was
completed using the R package, ‘ggplot2’. The list of USP18
co-expressed genes in CD4 and CD8 T cells was obtained from TISCH2
and the correlations between genes with an average logTPM of
>0.5 or a maximum logTPM of >2 were calculated.
Prognosis analysis
Kaplan-Meier (KM) curves and Cox regression analyses
were performed using the R package, ‘survival’ (23). The prognostic data used for KM
survival curve analysis were obtained from a previous study
(23) and samples were separated
into corresponding cohorts based on the median expression value of
USP18. The time-dependent receiver operating characteristic (ROC)
curve was constructed using the R package ‘timeROC’ (https://CRAN.R-project.org/package=timeROC). The two
aforementioned CGGA datasets served as validation sets in the KM
survival analysis. The nomogram was constructed with the R package,
‘regplot’ (https://CRAN.R-project.org/package=regplot). The
bootstrapping method was used and repeated 1,000 times to validate
the nomogram for 1, 3 and 5 years. All validation was conducted
using the R package, ‘rms’ (https://CRAN.R-project.org/package=rms).
Tumor immune profile analysis
The checkpoint inhibitors and major
histocompatibility complex (MHC) molecule genes were acquired from
the study by Charoentong et al (17). The relationship between the USP18
expression level and the score of 24 immune cells was assessed
using the R package, ‘GSVA’ (24).
The immune correlation analysis used data from 24 types of immune
cell markers (25). The immune
microenvironment analysis of the stromal score, estimate score,
immune score, tumor purity and USP18 expression was conducted with
the R package, ‘estimate’ and the score was calculated as
previously described by Yoshihara et al (26). The USP18 expression stratified gene
mutation files were differentiated using the Perl language and
visualized using the R package, ‘maftools’ (https://github.com/PoisonAlien/maftools).
Functional enrichment analyses
Gene Ontology (GO), KEGG and Gene Set Enrichment
Analysis (GSEA) functional analyses for the differentially
expressed genes (DEGs) were performed using the R packages,
‘enrichplot’ (https://github.com/YuLab-SMU/enrichplot),
‘org.Hs.eg.db’ (https://bioconductor.org/packages/org.Hs.eg.db/) and
‘clusterProfiler’ (https://guangchuangyu.github.io/software/clusterProfiler/)
(27-29).
The DEGs were selected with the R package, ‘DESeq2’ (https://github.com/thelovelab/DESeq2)
and the thresholds were |logFC|>1.5 and adjusted P<0.05. All
DEGs were included in the GSEA, using the USP18 low-expression
group as a reference.
Cell culture
U87MG ATCC (glioblastoma of unknown origin) and U251
cell lines were purchased from the Chinese National Infrastructure
of Cell Line Resource and cultured in DMEM supplemented with 10%
fetal bovine serum, 2 mM glutamine and 1% penicillin-streptomycin
(all from Gibco; Thermo Fisher Scientific, Inc.). The cell lines
were cultured in a 37˚C incubator containing 5% CO2.
U87MG ATCC was authenticated using STR profiling.
USP18 knockdown
Lipofectamine RNAiMAX (Invitrogen; Thermo Fisher
Scientific, Inc.) was used for transfecting USP18 targeting and
control (Ctrl; 40 nM) small interfering RNAs (siRNAs; 40 nM) into
U87MG ATCC and U251 cells, according to the manufacturer's
protocols and as previously described (30). The siRNA sequences used were as
follows: siUSP18#1, 5'-GGAAUUCACAGACGAGAAATT-3'; siUSP18#2,
5'-GGAAGAAGACAGCAACAUGTT-3'; and siCtrl, 5'-UUCUCCGAACGUGUCACGU-3'.
The USP18 knockdown efficiency was tested by reverse
transcription-quantitative PCR (RT-qPCR) analysis. The subsequent
experiments were conducted 48 h after transfection.
RNA isolation and RT-qPCR
Total RNA was extracted using TRIzol reagent (Thermo
Fisher Scientific, Inc.). The RNA was reverse transcribed using the
reverse transcription kit (cat. no. A3500; Promega Corp.) according
to the manufacturer's instructions, and qPCR assays were performed
with SYBR Green Master Mix (Takara Bio, Inc.) on a 7500 fast
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. The thermocycling conditions were as follows:
Pre-denaturation at 95˚C for 5 min and 40 cycles of denaturation at
95˚C for 15 sec, annealing at 60˚C for 20 sec and elongation at
72˚C for 30 sec. The following primers were used for qPCR: USP18
forward, 5'-CCTGAGGCAAATCTGTCAGTC-3' and reverse,
5'-CGAACACCTGAATCAAGGAGTTA-3'; and β-actin forward,
5'-CATGTACGTTGCTATCCAGGC-3' and reverse,
5'-CTCCTTAATGTCACGCACGAT-3'. The
2-ΔΔCq method was used to quantify
the fold changes in gene expression (31).
CCK-8 assay
Cell viability was assessed using CCK-8
(Sigma-Aldrich; Merck KGaA), following the manufacturer's protocols
as previously described (32).
Colony formation
The colony formation assay was used to examine the
cell colony formation ability of U87MG ATCC and U251 cells
following USP18 expression knockdown. The colony formation assay
was performed as previously described (32). Clusters of >50 cells were
regarded as colonies. The colonies were calculated using ImageJ
software (v.1.47; National Institutes of Health).
Apoptosis assay
The transfected U87MG ATCC and U251 cells were
cultured for 48 h at 37˚C. Subsequently, the cells were thoroughly
mixed with propidium iodide (5 µl) and binding buffer (500 µl), and
then incubated with fluorescein isothiocyanate-conjugated
anti-annexin V antibody (5 µl) at room temperature for 10 min,
according to the manufacturer's protocols. Finally, the cells were
evaluated using a CytoFLEX LX (Beckman Coulter, Inc.) and the data
were analyzed using CytTExpert software (v.2.4; Beckman Coulter,
Inc.).
Statistical analysis
The gene expression data were presented as the mean
± standard deviation and the survival hazard ratios (HRs) were
presented as HR (95% confidence interval). The counting and
statistical analyses were repeated three times. The cell line
experimental data were analyzed using GraphPad Prism 8.0 software
(Dotmatics). The age, isocitrate dehydrogenase (IDH) mutation and
1p19q co-deletion status of different groups were compared using
the Wilcoxon rank-sum test. The WHO grade and histological type
were compared using Kruskal-Wallis test with Dunn post hoc tests.
Survival analysis was conducted using the log-rank test. Gene
correlation analysis was conducted using Spearman's correlation
analysis. For multiple comparisons vs. the same control group,
including mRNA levels, cell viability, colony numbers and apoptosis
percentage, one-way ANOVA followed by Dunnett's test was used.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Differential expression and clinical
correlation analyses of USP18 in glioma
As indicated in Fig.
1, A pan-cancer analysis of USP18 expression using data from
TCGA was performed and it was found that USP18 was upregulated in
most cancer types (Fig. 1A). To
ensure TCGA samples were sufficient and comparable, the analysis
was repeated using data from the UCSC. USP18 was significantly
differentially expressed in various types of cancers in the UCSC
dataset, with high expression observed in LGG and GBM and low
expression observed in adrenocortical carcinoma and kidney
chromophobe (Fig. 1C).
Furthermore, the expression level of USP18 in the GBM dataset from
TCGA was examined and an upregulation of USP18 was observed
(Fig. 1B). The same result was
observed in the LGG and GBM datasets from the UCSC (Fig. 1D). Patients with LGG or GBM, aged
>60 years, with wild-type IDH and without the 1p/19q co-deletion
had a higher expression of USP18 (Fig.
1E-G). However, the expression level of USP18 revealed no
sex-specific differences (Fig.
1H). In more advanced glioma WHO grades, the USP18 expression
level was significantly higher (Fig.
1I). In addition, patients with GBM expressed notably higher
USP18 levels than patients with other histological types and the
expression of USP18 was not significantly different between
patients with oligoastrocytoma, astrocytoma or oligodendroglioma
(Fig. 1J).
