Introduction
Oligomeric proanthocyanidins (OPCs) are extensively
distributed in the plant kingdom and are present in high
concentrations in certain plant-based foods and beverages (1). Previous studies have suggested that
OPCs provide health benefits, such as a reduced risk of vascular
disease and certain types of cancer (2,3).
OPCs have also been shown to alleviate EtoH-induced liver steatosis
and injury in mice, while increasing superoxide dismutase (SOD)
activity (4). OPCs extracted from
apples have also been shown to exert antioxidant effects (5). However, the mechanisms through which
OPCs protect cells against oxidative stress require clarification.
All living organisms are constantly challenged by internal and
external oxidative stress, such as those from drugs, xenobiotics,
heavy metals and ionizing radiation, resulting in the increased
production of reactive oxygen species (ROS) (6). ROS include superoxide anion
(O2•−), hydrogen peroxide
(H2O2) and the extremely reactive hydroxyl
radical (OH−), with each species displaying a specific
range of action and distinct molecular targets (7,8).
ROS exert significant effects in cancer cells, such as stimulating
cellular growth, promoting mutations and inducing resistance to
anticancer agents (9). However, a
previous study reviewed the possible therapeutic effects of ROS,
suggesting that increased intrinsic ROS-induced stress may prove
beneficial as it may enhance the killing effect of therapeutic
drugs, as continuous ROS insults decrease the tolerance of cancer
cells (10). Others have reported
however, that ROS derived from circulating inflammatory cells
contribute to the induction of lung injury (11).
Nuclear factor erythroid 2-related factor 2 (Nrf2)
is commonly known to play a role in the transcriptional regulation
of genes encoding antioxidant proteins under stress conditions
(12,13). Target genes of Nrf2 are involved
in the glutathione synthesis, the elimination of ROS and xenobiotic
metabolism (14,15) and some of these enzymes include
SOD, glutathione peroxidase (GSH-Px), glutathione reductase,
catalase (CAT), heme oxygenase-1 (HO-1), NAD(P) H quinone
dehydrogenase 1 (NQO1) and thioredoxin reductase 1 (TXNRD1). Under
normal conditions, Nrf2 is constantly polyubiquitinated by the
Keap1-Cul3 E3 ligase and degraded by the 26S proteasome (16). In the presence of electrophiles or
ROS, the ability of the Keap1-Cul3 E3 ligase to target Nrf2 for
degradation becomes impaired, stabilized Nrf2 accumulates in the
nuclei, heterodimerizes with small Maf proteins and activates
target genes for cytoprotection through the antioxidant response
element (ARE) with a consensus sequence 5′-TGACNNNGC-3′ (17,18). The increased expression of Nrf2
target genes and the increased stability of Nrf2 caused by somatic
mutations in Nrf2 and Keap1 are well documented in human cancer
(19). These data indicate that
the Nrf2-ARE pathway plays an important role against oxidative
stress in cancer cells.
In this sudy, we investigated the anti-oxidative
potential of OPCs against H2O2-induced
oxidative stress in A549 non-small cell lung cancer cells. OPCs
prevent oxidative stress by the accumulation of Nrf2 protein in
A549 cells. Further research will be crucial in determining how
this inhibitor mediates lung protection and its use as a clinical
approach in lung disease.
Materials and methods
Chemicals and reagents
OPC (purity ≥98%) was purchased from Zelang
Biological Technology Co., Ltd. (Nanjing, China).
H2O2 was obtained from Sigma-Aldrich (St.
Louis, MO, USA). RPMI-1640 medium and fetal bovine serum (FBS) were
obtained from Gibco Industries, Inc. (Grand Island, NY, USA).
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT),
trypsin, L-glutamate, MK-801 and dimethyl sulfoxide (DMSO) were
purchased from Sigma-Aldrich. The DCFH-DA ROS assay kit was
purchased from the Beyotime Institute of Biotechnology (Jiangsu,
China). GSH-Px, CAT and SOD assay kits were procured from Nanjing
Jiancheng Bioengineering Institute (Jiangsu, China). The
bicinchoninic acid (BCA) protein reagent was from the Beyotime
Institute of Biotechnology Co. Ltd., Shanghai, China).
Cell culture and treatment
For this study, A549 cells were obtained from the
cell bank of the Chinese Academy of Science (Shanghai, China). The
A549 cells were cultured in RPMI-1640 medium supplemented with 10%
FBS 2 mM L-glutamine, penicillin (100 U/ml) and streptomycin (100
mg/ml) (Wako, Osaka, Japan). The cells were cultured in a
humidified incubator containing 5% CO2 and 95% air at
37°C. The cells were divided into 3 groups based on the treatments
as follows: the control group (cells treated with culture medium),
H2O2 group (cells exposed to
H2O2 for 12 h at a final concentration of 200
µM) and the H2O2 + group (cells
pre-treated with 50 mg/l OPC for 5 h and then exposed to 200
µM H2O2 for 12 h). The cells were
subjected to the different treatments and were then subjected to
serial analyses, including flow cytometry (FCM), MTT, RT-qPCR and
western blot analysis.
Assessment of the viable number of
cells
Cell viability was performed using the MTT
colorimetric assay. Following cell treatment, the medium was
removed, and the cells were incubated with 20 µl of 5 mg/ml
MTT solution for 4 h at 37°C. The dark blue formazan was dissolved
with 150 µl of DMSO, and the absorbance was measured at 570
nm using a microplate reader (Multiskan MK3, Thermo Fisher
Scientific, Waltham, MA, USA). Cell viability was expressed as a
percentage of the untreated controls.