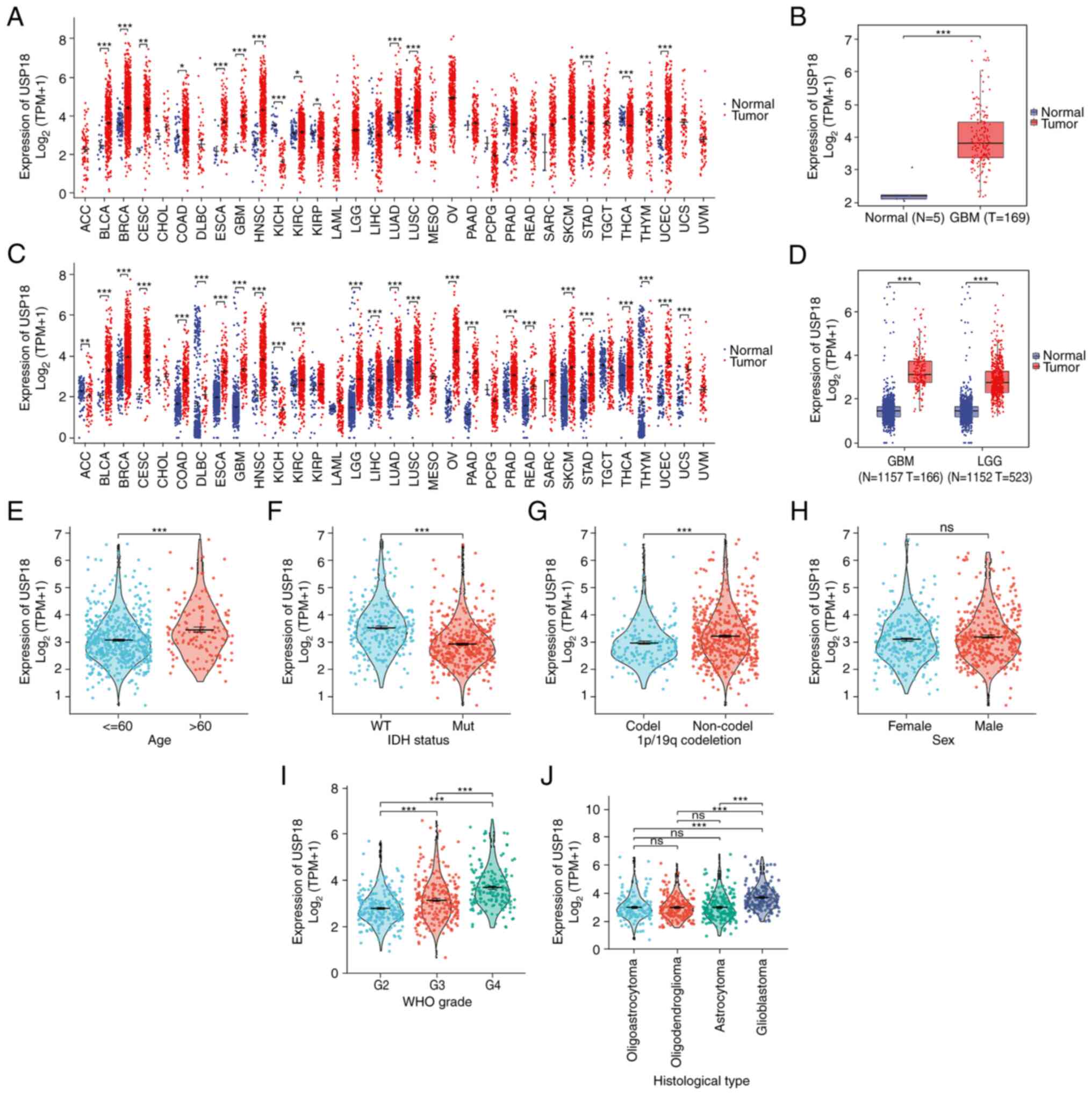 | Figure 1Differential expression and clinical
correlation analyses of USP18 in glioma. (A) USP18 expression in
pan-cancer using The Cancer Genome Atlas data. Red indicated tumor
samples and blue indicated normal samples. (B) USP18 expression in
GBM using TCGA data. (C) USP18 expression in pan-cancer using
University of California Santa Cruz data. (D) USP18 expression in
LGG and GBM using UCSC data. Clinical correlation analyses
comparing (E) age in years, (F) IDH status, (G) 1p/19q co-deletion,
(H) sex, (I) WHO grade and (J) histological type with USP18
expression in patients with LGG and GBM. Red indicates patients
aged >60 years, IDH mutation, 1p/19q non-co-deletion, male, WHO
grade 3 and oligodendroglioma. Blue represents patients aged <60
years, IDH wild-type, 1p/19q co-deletion, female, WHO grade 2 and
oligoastrocytoma. Green represents groups with WHO grade 4 and
astrocytoma, while purple indicates the GBM cohort.
*P<0.05, **P<0.01 and
***P<0.001; ns, no significance. The abbreviation for
cancers in A and C may be found at https://gdc.cancer.gov/resources-tcga-users/tcga-code-tables/tcga-study-abbreviations.
USP18, ubiquitin-specific protease 18; N, normal tissue; T, tumor
tissue; WT, wild-type; Mut, mutant; codel, codeletion; GBM,
glioblastoma; LGG, lower grade glioma; IDH, isocitrate
dehydrogenase; WHO, World Health Organization. |
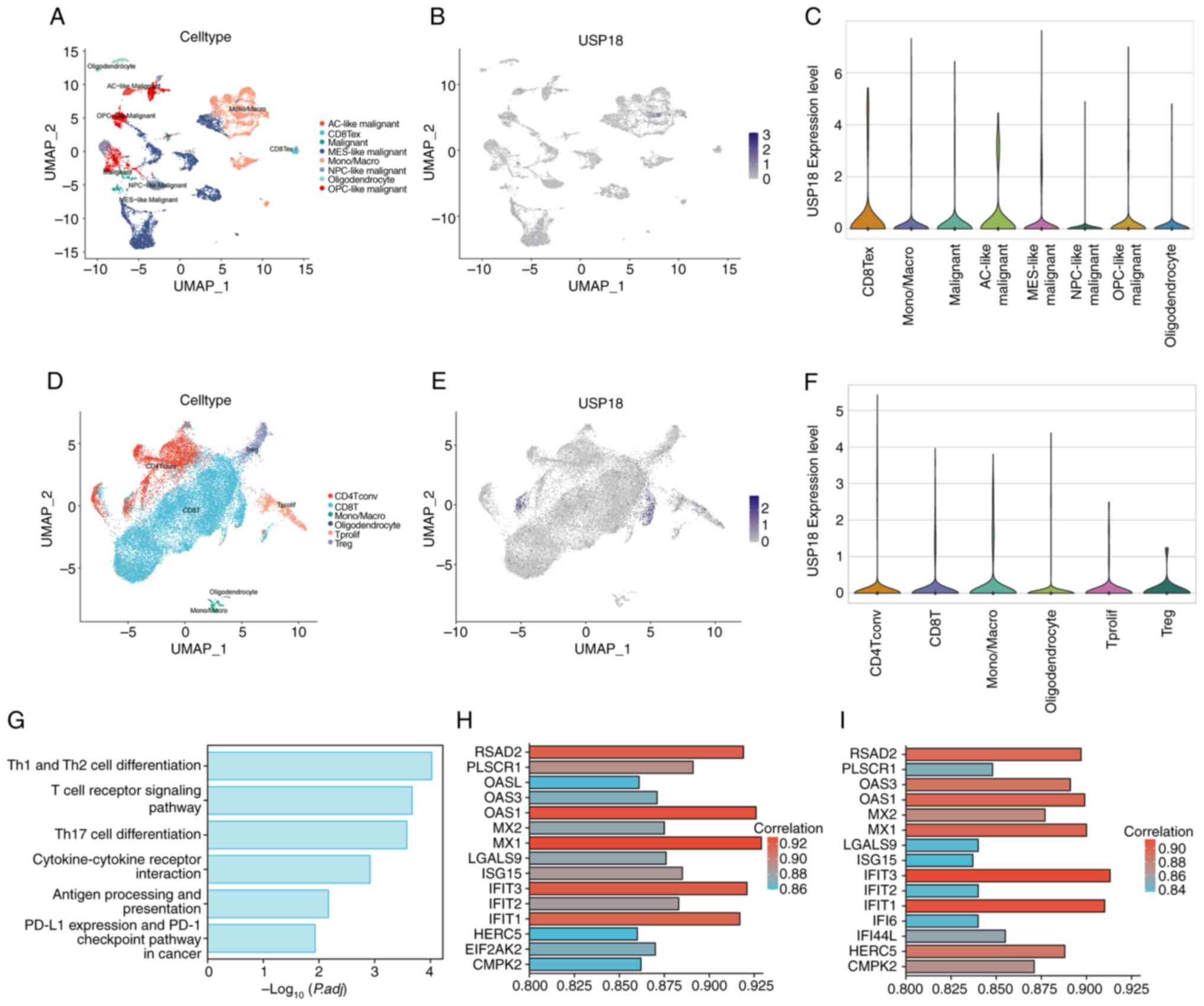
USP18 expression patterns in the
glioma single-cell landscape
The glioma tissue single-cell dataset was selected
for dimensionality reduction (Fig.
2A). The overall expression level of USP18 was low in this
dataset and no characteristic cell expression cluster was observed
(Fig. 2B). The USP18 expression
level in different cell clusters was further compared (Fig. 2C) and it was determined that USP18
was significantly upregulated in terminally exhausted CD8 T cells
(CD8 Tex). Since the USP18 negative regulation mechanism of the
IFN-I pathway in cancer has been demonstrated, it was hypothesized
that USP18 might play a specific role in glioma T cells. Therefore,
the glioma T cell dataset was selected for further analysis of the
USP18 expression pattern (Fig.
2D), and it was demonstrated that USP18 was highly expressed in
a small number of CD8 and CD4 T cells (Fig. 2E). However, there was no
significant difference in the USP18 expression level in each of the
cell clusters shown in the violin plot (Fig. 2F). The pathways enriched in the CD8
Tex subcluster of the glioma tissue dataset were the T cell subtype
differentiation and T cell receptor-related pathways, antigen
presentation and programmed cell death protein 1 in cancer
(Fig. 2G). The genes co-expressed
with USP18 in the CD4 (Fig. 2H)
and CD8 (Fig. 2I) T cell subsets
of the glioma T cell dataset included the IFN-induced protein with
tetratricopeptide repeats (IFIT) family, the 2'-5'-oligoadenylate
synthetase family and the MX dynamin-like GTPase family.
Upregulation of USP18 is related to a
worse survival outcome in patients with glioma
A KM survival curve analysis based on the USP18
expression levels in TCGA cohort was conducted and the results
demonstrated that patients with glioma and upregulated USP18
expression had a worse overall survival (OS) (Fig. 3A), disease-specific survival (DSS)
(Fig. 3B) and progression-free
interval (PFI) (Fig. 3C). The area
under the curve values at 1, 3 and 5 years for OS (Fig. 3D), DSS (Fig. 3E) and PFI (Fig. 3F) were all >0.6. A validation
analysis of the KM survival curves using the CGGA_325 (Fig. 3G) and CGGA_693 (Fig. 3H) datasets was then performed.
Patients with high USP18 expression in both validation sets had a
worse prognosis. The 5-year prognostic predictive power was the
best for OS, DSS and PFI. To build a prognostic model to predict
survival risk, a prognostic nomogram was constructed at 1, 3 and 5
years based on USP18 expression and the patient clinicopathological
parameters (Fig. 3I).
Consequently, the constructed calibration curves had a satisfactory
consistency at 1, 3 and 5 years (Fig.
3J).
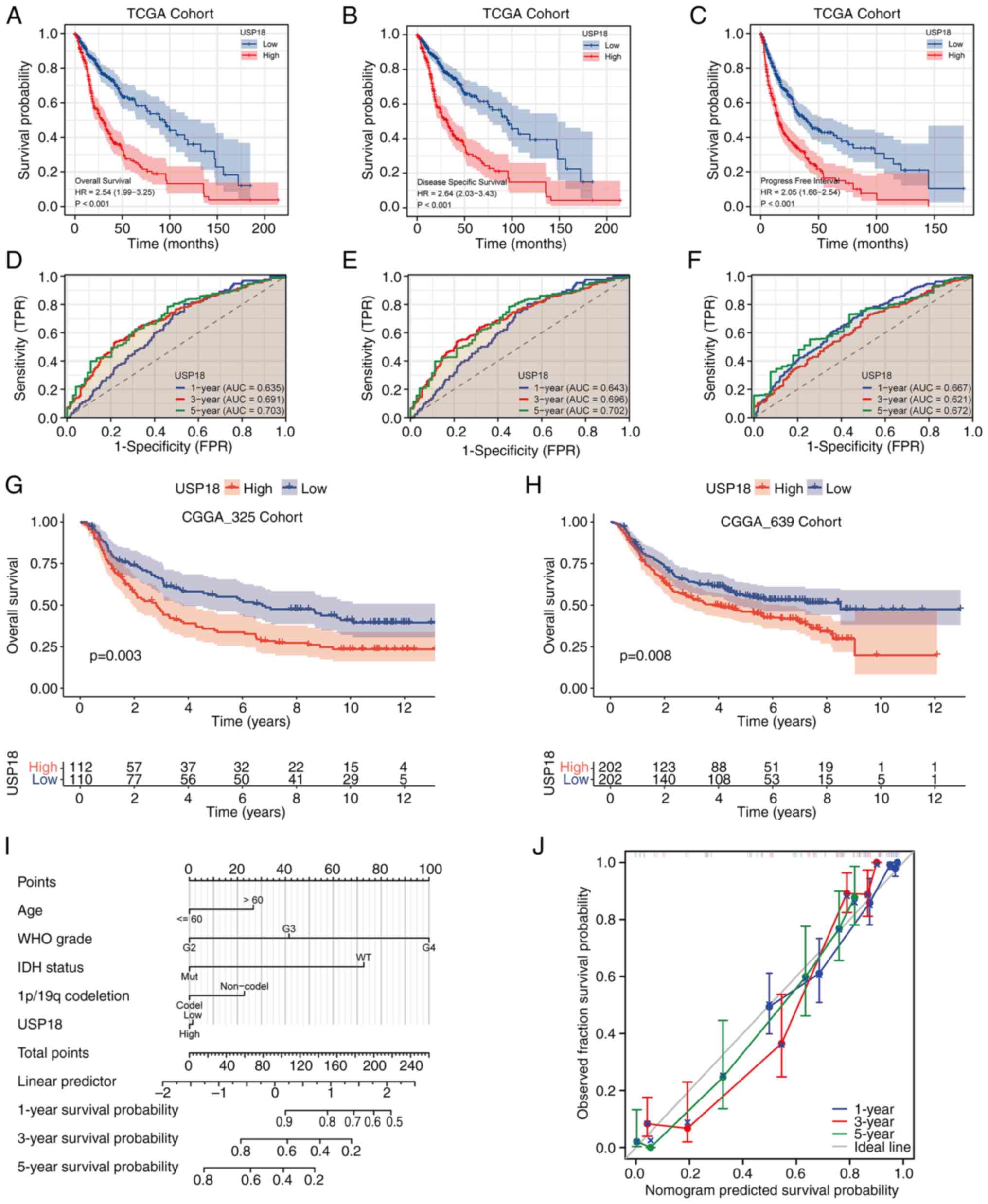 | Figure 3Survival and ROC analyses for
different USP18 expression levels. (A) OS, (B) DSS and (C) PFI
analyses of patients with GBM or LGG with high or low USP18
expression. Red indicates the high USP18 expression cohorts and
blue represents the low USP18 expression cohorts. Time-dependent
ROC curve analysis at 1, 3 and 5 years for (D) OS, (E) DSS and (F)
PFI. Blue indicates the ROC curve for USP18 expression at 1 year,
red indicates 3 years and green indicates 5 years. Kaplan-Meir
survival analysis according to the USP18 expression level in the
(G) CGGA_325 and (H) CGGA_639 test cohorts. Red indicates the high
USP18 expression cohorts and blue indicates the low USP18
expression cohorts. (I) Nomograms at 1, 3 and 5 years based on
USP18 expression and clinicopathological parameters. (J)
Calibration curve analysis at 1, 3 and 5 years. Blue indicates the
calibration curve of USP18 at 1 year, red indicates 3 years and
green indicates 5 years. The grey line indicates the ideal curve.
ROC, receiver operating characteristic; USP18, ubiquitin-specific
protease 18; OS, overall survival; DSS, disease-specific survival;
PFI, progression-free interval; GBM, glioblastoma; LGG, lower grade
glioma; CGGA, Chinese Glioma Genome Atlas; TCGA, The Cancer Genome
Atlas; WHO, World Health Organization; IDH, isocitrate
dehydrogenase. |
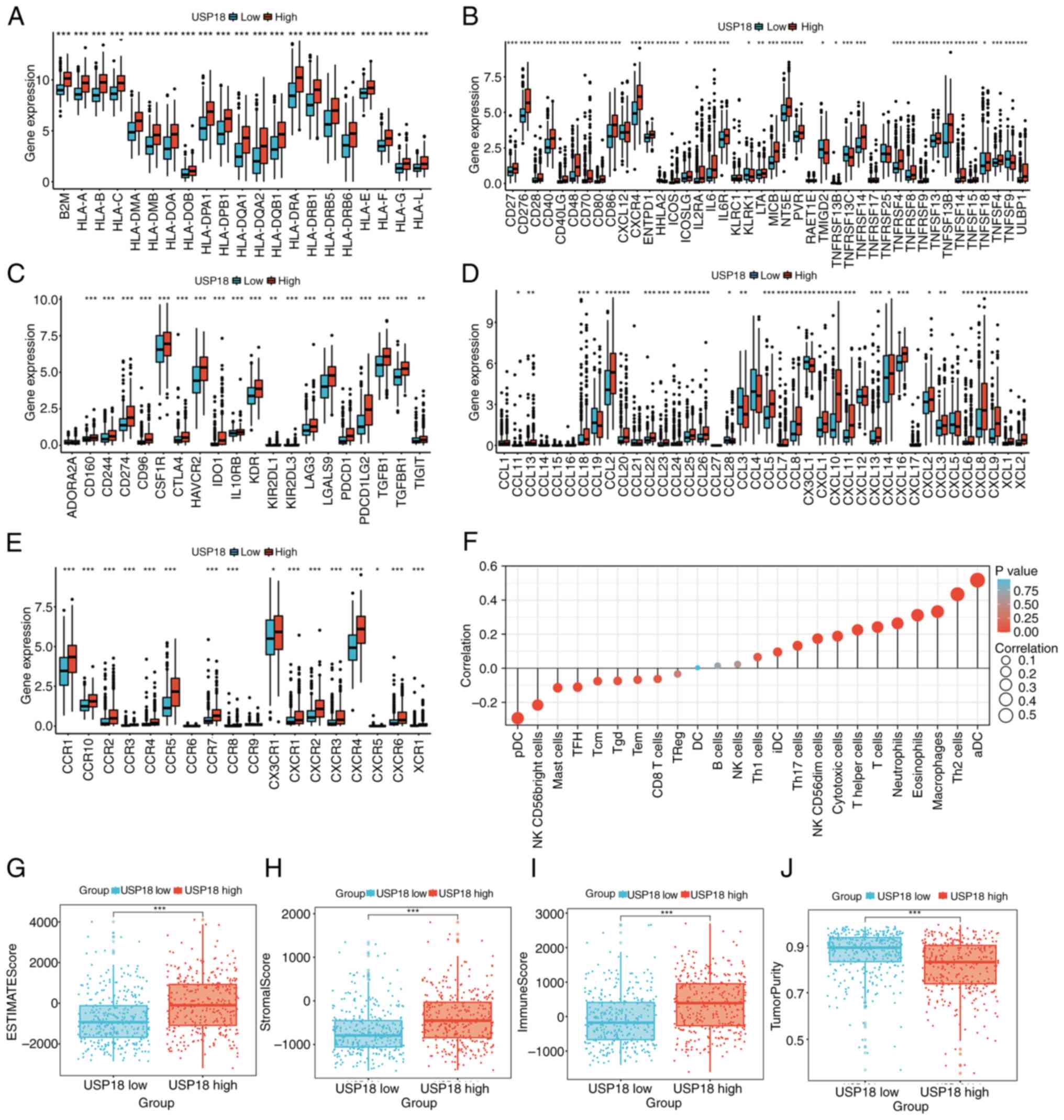
Crosstalk of USP18 expression on the
glioma immune microenvironment and immunotherapy
The MHC molecule (Fig.