Western blot analysis
The cells were lysed in RIPA buffer (150 mM NaCl, 1%
Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH
8.0) supplemented with a protease and phosphatase inhibitor
cocktail on ice for 15 min, sonicated and centrifuged at 13,000 × g
for 15 min. The protein content of the supernatants was measured by
BCA reagent (Pierce, Rockford, IL, USA). For the examination of
Nrf2 expression, nuclear proteins and cytoplasmic proteins were
separated using a Bioepitope Nuclear and Cytoplasmic Extraction kit
(Bioworld Technology, St. Louis Park, MN, USA) according to the
manufacturer's instructions. Cellular proteins were separated by
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE) and transferred onto PVDF membranes. The membranes were
probed with the following primary antibodies (Abs): anti-TXNRD1
(ab16847; 1:1,000; Abcam, Cambridge, MA, USA); anti-Nrf2 (#12721;
1:1,000; Cell Singaling Technology, Danvers, MA, USA); anti-tubulin
(ab7291; 1:5,000); anti-HO-1 (ab13248; 1:1,000); lamin B (ab8980;
1:1,000) and anti-NQO1 (ab34173; 1:1,000) (all from Abcam,
Cambridge, MA, USA). This was followed by detection using
HRP-conjugated secondary antibodies to rabbit IgG (GE Healthcare,
Piscataway, NJ, USA).
RT-qPCR
Total RNA was extracted from the A549 cells using
TRIzol reagent and then reverse transcribed into cDNA using the
QuantScript reverse transcription kit (Tiangen, Beijing, China)
according to the manufacturer's instructions. Amplification was
conducted using a SYBR-Green I PCR kit (Roche, Indianapolis, IN,
USA). The PCR reaction conditions were as follows: initial
denaturation at 95°C for 10 min, followed by 35 amplification
cycles of 95°C for 10 sec, 55°C for 10 sec, 72°C for 15 sec, and a
final extension at 72°C for 10 min. β-actin was used as an internal
reference gene. Relative values for mRNA levels were calculated
using the following formula: fold change = 2−ΔΔCq. The
primers used for each gene are listed in Table I.
 | Table ISequence of the primers used in
RT-qPCR. |
Table I
Sequence of the primers used in
RT-qPCR.
| Gene | Forward primer
(5′→3′) | Reverse primer
(5′→3′) |
|---|
| Nrf2 |
GAGACAGGTGAATTTCTCCCAAT |
GGGAATGTGGGCAACCTGGG |
| HO-1 |
CAGGCAGAGAATGCTGATTC |
GCTCTTCTGGGAAGTAGACAGG |
| NQO1 |
AAGAAAGGATGGGAGGTGGT |
GCTTCTTTTGTTCAGCCACA |
| TXNRD1 |
GGAACTAGATGGGGTCTCGG |
TCTTGCAGGGCTTGTCCTAA |
| β-actin |
GATCATTGCTCCTCCTGAGC |
ACTCCGCTTGCTGATCCAC |
Intracellular ROS staining
The cells were incubated with 300 nM
dichloro-dihydro-fluorescein diacetate (DCF-DA) (Sigma-Aldrich) for
10 min at 37°C. Following 2 washes in phosphate-buffered saline
(PBS)-1X, DCF-DA fluorescence was analyzed using a flow cytometer
(BD Biosciences, Franklin Lakes, NJ, USA) and FlowJo software.
Measurement of intracellular GSH-Px, CAT
and SOD levels
The A549 cells were plated in 6-well plates at a
density of 10×104 cells/ml in 2.5 ml of culture medium.
After the cells were treated in accordance with the experimental
design, the cells were then harvested, disrupted ultrasonically on
ice and centrifuged at 2,500 × g for 10 min at 4°C. The
supernatants were collected and stored −20°C for subsequent
analysis. The levels of SOD, GSH-Px and CAT were detected using a
Wallac 1420 microplate spectrophotometer (Perkin Elmer, Waltham,
MA, USA) using commercially available assay kits (Jiancheng
Bioengineering Institute) following the manufacturers'
instructions.
Determination of the activity of
malondialdehyde (MDA)
A reaction mixture containing 0.2 ml cell lysate and
4.2 ml reaction buffer was treated by ultrasound for a certain time
using a 22 kHz ultrasound generator (HN-1000Y; Shanghai Hanno
Instrument Corp., Shanghai, China) equipped with a tapered horn tip
(10-mm end diameter) and boiled for 10 min in water bath, then
cooled down and centrifuged at 4,000 rpm for 10 min. the
supernatants were measured using a kit purchased from the Nanjing
Jiancheng Bioengineering Institute according to the thiobarbituric
acid method, which is based on the fact that MDA reacts with
thiobarbituric acid to form thiobarbituric acid reactive substances
(TBARS) with a maximum absorbance at 530 nm. The experiments were
performed in triplicate.
Knockout of the Nrf2 gene using the
CRISPR/Cas9 mediated genome
Target sequences for CRISPR interference (20) were designed at CRISPR direct
(http://crispr.dbcls.jp/). The target sequences
for human Nrf2 were TAGTTCATGAGCGTGA TGAT (exon 3) and
TGCCATAATTGTTACACATT (exon 4). Two oligonucleotides with
Bbs1 restiction sites for guide RNAs (gRNAs) were
synthesized by Shenggong (Shanghai, China) and cloned into the
PX330 CRISPR/Cas9 vector (Addgene, Cambridge, MA, USA). The cells
were transfected the two PX330 guides and PMT-puro by using PolyJet
reagent (Invitrogen, Carlsbad, CA, USA). The cells were selected
with 1.5 µg/ml puromycin and stable clones were maintained
in 0.5 µg/ml puromycin. The allele-specific primers were
designed as follows: forward, GATTCTTGTGAAGCAGTCCAGC and reverse,
ATCCAGTGACTTCTTGAATGCTT. Lastly, the potential positive clones from
PCR results were selected and this was followed by electrophoresis
on a 1% agrose gel. The positive clones will loss about 1000 bp DNA
fragment.
DNA gel electrophoresis
First, the cells were spinned down in the PCR plate,
and the supernatant was discard, leaving 10 µl medium. The
cells were then lysed using single cell lysis buffer (1 mM EDTA,
10% Tween-20) and then boiled at 65 °C for 2 h; 95 °C for 30 min.
PCR was then performed with cell lysis buffer and allele-specific
primers followed by electrophoresis on a 1% agrose gel. The DNA gel
was scanned using a gel imaging system (Tannon 3500R; Tanon Science
& Technology Co., Ltd., Shanghai, China).
Luciferase assay
The NQO1-ARE promoter was cloned into pGL3-Basic
(Promega, Madison, WI, USA) vector by standard methods. For the
luciferase assay, the A549 cells were transiently transfected with
the luciferase construct and β-galactosidase (as an internal
control). The cells were transfected using PolyJet reagent
(Invitrogen) according to the manufacturer's instructions.
Luciferase activity was determined using a luminometer. The medium
was removed from the cells, and the cells were then lysed in 100