4A), Immunostimulator (Fig.
4B) immunoinhibitor (Fig. 4C),
chemokine (Fig. 4D) and chemokine
receptor (Fig. 4E) gene levels
were compared between the two USP18 expression cohorts and it was
revealed that most were upregulated in the high USP18 expression
cohort. However, the immunostimulatory gene, TNF superfamily member
9 and the chemokine receptor genes, C-C motif chemokine ligand
(CCL)3 and CCL19, were downregulated in patients with high USP18
expression. Overall, the proportions of different immune cell
types, such as activated dendritic cells (DCs), macrophages, T
cells, neutrophils and natural killer (NK) CD56dim cells, were
significantly positively correlated with the USP18 expression
level, while the other immune cell types were negatively
correlated, such as plasmacytoid DCs and NK CD56-bright cells
(Fig. 4F). Furthermore, the high
USP18 expression cohort was characterized by a high stromal score,
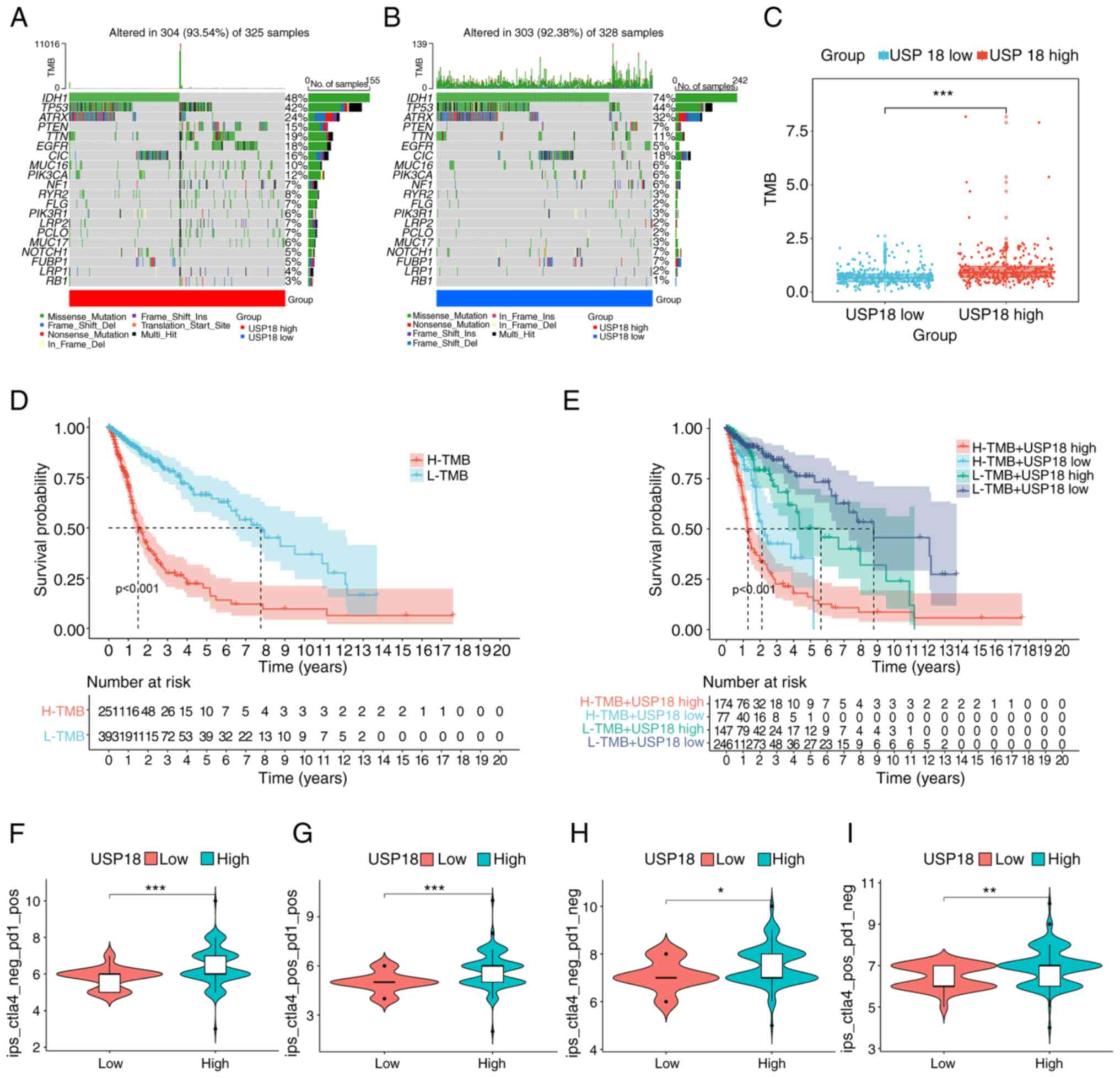
immune score and low tumor purity (Fig. 4G-J). The frequency of PTEN and EGFR
mutations in the high USP18 expression cohort was also higher
(Fig. 5A). However, the low USP18
expression cohort had a higher IDH1 and ATRX mutation frequency
(Fig. 5B). A higher tumor mutation
burden (TMB) was observed in the high USP18 expression group, which
indicated tumor neoantigen potential (Fig. 5C). It is also worth noting that TMB
had a prognostic effect on glioma (Fig. 5D) and patients with a high TMB and
high USP18 expression had a worse prognosis (Fig. 5E). Furthermore, patients with
glioma and upregulated USP18 expression and pos_PD1 had a higher
IPS, suggesting a higher immunotherapeutic potential (Fig. 5F-I).
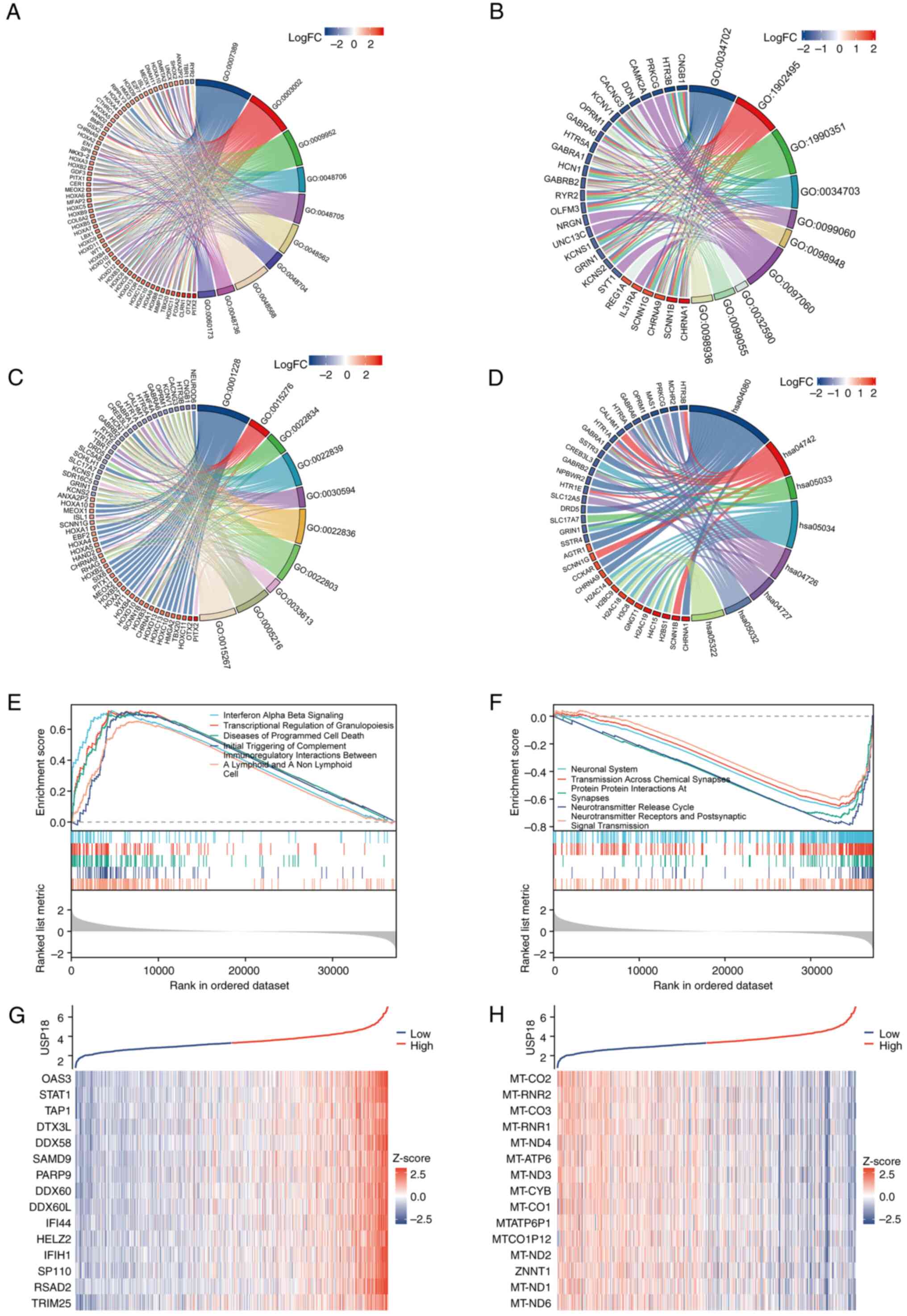
USP18 is associated with multiple
signaling pathways in glioma
Statistically significant DEGs between the USP18 low
and high expression cohorts in the LGG and GBM datasets were
identified, which included 462 upregulated and 175 downregulated
genes. GO analysis chord diagrams incorporated the 10 most
significant GO terms (P<0.001), while the KEGG pathway analysis
chord diagram incorporated the 8 most significant pathway terms
(P<0.001). The term, ‘biological process’, included genes
enriched in the ‘pattern specification process’ and the
‘development of the embryonic skeletal system’ (Fig. 6A). The term, ‘cellular component’,
included genes enriched in the ‘transmembrane transporter complex’,
an integral component of the postsynaptic membrane (Fig. 6B). While the ‘molecular function’
term included genes enriched in ‘ligand-gated ion channel activity’
and ‘ion-gated channel activity’ (Fig.
6C). The KEGG analysis revealed an association with
‘neuroactive ligand-receptor interaction’ and ‘taste transduction’
(Fig. 6D). Next, all DEGs were
incorporated into GSEA. The main enriched pathways in the high
USP18 expression group were ‘IFN alpha beta signaling’,
‘transcriptional regulation of granulopoiesis’ and ‘initial
triggering of complement’ (Fig.
6E). The pathways enriched in the low USP18 expression group
were the ‘neuronal system’, ‘neurotransmitter release cycle’ and
‘neurotransmitter receptors and postsynaptic signal transmission’
(Fig. 6F). Genes co-expressed with
USP18 included STAT1 and OSAS3 (Fig.
6G), and the genes negatively correlated with USP18 were mainly
mitochondrially encoded genes (Fig.
6H).
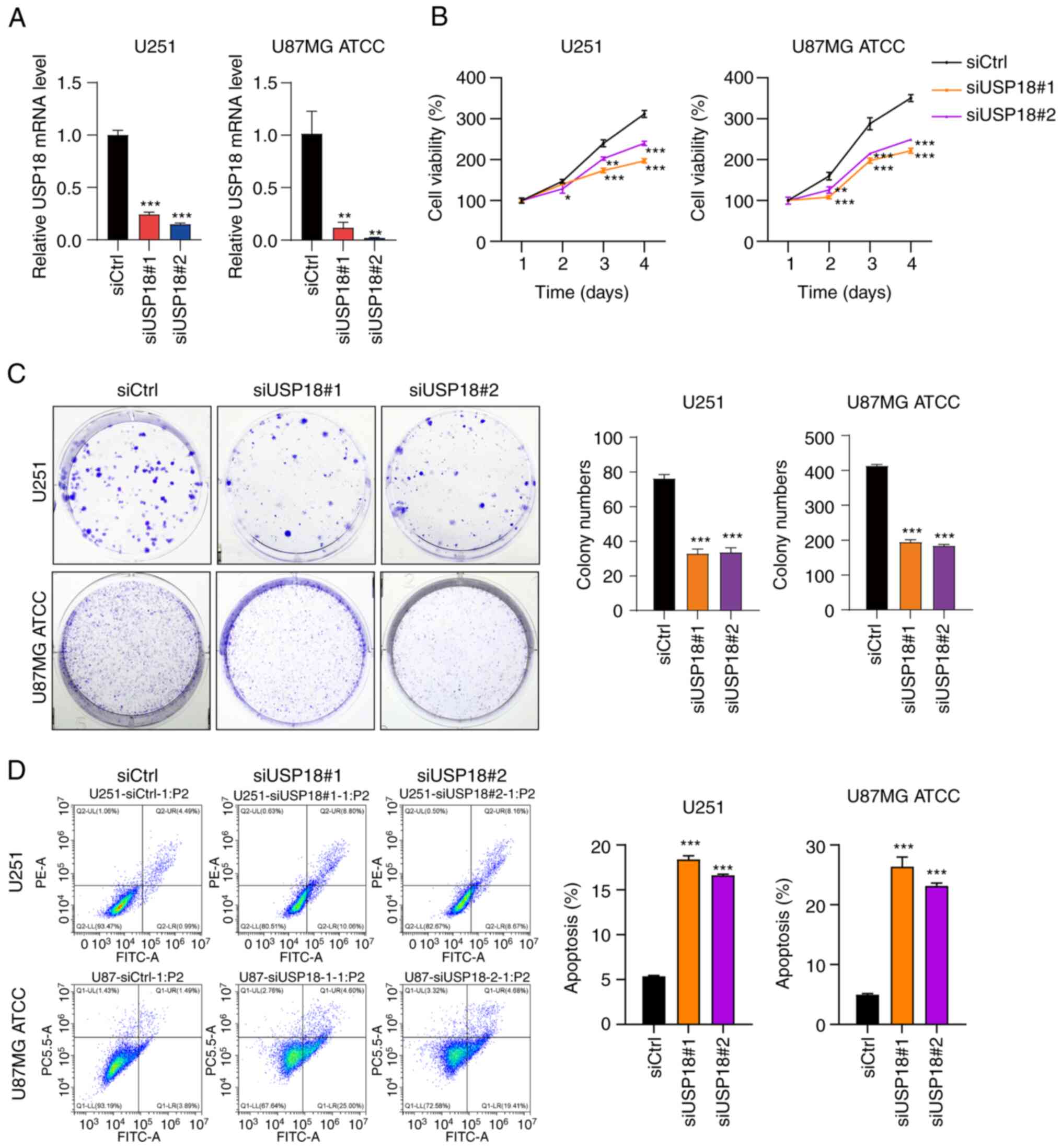
Knocking down USP18 expression
suppresses proliferation and colony formation and induces the
apoptosis of glioma cells
To investigate the biological role of USP18 in the
development of glioma, two siRNAs targeting USP18 expression
(siUSP18#1 and siUSP18#2) were transfected into glioma cells to
knockdown USP18 expression. The RT-qPCR validation assay confirmed
that USP18 expression was significantly reduced in U87MG ATCC and
U251 cells transfected with siUSP18 (Fig. 7A). Next, the CCK-8 assay was used
to evaluate the impact of USP18 knockdown on glioma cell
proliferation. The viability of U87MG ATCC and U251 cells following
USP18 knockdown was significantly reduced on days 3 and 4 (Fig. 7B). Analogous results were obtained
in the colony formation assay. Compared with the siCtrl group, the
number of colonies in the siUSP18#1 and siUSP18#2 groups decreased
significantly (Fig. 7C). Moreover,
it was also found that USP18 knockdown significantly induced the
apoptosis of U251 and U87MG ATCC cells (Fig. 7D). Collectively, these results
indicated that USP18 knockdown inhibited the proliferation and
induced the apoptosis of glioma cells.