µl of extraction buffer (100 mM KPO4, pH 7.4, 4.0
mM ATP, 1.5 mM MgSO4, 1.0 mM dithiothreitol and 0.1%
Triton X-100). Cell lysates (25 µl) were added to 350
µl of luciferase assay buffer (100 mM KPO4, pH
7.4, 4.0 mM ATP, and 1.5 mM MgSO4) and then 100
µl of 1.0 mM D-Luciferin was injected into each culture tube
using a luminometer that integrated the luminescence over 10 sec.
β-galactosidase activity was measured using
o-nitrophenyl-β-D-galactopyranoside (ONPG; Sigma-Aldrich) as
a substrate. Briefly, the cell lysates were incubated with ONPG
(0.4 mg/ml) in reaction buffer (0.1 M sodium phosphate pH 7.5, 10
mM KCl, 1 mM MgCl2) for 1 h and the absorbance was then
measured at 405 nm. The data are expressed as a ratio of luciferase
to β-galactosidase activity.
Statistical analysis
All data are presented as the means ± standard
deviation (SD). Statistical analysis was carried out using one-way
ANOVA followed by the Scheffe test using SPSS 17.0 software (SPSS
Inc., Chicago, IL, USA). Statistical significance was set at
p<0.05.
Results
OPC protects against cell death and
attenuates oxidative stress in A549 cells exposed to
H2O2
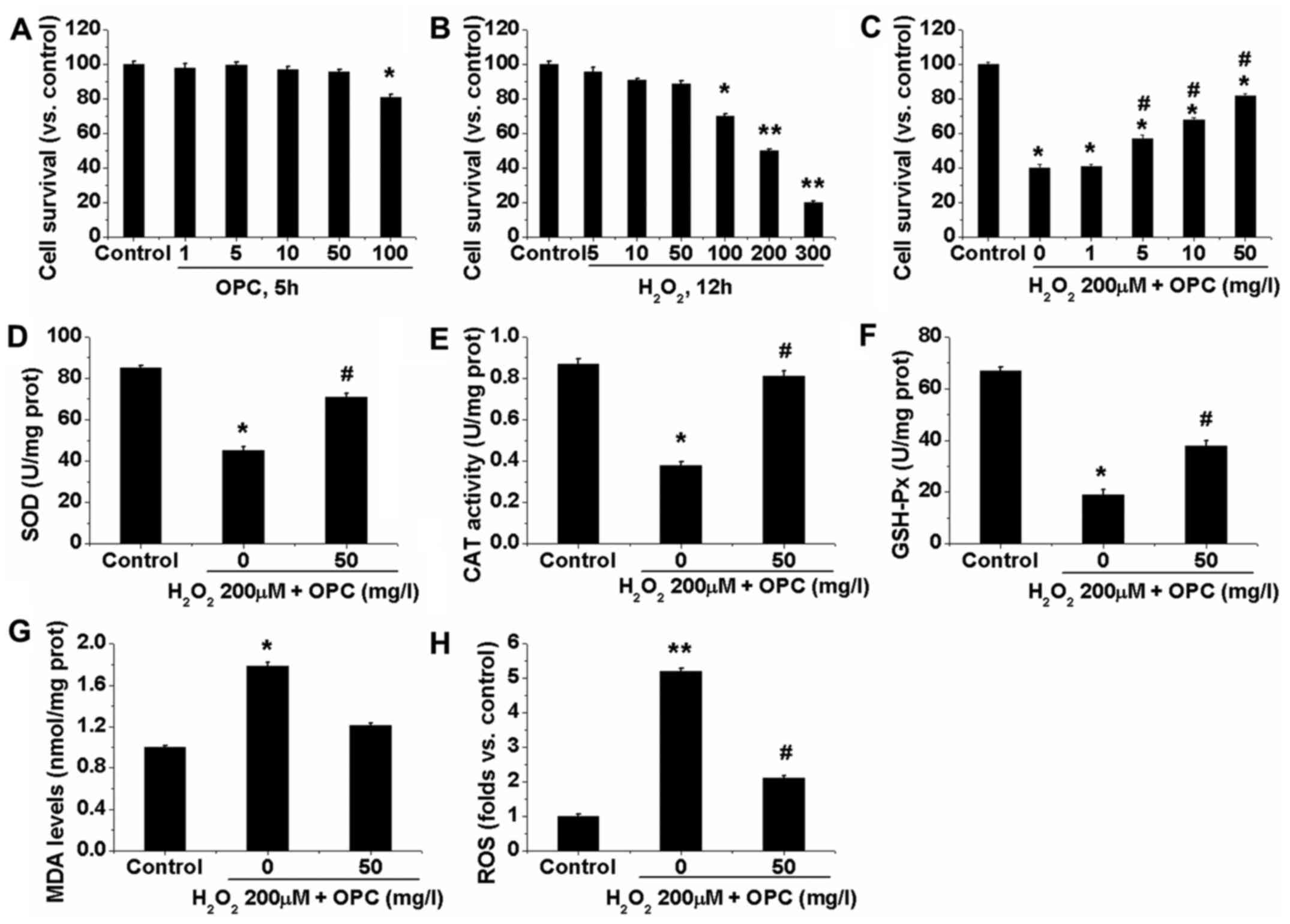
We first examined the potential effects of OPC in
A549 cells. The resutls of MTT assay for cell survival shown in
Fig. 1A demonstrated that the
viability of the A549 cells was not significantly decreased when
the cells were treated with OPC at the concentration of ≤50 mg/l.
However, at the concentration of ≥100 mg/l OPC, A549 cell viability
was slightly, but significantly decreased (Fig. 1A). Subsequently,
H2O2-induced oxidative damage was
investigated in the A549 cells by MTT assay. The viability of the
A549 cells decreased following exposure to
H2O2 in a dose-dependent manner. Following 12
h of exposure to 100 µM H2O2, cell
viability decreased to 70% of the control, whereas 200 µM
decreased cell viability to 50% and this concentration was utilized
for further experiments (Fig.
1B). Significantly, treatment with OPC at 1–50 mg/l