Discussion
The role of USP18 in viral infectious diseases,
particularly those related to the central nervous system (CNS), has
been the focus of previous research (33,34).
The role of USP18 in the IFN pathway has therefore encouraged
research in the field of malignant tumors (35). Since USP18 was first identified in
human melanoma cell lines in 2001, research on the effects of USP18
on cancer has increased (8). There
is also growing evidence that USP18 is upregulated in melanoma and
liver cancer (36-39).
A pan-cancer analysis of USP18 expression was also conducted in the
present study. Although the function of USP18 in the modulation of
signaling pathways in glioma has been previously described
(7), its role has not been
exhaustively elucidated. Therefore, in the present study, the high
expression of USP18 in glioma was verified using TCGA and UCSC
database and these findings were consistent with a previous study
(15). The upregulation of USP18
expression may be induced by the IFN-I response elicited by glioma
cells, which is similar to other IFN-induced genes (15). The mechanism may involve free ISG15
blocking S-phase kinase-associated protein-Cullin-F-box
protein-mediated ubiquitination and subsequent proteasomal
degradation of USP18(40). In the
present study, USP18 expression in glioma was further verified
using single-cell data and in the glioma microenvironment,
significantly high expression of USP18 in CD8 T cells was observed.
The CD8 T cell cluster is associated with IFN-I signaling and T
cell differentiation. Furthermore, USP18 has been revealed to
regulate T cell activation and T helper 17 cell differentiation
through deubiquitination of the TAK1-TGF-β activated kinase 1
complex (41). This suggests that
USP18 may play a similar role in regulating differentiation and
inhibiting IFN-I signaling in glioma T cells, thereby promoting
glioma progression. In the present study, USP18 co-expressed genes
were further identified in the glioma T cell subgroups and most of
these genes were IFN-inducible. For example, the IFIT genes are
important mediators of innate immunity (42), and the IFN-induced MX genes enhance
endoplasmic reticulum stress-related cell death (43). USP18 localizes to the nucleus, is
recruited to IFNAR2 by STAT2 and interferes with the assembly of
the ternary IFN-IFNAR1-IFNAR2 complex, thereby acting as a negative
regulator of the IFN-I signaling pathway (44).
Glioma is characterized by heterogeneity and
differences in glioma classification, prognosis and treatment based
on clinical features have been widely discussed (45,46).
To verify whether USP18 expression is associated with clinical
characteristics, a clinical correlation analysis was performed in
the present study and it was found that USP18 was highly expressed
in patients >60 years old, with wild-type IDH, non-co-deletion
of 1p19q, WHO grade 3 or 4 and GBM. A survival analysis of all
three dataset cohorts was also performed and it demonstrated that
USP18 upregulation was related to poor survival outcomes.
Since the tumor immune microenvironment has been
deemed a key element involved in tumor development, immune
infiltration and differences in immune-related gene expression were
evaluated in the present study (47). It was found that the proportion of
various immune cells was related to the USP18 expression level, and
the immuno-genes were mostly upregulated in the high USP18
expression subgroup. Since functional lymphangion was described in
the meninges in 2015, the tumor microenvironment (TME) in GBM has
been considered an immunologically distinct organ (48,49).
The TME of GBM consists of microglia (permanent macrophages of the
CNS), macrophages (peripheral bone marrow-derived macrophages),
tumor-infiltrating lymphocytes and tumor-infiltrating DCs (47,50,51).
Microglia and macrophages in glioma are tumor-associated
macrophages (TAMs) and represent distinctly different cell groups
(52). Furthermore, TAMs have been
demonstrated to promote GBM infiltration and GBM proliferation
(47). Neutrophils in GBM, or
tumor-associated neutrophils, excrete elastase to boost tumor
proliferation and angiogenesis (53,54).
The results of the present study demonstrated that the USP18
expression level was positively correlated with macrophage and
neutrophil infiltration, indicating that USP18 may promote GBM
progression through this infiltration. T cells engaged in the
adaptive immune response can restrict glioma growth (55). The nuclear factor-κ light chain
enhancer of activated B cells (NF-κB) pathway, a T cell activating
pathway, has also been reported (56). Regulatory T cells, which express
the Forkhead Box P3 transcription factor, downregulate the NF-κB
pathway to promote immunosuppression in the adaptive immune
response (57,58). In addition, USP18 has been revealed
to suppress the NF-κB pathway in innate immune cells (59). Thus, further research is necessary
to explore the USP18 and NF-κB pathways in the adaptive
immunological response. However, these results suggested that USP18
may crosstalk with immunotherapy in glioma. In the present study,
the potential impact of USP18 expression on immunotherapy was
explored. The results suggested that patients with glioma with high
USP18 expression may exhibit improved response to
immunotherapy.
GO, KEGG and GSEA pathway analyses of DEGs were
conducted in the present study and it was found that the GO terms,
CC and MF, were both enriched in intercellular signal transduction
processes, such as those involving the ion channel complex and
ligand-gated ion channel activity. KEGG analysis revealed a similar
result, with neuroactive ligand-receptor interactions being the
most significant term. The GSEA indicated that the DEGs were mainly
enriched in gene sets associated with the IFN, complement
activation signaling pathways. Further co-expression analysis
indicated that USP18 may play a role in the regulation of IFN
signaling and mitochondrial oxidative respiratory chain
transmission. Mitochondrial-encoded genes involved in the
regulation of apoptosis in glioma cells have also been reported
(60). The respiratory chain
transmission and oxidative phosphorylation metabolism associated
with these genes may provide the feasibility of an in-depth study
of the mechanism of USP18 in glioma.
IFN-I are the largest IFN class of the three types
(61). IFN-I is regularly used as
a biotherapeutic agent in clinical malignancy as it can directly
restrict tumor growth and progression and improves the immune
surveillance against cancer (62-64).
However, GBM is strongly resistant to IFN-I-induced apoptosis and
therefore IFN-I achieves little efficacy in GBM therapy (65-68).
TNF-related apoptosis-inducing ligand (TRAIL), which exerts
specific pro-apoptotic activity against transformed cells, belongs
to the TNF superfamily (69,70).
Downregulation of USP18 was initially reported to enhance apoptosis
induced by IFN-α or TRAIL in different cell lines, activating the
extrinsic apoptosis pathway and the associated upregulation of
TRAIL (71). In addition, USP18
can also maintain resistance against IFN-I treatment of GBM, which
is TRAIL-dependent (72). These
findings provided a unique perspective on the resistance of GBM to
IFN-I, and USP18 may play a key role. GBM currently lacks effective
therapies (73), and the
first-line drugs for the treatment of GBM are temozolomide and
bevacizumab. Research to identify targeted agents for future
immunotherapy, gene therapy and antiangiogenic strategies are
required to combat GBM tumor heterogeneity (74).
In the present study, to determine whether USP18
exerted an effect on glioma proliferation, RT-qPCR, CCK-8, colony
formation and apoptosis experiments were performed. Knockdown of
USP18 expression suppressed the proliferation capacity and colony
formation of glioma cells. USP18-mediated glioma proliferation may
be related to EMT. EMT maintains epithelial cells in an unstable
quasi-mesenchymal cell state (75,76).
Although the detailed mechanisms underlying EMT in GBM remain
unknown, evidence indicates that EMT is engaged in the invasion and
growth of GBM (77). Furthermore,
USP18 has been discovered to stabilize TWIST and its expression,
which may contribute to promoting EMT in GBM (15). A recent study has also indicated
that USP18 affects EMT and promotes glioma stem cell growth
(78).
This study still has limitations. For instance, the
specific mechanism by which USP18 affects glioma cell proliferation
was not elucidated and the experiments were only conducted at the
cellular level, requiring further verification by animal
experiments, such as in vivo tumorigenicity assay in athymic
nude mice.
In conclusion, the present study indicated that
USP18 may be involved in glioma invasion, growth, metastasis,
immune response and tolerance. USP18 may be valuable as a latent
outcome indicator and a latent therapy target in glioma.
Furthermore, the results of the present study provided additional
information regarding the molecular mechanisms involving USP18 in
glioma. Therefore, future studies should focus on the role of USP18
in glioma.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant no. 81201991), the Basic
Research Program of Shanxi Province (grant no. 202103021224400),
the Key Medical Scientific Research Projects of Shanxi (grant no.
2021XM35) and the Four ‘Batches’ Innovation Project of Invigorating
Medical through Science and Technology of Shanxi (grant no.
2022RC07).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in TCGA (https://portal.gdc.cancer.gov/), UCSC Xena database
(https://xenabrowser.net/datapages/),
CGGA (http://www.cgga.org.cn/) and TISCH2
(http://tisch.comp-genomics.org/)
repositories.
Authors' contributions
YC, RL and ZL conceived and designed the study and
conducted the experiments. ZL and PL acquired the data. JL, ZZ and
JH analyzed the results. YC, RL and BY operated software and
visualized the results. YW, YZ and GG supervised the experiments
and validated the results. YC and RL confirm the authenticity of
all the raw data. All authors read and approved the final version
of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Goodenberger ML and Jenkins RB: Genetics
of adult glioma. Cancer Genet. 205:613–621. 2012.PubMed/NCBI View Article : Google Scholar
|
|
2
|
WHO Classification of Tumours Editorial
Board. World Health Organization classification of tumours of the
central nervous system. 5th edition. Lyon: International Agency for
Research on Cancer, 2021.
|
|
3
|
Ostrom QT, Price M, Neff C, Cioffi G,
Waite KA, Kruchko C and Barnholtz-Sloan JS: CBTRUS statistical
report: Primary brain and other central nervous system tumors
diagnosed in the United States in 2015-2019. Neuro Oncol. 24 (Suppl
5):v1–v95. 2022.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Ohgaki H and Kleihues P: Epidemiology and
etiology of gliomas. Acta Neuropathol. 109:93–108. 2005.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Verhaak RGW, Hoadley KA, Purdom E, Wang V,
Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al:
Integrated genomic analysis identifies clinically relevant subtypes
of glioblastoma characterized by abnormalities in PDGFRA, IDH1,
EGFR, and NF1. Cancer Cell. 17:98–110. 2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Lee E, Yong RL, Paddison P and Zhu J:
Comparison of glioblastoma (GBM) molecular classification methods.
Semin Cancer Biol. 53:201–211. 2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Basters A, Knobeloch KP and Fritz G:
USP18-a multifunctional component in the interferon response.
Biosci Rep. 38(BSR20180250)2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Liu LQ, Ilaria R Jr, Kingsley PD, Iwama A,
van Etten RA, Palis J and Zhang DE: A novel ubiquitin-specific
protease, UBP43, cloned from leukemia fusion protein
AML1-ETO-expressing mice, functions in hematopoietic cell
differentiation. Mol Cell Biol. 19:3029–3038. 1999.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Zhang X, Shin J, Molitor TW, Schook LB and
Rutherford MS: Molecular responses of macrophages to porcine
reproductive and respiratory syndrome virus infection. Virology.
262:152–162. 1999.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kang D, Jiang H, Wu Q, Pestka S and Fisher
PB: Cloning and characterization of human ubiquitin-processing
protease-43 from terminally differentiated human melanoma cells
using a rapid subtraction hybridization protocol RaSH. Gene.
267:233–242. 2001.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Malakhov MP, Malakhova OA, Kim KI, Ritchie
KJ and Zhang DE: UBP43 (USP18) specifically removes ISG15 from
conjugated proteins. J Biol Chem. 277:9976–9981. 2002.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Malakhova OA, Yan M, Malakhov MP, Yuan Y,
Ritchie KJ, Kim KI, Peterson LF, Shuai K and Zhang DE: Protein
ISGylation modulates the JAK-STAT signaling pathway. Genes Dev.
17:455–560. 2003.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Malakhova OA, Kim KI, Luo J-K, Zou W,
Kumar KGS, Fuchs SY, Shuai K and Zhang DE: UBP43 is a novel
regulator of interferon signaling independent of its ISG15
isopeptidase activity. EMBO J. 25:2358–2367. 2006.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Arimoto KI, Miyauchi S, Troutman TD, Zhang
Y, Liu M, Stoner SA, Davis AG, Fan JB, Huang YJ, Yan M, et al:
Expansion of interferon inducible gene pool via USP18 inhibition
promotes cancer cell pyroptosis. Nat Commun. 14(251)2023.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Cai X, Feng S, Zhang J, Qiu W, Qian M and
Wang Y: USP18 deubiquitinates and stabilizes Twist1 to promote
epithelial-mesenchymal transition in glioblastoma cells. Am J
Cancer Res. 10:1156–1169. 2020.PubMed/NCBI
|
|
16
|
Vivian J, Rao AA, Nothaft FA, Ketchum C,
Armstrong J, Novak A, Pfeil J, Narkizian J, Deran AD,
Musselman-Brown A, et al: Toil enables reproducible, open source,
big biomedical data analyses. Nat Biotechnol. 35:314–316.
2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Charoentong P, Finotello F, Angelova M,
Mayer C, Efremova M, Rieder D, Hackl H and Trajanoski Z: Pan-cancer
immunogenomic analyses reveal genotype-immunophenotype
relationships and predictors of response to checkpoint blockade.
Cell Rep. 18:248–262. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ceccarelli M, Barthel FP, Malta TM,
Sabedot TS, Salama SR, Murray BA, Morozova O, Newton Y, Radenbaugh
A, Pagnotta SM, et al: Molecular profiling reveals biologically
discrete subsets and pathways of progression in diffuse glioma.
Cell. 164:550–563. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wang L, Babikir H, Müller S, Yagnik G,
Shamardani K, Catalan F, Kohanbash G, Alvarado B, Di Lullo E,
Kriegstein A, et al: The phenotypes of proliferating glioblastoma
cells reside on a single axis of variation. Cancer Discov.
9:1708–1719. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Sun D, Wang J, Han Y, Dong X, Ge J, Zheng
R, Shi X, Wang B, Li Z, Ren P, et al: TISCH: A comprehensive web
resource enabling interactive single-cell transcriptome
visualization of tumor microenvironment. Nucleic Acids Res. 49
(D1):D1420–D1430. 2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Butler A, Hoffman P, Smibert P, Papalexi E
and Satija R: Integrating single-cell transcriptomic data across
different conditions, technologies, and species. Nat Biotechnol.
36:411–420. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Liu J, Lichtenberg T, Hoadley KA, Poisson
LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee
AV, et al: An integrated TCGA pan-cancer clinical data resource to
drive high-quality survival outcome analytics. Cell.
173:400–416.e11. 2018.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14(7)2013.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Bindea G, Mlecnik B, Tosolini M,
Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T,
Lafontaine L, Berger A, et al: Spatiotemporal dynamics of
intratumoral immune cells reveal the immune landscape in human
cancer. Immunity. 39:782–795. 2013.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Yoshihara K, Shahmoradgoli M, Martínez E,
Vegesna R, Kim H, Torres-Garcia W, Treviño V, Shen H, Laird PW,
Levine DA, et al: Inferring tumour purity and stromal and immune
cell admixture from expression data. Nat Commun.
4(2612)2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Gene Ontology Consortium. Gene ontology
consortium: Going forward. Nucleic Acids Res. 43 (Database
Issue):D1049–D1056. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Li R, Wang YY, Wang SL, Li XP, Chen Y, Li
ZA, He JH, Zhou ZH, Li JY, Guo XL, et al: GBP2 as a potential
prognostic predictor with immune-related characteristics in glioma.
Front Genet. 13(956632)2022.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ren Y, Yang B, Guo G, Zhang J, Sun Y, Liu
D, Guo S, Wu Y, Wang X, Wang S, et al: GBP2 facilitates the
progression of glioma via regulation of KIF22/EGFR signaling. Cell
Death Discov. 8(208)2022.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Sun Q, Li J, Wang R, Sun T, Zong Y, Wang
C, Liu Y, Li X, Song Y and Zhang Y: Coxsackievirus A6 infection
causes neurogenic pathogenesis in a neonatal murine model. Viruses
15: 511, 223.
|
|
34
|
Selinger M, Věchtová P, Tykalová H,
Ošlejšková P, Rumlová M, Štěrba J and Grubhoffer L: Integrative RNA
profiling of TBEV-infected neurons and astrocytes reveals potential
pathogenic effectors. Comput Struct Biotechnol J. 20:2759–2777.
2022.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Ritchie KJ, Hahn CS, Kim KI, Yan M,
Rosario D, Li L, de la Torre JC and Zhang DE: Role of ISG15
protease UBP43 (USP18) in innate immunity to viral infection. Nat
Med. 10:1374–1378. 2004.PubMed/NCBI View
Article : Google Scholar
|
|
36
|
Hong B, Li H, Lu Y, Zhang M, Zheng Y, Qian
J and Yi Q: USP18 is crucial for IFN-γ-mediated inhibition of B16
melanoma tumorigenesis and antitumor immunity. Mol Cancer.
13(132)2014.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Mustachio LM, Kawakami M, Lu Y,
Rodriguez-Canales J, Mino B, Behrens C, Wistuba I, Bota-Rabassedas
N, Yu J, Lee JJ, et al: The ISG15-specific protease USP18 regulates
stability of PTEN. Oncotarget. 8:3–14. 2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Tan Y, Zhou G, Wang X, Chen W and Gao H:
USP18 promotes breast cancer growth by upregulating EGFR and
activating the AKT/Skp2 pathway. Int J Oncol. 53:371–383.
2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Tong HV, Hoan NX, Binh MT, Quyen DT, Meyer
CG, Hang DTT, Hang DTD, Son HA, Van Luong H, Thuan ND, et al:
Upregulation of Enzymes involved in ISGylation and ubiquitination
in patients with hepatocellular carcinoma. Int J Med Sci.