(non-cytotoxic concentrations) markedly attenuated the
H2O2-induced decrease in A549 cell viability
(Fig. 1C). We then examined the
effects of OPC on the activities of a series of phase II
metabolizing enzymes and antioxidants. The results demonstrated
that the enzyme activities of SOD, CAT and GSH-Px were
significantly decreased in the H2O2-treated
A549 cells, and that OPC at 50 mg/l enhanced their activities
(Fig. 1D–F). We also determined
the contents of ROS and MDA within the A549 cells. We found that
the levels of ROS and MDA were significantly elevated in the
H2O2-exposed cells. Similarly, OPC at 50 mg/l
significantly decreased the contents of ROS and MDA (Fig. 1G and H). These results indicated
that OPC exerted potent antioxidant effects on
H2O2-exposed A549 cells.
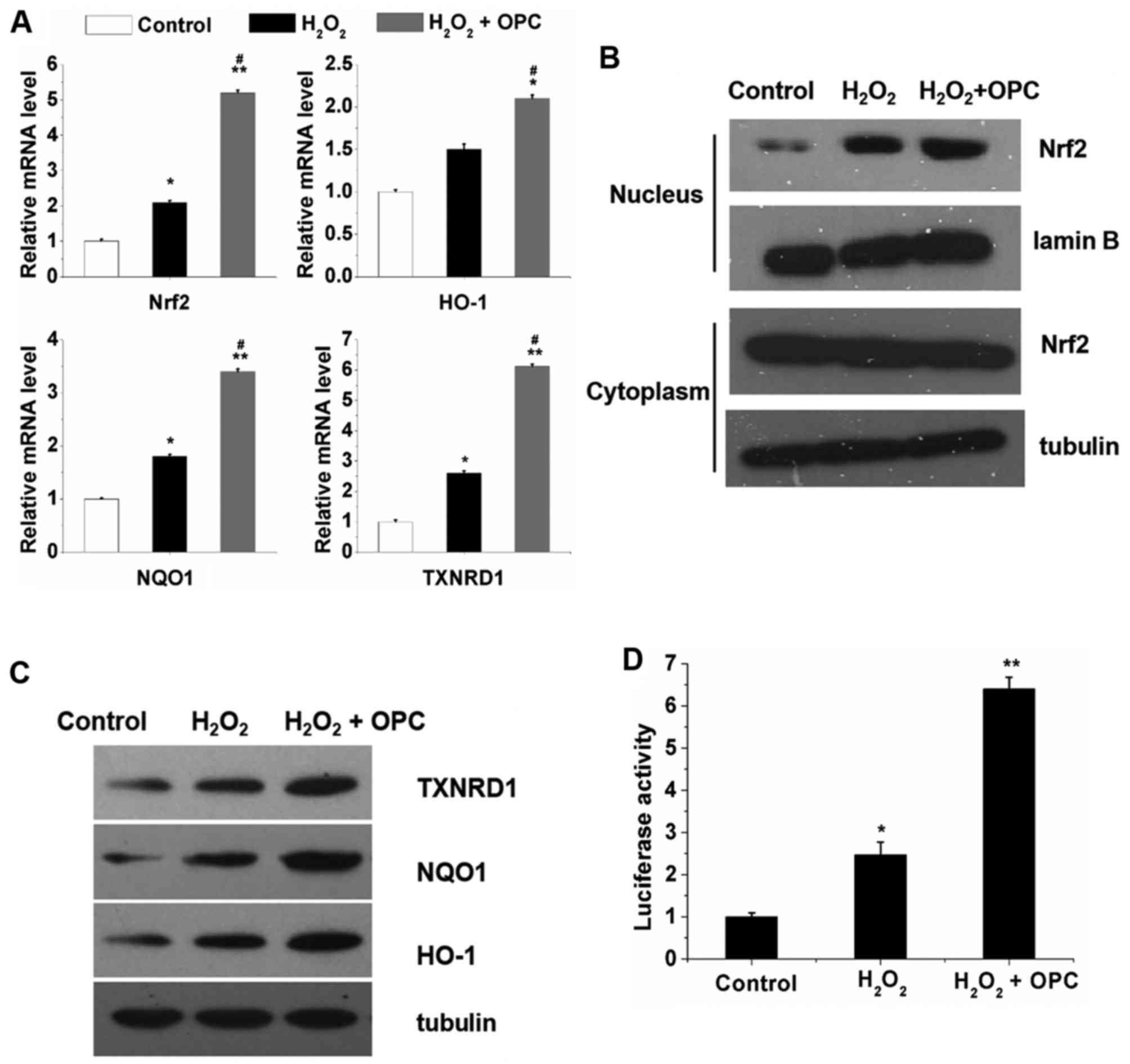
OPC increases the transcriptional
activity of Nrf2 and the Nrf2-dependent antioxidant response of
A549 cells exposed to H2O2
In order to elucidate the potential mechanisms
responsible for the protective effects of OPC against
H2O2-induced oxidative stress, the A549 cells
were exposed to H2O2 for 12 h following
treatment or not with OPC. We then examined the mRNA and protein
expression of Nrf2 and Nrf2 target genes. We found that
H2O2 increased the mRNA expression of Nrf2
and Nrf2 target genes and this increase was further enhanced by OPC
(Fig. 2A). Furthermore, the
results of western blot analyses revealed that the nuclear protein
expression of Nrf2 was significantly increased by
H2O2 and OPC; however, the cytoplasmic
protein expression of Nrf2 was not altered by
H2O2 and OPC (Fig. 2B). As regards HO-1, NQO1 and
TXNRD1, their protein expression was significantly increased by OPC
(Fig. 2C). Subsequently, to
investigate the effects of OPC on Nrf2 transcriptional activity
under conditions of H2O2- induced oxidative
stress, the enzymatic activity of NQO1 was also examined. As
observed with the mRNA expression of NQO1, which increased
following treatment with OPC in the A549 cells, the enzymatic
activity of NQO1 was also increased in the A549 cells following
treatment with OPC (Fig. 2D).
Taken together, these results indicate that OPC promotes the
nuclear translocation of Nrf2, leading to the enhanced expression
of its target genes implicated in the protective effects of OPC
against H2O2-induced oxidative stress in A549
cells.
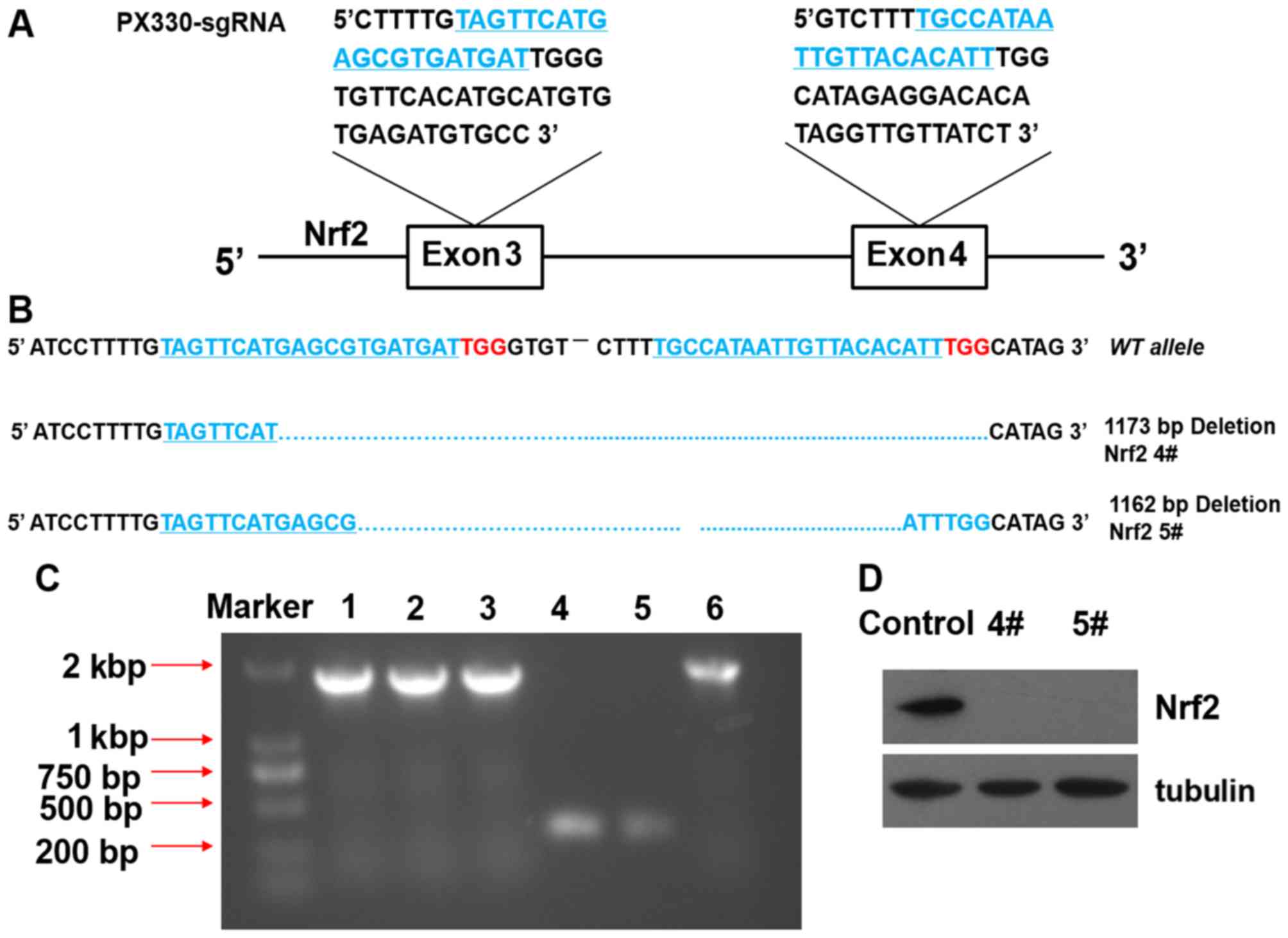
Effects of the knockout of Nrf2 in A549
using the CRISPR/Cas9 system
As Nrf2 was above shown to participate in the
protective effects of OPC against oxidative stress, we knocked out
Nrf2 in the A549 cells in order to further explore the overall
influence of the process. We used the CRISPR/Cas9 system that has
been reported to efficiently disrupt genes in cells (21). As a result, we obtained 2 Nrf2
knockout A549 clones, 4# and 5#. The DNA sequences of the 2 clones
revealed a deletion of the 1173 and 1162 bases in both alleles
(Fig. 3A and B). DNA gel-image
indicated that Nrf2 deletion >1,000 bases between exon 3 and
exon 4 (Fig. 3C). Western blot
analysis revealed that Nrf2 was completely knocked out in the 2
clone cells by the absence of Nrf2 expression (Fig. 3D).
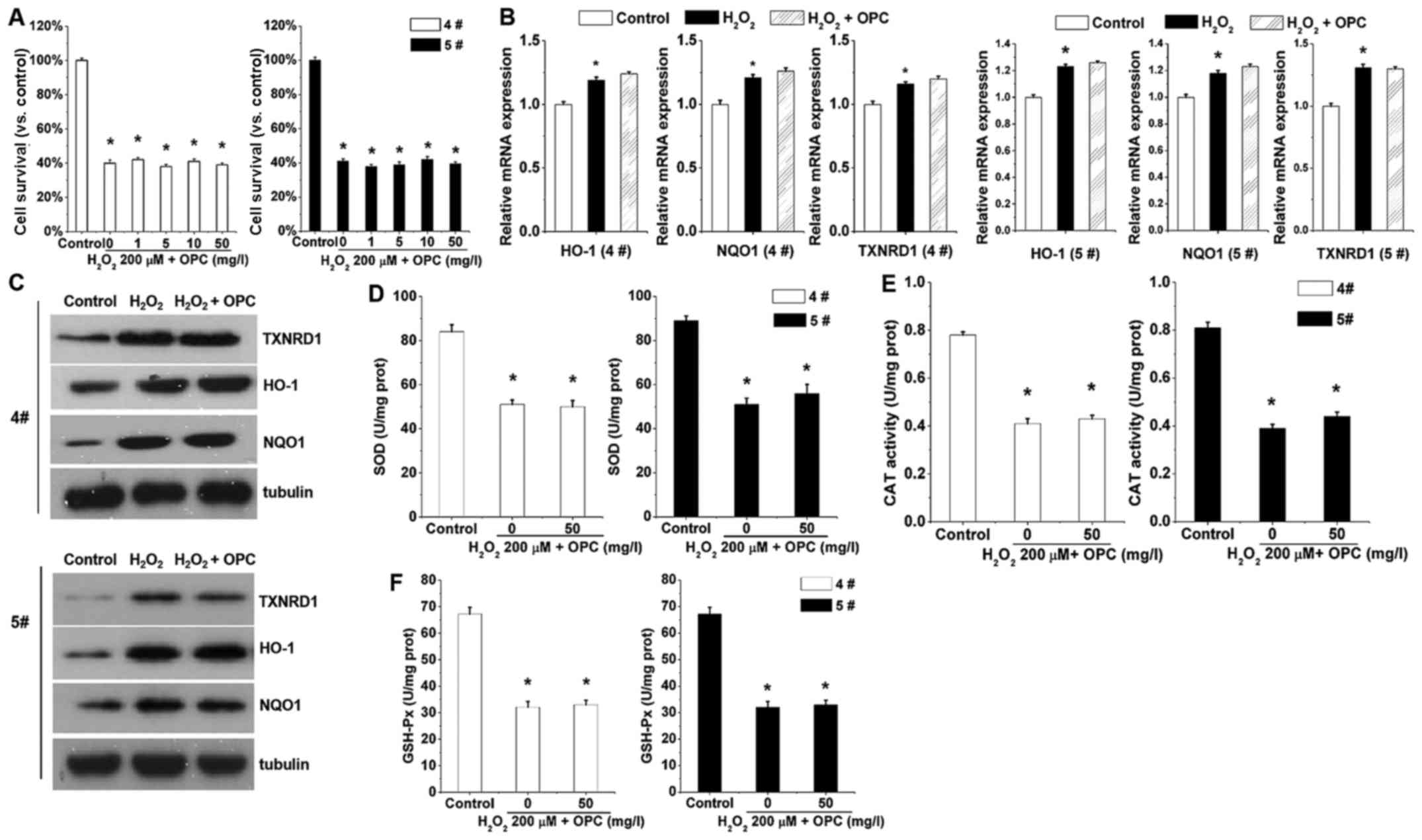
Nrf2 mediates
H2O2-induced antioxidant activity
We then characterized the function of OPC in A549
cells in which Nrf2 had been knocked out. Note that in the A549
cells in which Nrf2 was knocked out,
H2O2-induced cell death was exacerbated
(Fig. 4A), indicating that OPC
exerts its protective effects via basal Nrf2. The results shown in
Fig. 2A and C demonstrated that
the mRNA and protein expression levels of HO-1, NQO1 and TXNRD1
were significantly increased following treatment with OPC. However,
the antioxidant activity was almost abolished with Nrf2 knockout
(Fig. 4D–F). OPC was almost
ineffective against H2O2-mediated SOD, CAT
and GSH-Px activity when Nrf2 was knocked out (Fig. 4D–F). This finding suggested that
OPC protected the A549 cells against
H2O2-induced oxidative stress via the
Nrf2-ARE pathway.