17:347–353. 2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Zhang X, Bogunovic D, Payelle-Brogard B,
Francois-Newton V, Speer SD, Yuan C, Volpi S, Li Z, Sanal O,
Mansouri D, et al: Human intracellular ISG15 prevents
interferon-α/β over-amplification and auto-inflammation. Nature.
517:89–93. 2015.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Liu X, Li H, Zhong B, Blonska M,
Gorjestani S, Yan M, Tian Q, Zhang DE, Lin X and Dong C: USP18
inhibits NF-κB and NFAT activation during Th17 differentiation by
deubiquitinating the TAK1-TAB1 complex. J Exp Med. 210:1575–1590.
2013.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Zheng C, Zheng Z, Zhang Z, Meng J, Liu Y,
Ke X, Hu Q and Wang H: IFIT5 positively regulates NF-κB signaling
through synergizing the recruitment of IκB kinase (IKK) to
TGF-β-activated kinase 1 (TAK1). Cell Signal. 27:2343–2354.
2015.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Numajiri Haruki A, Naito T, Nishie T,
Saito S and Nagata K: Interferon-inducible antiviral protein MxA
enhances cell death triggered by endoplasmic reticulum stress. J
Interferon Cytokine Res. 31:847–856. 2011.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Arimoto KI, Löchte S, Stoner SA, Burkart
C, Zhang Y, Miyauchi S, Wilmes S, Fan JB, Heinisch JJ, Li Z, et al:
STAT2 is an essential adaptor in USP18-mediated suppression of type
I interferon signaling. Nat Struct Mol Biol. 24:279–289.
2017.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Molinaro AM, Taylor JW, Wiencke JK and
Wrensch MR: Genetic and molecular epidemiology of adult diffuse
glioma. Nat Rev Neurol. 15:405–417. 2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Nicholson JG and Fine HA: Diffuse glioma
heterogeneity and its therapeutic implications. Cancer Discov.
11:575–590. 2021.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Hernández A, Domènech M, Muñoz-Mármol AM,
Carrato C and Balana C: Glioblastoma: Relationship between
metabolism and immunosuppressive microenvironment. Cells.
10(3529)2021.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Louveau A, Smirnov I, Keyes TJ, Eccles JD,
Rouhani SJ, Peske JD, Derecki NC, Castle D, Mandell JW, Lee KS, et
al: Structural and functional features of central nervous system
lymphatic vessels. Nature. 523:337–341. 2015.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Quail DF and Joyce JA: The
microenvironmental landscape of brain tumors. Cancer Cell.
31:326–341. 2017.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Sprooten J, Agostinis P and Garg AD: Type
I interferons and dendritic cells in cancer immunotherapy. Int Rev
Cell Mol Biol. 348:217–262. 2019.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Pyfferoen L, Brabants E, Everaert C, De
Cabooter N, Heyns K, Deswarte K, Vanheerswynghels M, De Prijck S,
Waegemans G, Dullaers M, et al: The transcriptome of lung
tumor-infiltrating dendritic cells reveals a tumor-supporting
phenotype and a microRNA signature with negative impact on clinical
outcome. Oncoimmunology. 6(e1253655)2016.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Chen Z, Feng X, Herting CJ, Garcia VA, Nie
K, Pong WW, Rasmussen R, Dwivedi B, Seby S, Wolf SA, et al:
Cellular and molecular identity of tumor-associated macrophages in
glioblastoma. Cancer Res. 77:2266–228. 2017.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Dapash M, Hou D, Castro B, Lee-Chang C and
Lesniak MS: The interplay between glioblastoma and its
microenvironment. Cells. 10(2257)2021.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Wu L and Zhang XHF: Tumor-associated
neutrophils and macrophages-heterogenous but not chaotic. Front
Immunol. 11(553967)2020.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Broekman ML, Maas SLN, Abels ER, Mempel
TR, Krichevsky AM and Breakefield XO: Multidimensional
communication in the microenvirons of glioblastoma. Nat Rev Neurol.
14:482–495. 2018.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Blanchett S, Boal-Carvalho I, Layzell S
and Seddon B: NF-κB and extrinsic cell death pathways-entwined
do-or-die decisions for T cells. Trends Immunol. 42:76–88.
2021.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Kim CH: FOXP3 and its role in the immune
system. Adv Exp Med Biol. 665:17–29. 2009.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Lu L, Barbi J and Pan F: The regulation of
immune tolerance by FOXP3. Nat Rev Immunol. 17:703–717.
2017.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Yang Z, Xian H, Hu J, Tian S, Qin Y, Wang
RF and Cui J: USP18 negatively regulates NF-κB signaling by
targeting TAK1 and NEMO for deubiquitination through distinct
mechanisms. Sci Rep. 5(12738)2015.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Cui P, Wei F, Hou J, Su Y, Wang J and Wang
S: STAT3 inhibition induced temozolomide-resistant glioblastoma
apoptosis via triggering mitochondrial STAT3 translocation and
respiratory chain dysfunction. Cell Signal.
71(109598)2020.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Schneider WM, Chevillotte MD and Rice CM:
Interferon-stimulated genes: A complex web of host defenses. Annu
Rev Immunol. 32:513–545. 2014.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Bracci L, Sistigu A, Proietti E and
Moschella F: The added value of type I interferons to cytotoxic
treatments of cancer. Cytokine Growth Factor Rev. 36:89–97.
2017.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Parker BS, Rautela J and Hertzog PJ:
Antitumour actions of interferons: Implications for cancer therapy.
Nat Rev Cancer. 16:131–144. 2016.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Zitvogel L, Galluzzi L, Kepp O, Smyth MJ
and Kroemer G: Type I interferons in anticancer immunity. Nat Rev
Immunol. 15:405–414. 2015.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Furnari FB, Fenton T, Bachoo RM, Mukasa A,
Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al:
Malignant astrocytic glioma: Genetics, biology, and paths to
treatment. Genes Dev. 21:2683–2710. 2007.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Stewart LA: Chemotherapy in adult
high-grade glioma: A systematic review and meta-analysis of
individual patient data from 12 randomised trials. Lancet.
359:1011–1018. 2002.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Yung WK, Prados M, Levin VA, Fetell MR,
Bennett J, Mahaley MS, Salcman M and Etcubanas E: Intravenous
recombinant interferon beta in patients with recurrent malignant
gliomas: A phase I/II study. J Clin Oncol. 9:1945–1949.
1991.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Fine HA, Wen PY, Robertson M, O'Neill A,
Kowal J, Loeffler JS and Black PM: A phase I trial of a new
recombinant human beta-interferon (BG9015) for the treatment of
patients with recurrent gliomas. Clin Cancer Res. 3:381–387.
1997.PubMed/NCBI
|
|
69
|
Manini I, Sgorbissa A, Potu H, Tomasella A
and Brancolini C: The DeISGylase USP18 limits TRAIL-induced
apoptosis through the regulation of TRAIL levels: Cellular levels
of TRAIL influences responsiveness to TRAIL-induced apoptosis.
Cancer Biol Ther. 14:1158–1166. 2013.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Dimberg LY, Anderson CK, Camidge R,
Behbakht K, Thorburn A and Ford HL: On the TRAIL to successful
cancer therapy? Predicting and counteracting resistance against
TRAIL-based therapeutics. Oncogene. 32:1341–1350. 2013.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Potu H, Sgorbissa A and Brancolini C:
Identification of USP18 as an important regulator of the
susceptibility to IFN-alpha and drug-induced apoptosis. Cancer Res.
70:655–665. 2010.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Sgorbissa A, Tomasella A, Potu H, Manini I
and Brancolini C: Type I IFNs signaling and apoptosis resistance in
glioblastoma cells. Apoptosis. 16:1229–1244. 2011.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Zong H, Verhaak RGW and Canoll P: The
cellular origin for malignant glioma and prospects for clinical
advancements. Expert Rev Mol Diagn. 12:383–394. 2012.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Omuro A and DeAngelis LM: Glioblastoma and
other malignant gliomas: A clinical review. JAMA. 310:1842–1850.
2013.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Nieto MA, Huang RYJ, Jackson RA and Thiery
JP: EMT: 2016. Cell. 166:21–45. 2016.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Nieto MA: Epithelial-mesenchymal
transitions in development and disease: Old views and new
perspectives. Int J Dev Biol. 53:1541–1547. 2009.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Noronha C, Ribeiro AS, Taipa R, Castro DS,
Reis J, Faria C and Paredes J: Cadherin expression and EMT: A focus
on gliomas. Biomedicines. 9(1328)2021.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Li L, Yin Y, Zhang J, Wu X, Liu J, Chai J,
Yang Y, Li M, Jia Q and Liu Y: USP18 regulates the malignant
phenotypes of glioblastoma stem cells. Pathol Res Pract.
247(154572)2023.PubMed/NCBI View Article : Google Scholar
|