Discussion
The increased production of ROS (22,23) is frequently observed in cancer
cells compared to normal cells, and this may be selectively used to
promote tumor cell death. Cellular exposure to xenobiotics, drugs,
heavy metals and ionizing radiation is able to generate ROS that
leads to oxidative stress and has a profound impact on the survival
and evolution of all living organisms (24). OPC, as a natural compound, has
high bioactivity and provides significantly greater protection
against free radicals, and free radical-induced lipid peroxidation
and DNA damage than vitamins C and E, and β-carotene both in in
vitro and in vivo models (25). In this study, we induced oxidative
stress in A549 cells using H2O2 to uncover
the underlying mechanisms of H2O2-induced
oxidative toxicity. Cells have internal antioxidant defense
enzymes, such as GSH-Px, CAT and SOD, which are critical protective
measures against oxidative stress-related disorders. It has been
previously suggested that antioxidants protect cells against
oxidative damage by reducing lipid peroxidation products and
elevating the levels of GSH-Px and SOD activities (26). In this study, the activities of
these antioxidant defense enzymes were observed to be significantly
reduced in A549 cells following exposure to
H2O2, and pre-treatment of the cells with OPC
for 5 h significantly attenuated the effects of
H2O2 on these enzymes, thereby boosting the
protective effects of the enzymes for the cells (Fig. 1D–F).
Nrf2 is a transcription factor that is activated by
increased ROS production, which induces the transcription of
several antioxidant and detoxification enzymes, including HO-1,
NQO1 and TXNRD1 (27). In this
study, we present our findings that OPC is able to upregulate the
Nrf2 signaling pathway under conditions of
H2O2-induced oxidatve stress by enhancing the
activation of ARE. Based on the molecular data obtained, we used
CRISPR/gRNA technology to knockout Nrf2 and demonstrated that OPC
is able to upregulate the Nrf2-dependent antioxidant response under
conditions of H2O2-induced oxidative stress.
Previous studies have shown that an advantage of the constitutive
activation of Nrf2 for cancer cells is the enhancement of
proliferation. This enhanced proliferation enhances the ability of
the cancer cells to detoxify the elevated levels of ROS associated
with constant division and growth (28,29).
In conclusion, the results obtained in this study
demonstrate that the protective effects of OPC against
H2O2-induced oxidative stress are dependent
on the enhanced activity of Nrf2, which facilitates cells viability
even in a highly oxidative environment.
References
|
1
|
Tatsuno T, Jinno M, Arima Y, Kawabata T,
Hasegawa T, Yahagi N, Takano F and Ohta T: Anti-inflammatory and
anti-melanogenic proanthocyanidin oligomers from peanut skin. Biol
Pharm Bull. 35:909–916. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
García-Conesa MT, Tribolo S, Guyot S,
Tomás-Barberán FA and Kroon PA: Oligomeric procyanidins inhibit
cell migration and modulate the expression of migration and
proliferation associated genes in human umbilical vascular
endothelial cells. Mol Nutr Food Res. 53:266–276. 2009. View Article : Google Scholar
|
|
3
|
Ray SD, Parikh H and Bagchi D:
Proanthocyanidin exposure to B6C3F1 mice significantly attenuates
dimethylnitrosamine-induced liver tumor induction and mortality by
differentially modulating programmed and unprogrammed cell deaths.
Mutat Res. 579:81–106. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang Z, Su B, Fan S, Fei H and Zhao W:
Protective effect of oligomeric proanthocyanidins against
alcohol-induced liver steatosis and injury in mice. Biochem Biophys
Res Commun. 458:757–762. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Eberhardt MV, Lee CY and Liu RH:
Antioxidant activity of fresh apples. Nature. 405:903–904.
2000.PubMed/NCBI
|
|
6
|
Forkink M, Basit F, Teixeira J, Swarts HG,
Koopman WJ and Willems PH: Complex I and complex III inhibition
specifically increase cytosolic hydrogen peroxide levels without
inducing oxidative stress in HEK293 cells. Redox Biol. 6:607–616.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Collins Y, Chouchani ET, James AM, Menger
KE, Cochemé HM and Murphy MP: Mitochondrial redox signalling at a
glance. J Cell Sci. 125:801–806. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
D'Autréaux B and Toledano MB: ROS as
signalling molecules: Mechanisms that generate specificity in ROS
homeostasis. Nat Rev Mol Cell Biol. 8:813–824. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zubovych IO, Straud S and Roth MG:
Mitochondrial dysfunction confers resistance to multiple drugs in
Caenorhabditis elegans. Mol Biol Cell. 21:956–969. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pelicano H, Carney D and Huang P: ROS
stress in cancer cells and therapeutic implications. Drug Resist
Updat. 7:97–110. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shasby DM, Vanbenthuysen KM, Tate RM,
Shasby SS, McMurtry I and Repine JE: Granulocytes mediate acute
edematous lung injury in rabbits and in isolated rabbit lungs
perfused with phorbol myristate acetate: Role of oxygen radicals.
Am Rev Respir Dis. 125:443–447. 1982.PubMed/NCBI
|
|
12
|
Kobayashi A, Ohta T and Yamamoto M: Unique
function of the Nrf2-Keap1 pathway in the inducible expression of
antioxidant and detoxifying enzymes. Methods Enzymol. 378:273–286.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Itoh K, Ishii T, Wakabayashi N and
Yamamoto M: Regulatory mechanisms of cellular response to oxidative
stress. Free Radic Res. 31:319–324. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Okawa H, Motohashi H, Kobayashi A,
Aburatani H, Kensler TW and Yamamoto M: Hepatocyte-specific
deletion of the keap1 gene activates Nrf2 and confers potent
resistance against acute drug toxicity. Biochem Biophys Res Commun.
339:79–88. 2006. View Article : Google Scholar
|
|
15
|
Niture SK, Khatri R and Jaiswal AK:
Regulation of Nrf2-an update. Free Radic Biol Med. 66:36–44. 2014.
View Article : Google Scholar
|
|
16
|
Villeneuve NF, Tian W, Wu T, Sun Z, Lau A,
Chapman E, Fang D and Zhang DD: USP15 negatively regulates Nrf2
through deubiquitination of Keap1. Mol Cell. 51:68–79. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Papaiahgari S, Zhang Q, Kleeberger SR, Cho
HY and Reddy SP: Hyperoxia stimulates an Nrf2-ARE transcriptional
response via ROS-EGFR-PI3K-Akt/ERK MAP kinase signaling in
pulmonary epithelial cells. Antioxid Redox Signal. 8:43–52. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Min KJ, Lee JT, Joe EH and Kwon TK: An
IκBα phosphorylation inhibitor induces heme oxygenase-1(HO-1)
expression through the activation of reactive oxygen species
(ROS)-Nrf2-ARE signaling and ROS-PI3K/Akt signaling in an
NF-κB-independent mechanism. Cell Signal. 23:1505–1513. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
DeNicola GM, Karreth FA, Humpton TJ,
Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES,
et al: Oncogene-induced Nrf2 transcription promotes ROS
detoxification and tumorigenesis. Nature. 475:106–109. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cong L, Ran FA, Cox D, Lin S, Barretto R,
Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al: Multiplex
genome engineering using CRISPR/Cas systems. Science. 339:819–823.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hsu PD, Lander ES and Zhang F: Development
and applications of CRISPR-Cas9 for genome engineering. Cell.
157:1262–1278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ogrunc M, Di Micco R, Liontos M,
Bombardelli L, Mione M, Fumagalli M, Gorgoulis VG and d'Adda di
Fagagna F: Oncogene-induced reactive oxygen species fuel
hyperproliferation and DNA damage response activation. Cell Death
Differ. 21:998–1012. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jaiswal AK: Nrf2 Signaling in coordinated
activation of antioxidant gene expression. Free Rad Biol Med.
36:1199–1207. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hamilton JW and Wetterhahn KE:
Differential effects of chromium(VI) on constitutive and inducible
gene expression in chick embryo liver in vivo and correlation with
chromium(VI)-induced DNA damage. Mol Carcinog. 2:274–286. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bagchi D, Bagchi M, Stohs SJ, Das DK, Ray
SD, Kuszynski CA, Joshi SS and Pruess HG: Free radicals and grape
seed proanthocyanidin extract: Importance in human health and
disease prevention. Toxicology. 148:187–197. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hong H and Liu GQ: Scutellarin attenuates
oxidative glutamate toxicity in PC12 cells. Planta Med. 70:427–431.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kobayashi E, Suzuki T and Yamamoto M:
Roles nrf2 plays in myeloid cells and related disorders. Oxid Med
Cell Longev. 2013:5292192013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Reddy NM, Kleeberger SR, Bream JH, Fallon
PG, Kensler TW, Yamamoto M and Reddy SP: Genetic disruption of the
Nrf2 compromises cell-cycle progression by impairing GSH-induced
redox signaling. Oncogene. 27:5821–5832. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Homma S, Ishii Y, Morishima Y, Yamadori T,
Matsuno Y, Haraguchi N, Kikuchi N, Satoh H, Sakamoto T, Hizawa N,
et al: Nrf2 enhances cell proliferation and resistance to
anticancer drugs in human lung cancer. Clin Cancer Res.
15:3423–3432. 2009. View Article : Google Scholar : PubMed/NCBI
|